Abstract
Acrolein (Acr), an α,β-unsaturated aldehyde, is abundant in tobacco smoke and cooking and exhaust fumes. Acr induces mutagenic α- and γ- hydroxy-1,N2-cyclic propano-deoxyguanosine adducts in normal human bronchial epithelial cells. Our earlier work has found that Acr-induced DNA damage preferentially occurs at lung cancer p53 mutational hotspots that contain CpG sites and that methylation at CpG sites enhances Acr-DNA binding at these sites. Based on these results, we hypothesized that this enhancement of Acr-DNA binding leads to p53 mutational hotspots in lung cancer. In this study, using a shuttle vector supF system, we tested this hypothesis by determining the effect of CpG methylation on Acr-DNA binding and the mutations in human lung fibroblasts. We found that CpG methylation enhances Acr-induced mutations significantly. Although CpG methylation enhances Acr-DNA binging at all CpG sites, it enhances mutations at selective—TCGA—sites. Similarly, we found that CpG methylation enhances benzo(a)pyrene diol epoxide binding at all –CpG- sites. However, the methylated CpG sequences in which benzo(a)pyrene diol epoxide-induced mutations are enhanced are different from the CpG sequences in which Acr-induced mutations are enhanced. CpG methylation greatly increases Acr-induced G to T and G to A mutation frequency to levels similar to these types of mutations found in the CpG sites in the p53 gene in tobacco smoke–related lung cancer. These results indicate that both CpG sequence context and the chemical nature of the carcinogens are crucial factors for determining the effect of CpG methylation on mutagenesis.
Introduction
Acrolein (Acr), an α,β−unsaturated aldehyde, is abundant in tobacco smoke (TS), cooking fumes and automobile exhaust fumes (1). Acr is also a by-product of lipid peroxidation generated endogenously in cells under oxidative stress (2). Acr, a major metabolite of the antitumor drugs cyclophosphamide and ifosfamide, is excreted in urine and accumulated in the bladder (3–5). It has been shown that intraperitoneal injection of Acr causes bladder tumors in rat models (6). Therefore, it has been concluded that Acr is the culprit of bladder cancer in patients who have been administered cyclophosphamide and ifosfamide in long-term treatment regimens (3,4,6,7).
Acr induces α- and γ-hydroxy-1,N2-cyclic propano-deoxyguanosine (α-OH-Acr-dG and γ-OH-Acr-dG) adducts in human cells (8–10). It has been found that both isomers of Acr-dG adducts are mutagenic and can induce mainly G to T and G to A mutations (9–20). By mapping Acr-dG adduct distribution at the nucleotide level in Acr-treated normal human bronchial epithelial cells, we have found that the Acr-DNA binding spectrum in the p53 gene coincides with the p53 mutational spectrum in lung cancer (21). Because Acr is abundant in TS and its level is to 10,000-fold that of benzo(a)pyrene (BP) in TS, we concluded that Acr is one of the major lung carcinogens in TS-induced lung cancer (21–23). Recent epidemiological studies suggested cooking fumes are also involved in the high incidence of lung cancer in non-smoking women in Asian countries (24). It has been found that Acr is the most abundant aldehyde in the urine of Chinese women who cook (25). These results together suggest that Acr contributes to lung carcinogenesis in non-smokers.
Our early work has shown that the major preferential sites of Acr-dG binding at p53 gene are guanines within CpG and –GAGG– sites and that the strong binding of Acr at CpG sites in the p53 gene in human lung cells is due to C5 cytosine methylation at these sequences (21). These results raise the possibilities that different DNA methylation patterns among individuals affect the Acr-DNA binding, as well as their susceptibilities toward Acr-induced DNA damages and mutations.
The biological outcome of the preferential Acr binding at methylated CpG sequences, however, has not yet been elucidated. In this study, using a shuttle vector pSP189 containing the supF gene as a target gene, we determined the effect of C5 cytosine CpG methylation on Acr- and benzo(a)pyrene diol epoxide (BPDE)-induced DNA binding and mutagenesis in human lung fibroblasts. BP is a potent carcinogen, which induces cancer in the aerodigestive system in hamster (26). Previously we have found that BPDE, the metabolite of BP, preferentially binds at lung cancer p53 mutational hotspots, which contain CpG sequences (27–29).
Herein, we report that CpG methylation enhances Acr-induced mutations significantly. Although CpG methylation enhances Acr-DNA binding at all CpG sites, it enhances Acr-induced mutations at selective –TCGA– sites. Although CpG methylation also enhances BPDE binding at all CpG sites, BPDE and Acr enhance mutations in different CpG sequence contexts. However, both Acr- and BPDE-induced mutations at methylated CpG sites are mainly G to T and G to A. These results indicate that both Acr and BP can contribute to lung cancer p53 mutations, and that sequence context, as well as chemical nature of the carcinogens, plays crucial roles in determining CpG methylation–enhanced mutagenesis.
Materials and methods
Cell culture and chemicals
Normal human lung fibroblasts (NHLF; American Type Culture Collection, Manassas, VA) were grown in modified Eagle's medium supplemented with 10% fetal bovine serum (Gibco/Invitrogen, Grand Island, NY). Stock solutions of Acr (Sigma, St. Louis, MO) and BPDE (Chemsyn Science Laboratories, Lenexa, KS) were prepared immediately before use.
Methylation and Acr modification of supercoiled pSP189 plasmids
The shuttle vector pSP189, which contains the tyrosine suppressor tRNA coding gene supF as a mutation target, was kindly provided by M. Seidman (National Institute on Aging, NIH, Baltimore, MD) and purified as described previously (30). The supercoiled pSP189 plasmids and 32P-end-labeled supF DNA fragments, purified as described previously (16,21) were subjected to Sss I methylase (New England Biolabs, Beverly, MA) treatment in the presence of S-adenosylmethionine according to the manufacturer’s instructions to methylate all cytosines at CpG sites. The methylation status of pSP189 was confirmed by restriction enzyme, Msp I and Hpa II, digestion (Supplementary Figure S1, available at Carcinogenesis Online). Methylated and unmethylated pSP189 plasmids were modified with Acr (0–1mM) for 24h at 37°C or with BPDE (2 µM) for 1h at 37°C, and then purified by repeated phenol and diethyl ether extraction. The DNA was then precipitated with ethanol and dissolved in TE buffer (10mM Tris-HCl, pH 8.0, 1mM ethylenediaminetetraacetic acid).
