A repertoire of mechanisms in the autophagy system combats viral infections.
Keywords: autophagy, dendritic cells, innate immunity, T-cell responses, virus infection
Abstract
Autophagy is an evolutionarily ancient process eukaryotic cells utilize to remove and recycle intracellular material in order to maintain cellular homeostasis. In metazoans, the autophagy machinery not only functions in this capacity but also has evolved to perform a diverse repertoire of intracellular transport and regulatory functions. In response to virus infections, the autophagy machinery degrades viruses, shuttles viral pathogen-associated molecular patterns to endosomes containing Toll-like receptors, facilitates viral-antigen processing for major histocompatibility complex presentation and transports antiviral proteins to viral replication sites. This is accomplished through canonical autophagy or through processes involving distinct subsets of the autophagy-related genes (Atgs). Herein, we discuss how the variable components of the autophagy machinery contribute to antiviral defense and highlight three emerging themes: first, autophagy delivers viral cytosolic components to several distinct endolysosomal compartments; second, Atg proteins act alone, as subgroups or collectively; and third, the specificity of autophagy and the autophagy machinery is achieved by recognition of triggers and selective targeting by adaptors.
Introduction
Autophagy is a highly conserved process of degradation of intracellular components via the lysosomal machinery. Autophagy plays a crucial role in normal cell growth, development, repair, and survival during cellular starvation. It can also be selective—for example, in degradation of mitochondria (mitophagy) or foreign bodies (xenophagy). Autophagy includes several mechanistically distinct processes, including macroautophagy (in which targets are sequestered in a double-membrane structure), microautophagy and chaperone-mediated autophagy. Here we focus our discussion to the roles of macroautophagy (hereafter referred to as autophagy) and the associated (Atg) proteins in viral infection.
Classical autophagy initiation begins with the complex involving Beclin-1 (Atg6) and the class III phosphoinositide 3-kinase (PI3K), vacuolar protein sorting 34 (Vps34), which initiate phagophore membrane formation (1). Upon initiation, two different ubiquitin-like protein conjugation systems mediate the elongation of the autophagosome membrane (2). The Atg5–Atg12 conjugation system first conjugates Atg12 to Atg7, and this is followed by the transfer of Atg12 to Atg10 (3). After transfer of Atg12 to Atg10, Atg12 is transferred to Atg5 via a covalent bond. The Atg5–Atg12 conjugate forms a functional complex with Atg16, and this multimeric complex is crucial in autophagosome formation. The second conjugation system is initiated with the cleavage of microtubule-associated protein 1 light chain 3 (LC3) (Atg8) by Atg4b to LC3-I. Next, LC3-I is bound and activated by Atg7 and is transferred to Atg3. LC3-I is subsequently covalently linked to the lipid phosphatidylethanolamine (PE) (4). The Atg5–Atg12–Atg16 complex acts as the E3 enzyme on the LC3 conjugation reaction to generate LC3–PE (LC3-II), which is incorporated into both the cytoplasmic and luminal faces of the elongating autophagosomal membrane, and facilitates closure of the double-membrane autophagosome (5).
During viral infection, many cellular processes are either bypassed or subverted such that they are controlled by the virus. In response, metazoan hosts have developed complex antiviral systems to attempt to both counter viral infection and restore host autonomy (6). The Atg proteins and autophagy machinery function at this crux—playing key roles in both positively and negatively regulating antiviral immune responses. In the past decade, additional levels of detail have been uncovered in the relationship between the autophagy machinery and host antiviral immune responses.
Here, we focus on how autophagy contributes to antiviral defense and restoration of cellular homeostasis during viral infection while highlighting three emerging themes. First, it has become clear that the autophagy machinery does not function exclusively as a cytosol-to-lysosome degradation mechanism in the context of the metazoan antiviral immune response. In addition, the Atg proteins serve as a highly adaptable intracellular transport system. Second, Atg proteins contribute to antiviral immune responses via three distinct mechanisms: by operating independently, in combination, and collectively via canonical autophagy. Third, the autophagy machinery achieves temporal, spatial and mechanical specificity via utilization of a diverse set of triggers, sensors and adaptors (7, 8) (Fig. 1). We propose that an Atg-dependent mechanism of viral defense is initiated by a trigger that activates a sensor, which induces signaling that mobilizes Atg proteins to engulf substrates marked for destruction. Well-known sensors include the Toll-like receptors (TLRs) and cytokine receptors which, upon detection of their ligands, stimulate autophagy (Fig. 1). Selectivity of such substrates is determined by adaptors that couple membranes containing LC3 and target substrates through specific binding motifs. The LC3-interacting motif (LIR) is one such motif commonly found in adaptors, and consists of a linear tetrapeptide sequence that binds directly to LC3 (9). Known adaptors include p62 (10), Sma and Mad related family (SMAD) ubiquitin regulatory factor 1 (SMURF-1) (11), NDP52 (12), NBR1 (13), NIX/Bnip3L (14, 15) and optineurin (16). The role of triggers, sensors, adaptors and effector functions that are mediated by various sets of Atg proteins is discussed below.
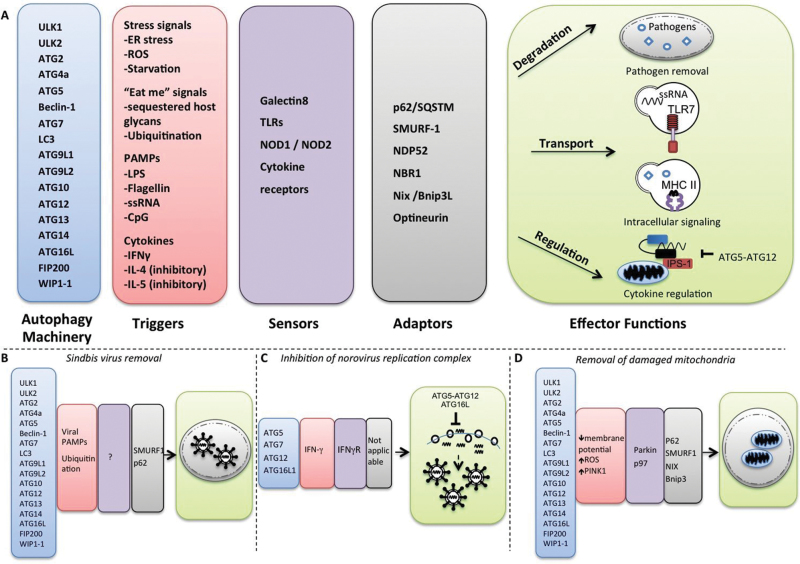
Fig. 1.
