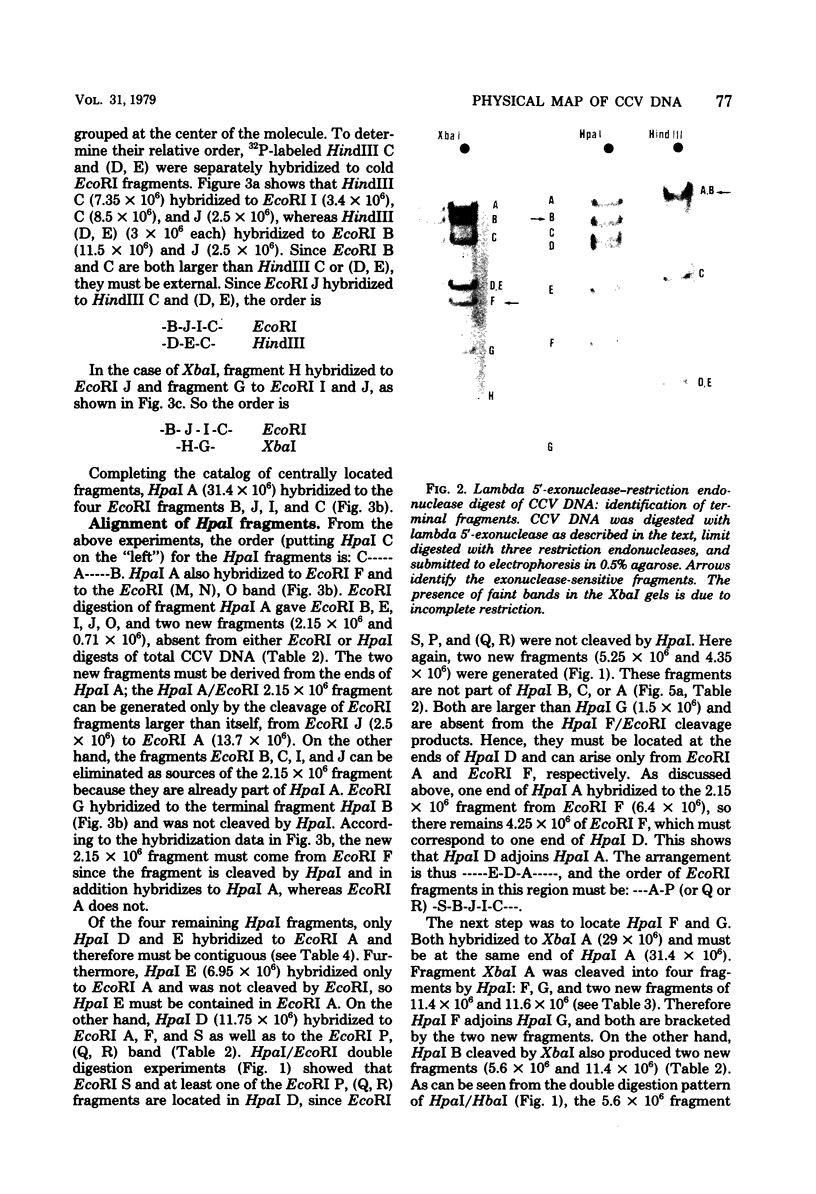
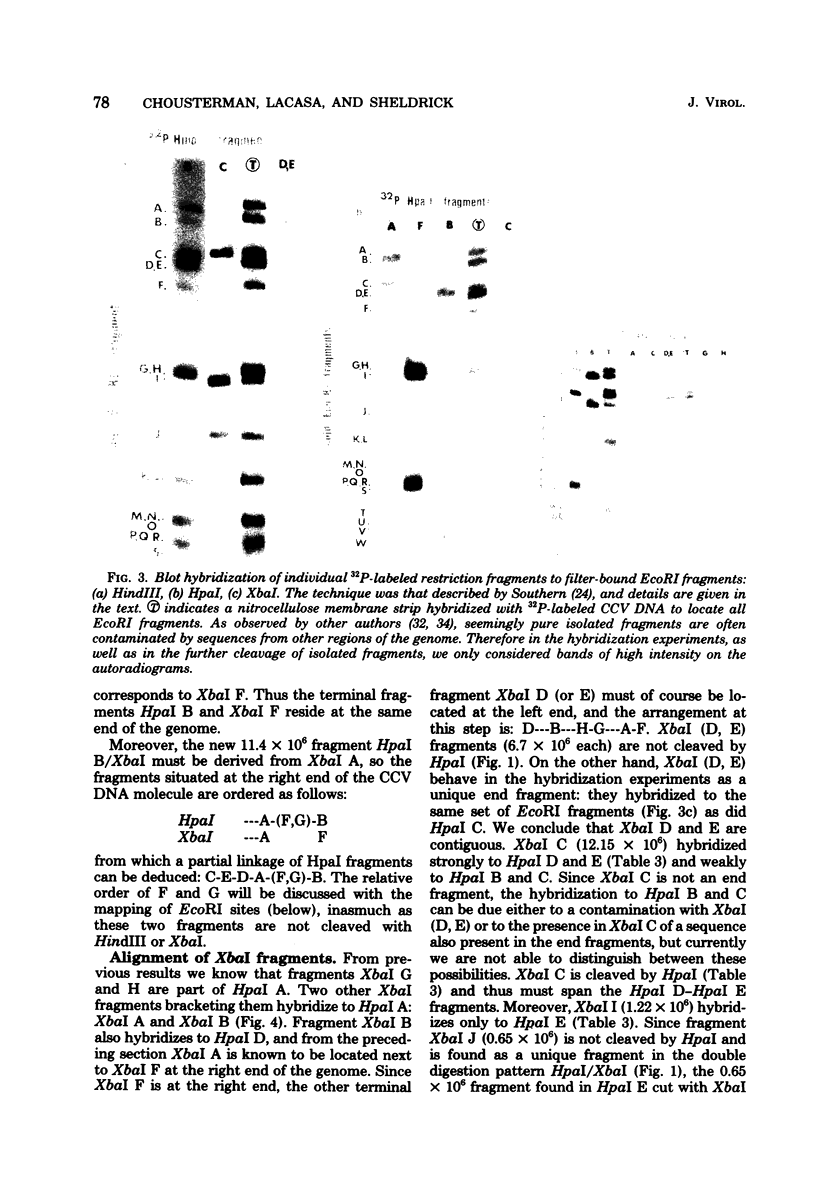
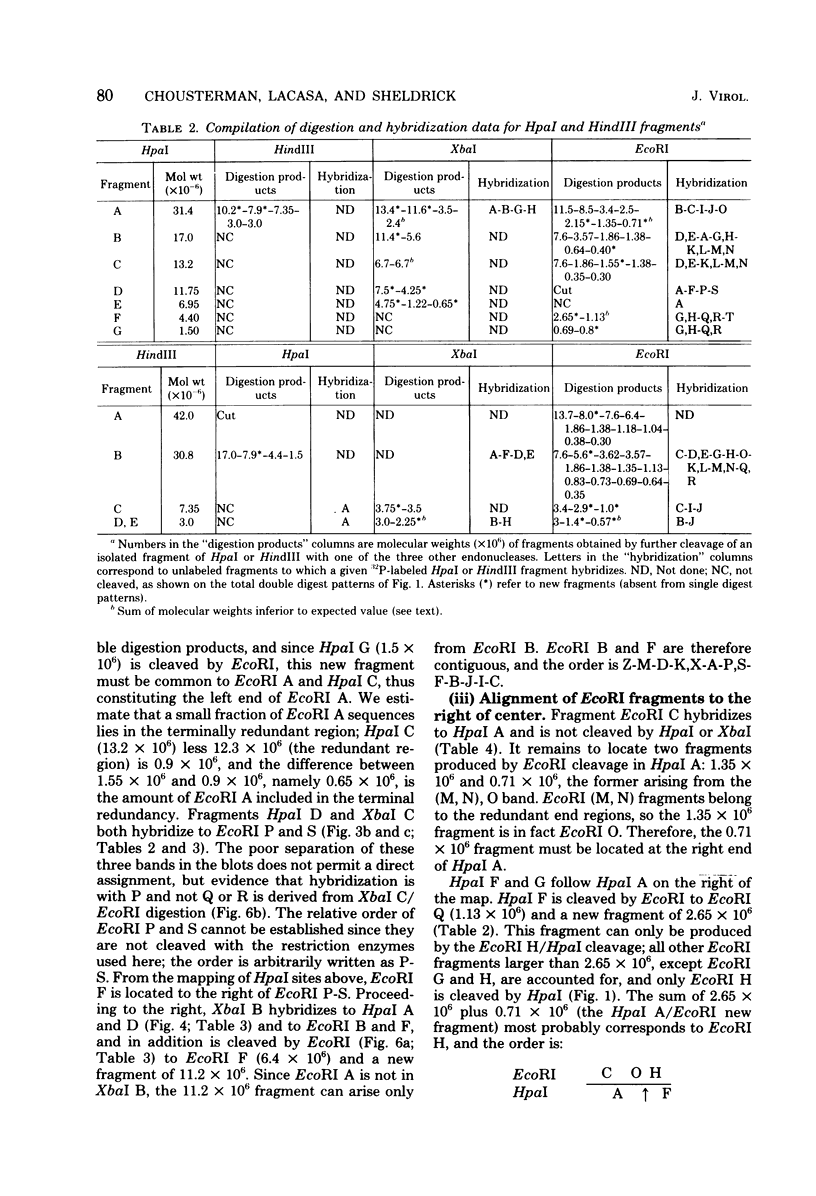
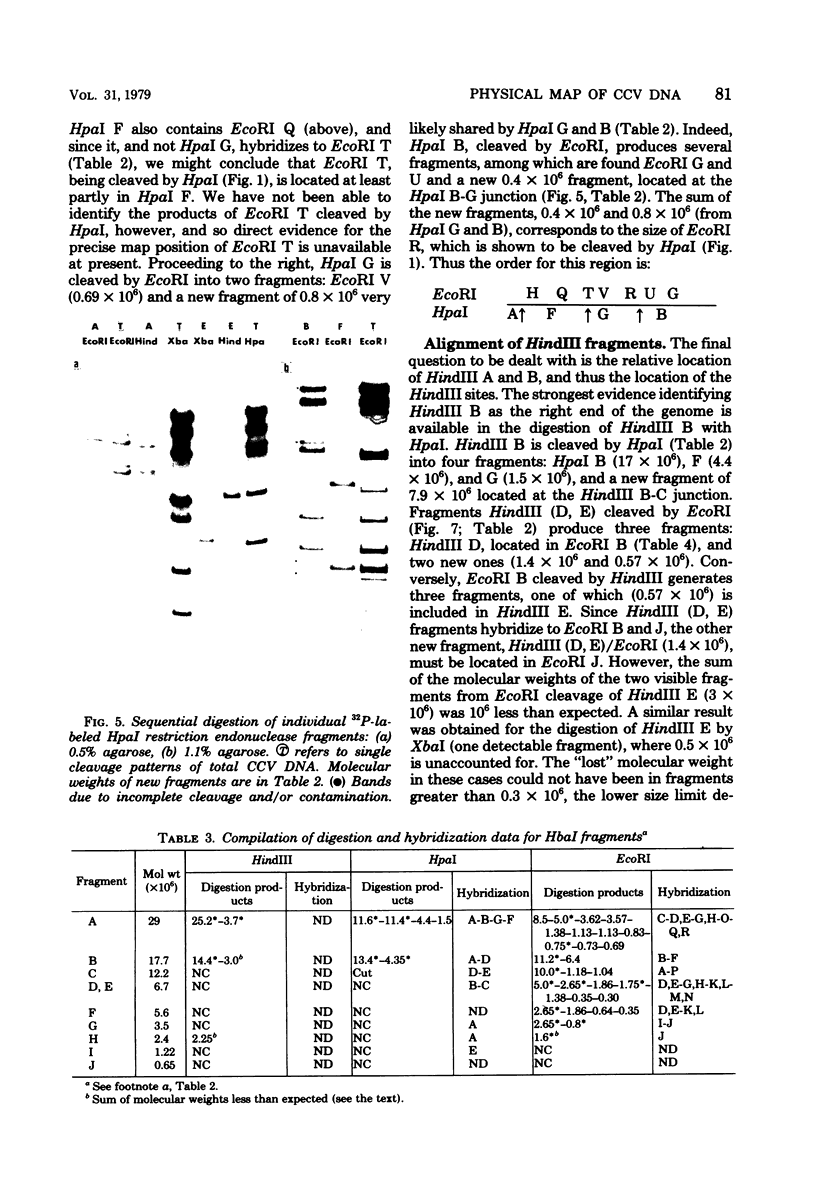
Abstract
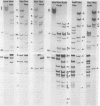
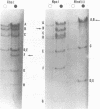
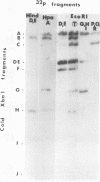
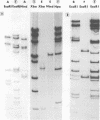
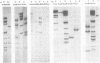
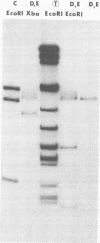
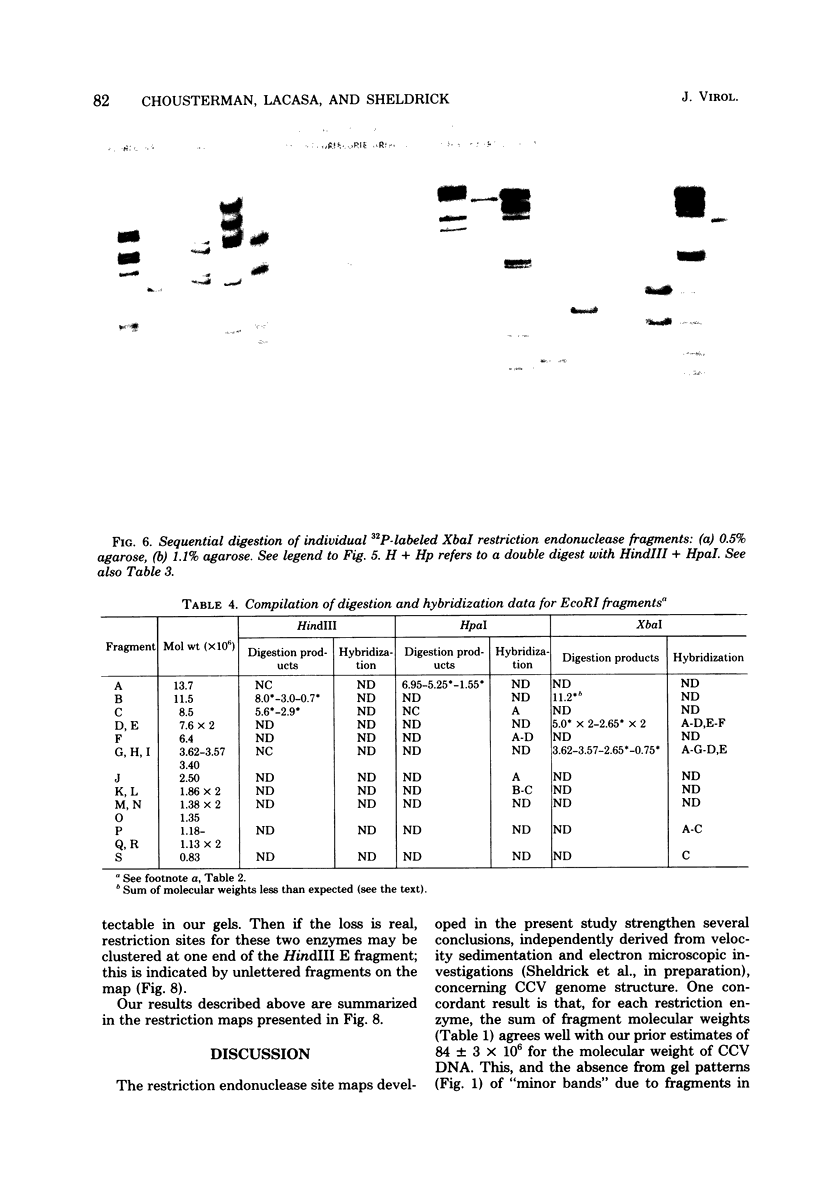
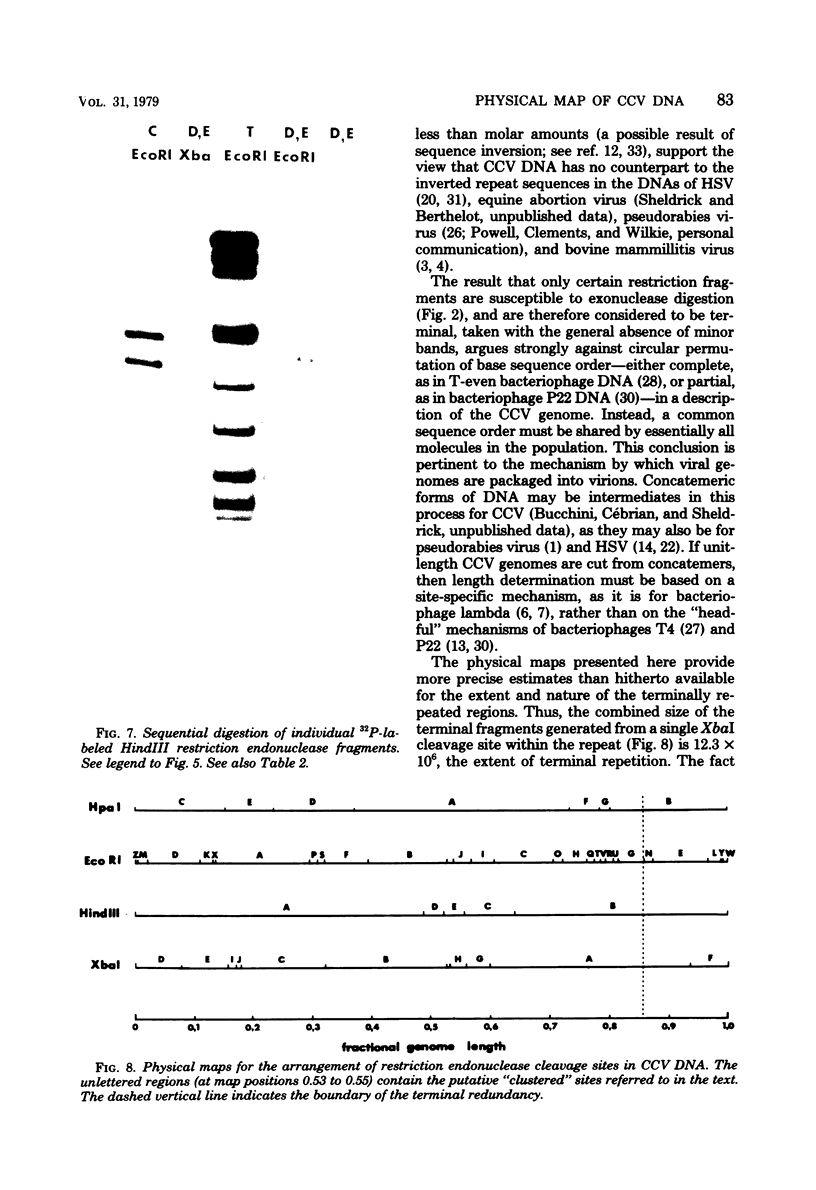
The overall arrangement of nucleotide sequences in the DNA of channel catfish virus has been studied by cleavage with four restriction endonucleases. Physical maps have been developed for the location of sites for EcoRI, HindIII, HpaI, and XbaI. The sum of the molecular weights of fragments generated by each restriction enzyme indicates a molecular weight of approximately 86 × 106 for the channel catfish virus genome. Fragments corresponding to the molecular ends of channel catfish virus DNA have been identified by their sensitivity to exonuclease treatment. The distribution of restriction sites in the genome shows that sequences included in a 12 × 106-molecular weight region at one end are repeated with direct polarity at the other end, and that the overall genomic sequence order is nonpermuted.