Immunochemical method for Acr-dG adduct detection.
Acr-modified DNA (1 µg) was denatured at 95°C for 10min, loaded onto nylon membranes using a Bio-Dot SF microfiltration apparatus (Bio-Rad, Hercules, CA) and cross-linked onto the membrane with UV. After blocking for 1h at room temperature in phosphate-buffered saline (137mM NaCl, 2.7mM KCl, 8mM Na2HPO4, 1.46mM KH2PO4, pH 7.0) -1% Tween-20 phosphate- buffered saline (PBS) with 1% Tween-20 (PBST) containing 5% (w/v) non-fat milk, the membrane was probed overnight at 4°C with anti-Acr-dG mouse monoclonal antibody (raised in-house). After washing with phosphate-buffered saline (PBS) with 1% Tween-20 to remove unbound primary antibody, horseradish peroxidase (HRP)-conjugated secondary antibodies (1:5000 in phosphate-buffered saline (PBS) with 1% Tween-20 with 5% non-fat milk; Santa Cruz Biotechnology, Santa Cruz, CA) were added for 2h at room temperature. Ultimately, Acr-dG adducts were detected using an ECL Plus chemiluminescence kit (PerkinElmer, Boston, MA) and medical X-ray films (Fuji, Düsserldorf, Germany) that were scanned with a CanoScan 8800F Laser Scanning Densitometer (Eastman Kodak, New York, NY). After antibody detection, the same membrane was stained with methylene blue (Molecular Research Center, Cincinnati, OH) to provide an estimated amount of DNA present.
Acr-DNA adduct analysis by two-dimensional thin layer chromatography/high-performance liquid chromatography.
Acr-DNA adducts were analyzed by thin layer chromatography/high-performance liquid chromatography (2D TLC/HPLC) as described previously (9,10,16,21).
Mutation assaysThe methods used for supF mutation detection were the same as described previously (9,10,16,21).
UvrABC nuclease incision on Acr-modified DNA and determination of Acr-DNA adduct distribution in the supF gene
It is well established that UvrA, UvrB and UvrC proteins function in concert (termed UvrABC nuclease) can incise bulky DNA damage (31). We have found that UvrABC nuclease can incise Acr-dG quantitatively and specifically (16,21); therefore, we used UvrABC incision method to identify and quantify the Acr-dG distribution in the supF sequence. Methods for the UvrA, UvrB and UvrC protein preparations and the UvrABC nuclease incision conditions were the same as described previously (31). Standard UvrABC incision assays were carried out in a 25 μl solution containing 100mM KCl, 1mM adenosine triphosphate, 10mM MgCl2, 10mM Tris (pH 7.5) and 1mM ethylenediaminetetraacetic acid. UvrABC nuclease was added to Acr- or BPDE-modified DNA at a molar ratio of 6:1 (UvrABC nuclease: plasmid DNA).
To map the Acr-dG distribution in the supF sequence, the supF DNA fragments were 32P-5′-single-end-labeled by two methods. Method I, the supF sequence was PCR amplified using EcoR I forward primer (5′-GAATTCGAGACCCTGCTC-) and BamH I reverse primer (5′-TAAGGATCCGGGTACCGAA-). Only forward or reverse primer was 32P-5′-end labeled. Method II, the pSP189 DNA was first digested with restriction enzyme BamH I, 5′-end-labeled with 32P- γ-adenosine triphosphate and further digested with restriction enzyme EcoR I to generate a single 5′-end-[32P]-labeled DNA fragment containing the supF sequence (non-transcribed [NT] strand). To generate a single 5′-end-32P-labeled DNA fragment containing the supF sequence in the opposite strand (transcribed [T] strand), pSP189 DNA was first digested with restriction enzyme EcoR I, 5′-end-labeled with 32P- γ-adenosine triphosphate and further digested with restriction enzyme BamH I. Both methods generate the same 131bp (EcoR I-BamH I) supF fragments. These methylated or unmethylated DNA fragments were modified with Acr (0–1mM, at 37°C for 24h) or BPDE (1 × 10−3 and 1 × 10−4 µM at 37°C for 1h) and then reacted with UvrABC nuclease. The resultant DNA samples were separated by electrophoresis over 8% denaturing polyacrylamide gels in parallel with Maxam and Gilbert sequencing reaction products as described previously (32).
Statistical analysis
The Student’s t-test was used to determine the significance of Acr-dG adduct analysis and mutation assay results. The differences were considered to be significant at P value less than or equal to 0.05.