Cooperation between Atg proteins, triggers and adaptors in antiviral defense. (A) Distinct triggers (in red) and set subsets of Atg proteins (in blue) interact with sensors and adaptors (in gray) in order to perform a diverse set of effector functions (in green). In (B), the canonical autophagy machinery selectively removes Sindbis viral particles in infected neurons via the adaptor proteins SMURF1 and p62. The p62 interacts with Sindbis viral capsids, facilitating efficient targeting and clearance of viral proteins. (C) In MNV infection, a subset of Atg proteins including Atg5, Atg7, Atg12 and Atg16L1 disrupt viral replication complexes via an IFN-γ-dependent mechanism. In this case, the term ‘adaptor’ may not be relevant because this Atg-dependent antiviral mechanism does not involve autophagosomes. (D) The canonical autophagy machinery also facilitates removal of damaged mitochondria. This depends on mitochondrial adaptors as well as other unknown triggers and sensors.
The Atg machinery functions as a general and highly adaptable intracellular transport system
It is now clear that the autophagy machinery functions in the transport of cytosolic components to various endolysosomal compartments. In the context of viral infection, known transport functions include transport from the cytosol to lysosomes, transport from the cytosol to endosomes that contain TLRs (i.e. TLR-signaling endosomes) and transport from the cytosol to major histocompatibility complex (MHC)-loading endosomes. Such processes maintain order in the cytoplasmic domain, deliver viral ligands to topographically restricted compartments and facilitate interactions without relying on simple diffusion.
Delivery of viral particles to lysosomes
The most direct mechanism by which the autophagy process controls viral replication is direct degradation of viral particles, proteins or replication complexes via delivery to the lysosome. Indeed, several independent lines of evidence have demonstrated that the autophagy machinery sequesters and eliminates both bacterial and viral particles and proteins. This process, termed xenophagy (virophagy), was first demonstrated in Sindbis virus infection. Genetic deletion or impairment of host Atg genes results in increased Sindbis viral protein levels, neuron death and mortality (17, 18). Interestingly, deletion of host Atg proteins had little impact on Sindbis virus replication and facilitated neuronal survival primarily by mediating clearance of accumulated viral proteins (18).
Atg proteins have also been shown to control replication of vesicular stomatitis virus (VSV) in Drosophila (19) but not in its natural, vertebrate hosts (20), implying that many viral pathogens have evolved mechanisms to neutralize the Atg machinery of their natural hosts. Further support of such a model has come from studies in the herpes family of viruses. Infection with a mutant strain of HSV-1 lacking a domain of the infected cell protein 34.5 (ICP34.5) involved in inhibition of autophagy results in decreased neurovirulence and mortality in vivo (21, 22) but no difference in replication in cell lines in vitro (23).
Interestingly, several of the viruses in which the degradative capacity of the autophagy machinery has been shown to decrease morbidity and mortality in animal models are neurotropic viruses (24). We have recently found that upon vaginal infection, autophagy is required to restrict HSV-1 replication in peripheral nervous system neurons but not in the primary target of virus infection, the vaginal keratinocytes (25). Together, these results suggest that autophagy, a non-lytic defense mechanism, is preferentially utilized as a form of innate antiviral defense in post-mitotic, irreplaceable cell types such neurons in HSV-1 infection.
Although it is not the focus of this review, it is important to note that the Atg proteins and autophagy machinery are subverted by numerous viral pathogens to perform functions that assist viral replication. The multifunctional, highly adaptable nature of the Atg machinery likely makes it an ‘easy’ target for such subversion tactics. Numerous viral pathogens, including human immunodeficiency virus-1 (HIV-1), gamma herpes virus-68, hepatitis B virus, hepatitis C virus and poliovirus have evolved mechanisms to abrogate or subvert the autophagy machinery to augment viral replication (26).
Delivery to TLR-signaling compartments
The autophagy machinery has also been shown to play a crucial role in transporting pathogens and their associated pathogen-associated molecular patterns (PAMPs) to innate signaling compartments. In plasmacytoid dendritic cells (pDCs), autophagy plays a critical role in delivery of viral ligands into the TLR7-signaling endosomes, which results in the production of type I interferons and cytokines (Figs 2 and 3).
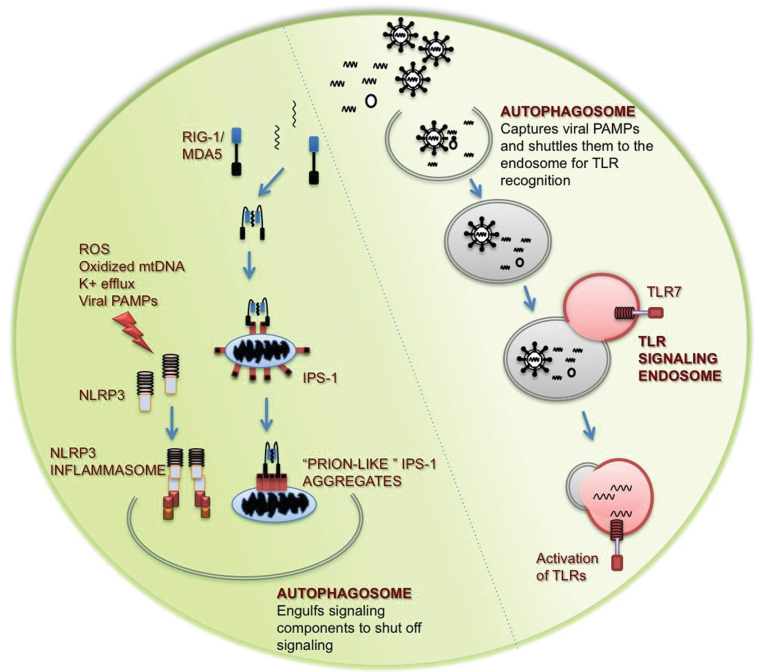
Fig. 2.
Role of the autophagy machinery in innate immune signaling. In endosomal viral recognition through TLRs, autophagy facilitates the transport of viral ligands to the signaling endosome where TLRs recognize viral PAMPs and initiate production of pro-inflammatory cytokines and type I interferons. In contrast, autophagy plays an important role in negatively regulating cytosolic viral recognition. Autophagy indirectly regulates innate antiviral signaling by removal of innate immune signaling complexes.