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ben-Porat T., Kaplan A. S., Stehn B., Rubenstein A. S. Concatemeric forms of intracellular herpesvirus DNA. Virology. 1976 Feb;69(2):547–560. doi: 10.1016/0042-6822(76)90484-0. [DOI] [PubMed] [Google Scholar]
- Bornkamm G. W., Delius H., Fleckenstein B., Werner F. J., Mulder C. Structure of Herpesvirus saimiri genomes: arrangement of heavy and light sequences in the M genome. J Virol. 1976 Jul;19(1):154–161. doi: 10.1128/jvi.19.1.154-161.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman T. G., Roizman B. Anatomy of bovine mammillitis DNA II. Size and arrangements of the deoxynucleotide sequences. J Virol. 1978 Jul;27(1):239–254. doi: 10.1128/jvi.27.1.239-254.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman T. G., Roizman B. Anatomy of bovine mammillitis DNA. I Restriction endonuclease maps of four populations of molecules that differ in the relative orientation of their long and short components. J Virol. 1978 Jan;25(1):395–407. doi: 10.1128/jvi.25.1.395-407.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements J. B., Cortini R., Wilkie N. M. Analysis of herpesvirus DNA substructure by means of restriction endonucleases. J Gen Virol. 1976 Feb;30(2):243–256. doi: 10.1099/0022-1317-30-2-243. [DOI] [PubMed] [Google Scholar]