Results
C5 cytosine methylation at CpG sequences enhances Acr-dG adduct formation
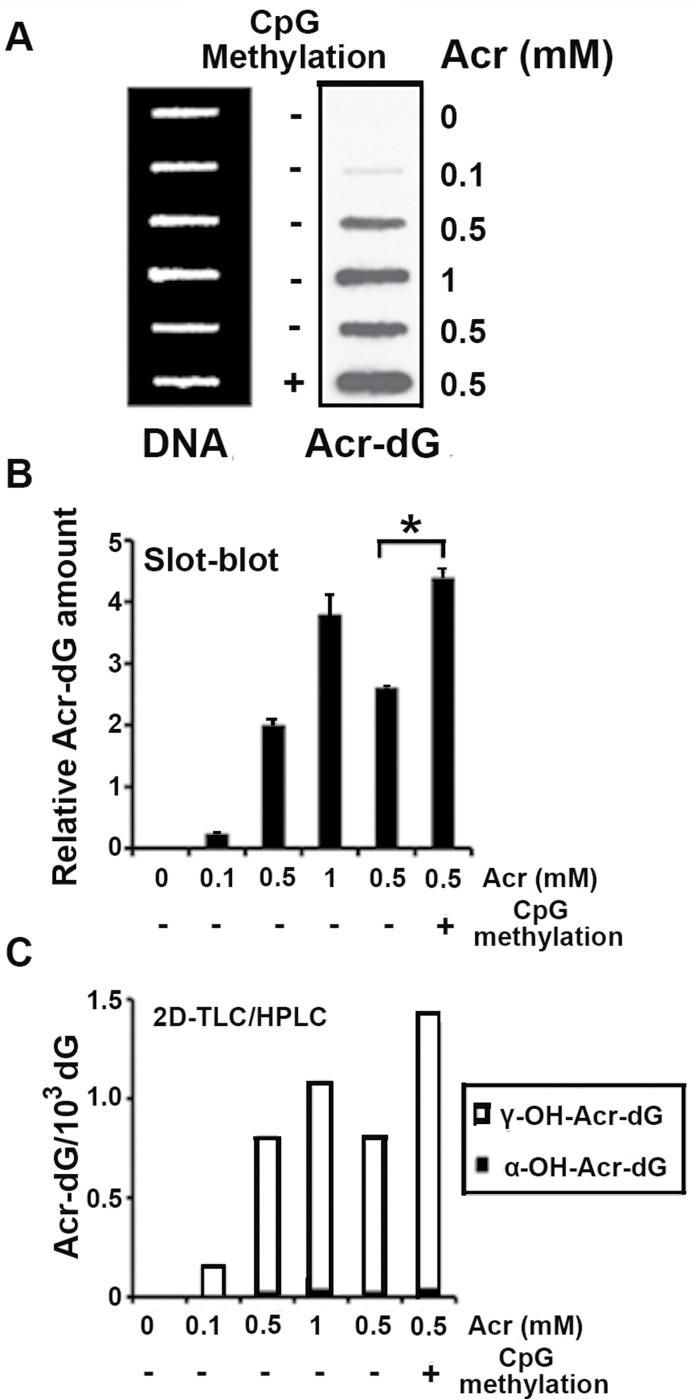
Using the UvrABC incision method to quantify Acr-dG adduct formation in the p53 gene sequences, our earlier work found that C5 methylation at CpG sequences of p53 gene enhances the binding of Acr at CpG sites (21). In order to determine the effect of this methylation on mutation induction, we used the supF gene in the pSP189 shuttle vector as a mutation target for mutation assay. The pSP189 plasmid DNA was subjected to methylation at C5 cytosine at CpG sites with SssI methylase, and the methylation status was confirmed by isoschizomers, HpaII and MspI digestion (Supplementary Figure S1, available at Carcinogenesis Online). Plasmid DNA with or without C5 cytosine methylation was then modified with different concentrations of Acr. The Acr-dG adducts formed in these plasmid DNAs were then determined by two methods: 32P postlabeling 2D TLC/HPLC and the slot-blot method using monoclonal antibody against Acr-dG adducts (Supplementary Figure S2, available at Carcinogenesis Online). The results in Figure 1 show that the relative amount of Acr-dG formed in methylated pSP189 plasmids is 1.7-fold (P = 0.0262) higher than Acr-dG formed in ummethylated plasmids determined by both Acr-dG-specific antibody assay (Figure 1A and 1B) and 2D TLC/HPLC analysis (Figure 1C)
Fig. 1.
DNA methylation enhanced Acr-DNA adduct formation. Plasmid pSP189 DNA was methylated by the CpG methylase SssI, and the methylated and unmethylated pSP189 plasmids were modified with Acr (0–1mM). The amount of Acr-DNA adducts formed in the DNA was quantified by a slot-blot method with monoclonal Acr-dG antibody (A and B), and by the 32P postlabeling 2D TLC/HPLC method (C). (A) Acr-modified DNA (1 µg) was loaded onto nylon membrane using a slot-blot apparatus, the relative levels of Acr-dG adducts were detected using anti-Acr-dG antibody (right lane) and the relative amount of DNA on the same membrane was detected by methylene blue staining (left lane). (B) Quantifications of Acr-dG adducts from (A) using Acr-dG antibody signals, which were normalized by the amount of DNA. (C) Quantifications of the amount of Acr-dG formed in the pSP189 plasmid DNA by the 32P -postlabeling 2D TLC/HPLC method (* indicates P < 0.05).
C5 cytosine methylations at CpG sites enhance Acr-induced supF mutations
Previous results have shown that Acr-dG adducts are mutagenic and that the mutation frequency increases proportionally to the amount of Acr-dG adducts (10,16,21). Because Acr induced more Acr-dG adducts in methylated pSP189 DNA than in unmethylated pSP189 DNA (Figure 1), we expected that the former would induce more mutations than the latter and that the fold of increase in mutation frequency would reflect the fold of increase of Acr-dG adducts. The results in Table I showed that CpG methylation enhanced spontaneous mutation frequency by 1.5-fold (P < 0.05). The results also showed that Acr modification induced more mutations in methylated pSP189 than in unmethylated pSP189 and that fold of mutation frequency increase (2.3-fold) was in fact similar to the fold of increase in Acr-dG adducts (1.7-fold).
Table I.
Effect of CpG methylation on Acr-modification-induced mutations in the supF gene in normal human lung fibroblasts
| Exp. | Acra | CpG Methylationb | White colonies/total colonies | Mutation frequency (×104) | Fold changec | P valued |
|---|---|---|---|---|---|---|
| 1 | − | − | 5/20720 | 2.4 | ||
| 2 | − | − | 4/17660 | 2.3 | ||
| 1 | − | + | 5/14480 | 3.5 | 1.4 | |
| 2 | − | + | 6/17920 | 3.3 | 1.5 | |
| 1 | + | − | 18/8350 | 21.6 | ||
| 2 | + | − | 21/9560 | 22.0 | ||
| 1 | + | + | 38/7280 | 52.2 | 2.4 | |
| 2 | + | + | 43/8500 | 50.6 | 2.3 | 0.03522 |
aAcr: pSP189 plasmids were treated with Acr (0.5mM) at 37°C for 24h.
bCpG methylation: pSP189 plasmids were treated wit SssI methyltransterase at 37°C for 1h.
cFold change of mutation frequency between unmethylation and methylation in each experiment.
dP value: statistical significance was tested by Student’s t-test.