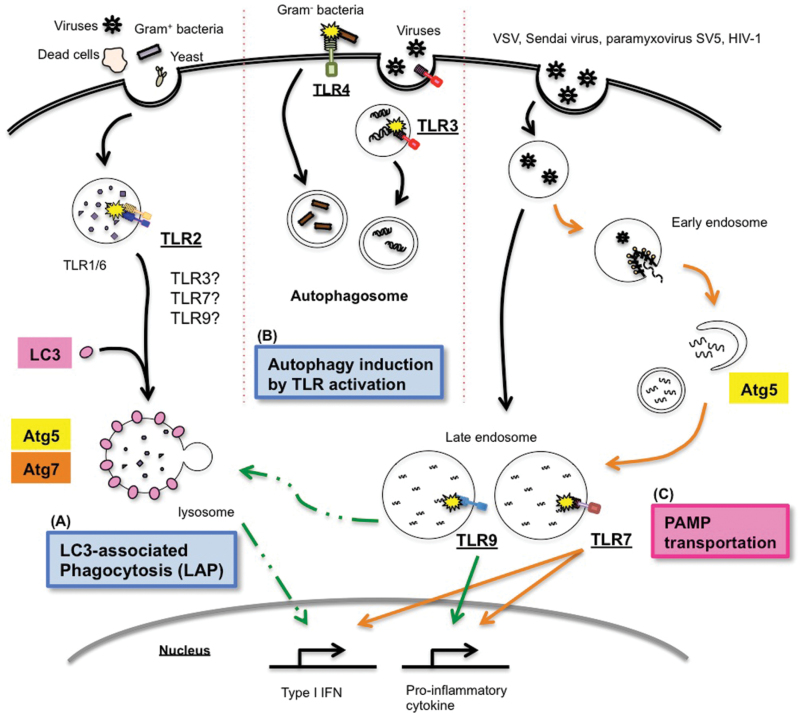
Fig. 3.
TLRs and autophagy in innate immunity. (A) Cell wall components of Gram-positive bacteria and yeast are recognized by TLR2 whose activation triggers the association of LC3 with the phagosomal membrane and promotes phagolysosomal fusion. Phagocytosis of dead cells through recognition of surface PtdSr by Tim4 also utilizes LAP. LAP is also critical for processing and presentation of HSV antigens for MHC II by DCs. (B) Several TLRs are known to induce autophagy. (C) In pDCs, autophagy enables TLR7 recognition of cytosolic virus replication products or cytosolic viral genomes, resulting in both type I interferons and proinflammatory cytokine production (orange lines). In addition, TLR9 signaling for type I interferon production requires Atg5, possibly through the LAP-dependent signaling pathway (dotted green lines). Cytokine induction downstream of TLR9 occurs independently of Atg5 (solid line).
This link was first demonstrated for VSV, where Atg5-deficient pDCs were incapable of recognizing VSV infection through TLR7 (27). In contrast to influenza virus, replication of VSV was essential for TLR7 stimulation. Although viral infection did not change the overall incidence of autophagy in pDCs, pharmacological inhibition of autophagy also abolished IFN-α secretion from wild-type pDCs, suggesting that the autophagy machinery, and not Atg5 per se, is required for TLR7-dependent recognition of VSV. This study thus indicated that autophagy is required for delivery of cytosolic VSV replication intermediates to the TLR7-signaling endosome for recognition.
Additionally, TLR7-dependent IFN-α production requires autophagy in human pDCs following infection with HIV-1 or transfection of HIV-1 RNA (28). Consistent with these findings, Manuse et al. (29) have demonstrated that TLR7-dependent secretion of IFN-α by paramyxovirus Simian virus 5 (SV5) in pDCs depends on autophagy. Curiously, SV5 infection of pDCs did not result in IL-6 secretion, and replication of SV5 was not a prerequisite for TLR7 stimulation. As discussed below, these data are consistent with the role of autophagy in promoting TLR7 and TLR9 signaling for type I interferon production (30) (Fig. 3). These data indicate that autophagy is needed for viral recognition through endosomal TLRs for certain ssRNA viruses.
Delivery to MHC-loading compartments
Viral antigenic peptides are directed into the MHC antigen-presentation pathway through two distinct classical mechanisms: intracellular and extracellular antigens are transported to MHC class I and MHC class II compartments, respectively [reviewed extensively in (31, 32)]. In addition, in certain cell types such as dendritic cells, extracellular antigens can be transported for presentation by MHC I (cross-presentation). Intriguingly, autophagy has also been shown to traffic cytosolic and nuclear antigens into the MHC II antigen-presentation compartments (MIIC) (33–37).
An important example is the Epstein–Barr virus (EBV) nuclear antigen-1 (EBNA-1), which was shown to accumulate in autophagosomes following inhibition of lysosomal acidification by chloroquine treatment (33). Cytosolic antigens captured by autophagosomes may gain access into MIIC either through direct fusion of autophagosomes with MIIC (34) or through fusion of autophagosomes with endosomes (38). In support of the first notion, Schmid et al. (34) demonstrated that under nutrient-rich conditions, autophagosomes constitutively form and fuse with MIIC compartments in antigen-presenting cells (APCs). Furthermore, targeting of influenza matrix protein 1 (MP1) to autophagosomes by fusion of MP1 with LC3 results in increased presentation of MP1 on MHC II. In addition to the role of autophagy in delivering cytosolic antigens to MIIC, LC3-associated phagocytosis (LAP) of extracellular antigens contributes to enhanced processing for presentation on MHC II, as discussed in more detail below.
Unlike the MHC II pathway, the contribution of the Atg machinery to antigen presentation on MHC I is less well understood. Emerging evidence suggests that enhanced autophagy in antigen-donor cells may facilitate antigen cross-presentation and hence antigen presentation on MHC I in DCs following phagocytosis of such cells (39, 40). Of interest, in vivo infection with an HSV-1 mutant strain that has a deletion in the Beclin-1 binding domain of ICP34.5 resulted in enhanced antigen presentation on MHC II and better priming of antiviral CD4+ T cells (41). Thus, intact autophagy in virally infected dying cells may also play an important role in facilitating presentation on MHC II of bystander DCs. In addition, HSV-1 infection induces the formation of distinct four-membrane autophagosome-like structures from the nuclear membrane, which contribute to antigen presentation to CD8+ T cells (42). Interestingly, in this case, antigens processed in autophagosomes still require additional proteasomal processing for efficient presentation. Autophagy thus contributes to presentation on both MHC I and MHC II in the antigen-donor cells and in APCs.
Atg proteins contribute to antiviral immune responses via distinct mechanisms
Atg proteins contribute to antiviral immune responses by three distinct mechanisms: individually, in subgroups and collectively via canonical autophagy. Here, we discuss and provide several illustrative examples of these mechanisms.