- Emmons S. W. Bacteriophage lambda derivatives carrying two copies of the cohesive end site. J Mol Biol. 1974 Mar 15;83(4):511–525. doi: 10.1016/0022-2836(74)90511-7. [DOI] [PubMed] [Google Scholar]
- Feiss M., Bublitz A. Polarized packaging of bacteriophage lambda chromosomes. J Mol Biol. 1975 Jun 5;94(4):583–594. doi: 10.1016/0022-2836(75)90323-x. [DOI] [PubMed] [Google Scholar]
- Fleckenstein B., Bornkamm G. W., Mulder C., Werner F. J., Daniel M. D., Falk L. A., Delius H. Herpesvirus ateles DNA and its homology with Herpesvirus saimiri nucleic acid. J Virol. 1978 Jan;25(1):361–373. doi: 10.1128/jvi.25.1.361-373.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Given D., Kieff E. DNA of Epstein-Barr virus. IV. Linkage map of restriction enzyme fragments of the B95-8 and W91 strains of Epstein-Barr Virus. J Virol. 1978 Nov;28(2):524–542. doi: 10.1128/jvi.28.2.524-542.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodheart C. R., Plummer G. The densities of herpesviral DNAs. Prog Med Virol. 1975;19:324–352. [PubMed] [Google Scholar]
- Hayward G. S., Jacob R. J., Wadsworth S. C., Roizman B. Anatomy of herpes simplex virus DNA: evidence for four populations of molecules that differ in the relative orientations of their long and short components. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4243–4247. doi: 10.1073/pnas.72.11.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson E. N., Jackson D. A., Deans R. J. EcoRI analysis of bacteriophage P22 DNA packaging. J Mol Biol. 1978 Jan 25;118(3):365–388. doi: 10.1016/0022-2836(78)90234-6. [DOI] [PubMed] [Google Scholar]
- Jacob R. J., Roizman B. Anatomy of herpes simplex virus DNA VIII. Properties of the replicating DNA. J Virol. 1977 Aug;23(2):394–411. doi: 10.1128/jvi.23.2.394-411.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KIRBY K. S. A new method for the isolation of deoxyribonucleic acids; evidence on the nature of bonds between deoxyribonucleic acid and protein. Biochem J. 1957 Jul;66(3):495–504. doi: 10.1042/bj0660495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little J. W., Lehman I. R., Kaiser A. D. An exonuclease induced by bacteriophage lambda. I. Preparation of the crystalline enzyme. J Biol Chem. 1967 Feb 25;242(4):672–678. [PubMed] [Google Scholar]
- RICHARDSON C. C., KORNBERG A. A DEOXYRIBONUCLEIC ACID PHOSPHATASE-EXONUCLEASE FROM ESCHERICHIA COLI. I. PURIFICATION OF THE ENZYME AND CHARACTERIZATION OF THE PHOSPHATASE ACTIVITY. J Biol Chem. 1964 Jan;239:242–250. [PubMed] [Google Scholar]