Analysis of Acr modification induced mutation signature and spectrum in methylated and unmethylated supF gene
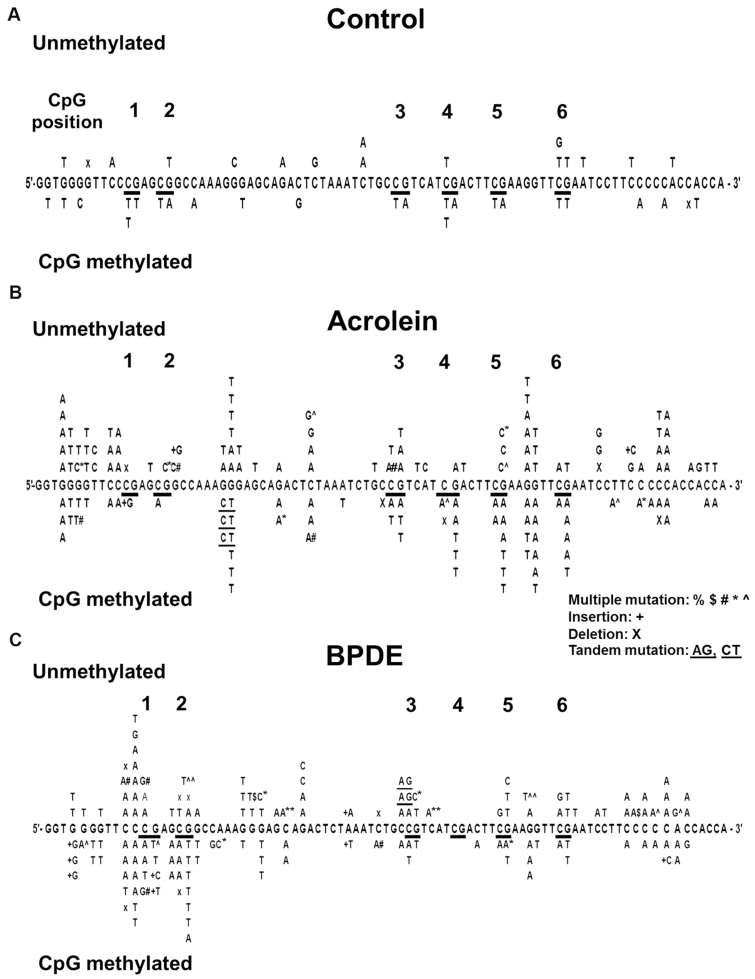
In order to understand the methylation effect on mutagenesis at the DNA sequence level, we sequenced spontaneous supF mutants in methylated and unmethylated pSP189 plasmid DNA and supF mutants induced by Acr modifications in methylated and unmethylated pSP189 plasmid DNA. Results in Figure 2A show that although supF spontaneous mutations occur randomly at -G-, -C- and -A- sites in unmethylated plasmid DNA, supF mutations occur mainly at -CpG- sites in methylated plasmid DNA. Furthermore, 12 out of 14 mutations that occur at CpG sites are G to A or C to T mutations. These results suggest that these mutations are due to deamination of methylated cytosines at -CpG- sites, which will result in C to T mutations or G to A mutations (Figure 2A).
Fig. 2.
Effect of cytosine methylation at CpG sites on spontaneous, Acr-dG- and BPDE-dG-induced mutational spectrum in the supF gene. Plasmid pSP189 DNA was methylated at CpG sites by SssI CpG methylase. Methylated and unmethylated pSP189 plasmids were (A) mock modified, (B) modified with Acr (0.5mM at 37°C for 24h) and (C) BPDE (2 µM at 37°C for 1h) and transfected into NHLF. After 72h incubation, the replicated plasmid DNA was recovered, treated with Dpn I, and transformed into indicator Escherichia coli cells according to the method described previously (10,16,21). The supF gene in the plasmid pSP189 DNA isolated from white mutant colonies was sequenced. The supF mutations were depicted on upper panel (unmethylated DNA) and lower panel (methylated DNA) of the supF gene sequence. Note: (i) CpG methylation enhances C to T and G to A mutations (12 out of 14) at CpG sites. (ii) CpG methylation enhanced Acr-induced mutations at -TCGA- (-CpG- positions 4, 5 and 6) but not at other CpG sequences such as -CCGA- (positions 1), -GCGG- (position 2) and -CCGT- (position 3). (iii) CpG methylation enhances BPDE-induced mutations at -CCGA- (positions 1), -GCGG- (position 2) but not at other CpG sequences such as -CCGT- (position 3) and -TCGA- (-CpG- positions 4, 5 and 6).
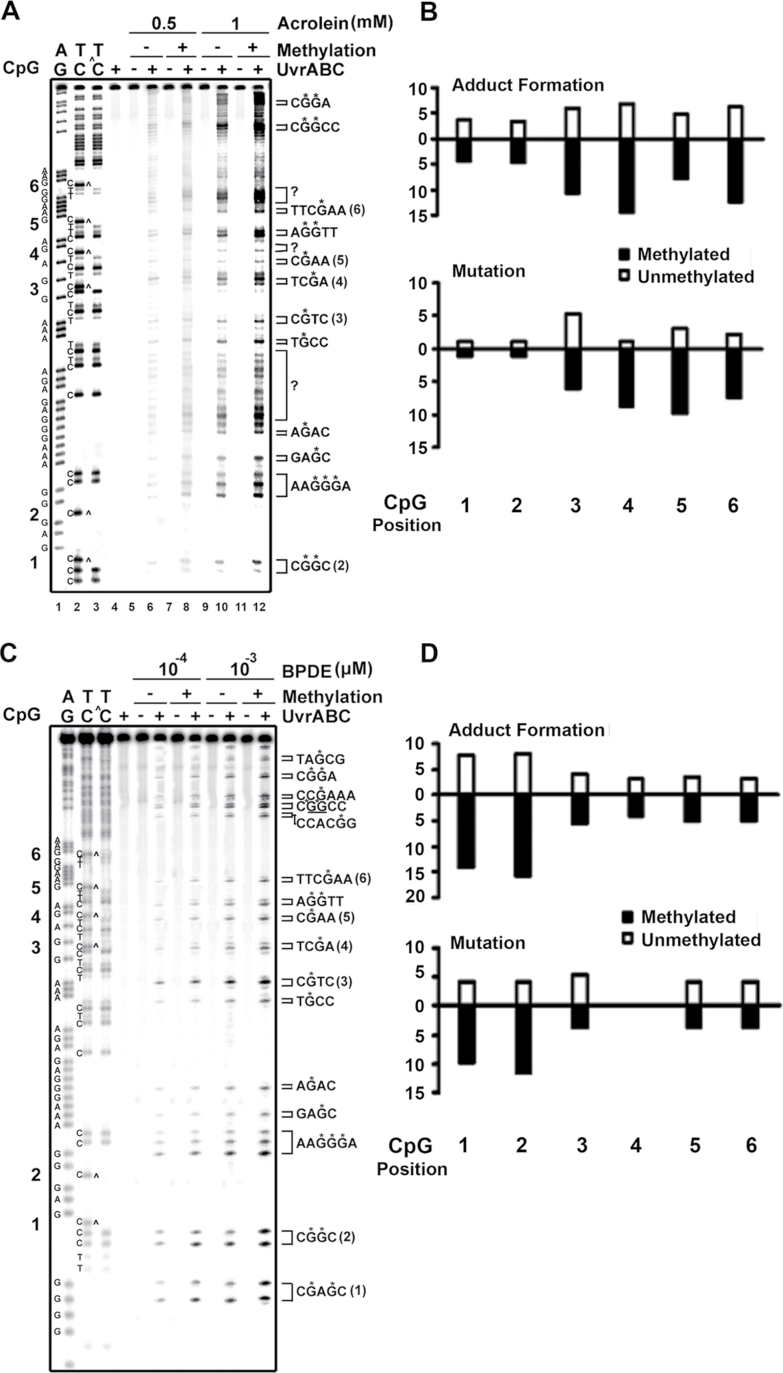
Results in Figure 2B showed that Acr modifications induced a high mutation frequency at a run of 3 G’s and -AGG- sequences in both methylated and unmethylated plasmid DNA, indicating these sequences were Acr mutation hotspots and that cytosine methylation at -CpG- sites did not affect Acr mutation induction at these sites. However, CpG methylations enhanced mutation frequency at 3 (positions 4, 5 and 6) out of 6 -CpG- sites, which have identical flanking sequences -TCGA-. There are two possibilities that can result in this dichotomous effect of methylation on Acr-induced mutagenesis at different CpG sequences: one, not all C5 cytosine methylation at different -CpG- sequences will enhance Acr-dG adduct formation, and two, Acr-dG formed at different CpG sites have different susceptibility to DNA repair and effect on the fidelity of translesion synthesis. To test the first possibility, we mapped the Acr-dG adduct distribution in the non-transcribed strand of supF sequences from methylated and unmethylated pSP189 DNA modified with Acr. Results in Figure 3A and Supplementary Figure S3, available at Carcinogenesis Online, showed that compared with unmethylated DNA fragments, Maxam and Gilbert sequencing reaction products of methylated DNA fragments lack C ladder bands, indicating that all the cytosines at -CpG- sites are methylated (cf lane 2 versus lane 3). Figure 3A and Supplementary Figure S3, available at Carcinogenesis Online, also showed that C5 methylation at CpG sequences enhanced Acr-DNA binding; these results are consistent with results in Figure 1, which show CpG