Regulation of innate antiviral signaling through canonical autophagy
Canonical autophagy mediates lysosomal degradation of intracellular components and contributes to viral responses via two broad categories: directly via degradation of viruses or viral components (xenophagy, discussed above) and indirectly via regulation of immune signaling. Intracellular antiviral defense relies heavily on the activation of cell-intrinsic viral-recognition pathways. Here, we focus on the retinoic-acid-inducible gene I (RIG-I)-like receptor (RLR) family and the nucleotide-binding oligomerization domain-containing protein (NOD)-like receptors (NLRs), which play an important role in initiating innate and adaptive antiviral immunity (43). These two pathways converge at the mitochondria, which play a key role in shaping the innate response to intracellular pathogens. Mitochondria serve as a signaling platform for innate immune complexes, produce reactive oxygen species (ROS), which further modulate cytokine production, and can initiate cell death pathways (44). Work within the past 5 years has demonstrated that mitochondrial function and innate-immune signaling reciprocally regulate each other (45–49). Importantly, autophagy is the only known mechanism for degradation of whole mitochondria. Perturbations of the core autophagy machinery or deficiencies in selective clearance of mitochondria (mitophagy) thus have important ramifications for RLR and NLR signaling.
The RLR family consists of RIG-I (50, 51) and melanoma differentiation-associated gene 5 (MDA5) (52, 53), which recognize RNA viral PAMPs in the cytosol. Activated RLRs bind to the adaptor protein, IFN-β promoter stimulator 1 (IPS-1) (54–57), located on the mitochondria to induce production of pro-inflammatory cytokines and type I interferons. We have found that autophagy contributes to homeostatic regulation of innate antiviral defense through the clearance of dysfunctional mitochondria, and demonstrated that ROS associated with mitochondria play a key role in potentiating RLR signaling (Figures 1 and 2). Cells defective in autophagy exhibit enhanced RLR signaling and resistance to infection by VSV, mostly because of enhanced mitochondrial ROS-dependent activation of the RIG-I pathway (20). IPS-1 has also been reported to form large prion-like fibrils after viral infection, which potently activate cytokine production and are self-propagating (58). Autophagy may be the only mechanism capable of clearing these prion-like aggregates on the mitochondria and allow a cell to control RLR stimulation (Fig. 2).
NLRs comprise a large family of intracellular pattern-recognition receptors (PRRs) that regulate innate immunity in response to recognition of various PAMPs and stress signals (59). NLRP3 (60) forms an inflammasome with an adaptor, apoptosis-associated speck-like protein containing a caspase-recruitment domain (ASC) and activates Caspase-1-dependent cleavage of potent pro-inflammatory cytokines such as IL-1β. Atg16L1-deficient macrophages and DCs have enhanced production of IL-1β and IL-18 upon LPS stimulation, highlighting the role of macroautophagy in inhibiting NLRP3 inflammasomes (61, 62). NLRP3 infla-mmasomes that can detect viral damage require two signals for activation: signal 1 mediated by TLR or RLR stimulation and signal 2 mediated by damage signals (63). It was recently shown that several mitochondrial components can function as signal 2 (64–66). Three additional key reports have shown that mitochondria play a key role in the activation of NLRP3 inflammasomes through their ROS, mitochondrial DNA (mtDNA) and oxidized mtDNA (65–67). These studies confirmed the accumulation of damaged mitochondria and mitochondrial ROS when autophagy is deficient (65–67) and showed autophagy-mediated regulation of NLRP3 inflammasome-dependent cytokine secretion. Thus, the increased activation of caspase-1 and secretion of IL-1β seen in Atg16L1-deficient cells (61, 62) may be in part explained by the lack of mitophagy.
Activation of NLRP3 stimulates re-localization of NLRP3 to close proximity of the mitochondria, likely for sensing of mitochondrial ROS (66). Further, IPS-1 localization to the mitochondria facilitates RLR signaling at the mitochondrial surface. We suggest that our current understanding of the role of mitochondrial function in antiviral immunity implies the existence of a ‘mitoxosome’, a mitochondrial oxidative signalosome where multiple pathways of viral recognition and cellular stress signals converge on the surface of the mitochondria and where they are integrated to form a coordinated antiviral response (44). This not only allows for the coordination of antiviral signals but also provides a centralized location from which signals can be readily terminated by autophagy to restore homeostasis.
Atg subgroups and LAP
Recent data demonstrated that distinct subgroups of the Atg genes contribute to antiviral immune responses. In vitro and in vivo replication of murine norovirus (MNV) was restricted by a subset of Atg proteins including Atg5, Atg7, Atg12 and Atg16L1, but not Atg4b, implying that induction of canonical autophagy is not required for this process (68) (Fig. 1C). Interestingly, a specific trigger (i.e. IFN-γ) was required for this Atg-dependent control of MNV replication. Atg genes have also been shown to play an important role in the control of bacterial and parasitic intracellular pathogens (69), and a direct inhibitory role for the Atg5–Atg12 conjugate of RLR signaling has also been described (70). This segmentation of the Atg machinery may represent a host countermeasure to circumvent pathogens’ attempt to abrogate autophagy.
Components of the autophagy machinery have also been linked to phagosome maturation, resulting in TLR-dependent pathogen destruction (71). This seminal work demonstrated that macrophages utilize Atg proteins to recruit LC3 to the phagosomal membrane for efficient fusion with lysosomes (Fig. 3). Zymosan-treated RAW264.7 cells showed rapid recruitment of LC3 to zymosan-containing phagosomes and induced phagosome fusion with lysosomes. This was inhibited by genetic deletion of TLR (Tlr2 -/-) or Atg5/Atg7. Intriguingly, no double-membrane structures were observed proximal to LC3+ phagosomes in zymosan-treated cells, suggesting that the TLR-induced LC3 localization to phagosomes occurs independently of canonical autophagy. Furthermore, a recent report indicates that LAP is also associated with dead-cell clearance via Tim4 engagement by phosphatidylserine (PtdSr) (72). Atg5 and Atg7, but not unc51-like kinase 1 (ULK1), which is critical in autophagosome biogenesis, were shown to be involved in phagosomal acidification and accelerated digestion of dead-cell corps upon phagocytosis of dead cells. No double-membrane structure was observed, suggesting that this process resulted from the direct recruitment of Atg proteins to the phagosome membrane. LAP-dependent engulfment and destruction also applies to entosis, which is a live-cell engulfment program between epithelial cells (73). During entosis, the autophagy lipidation machinery and Vps34, but not the mTOR-regulated ULK–ATG13–FIP200 complex, was required for lysosome fusion and the degradation of internalized cargo.
