Abstract
The heritability of most common, multifactorial diseases is rather modest and known genetic effects account for a small part of it. The remaining portion of disease aetiology has been conventionally ascribed to environmental effects, with an unknown part being stochastic. This review focuses on recent studies highlighting stochastic events of potentially great importance in human disease—the accumulation of post-zygotic structural aberrations with age in phenotypically normal humans. These findings are in agreement with a substantial mutational load predicted to occur during lifetime within the human soma. A major consequence of these results is that the genetic profile of a single tissue collected at one time point should be used with caution as a faithful portrait of other tissues from the same subject or the same tissue throughout life. Thus, the design of studies in human genetics interrogating a single sample per subject or applying lymphoblastoid cell lines may come into question. Sporadic disorders are common in medicine. We wish to stress the non-heritable genetic variation as a potentially important factor behind the development of sporadic diseases. Moreover, associations between post-zygotic mutations, clonal cell expansions and their relation to cancer predisposition are central in this context. Post-zygotic mutations are amenable to robust examination and are likely to explain a sizable part of non-heritable disease causality, which has routinely been thought of as synonymous with environmental factors. In view of the widespread accumulation of genetic aberrations with age and strong predictions of disease risk from such analyses, studies of post-zygotic mutations may be a fruitful approach for delineation of variants that are causative for common human disorders.
Keywords: Genetics, Genome-wide, Clinical genetics, Complex traits, Copy-number
Introduction
Over the past three decades, projects in human genetics searching for genotype–phenotype correlations have mostly focused on analyses of the inherited genome. These include studies of genes causing monogenic disorders and more recent analyses of the association of complex diseases with single nucleotide polymorphisms (SNP) in genome-wide association studies (GWAS). The prevailing approach has been analysis of DNA from a single tissue (usually blood) sampled at a single time point (non-longitudinal sampling). The general foundation and rationale for these studies has been the assumption that the vast majority of cells in the human soma are genetically identical; in other words, that the genome of somatic cells is stable across the human lifespan. In this review we discuss recent findings that challenge this assumption1–3 and argue that post-zygotic changes represent an underestimated source of variation responsible for the development of human phenotypes. In recent years, the GWAS have dominated the human medical genetic landscape of complex diseases and have, notwithstanding their shortcomings, contributed to our knowledge of human genetics.4 They have improved our understanding of the genetic basis of many human traits, as >1200 variants associated with >165 different human traits and diseases have been described.4–8 However, to the chagrin of the field, the portion of the estimated heritability explained by the GWAS findings has been unexpectedly low. Many explanations have been proposed for the ‘missing heritability’ of complex traits, including human disease.4–8 Faced with the inefficiency with which inherited biology explains and predicts disease, we argue that the weight should shift to the non-inherited component which, until now, has routinely been thought of as synonymous with environmental factors.
Post-zygotic DNA sequence mutations, although known to occur in normal cells, were not considered to be a major factor behind common diseases, but recent evidence seriously challenges this belief.1–3 This review has been inspired by our results1 and two other papers supporting and extending our conclusions,2 3 showing an age dependent accumulation of post-zygotic mutations in non-tumoral cell lines constituting the human soma. Our focus is to highlight the importance of somatic mosaicism as a potentially crucial factor causing complex human diseases. According to a common metaphor ‘A beloved child is called many things’; the phenomenon that is discussed here has many names—for example, somatic mosaicism, somatic variation, post-zygotic changes, de novo variants, aberrations acquired during lifetime, and detectable clonal mosaicism. All these terms fall into a definition of mosaicism as the presence of genetically distinct lineages of cells in a single organism that is derived from the same zygote. We use here ‘post-zygotic variation’ or ‘post-zygotic mosaicism’ as unifying terms for all DNA changes acquired during life, from single base pair mutations to aberrations at the chromosomal level. The term ‘mosaicism’ was first used in biology in the end of the 19th century by W Roux and A Weismann to describe differential usage of genetic information during development. This incorrect explanation of mosaic development and ontogenetic differentiation became later known as the Roux-Weismann theory of qualitative nuclear division.9 More recently, in 1956 CW Cotterman used the term ‘somatic mosaicism’ to define antigenic variation.10