methylation enhances overall Acr-DNA adduct formation by 1.7-fold. CpG methylation enhances Acr-DNA binding at all CpG sites (cf lane 6 versus lane 8 and lane 10 versus lane 12). However, the enhancements of Acr-DNA binding were much more pronounced at positions 4–6, which had a -TCGA- sequence (3- to 4-fold), than in positions 1 and 2, in which the CpG sites were in different sequence context (<2-fold; Figure 3B and Supplementary Figure S3, available at Carcinogenesis Online). Nonetheless, these results indicated that enhancement of Acr-dG adduct formation contributes to mutation enhancement at CpG sites.
Fig. 3.
Effect of CpG methylation on Acr-DNA, and BPDE-DNA binding spectrum in supF gene. PCR DNA fragments (131bp) containing the supF gene, which were 5′-end-32P-labeled at the non-transcribed strand, were methylated at CpG sites in the same manner as described in Figure 1, and then modified with different concentrations of (A and B) Acr (0–1mM), and (C and D) BPDE (1×10−3 to 1×10−4 µM at 37°C for 1h). The Acr-dG and BPDE-dG DNA adduct distributions in the supF sequences were mapped by the UvrABC incision method (10,16,21) (*G indicates the UvrABC incision G site). (A and C) typical autoradiographs. Symbols: GA and TC are Maxam and Gilbert sequencing reaction products. TC^ represents Maxam and Gilbert sequencing reaction products from methylated DNA fragments. Note: CpG methylation caused missing C ladders at CpG sites. Brackets with question markers indicate that we are unable to assign these UvrABC incision bands are corresponding to Acr-modified G residues in the 32P-labeled strand. These bands could be resulted from UvrABC incisions of Acr modifications of bases other than G residues or Acr-dG adducts in the opposite strand. (B and D) Quantifications. The relative amount of Acr-DNA and BPDE-DNA adducts formed at six CpG sites (positions 2–6 from (A and C) and position 1 from Supplementary Figure S3, available at Carcinogenesis Online) in the supF gene (upper panel) and the relative mutation frequency induced by Acr and BPDE modifications (lower panel) at these CpG sites are shown. Note: (i) CpG methylation enhances Acr binding at both CpG and non-CpG sites. (ii) CpG methylation enhances BPDE-DNA binding at all CpG sites.
We further analyzed the Acr-dG adduct induced mutation signatures at CpG sites in the unmethylated and methylated supF genes. The results in Figure 2B and Table II showed that CpG methylation enhanced Acr-dG-induced G to T transversions (36% to 54%) and that the signatures of the mutations induced by Acr-dG adducts at both unmethylated CpG sites (36% G to T and 28% G to A) and methylated CpG sites (54% G to T and 39% G to A) were similar to the mutation signatures at CpG sites of the p53 gene (49% G to T and 36% G to A) in lung cancer. These results were consistent with the interpretation that Acr is a lung cancer etiological agent (21).
Table II.
Identification of types of mutations induced by Acr or BPDE in the CpG methylated and unmethylated supF gene compared with types of mutations that occur at CpG mutational hotspots in the p53 gene in tobacco smoke–related lung cancer
| Mutations at CpG site | Control | Acrb | BPDEc | p53d | |||
|---|---|---|---|---|---|---|---|
| UMa | Ma | UM | M | UM | M | ||
| G to T | 2 (40%) | 1 (6%) | 9 (36%) | 15 (54%) | 10 (48%) | 20 (71%) | 296 (49%) |
| G to A | 2 (40%) | 17 (94%) | 7 (28%) | 11 (39%) | 1 (4%) | 3 (11%) | 219 (36%) |
| Otherse | 1 (20%) | 0 (0%) | 9 (36%) | 2 (7%) | 10 (48%) | 5 (18%) | 96 (15%) |
aUM: unmethylated supF; M: methylated supF.
bAcr: 0.5mM at 37°C for 24h.
cBPDE: 2 µM at 37°C for 1h.
dCpG sites at codons 157, 158, 175, 245, 248, 273 and 282 of p53 in TS-related lung cancer.
eOther types of mutations including G to C, insertion, deletion or tandem mutations.