The importance of LAP in viral infection has been shown in two distinct arms: in processing of microbial antigens for MHC II by DCs and in TLR signaling. Mice that have Atg5 deficiency in conventional DCs (cDCs) are impaired in their ability to efficiently prime CD4+ T cells to HSV infection in vivo, because of a decrease in capacity of the DCs to process and present phagocytosed antigen on MHC II (74). This process is likely dependent on LAP but not canonical autophagy, since rapamycin treatment (which induces canonical autophagy) did not enhance antigen presentation on MHC II, whereas TLR engagement within the phagosome was required for the Atg5-dependent enhancement of antigen processing and presentation on MHC II (74). However, the involvement of LAP in this process has not been formally demonstrated.
In addition, we demonstrated that Atg5 is required for the production of IFN-α but not IL-12p40 in pDCs following TLR9 engagement by CpG or HSV-2 (27). Further, recent unpublished data from our laboratory suggest that the LC3 recruitment to TLR9-containing endosomes facilitates TLR9-dependent type I interferon synthesis independently of canonical autophagy. Collectively, LAP plays a key role in phagosome maturation and degradation of internalized cargo, and possibly in phagosomal processing of microbial antigens for MHC II, and in endosomal TLR9 signaling for type I interferon production (Fig. 3).
Singular Atg protein regulation of antiviral immunity
Aside from their function as autophagy machinery components, Atg proteins are independently involved in regulating the magnitude of the host antiviral innate immune response. Upon dsDNA-dependent innate-immune stimulation, stimulator of interferon genes (STING) translocates from the endoplasmic reticulum (ER) to the Golgi apparatus, assembles with TBK1 in cytoplasmic punctate structures and induces type I interferon production. Atg9a and LC3 have recently been shown to co-localize with STING following dsDNA stimulation, thereby inhibiting its association with TBK1 and limiting dsDNA-induced innate immune responses (75). Importantly, Atg9a deficiency significantly increased both STING–TBK1 co-localization and downstream type I interferon production. No double-membrane structures were observed, however, and Atg9a regulation was Atg7-independent, indicating that Atg9a functions as a negative regulator of STING–TBK1-mediated immune responses independent of canonical autophagy.
Triggers, sensors and adaptors mediate antiviral functions of Atg machinery
The Atg machinery achieves temporal, spatial and mechanical specificity via utilization of a diverse set of triggers, sensors and adaptors. Known viral triggers of the autophagy machinery include viral PAMPs, cell stress and cytokines. These triggers are detected by sensors, which facilitate the appropriate activation of a select set of Atg proteins or the entire autophagy machinery. Further specificity is achieved via employment of adaptors that interact with conserved host or viral targets and the Atg proteins to direct viral components to autophagosomes. Autophagy triggers, sensors and adaptors thus operate cooperatively during virus infection to orchestrate efficient and selective targeting, transport and regulation necessary for antiviral defense.
Stress signals as triggers of autophagy
Viruses induce numerous cellular stresses, including oxidative and ER stress. Importantly, both impact innate immune signaling and induce autophagy. As discussed above, ROS not only amplify cytosolic antiviral signaling but also induce autophagy (76). In mammalian cells, ER stress has been shown to amplify innate antiviral signaling through the transcription factor X-box binding protein 1 (77) and has also been shown to induce autophagy (78). It is intriguing to postulate an integrated viral-sensing mechanism, whereby a cell incorporates recognition of viral presence through PRRs with other non-PRR-based indications of virus-induced cellular stress in order to initiate a robust response. Integration of such signals may provide a mechanism to tailor a response towards combating imminent danger (by turning on antiviral genes) versus directing a response towards irreversible damage that marks a point of no return for the cell (by inducing apoptosis).
TLR ligands and cytokines as triggers of autophagy
Stimulation of certain TLRs induces autophagy. TLR-dependent autophagy induction was first reported by Xu et al. (79), who demonstrated that TLR4 signaling induces autophagy in a TRIF-dependent, MyD88-independent manner in RAW264.7 cells. Further mechanistic insight was provided by Delgado et al. (80), who reported that TLR7 activation can also induce autophagy via a MyD88-dependent mechanism in RAW264.7 cells (Fig. 3). Shi and Kehrl (81) demonstrated that LPS stimulation of TLR4 indeed induces autophagy in macrophage cell lines, but observed a MyD88-dependent mechanism, whereas TLR1 and TLR3 stimulation induced autophagy through MyD88- and TRIF-dependent mechanisms, respectively. This report further demonstrated that MyD88 and TRIF engagement facilitates their interaction with Beclin-1, a key initiator of autophagy. This interaction induces autophagy by relieving Beclin-1 from its association with Bcl2. In addition, other immune receptors including CD40 and the B-cell receptor have been shown to induce autophagy (82).
Certain cytokine receptor signaling leads to the induction of autophagy. IFN-γ has been shown to induce autophagy in several contexts (83, 84) whereas the Th2 cytokines IL-4 and IL-5 have been shown to reduce autophagy upon starvation or upon IFN-γ stimulation (85). TNF-α has also been shown to induce autophagy in skeletal muscle cells as well as enhance antigen presentation on MHC class II (86, 87). In addition to cytokines, chemokine-receptor signaling induces autophagy. The HIV-1 envelope binds to CXCR4 and induces autophagy and cell death in bystander CD4+ T cells (88). Collectively, these findings indicate that different cytokine and chemokine receptors can induce or block autophagy. It is, however, important to note that immune receptor-dependent induction of autophagy appears to be dependent on both cell type and context (89). More studies are thus needed to delineate the molecular mechanism of autophagy induction by immune and cytokine-receptor engagement.
Adaptors for mitochondria
Several lines of evidence have demonstrated that mitochondria are specifically recognized and degraded through autophagy (mitophagy). There are developmental stages, such as mammalian erythroid and reticulocyte maturation, where Bnip3L (NIX) facilitates targeting all of the mitochondria for degradation (14, 15). Beyond these extreme examples of mitophagy, selective mitochondrial turnover is an essential part of intracellular quality control. Mitophagy of damaged mitochondria has been reported for depolarized mitochondria, which are rapidly sequestered and degraded (90). Mitochondrial dynamics are also important in facilitating selective mitophagy, where the mitochondria undergo asymmetrical fission, followed by a selective fusion of damaged components that specifically target the damaged mitochondria for autophagy (91).
Exciting developments of late have begun to reveal the mechanism(s) by which mitochondria are selectively removed by autophagy. In neurons and several other cell types, proteins that are critical in targeting selective clearance of damaged mitochondria include Parkin and PTEN-induced kinase 1 (PINK1) (92). Additional proteins that have been identified in mediating mitophagy include Atg3 and p62 as well as a number of recently identified proteins that are involved in one or more types of selective autophagy, including mitophagy (11). Of note, of the 141 genes identified for virophagy, 96 genes were also required for mitophagy (11), indicating a major overlap between molecules utilized for these two types of selective autophagy.