- Rhoades M., Rhoades E. A. Terminal repetition in the DNA of bacteriophage T5. J Mol Biol. 1972 Aug 21;69(2):187–200. doi: 10.1016/0022-2836(72)90224-0. [DOI] [PubMed] [Google Scholar]
- Rymo L., Forsblom S. Cleavage of Epstein-Barr virus DNA by restriction endonucleases EcoRI, HindIII and BamI. Nucleic Acids Res. 1978 Apr;5(4):1387–1402. doi: 10.1093/nar/5.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick P., Berthelot N. Inverted repetitions in the chromosome of herpes simplex virus. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):667–678. doi: 10.1101/sqb.1974.039.01.080. [DOI] [PubMed] [Google Scholar]
- Sheldrick P., Laithier M., Lando D., Ryhiner M. L. Infectious DNA from herpes simplex virus: infectivity of double-stranded and single-stranded molecules. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3621–3625. doi: 10.1073/pnas.70.12.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlomai J., Friedmann A., Becker Y. Replication intermediates of herpes simplex virus DNA. Virology. 1976 Feb;69(2):647–659. doi: 10.1016/0042-6822(76)90493-1. [DOI] [PubMed] [Google Scholar]
- Skare J., Summers W. C. Structure and function of herpesvirus genomes. II. EcoRl, Sbal, and HindIII endonuclease cleavage sites on herpes simplex virus. Virology. 1977 Feb;76(2):581–595. doi: 10.1016/0042-6822(77)90240-9. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Steinhart W. L., Grafstrom R. H., Hill C. W. Terminal fragments of herpes simplex virus DNA produced by restriction endonuclease. Biochem Biophys Res Commun. 1975 Nov 17;67(2):556–561. doi: 10.1016/0006-291x(75)90848-7. [DOI] [PubMed] [Google Scholar]
- Stevely W. S. Inverted repetition in the chromosome of pseudorabies virus. J Virol. 1977 Apr;22(1):232–234. doi: 10.1128/jvi.22.1.232-234.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streisinger G., Emrich J., Stahl M. M. Chromosome structure in phage t4, iii. Terminal redundancy and length determination. Proc Natl Acad Sci U S A. 1967 Feb;57(2):292–295. doi: 10.1073/pnas.57.2.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- THOMAS C. A., Jr, MACHATTIE L. A. CIRCULAR T2 DNA MOLECULES. Proc Natl Acad Sci U S A. 1964 Nov;52:1297–1301. doi: 10.1073/pnas.52.5.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M., Davis R. W. Studies on the cleavage of bacteriophage lambda DNA with EcoRI Restriction endonuclease. J Mol Biol. 1975 Jan 25;91(3):315–328. doi: 10.1016/0022-2836(75)90383-6. [DOI] [PubMed] [Google Scholar]
- Tye B. K., Huberman J. A., Botstein D. Non-random circular permutation of phage P22 DNA. J Mol Biol. 1974 Jan 5;85(4):501–528. doi: 10.1016/0022-2836(74)90312-x. [DOI] [PubMed] [Google Scholar]