Effect of CpG methylation on BPDE-dG adduct induced mutagenesis
It has been well established that CpG methylation enhances bulky chemical adduction at CpG sites (33–37). However, the results presented above show that CpG methylation manifested a sequence context–dependent differential effect on Acr-dG adduction and mutation induction. This differential effect could be Acr-dG adduct specific and/or the DNA sequence context specific. To test these two possibilities, we modified methylated and unmethylated pSP189 with BPDE and then determined BPDE-induced mutations and BPDE-dG adduct distribution in the supF gene sequences. Results in Figure 2C and Table III showed that (i) CpG methylation enhanced mutation frequency at sites 1 (-CCGA-) and 2 (-GCGG-) and (ii) CpG methylation did not enhance total mutation frequency significantly. These results showed that CpG methylation had different effects on BPDE-induced mutagenesis than it had on Acr-induced mutagenesis. Two possibilities can account for this difference: one, CpG methylation affected Acr and BPDE binding differently, and two, Acr-dG and BPDE-dG formed at different CpG sites had a different susceptibility to DNA repair and effect on the fidelity of translesion synthesis. To differentiate these possibilities, we mapped BPDE-dG adduct distribution in methylated and unmethylated supF gene DNA fragments modified with different concentrations of BPDE. The results in Figure 3C and 3D showed that (i) BPDE preferentially bound to G at -CCC*GA- or GC*GG sequences and CpG methylation greatly enhanced BPDE binding at these two sites and (ii) CpG methylation enhanced BPDE binding at all CpG sites. These results indicated that CpG methylation had distinct different effect on Acr- and BPDE-induced DNA adduct formation and mutagenesis.
Table III.
Effect of CpG methylation on BPDE modification–induced mutations in supF sequences in NHLF
| Exp. | BPDEa | CpG methylationb | White colonies/total colonies | Mutation frequency (×104) | Fold changec | P valued |
|---|---|---|---|---|---|---|
| 1 | − | − | 3/23960 | 1.3 | ||
| 2 | − | − | 4/25660 | 1.6 | ||
| 1 | − | + | 6/27480 | 2.2 | 1.7 | |
| 2 | − | + | 7/27920 | 2.5 | 1.6 | |
| 1 | + | − | 45/10350 | 43.5 | ||
| 2 | + | − | 43/9560 | 45.0 | ||
| 1 | + | + | 75/12880 | 58.2 | 1.3 | |
| 2 | + | + | 62/11200 | 55.4 | 1.2 | 0.00816 |
aBPDE: pSP189 plasmids were treated with BPDE (2 µM) at 37°C for 1h.
bCpG methylation: pSP189 plasmids were treated wit SssI methyltransterase at 37°C for 1h.
cFold change of mutation frequency between unmethylation and methylation in each experiment.
dP value: statistical significance was tested by Student’s t-test.
The results in Table II showed that CpG methylation enhanced BPDE-dG-induced G to T transversions (48% to 71%) and the signatures of mutations induced by BPDE-dG adducts at both unmethylated CpG sites (48% G to T and 4% G to A) and methylated CpG sites (71% G to T and 11% G to A) were different from the mutation signatures at CpG sites of the p53 gene (49% G to T and 36% G to A) in lung cancer.
Discussion
It has been long recognized that C5 cytosine methylation at CpG sequences greatly enhances bulky chemical-deoxyguanosine adduct formation at the methylated CpG sites (33–37). However, the mechanism that leads to this enhancement of chemical-DNA adduction remains unclear. It has been proposed that cytosine methylation increases the electron density at exocyclic amino group guanine that paired with the methylated cytosine, consequently enhancing its affinity with electrophiles such as BPDE, N-acetoxy-2-acetylaminofluorene, aflatoxin B1 diol epoxide and mitomycin C (18,27,28,33–37). However, this explanation is difficult to reconcile with the facts that not only the exocyclic amino group but also the C8 and N7 moieties of the guanine at the methylated CpG are more prone to be adducted by bulky chemicals than their counterparts at unmethylated CpG sites (36). 5C cytosine methylation has been shown to enhance the probability of base flipping of the paired guanine into the major groove (37). It is foreseeable that once the guanine is flipped to the major groove without pairing with cytosine, it has fewer constraints to its interaction with electrophiles at its electron centers at N2, N7 and C8 positions. However, the current results show that Acr binding affinity toward the methylated CpG sites was sequence dependent and that TmCGA was the preferential sequence. The enhanced Acr adduction at TmCGA sites led to enhanced mutagenesis at these sites. On the other hand, the enhancement of Acr-DNA binding at other CpG sites (positions 1–3) was much less than at the TmCGA site and no significant enhancement of mutagenesis was observed at these sites. These results raise the possibility that the Acr-dG adducts formed at CpG sequences other than TmCGA sequence are more susceptible to repair mechanisms, such as nucleotide excision repair, and/or cause less error during translesion synthesis.
We observed that Acr modifications induce a high mutation frequency at –GAGG- and –GGG/-CCC- sites. We also found that Acr preferentially bound at GAGG and GGG sites and that CpG methylation affected neither Acr-DNA binding (Figure 3A) nor mutation frequency at these sites (Figure 2B). These results indicate that the mutation frequency at GAGG and GGG reflects the level of DNA damage.
It is worth noting that CpG-methylated pSP189 plasmid DNA manifests a higher spontaneous mutation frequency in supF than unmethylated controls and that methylation-enhanced mutations are almost exclusively G to A or C to T mutations at the CpG sites (Figure 2A; Tables I and III). These results are consistent with the well-established finding that C5 cytosine methylation greatly enhances deamination, which leads to C to T base transformation (38).