Adaptors for viruses
As discussed above, autophagy has been shown to degrade viral particles and proteins by a process termed xenophagy or virophagy. Such clearance could occur by two distinct mechanisms: bulk (non-selective) autophagy and selective autophagy mediated by specific recognition of viral components or patterns. Although both mechanisms could remove viral particles, rapid and efficient removal would require the latter. Earlier studies implied that non-selective viral clearance might predominate, as autophagosomes observed by EM were found to contain viral particles as well as other cytoplasmic components such as mitochondria (21). Several recent intriguing studies have, however, demonstrated that at least in the case of Sindbis virus and HSV-1, viral particles are selectively targeted to autophagosomes for degradation by the adaptor proteins p62 (Sindbis) (18) and SMURF-1 (HSV-1 and Sindbis) (11) (Fig. 1B). p62 is a polyubiquitin-binding protein that is degraded by autophagy. It binds directly to LC3, which is covalently coupled to the autophagosomal membrane, and facilitates engulfment of a variety of polyubiquitinated substrates (10), including Sindbis virus capsids. Of note, SMURF-1 was identified as one of many key facilitators of selective autophagy, providing a wealth of targets for future studies aimed at understanding the mechanism of targeting and degradation of viral pathogens (93). Whether other adaptors (Fig. 1) are directly involved in virophagy and whether specific adaptors exist for different classes of viruses will be important to determine in future studies.
Conclusions
Recent studies have revealed molecular details of interactions between autophagy and innate virus-recognition pathways. The role of autophagy in the virus–host relationship is precisely tuned by contextual activation of triggers and adaptors. Consequently, gross induction (or conversely, inhibition) of canonical autophagy is unlikely to make significant headway as a treatment for most viral infections. Rapidly increasing understanding of the underpinning molecular pathways can, nevertheless, be leveraged into effective antiviral therapeutics in the future.
In this regard, HSV-1 provides an informative example of the promise and challenges of translating basic understanding of autophagy–virus interactions into novel therapeutics. The initial discovery that HSV-1 has at least two strategies to abrogate host autophagy (22) strongly implied that autophagy plays a critical role in anti-HSV immune responses. Subsequent studies have revealed that autophagy plays key roles in direct control of HSV-1 replication in neurons (16) and in activating CD4+ T cells (41). Further work has identified SMURF-1 as a key adaptor involved in targeting HSV-1 to autophagosomes for degradation (93). Further understanding of the molecular mechanisms governing interactions between autophagy and viruses and the development and delivery of specific agonists/antagonists are both necessary to translate our understanding into viable therapies.
Funding
This review was supported by grants from the National Institutes of Health to A.I. (AI081884, AI054359, AI062428 and AI064705) and to M.T. (F31 AG039163).
Acknowledgements
B.Y. was supported by National Institutes of Health Predoctoral Virology Training grant T32 AI055403, and O.A. is supported by the National Institutes of Health (NIH) National Research Service Award (T32AI07019) from the Interdisciplinary Immunology Training Program at Yale University. K.H. is supported by a Nakajima Foundation fellowship. A.I. is a recipient of the Burroughs Wellcome Investigator in Pathogenesis of Infectious Diseases. Research in the laboratory is supported by the NIH (grant numbers AI081884, AI054359, AI062428, and AI064705), and under Award Number U54AI057160 to the Midwest Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research (MRCE). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The authors declare no conflicts of interest.
References
- 1. Levine B., Klionsky D. J. 2004. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell 6: 463 [DOI] [PubMed] [Google Scholar]
- 2. Mizushima N., Levine B., Cuervo A. M., Klionsky D. J. 2008. Autophagy fights disease through cellular self-digestion. Nature 451: 1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mizushima N. 2007. Autophagy: process and function. Genes Dev. 21: 2861 [DOI] [PubMed] [Google Scholar]
- 4. Klionsky D. J., Emr S. D. 2000. Autophagy as a regulated pathway of cellular degradation. Science 290: 1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Levine B., Klionsky D. J. 2004. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell 6: 463 [DOI] [PubMed] [Google Scholar]
- 6. Stetson D. B., Medzhitov R. 2006. Type I interferons in host defense. Immunity 25: 373 [DOI] [PubMed] [Google Scholar]
- 7. Yu C., Chiang R., Chang T., Liao C., Lin Y. 2010. The interferon stimulator mitochondrial antiviral signaling protein facilitates cell death by disrupting the mitochondrial membrane potential and by activating caspases. J. Virol. 84: 2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sumpter R., Levine B. 2010. Autophagy and innate immunity: triggering, targeting and tuning. Semin. Cell Dev. Biol. 21: 699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Noda N. N., Ohsumi Y., Inagaki F. 2010. Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 584: 1379 [DOI] [PubMed] [Google Scholar]
- 10. Pankiv S., Clausen T. H., Lamark T., et al. 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy*[S]. J. Biol. Chem. 282: 24131 [DOI] [PubMed] [Google Scholar]
- 11. Orvedahl A., Sumpter R., Jr, Xiao G., et al. 2011. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature 480: 113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Thurston T. L., Ryzhakov G., Bloor S., von Muhlinen N., Randow F. 2009. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat. Immunol. 10: 1215 [DOI] [PubMed] [Google Scholar]
- 13. Kirkin V., Lamark T., Sou Y., et al. 2009. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell 33: 505 [DOI] [PubMed] [Google Scholar]
- 14. Schweers R. L., Zhang J., Randall M. S., et al. 2007. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc. Natl. Acad. Sci. U S A 104: 19500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sandoval H., Thiagarajan P., Dasgupta S. K., et al. 2008. Essential role for Nix in autophagic maturation of erythroid cells. Nature 454: 232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wild P., Farhan H., McEwan D. G., et al. 2011. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 333: 228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liang X. H., Kleeman L. K., Jiang H. H., et al. 1998. Protection against fatal sindbis virus encephalitis by Beclin, a novel Bcl-2-interacting protein. J. Virol. 72: 8586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Orvedahl A., MacPherson S., Sumpter R., Jr, Tallóczy Z., Zou Z., Levine B. 2010. Autophagy protects against sindbis virus infection of the central nervous system. Cell Host Microbe 7: 115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shelly S., Lukinova N., Bambina S., Berman A., Cherry S. 2009. Autophagy is an essential component of drosophila immunity against vesicular stomatitis virus. Immunity 30: 588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tal M. C., Sasai M., Lee H. K., Yordy B., Shadel G. S., Iwasaki A. 2009. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc. Natl. Acad. Sci. U S A 106: 2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Talloczy Z., Virgin H. W., IV, Levine B. 2006. PKR-dependent autophagic degradation of herpes simplex virus type 1. Autophagy 2: 24 [DOI] [PubMed] [Google Scholar]