- Wadsworth S., Jacob R. J., Roizman B. Anatomy of herpes simplex virus DNA. II. Size, composition, and arrangement of inverted terminal repetitions. J Virol. 1975 Jun;15(6):1487–1497. doi: 10.1128/jvi.15.6.1487-1497.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie N. M., Cortini R. Sequence arrangement in herpes simplex virus type 1 DNA: identification of terminal fragments in restriction endonuclease digests and evidence for inversions in redundant and unique sequences. J Virol. 1976 Oct;20(1):211–221. doi: 10.1128/jvi.20.1.211-221.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie N. M. Physical maps for Herpes simplex virus type 1 DNA for restriction endonucleases Hind III, Hpa-1, and X. bad. J Virol. 1976 Oct;20(1):222–233. doi: 10.1128/jvi.20.1.222-233.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittek R., Menna A., Schümperli D., Stoffel S., Müller H. K., Wyler R. HindIII and Sst I restriction sites mapped on rabbit poxvirus and vaccinia virus DNA. J Virol. 1977 Sep;23(3):669–678. doi: 10.1128/jvi.23.3.669-678.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K., Darlington R. W. Channel catfish virus: a new herpesvirus of ictalurid fish. J Virol. 1971 Oct;8(4):525–533. doi: 10.1128/jvi.8.4.525-533.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]