It has been argued that CpG mutational hotspots in the p53 gene in human lung cancer are mainly due to deamination at CpG sites rather than due to these sites being preferential sites for carcinogen binding (39–41). If this argument were correct, then based on our current results one would expect that the kinds of mutations occurring at CpG sites in the p53 gene in human lung cancer should be G to A and C to T mutations. However, more than 49% of the mutations occurring at CpG sites of the p53 gene in TS-related lung cancers are G to T and C to A mutations (Table II). Results from this study also show that 50–70% of mutations induced by both BPDE and Acr at methylated CpG sites are G to T and C to A mutations, whereas only 14% of spontaneous mutations at methylated CpG sites are G to T; these results further strengthen our conclusion that the mutational hotspots in TS-related lung cancer are due to preferential binding of cigarette carcinogens at these mutational hotspots (27,28,36).
We found that although CpG methylation enhanced BPDE binding at all CpG sites, it enhanced mutation frequency selectively at positions 1 (-CCGA-) and 2 (-GCGG-), but not at TCGA sites, which are the mutational hotspots for Acr modifications. CpG methylation at the -CCGT- site (position 3) does not enhance either BPDE or Acr-induced mutation frequency. These results, which were consistent with results from a previous report (42), indicate that the impact on DNA structure by CpG methylation is influenced by its neighboring sequence context, which may affect chemical binding, DNA repair efficiency, and/or fidelity of translesion synthesis.
DNA damage occurring at CpG sites is more resistant to repair as we have found previously (28). It is possible that CpG methylation enhances the susceptibility of guanines at both strands to chemical adduction, as the results shown in Figure 3 and Supplementary Figure S3, available at Carcinogenesis Online. It is also possible that if repair occurs at both DNA strands simultaneously, it will result in inducing DNA double-strand breaks, consequently increasing either mutations or lethality. Our results suggest that Acr and bulky chemicals may preferentially bind at methylated CpG islands. If this occurs at the promoter region of genes, the chemical binding and/or the repair process may affect protein interactions and methylation status. In this case, the activity of the downstream genes of the promoter may be affected. This effect may contribute to Acr antitumor activity, as well as to its carcinogenicity.
In summary, we found that CpG methylation enhances spontaneous mutations at CpG sites, which are almost exclusively G to A mutations indicating that cytosine methylation enhances deamination. CpG methylation enhances Acr-DNA adduct formation at all CpG sites; however, it enhance mutations at selective -TCGA- sites. CpG methylation greatly increases Acr-induced G to T and G to A mutation frequency to levels similar to these types of mutations found in the CpG sites in the p53 gene in TS-related lung cancer. CpG methylation also enhances BPDE-DNA adduct formation at all CpG sites; however, it enhances mutations at CpG sites with sequence contexts that are different from those enhanced by Acr modifications and mainly G to T mutations. These results indicate that both CpG sequence context and the chemical nature of the carcinogens are crucial factors for determining the effect of CpG methylation on mutagenesis.
Supplementary material
Supplementary Figures S1––S3 can be found at http://carcin.oxfordjournals.org/
Funding
National Institutes of Health (CA114541, ES014641, CA99007 and ES00260); UO1 CA086137 to Dr W.R.
Supplementary Material
Acknowledgement
We thank Drs. Catherine G. Klein and Gerd Pfeifer for critical review of this manuscript.
H.T.W., M.W.W., W.E.C. and M.Y. conducted the experiments and participated in writing; F.L.C., X.R.W. and W.R. participated in designing the experiments and writing; M.S.T. was in charge of designing the experiments, interpreting the results and manuscript writing Dr. Rom's efforts in this research (UO1 CA086137).
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- 2D TLC
two-dimensional thin layer chromatography
- α-OH-Acr-dG and γ-OH-Acr-dG
α-and γ-hydroxy-1,N2-cyclic propano-deoxyguanosine
- Acr
acrolein
- BP
benzo(a)pyrene
- BPDE
benzo(a)pyrene diol epoxide
- HPLC
high-performance liquid chromatography
- NHLF
normal human lung fibroblasts
- TS
tobacco smoke.
References
- 1.Stevens J.F., et al. (2008)Acrolein: sources, metabolism, and biomolecular interactions relevant to human health and disease. Mol. Nutr. Food Res. 527–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chung F.L., et al. (1996)Lipid peroxidation as a potential endogenous source for the formation of exocyclic DNA adducts. Carcinogenesis 172105–2111 [DOI] [PubMed] [Google Scholar]
- 3.Kehrer J.P., et al. (2000)The molecular effects of acrolein. Toxicol. Sci. 576–15 [DOI] [PubMed] [Google Scholar]
- 4.Ghilarducci D.P., et al. (1995)Fate and effects of acrolein. Rev. Environ. Contam. Toxicol. 14495–146 [DOI] [PubMed] [Google Scholar]
- 5.Sladek N.E.(1972)Therapeutic efficacy of cyclophosphamide as a function of inhibition of its metabolism. Cancer Res. 321848–1854 [PubMed] [Google Scholar]
- 6.Cohen S.M., et al. (1992)Acrolein initiates rat urinary bladder carcinogenesis. Cancer Res. 523577–3581 [PubMed] [Google Scholar]
- 7.Comes R., et al. Concise International Chemical Assessment Document No 43. World Health Organization; Geneva: (2002). [Google Scholar]
- 8.Chung F.L., et al. (1984)Formation of cyclic 1,N2-propanodeoxyguanosine adducts in DNA upon reaction with acrolein or crotonaldehyde. Cancer Res. 44990–995 [PubMed] [Google Scholar]
- 9.Wang H.T., et al. (2012)Effect of carcinogenic acrolein on DNA repair and mutagenic susceptibility. J. Biol. Chem. 28712379–12386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang M.S., et al. (2011)Acrolein induced DNA damage, mutagenicity and effect on DNA repair. Mol. Nutr. Food Res. 551291–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanuri M., et al. (2002)Error prone translesion synthesis past gamma-hydroxypropano deoxyguanosine, the primary acrolein-derived adduct in mammalian cells. J. Biol. Chem. 27718257–18265 [DOI] [PubMed] [Google Scholar]