- 22. Orvedahl A., Alexander D., Tallóczy Z., et al. 2007. HSV-1 ICP34.5 confers neurovirulence by targeting the beclin 1 autophagy protein. Cell Host Microbe 1: 23 [DOI] [PubMed] [Google Scholar]
- 23. Alexander D. E., Ward S. L., Mizushima N., Levine B., Leib D. A. 2007. Analysis of the role of autophagy in replication of herpes simplex virus in cell culture. J. Virol. 81: 12128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Orvedahl A., Levine B. 2008. Autophagy and viral neurovirulence. Cell Microbiol 10: 1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yordy B., Iijima N., Huttner A., Leib D., Iwasaki A. 2012. A neuron-specific role for autophagy in antiviral defense against herpes simplex virus. Cell Host & Microbe, 12::334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yordy B., Iwasaki A. 2011. Autophagy in the control and pathogenesis of viral infection. Curr. Opin. Virol. 1: 196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee H. K., Lund J. M., Ramanathan B., Mizushima N., Iwasaki A. 2007. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science 315: 1398 [DOI] [PubMed] [Google Scholar]
- 28. Zhou D., Kang K. H., Spector S. A. 2012. Production of interferon α by human immunodeficiency virus type 1 in human plasmacytoid dendritic cells is dependent on induction of autophagy. J. Infect. Dis. 205: 1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Manuse M. J., Briggs C. M., Parks G. D. 2010. Replication-independent activation of human plasmacytoid dendritic cells by the paramyxovirus SV5 requires TLR7 and autophagy pathways. Virology 405: 383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Iwasaki A. 2007. Role of autophagy in innate viral recognition. Autophagy 3: 354 [DOI] [PubMed] [Google Scholar]
- 31. Pamer E., Cresswell P. 1998. Mechanisms of MHC class I-restricted antigen processing. Annu. Rev. Immunol. 16: 323 [DOI] [PubMed] [Google Scholar]
- 32. Germain R. N., Margulies D. H. 1993. The biochemistry and cell biology and antigen processing and presentation. Annu. Rev. Immunol. 11: 403 [DOI] [PubMed] [Google Scholar]
- 33. Paludan C., Schmid D., Landthaler M., et al. 2005. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science 307: 593 [DOI] [PubMed] [Google Scholar]
- 34. Schmid D., Pypaert M., Munz C. 2007. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity 26: 79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Morris S., Swanson M. S., Lieberman A., et al. 2011. Autophagy-mediated dendritic cell activation is essential for innate cytokine production and APC function with respiratory syncytial virus responses. J. Immunol. 187: 3953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Crotzer V. L., Blum J. S. 2009. Autophagy and its role in MHC-mediated antigen presentation. J. Immunol. 182: 3335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Munz C. 2012. Antigen processing for MHC class II presentation via autophagy. Front. Immunol. 3: 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Berg T. O., Fengsrud M., Stromhaug P. E., Berg T., Seglen P. O. 1998. Isolation and characterization of rat liver amphisomes. Evidence for fusion of autophagosomes with both early and late endosomes. J. Biol. Chem. 273: 21883 [DOI] [PubMed] [Google Scholar]
- 39. Li Y., Wang L. X., Yang G., Hao F., Urba W. J., Hu H. M. 2008. Efficient cross-presentation depends on autophagy in tumor cells. Cancer Res 68: 6889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Uhl M., Kepp O., Jusforgues-Saklani H., Vicencio J. M., Kroemer G., Albert M. L. 2009. Autophagy within the antigen donor cell facilitates efficient antigen cross-priming of virus-specific CD8+ T cells. Cell Death Differ 16: 991 [DOI] [PubMed] [Google Scholar]
- 41. Leib D. A., Alexander D. E., Cox D., Yin J., Ferguson T. A. 2009. Interaction of ICP34.5 with Beclin 1 modulates herpes simplex virus type 1 pathogenesis through control of CD4+ T-cell responses. J. Virol. 83: 12164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. English L., Chemali M., Duron J., et al. 2009. Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat. Immunol. 10: 480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Iwasaki A., Medzhitov R. 2010. Regulation of adaptive immunity by the innate immune system. Science 327: 291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tal M. C., Iwasaki A. 2011. Mitoxosome: a mitochondrial platform for cross-talk between cellular stress and antiviral signaling. Immunol. Rev. 243: 215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ohman T., Rintahaka J., Kalkkinen N., Matikainen S., Nyman T. A. 2009. Actin and RIG-I/MAVS signaling components translocate to mitochondria upon influenza A virus infection of human primary macrophages. J. Immunol. 182: 5682 [DOI] [PubMed] [Google Scholar]
- 46. Castanier C., Garcin D., Vazquez A., Arnoult D. 2010. Mitochondrial dynamics regulate the RIG-I-like receptor antiviral pathway. EMBO Rep 11: 133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Koshiba T., Yasukawa K., Yanagi Y., Kawabata S. 2011. Mitochondrial membrane potential is required for MAVS-mediated antiviral signaling. Sci. Signal. 4::7. [DOI] [PubMed] [Google Scholar]
- 48. Yasukawa K., Oshiumi H., Takeda M., et al. 2009. Mitofusin 2 inhibits mitochondrial antiviral signaling. Sci. Signal. 2::47. [DOI] [PubMed] [Google Scholar]
- 49. Onoguchi K., Onomoto K., Takamatsu S., et al. 2010. Virus-infection or 5′ppp-RNA activates antiviral signal through redistribution of IPS-1 mediated by MFN1. PLoS Pathog 6: 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pichlmair A., Schulz O., Tan C. P., et al. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5'-phosphates. Science 314: 997 [DOI] [PubMed] [Google Scholar]
- 51. Hornung V., Ellegast J., Kim S., et al. 2006. 5'-Triphosphate RNA is the ligand for RIG-I. Science 314: 994 [DOI] [PubMed] [Google Scholar]
- 52. Kato H., Takeuchi O., Sato S., et al. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441: 101 [DOI] [PubMed] [Google Scholar]
- 53. Gitlin L., Barchet W., Gilfillan S., et al. 2006. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl. Acad. Sci. U S A 103: 8459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kawai T., Takahashi K., Sato S., et al. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6: 981 [DOI] [PubMed] [Google Scholar]
- 55. Seth R. B., Sun L., Ea C., Chen Z. J. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF3. Cell 122: 669 [DOI] [PubMed] [Google Scholar]
- 56. Meylan E., Curran J., Hofmann K., et al. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437: 1167 [DOI] [PubMed] [Google Scholar]
- 57. Xu L. G., Wang Y. Y., Han K. J., Li L. Y., Zhai Z., Shu H. B. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell 19: 727 [DOI] [PubMed] [Google Scholar]
- 58. Hou F., Sun L., Zheng H., Skaug B., Jiang Q. X., Chen Z. J. 2011. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell 146: 448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Martinon F., Mayor A., Tschopp J. 2009. The inflammasomes: guardians of the body. Annu. Rev. Immunol. 27: 229 [DOI] [PubMed] [Google Scholar]
- 60. Ting J. P., Lovering R. C., Alnemri E. S., et al. 2008. The NLR gene family: a standard nomenclature. Immunity 28: 285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Saitoh T., Fujita N., Jang M. H., et al. 2008. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature 456: 264 [DOI] [PubMed] [Google Scholar]
- 62. Harris J., Hartman M., Roche C., et al. 2011. Autophagy controls IL-1β secretion by targeting pro-IL-1β for degradation. J. Biol. Chem. 286: 9587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ichinohe T. 2010. Respective roles of TLR, RIG-I and NLRP3 in influenza virus infection and immunity: impact on vaccine design. Expert Rev. Vaccines 9: 1315 [DOI] [PubMed] [Google Scholar]
- 64. Van Bruggen R., Köker M. Y., Jansen M., et al. 2010. Human NLRP3 inflammasome activation is Nox1-4 independent. Blood 115: 5398 [DOI] [PubMed] [Google Scholar]
- 65. Nakahira K., Haspel J. A., Rathinam V. A. K., et al. 2011. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat. Immunol. 12: 222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhou R., Yazdi A. S., Menu P., Tschopp J. 2011. A role for mitochondria in NLRP3 inflammasome activation. Nature 469: 221 [DOI] [PubMed] [Google Scholar]
- 67. Shimada K., Crother T. R., Karlin J., et al. 2012. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity 36: 401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hwang S., Maloney N. S., Bruinsma M. W., et al. 2012. Nondegradative role of Atg5-Atg12/Atg16L1 autophagy protein complex in antiviral activity of interferon gamma. Cell Host Microbe 11: 397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhao Z., Fux B., Goodwin M., et al. 2008. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe 4: 458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ponpuak M., Davis A. S., Roberts E. A., et al. 2010. Delivery of cytosolic components by autophagic adaptor protein p62 endows autophagosomes with unique antimicrobial properties. Immunity 32: 329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sanjuan M. A., Dillon C. P., Tait S. W. G., et al. 2007. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature 450: 1253 [DOI] [PubMed] [Google Scholar]
- 72. Martinez J., Almendinger J., Oberst A., et al. 2011. Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc. Natl. Acad. Sci. U S A 108: 17396 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 73. Florey O., Kim S. E., Sandoval C. P., Haynes C. M., Overholtzer M. 2011. Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes. Nat. Cell Biol. 13: 1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lee H. K., Mattei L. M., Steinberg B. E., et al. 2010. In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity 32: 227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Saitoh T., Fujita N., Hayashi T., et al. 2009. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc. Natl. Acad. Sci. U S A 106: 20842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Scherz-Shouval R., Shvets E., Fass E., Shorer H., Gil L., Elazar Z. 2007. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J 26: 1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Smith J. A., Turner M. J., DeLay M. L., Klenk E. I., Sowders D. P., Colbert R. A. 2008. Endoplasmic reticulum stress and the unfolded protein response are linked to synergistic IFN-beta induction via X-box binding protein 1. Eur. J. Immunol. 38: 1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Verfaillie T., Salazar M., Velasco G., Agostinis P. 2010. Linking ER stress to autophagy: potential implications for cancer therapy. Int. J. Cell Biol. 2010: 930509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Xu Y., Jagannath C., Liu X., Sharafkhaneh A., Kolodziejska K. E., Eissa N. T. 2007. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity 27: 135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Delgado M. A., Elmaoued R. A., Davis A. S., Kyei G., Deretic V. 2008. Toll-like receptors control autophagy. EMBO J. 27: 1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Shi C., Kehrl J. H. 2008. MyD88 and Trif target Beclin 1 to trigger autophagy in macrophages. J. Biol. Chem. 283: 33175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Watanabe K., Tsubata T. 2009. Autophagy connects antigen receptor signaling to costimulatory signaling in B lymphocytes. Autophagy 5: 108 [DOI] [PubMed] [Google Scholar]
- 83. Gutierrez M. G., Master S. S., Singh S. B., Taylor G. A., Colombo M. I., Deretic V. 2004. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119: 753 [DOI] [PubMed] [Google Scholar]
- 84. Deretic V., Levine B. 2009. Autophagy, immunity, and microbial adaptations. Cell Host Microbe 5: 527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Harris J., De Haro S. A., Master S. S., et al. 2007. T helper 2 cytokines inhibit autophagic control of intracellular Mycobacterium tuberculosis. Immunity 27: 505 [DOI] [PubMed] [Google Scholar]
- 86. Keller C. W., Fokken C., Turville S. G., et al. 2011. TNF-α induces macroautophagy and regulates MHC class II expression in human skeletal muscle cells. J. Biol. Chem. 286: 3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Djavaheri-Mergny M., Amelotti M., Mathieu J., et al. 2006. NF-κB activation represses tumor necrosis factor-α-induced autophagy. J. Biol. Chem. 281: 30373 [DOI] [PubMed] [Google Scholar]
- 88. Espert L., Denizot M., Grimaldi M., et al. 2006. Autophagy is involved in T cell death after binding of HIV-1 envelope proteins to CXCR4. J. Clin. Invest. 116: 2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Saitoh T., Akira S. 2010. Regulation of innate immune responses by autophagy-related proteins. J. Cell. Biol. 189: 925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Rodriguez-Enriquez S., Kim I., Currin R. T., Lemasters J. J. 2006. Tracker dyes to probe mitochondrial autophagy (mitophagy) in rat hepatocytes. Autophagy 2: 39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Twig G., Elorza A., Molina A. J. A., et al. 2008. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J 27: 433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Youle R. J., Narendra D. P. 2011. Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 12: 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Orvedahl A., Sumpter R., Jr, Xiao G., et al. 2011. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature 480: 113 [DOI] [PMC free article] [PubMed] [Google Scholar]