- 12.Kawanishi M., et al. (1998)Molecular analysis of mutations induced by acrolein in human fibroblast cells using supF shuttle vector plasmids. Mutat. Res. 41765–73 [DOI] [PubMed] [Google Scholar]
- 13.Minko I.G., et al. (2003)Translesion synthesis past acrolein-derived DNA adduct, gamma -hydroxypropanodeoxyguanosine, by yeast and human DNA polymerase eta. J. Biol. Chem. 278784–790 [DOI] [PubMed] [Google Scholar]
- 14.Sanchez A.M., et al. (2003)Comparative evaluation of the bioreactivity and mutagenic spectra of acrolein-derived alpha-HOPdG and gamma-HOPdG regioisomeric deoxyguanosine adducts. Chem. Res. Toxicol. 161019–1028 [DOI] [PubMed] [Google Scholar]
- 15.VanderVeen L.A., et al. (2001)Evaluation of the mutagenic potential of the principal DNA adduct of acrolein. J. Biol. Chem. 2769066–9070 [DOI] [PubMed] [Google Scholar]
- 16.Wang H.T., et al. (2009)Mutagenicity and sequence specificity of acrolein-DNA adducts. Chem. Res. Toxicol. 22511–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang I.Y., et al. (2002)Mutagenesis by acrolein-derived propanodeoxyguanosine adducts in human cells. Biochemistry 4113826–13832 [DOI] [PubMed] [Google Scholar]
- 18.Yang I.Y., et al. (2001)Responses to the major acrolein-derived deoxyguanosine adduct in Escherichia coli. J. Biol. Chem. 2769071–9076 [DOI] [PubMed] [Google Scholar]
- 19.Yang I.Y., et al. (2002)Genotoxic mechanism for the major acrolein-derived deoxyguanosine adduct in human cells. Chem. Res. Toxicol. 15160–164 [DOI] [PubMed] [Google Scholar]
- 20.Yang I.Y., et al. (2003)Mammalian translesion DNA synthesis across an acrolein-derived deoxyguanosine adduct. Participation of DNA polymerase eta in error-prone synthesis in human cells. J. Biol. Chem. 27813989–13994 [DOI] [PubMed] [Google Scholar]
- 21.Feng Z., et al. (2006)Acrolein is a major cigarette-related lung cancer agent: Preferential binding at p53 mutational hotspots and inhibition of DNA repair. Proc. Natl. Acad. Sci. U.S.A. 10315404–15409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujioka K., et al. (2006)Determination of toxic carbonyl compounds in cigarette smoke. Environ. Toxicol. 2147–54 [DOI] [PubMed] [Google Scholar]
- 23.Hoffman D., et al. (1990)The Handbook of Experimental Pharmacology Springer; Heidelberg: 70–74 [Google Scholar]
- 24.Kligerman S., et al. (2011)Epidemiology of lung cancer in women: risk factors, survival, and screening. AJR. Am. J. Roentgenol. 196287–295 [DOI] [PubMed] [Google Scholar]
- 25.Hecht S.S., et al. (2010)Elevated levels of volatile organic carcinogen and toxicant biomarkers in Chinese women who regularly cook at home. Cancer Epidemiol. Biomarkers Prev. 191185–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saffiotti U., et al. (1968)A method for the experimental induction of bronchogenic carcinoma. Cancer Res. 28104–124 [PubMed] [Google Scholar]
- 27.Denissenko M.F., et al. (1996)Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in P53. Science 274430–432 [DOI] [PubMed] [Google Scholar]
- 28.Denissenko M.F., et al. (1998)Slow repair of bulky DNA adducts along the nontranscribed strand of the human p53 gene may explain the strand bias of transversion mutations in cancers. Oncogene 161241–1247 [DOI] [PubMed] [Google Scholar]
- 29.Smith L.E., et al. (2000)Targeting of lung cancer mutational hotspots by polycyclic aromatic hydrocarbons. J. Natl. Cancer Inst. 92803–811 [DOI] [PubMed] [Google Scholar]
- 30.Canella K.A., et al. (2000)Mutation spectra in supF: approaches to elucidating sequence context effects. Mutat. Res. 45061–73 [DOI] [PubMed] [Google Scholar]
- 31.Tang M.S. (ed) (1996)Technologies for Detection of DNA Damage and Mutation Plwnum; New York: 139–152 [Google Scholar]
- 32.Maxam A.M., et al. (1980)Sequencing end-labeled DNA with base-specific chemical cleavages. Meth. Enzymol. 65499–560 [DOI] [PubMed] [Google Scholar]
- 33.Johnson W.S., et al. (1995)Selective recognition of the m5CpG dinucleotide sequence in DNA by mitomycin C for alkylation and cross-linking. Bioorg. Med. Chem. 3851–860 [DOI] [PubMed] [Google Scholar]
- 34.Tornaletti S., et al. (1995)Complete and tissue-independent methylation of CpG sites in the p53 gene: implications for mutations in human cancers. Oncogene 101493–1499 [PubMed] [Google Scholar]
- 35.Denissenko M.F., et al. (1997)Cytosine methylation determines hot spots of DNA damage in the human P53 gene. Proc. Natl. Acad. Sci. U.S.A. 943893–3898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen J.X., et al. (1998)Carcinogens preferentially bind at methylated CpG in the p53 mutational hot spots. Cancer Res. 582070–2075 [PubMed] [Google Scholar]
- 37.Li V.S., et al. (2001)The effect of C(5) cytosine methylation at CpG sequences on mitomycin-DNA bonding profiles. Bioorg. Med. Chem. 9863–873 [DOI] [PubMed] [Google Scholar]
- 38.Gonzalgo M.L., et al. (1997)Mutagenic and epigenetic effects of DNA methylation. Mutat. Res. 386107–118 [DOI] [PubMed] [Google Scholar]
- 39.Krawczak M., et al. (1998)p53 mutations, benzo[a]pyrene and lung cancer. Mutagenesis 13319–320 [DOI] [PubMed] [Google Scholar]
- 40.Cooper D.N., et al. Human Gene Mutation. BIOS Scientific Publishers; Oxford, UK: (1993). [Google Scholar]
- 41.Krawczak M., et al. (1995)Somatic spectrum of cancer-associated single basepair substitutions in the TP53 gene is determined mainly by endogenous mechanisms of mutation and by selection. Hum. Mutat. 548–57 [DOI] [PubMed] [Google Scholar]
- 42.Yoon J.H., et al. (2001)Methylated CpG dinucleotides are the preferential targets for G-to-T transversion mutations induced by benzo[a]pyrene diol epoxide in mammalian cells: similarities with the p53 mutation spectrum in smoking-associated lung cancers. Cancer Res. 617110–7117 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.