Post-zygotic mosaicism has been studied in human embryos,11 12 fetuses from spontaneous abortions,13 and children with birth defects or developmental delay.14 15 However, until recently,1–3 little has been known about post-zygotic mosaicism in human adult and aging but otherwise healthy individuals. This review does not focus on de novo mutations in the germline that are known to cause monogenic autosomal dominant and X-linked diseases, or those recently found to be part of the aetiology of neurodevelopmental diseases. For the latter, we refer to a recent review on this topic.16 Likewise, we do not discuss paternal age effect mutations and selfish spermatogonial selection in relation to various human disorders.17 There are two well known examples of physiological and locus specific post-zygotic variation in the nuclear genome. The first are somatic rearrangements of immunoglobulin (Ig) and T cell receptor (TCR) genes in B and T lymphocytes. The Ig and TCR genes are inactive in most cells, but undergo a tightly regulated reshuffling in order to become activated, which leads to individual B or T lymphocytes producing a mono-specific antibody or TCR, respectively.18 The second example is the variation of telomere length; a special case of structural post-zygotic change. The length of telomeres functions as a clock for the number of cell divisions, limiting the replicative capacity of cells, which is important for cell senescence, aging, and cancer.19–23 All other known examples of post-zygotic variation, which is a focus of this review, are apparently a result of stochastic, random processes.
An adult human body has been estimated to contain 1013–1014 cells and the number of cells produced during a human lifetime is assessed as more than 1016. Each somatic cell division is inherently coupled with a risk for mutations and there are estimates of the number of mutations that could be expected to arise during human life.24–26 We quote from Lynch 201026: “… with a human germ-line mutation rate of ∼10−8 base substitutions/site/generation, a site in a somatic nucleus will be mutated with a probability of 10−7 to 10−6 by the average age of reproduction, with the burden being higher in older individuals. With a diploid genome size of 6×109 sites and ∼1013 cells per soma, the body of a middle-aged human might then contain >1016 mutations (not including insertions, deletions, or other larger scale mutations). Only about 1% of the human genome consists of coding DNA, so a substantial fraction of somatic mutations will be inconsequential, but even if just 1% of coding mutations had significant fitness effects, the total body burden of mutations would be of order 1012”. The above numbers have been calculated based on studies of single nucleotide variants. It should be stressed that structural variants, although less well studied than single nucleotide polymorphisms (SNPs), are estimated to be more common. Comparisons of germline mutation frequencies of SNPs versus copy number variations (CNVs) indicate that the latter are more common by a few orders of magnitude.27 28 Furthermore, the base substitutional mutation rate per cell division in somatic cells is 4–25 times greater than the corresponding rate for germline (reviewed in Lynch25). Thus, the predicted burden of post-zygotic mutations in the human soma during a single lifetime is overwhelming.
Given this vast amount of expected variation, it is likely that a considerable part of these events have consequences for cellular phenotypes. However, for a phenotype to occur at the level of an organism, a mutation should strike a substantial number of cells, which are in an appropriate spatial and temporal window of development. It might be helpful to consider the above numbers using an analogy with Darwinian selection. During evolution of species, most new mutations are either disadvantageous to the organism (eliminated from the gene pool because of their negative effect on fitness) or are neutral passengers, not providing an advantage or disadvantage, and are therefore not leading to their relative increase in the gene pool. Only a minority of new mutations are propagated in following generations, by increasing the fitness of the affected organism and its progeny. Similar reasoning might be applied to the post-zygotic mutations within a human soma. It is likely that a large group of post-zygotic mutations are never detected because of their detrimental effect on the affected cell and its elimination by apoptosis/growth arrest. The phenotypically neutral passenger mutations are not easily studied either, since they are not increasing in the relative frequency of the affected cell clone over all other cells. The only mutations that are readily detectable are those providing the affected cell with a proliferative advantage and this has been known to be the main mechanism of tumorigenesis. The three recent studies1–3 show that this can also occur in lineages of normal cells in healthy individuals.
Recent findings on post-zygotic variation in phenotypically normal human cells
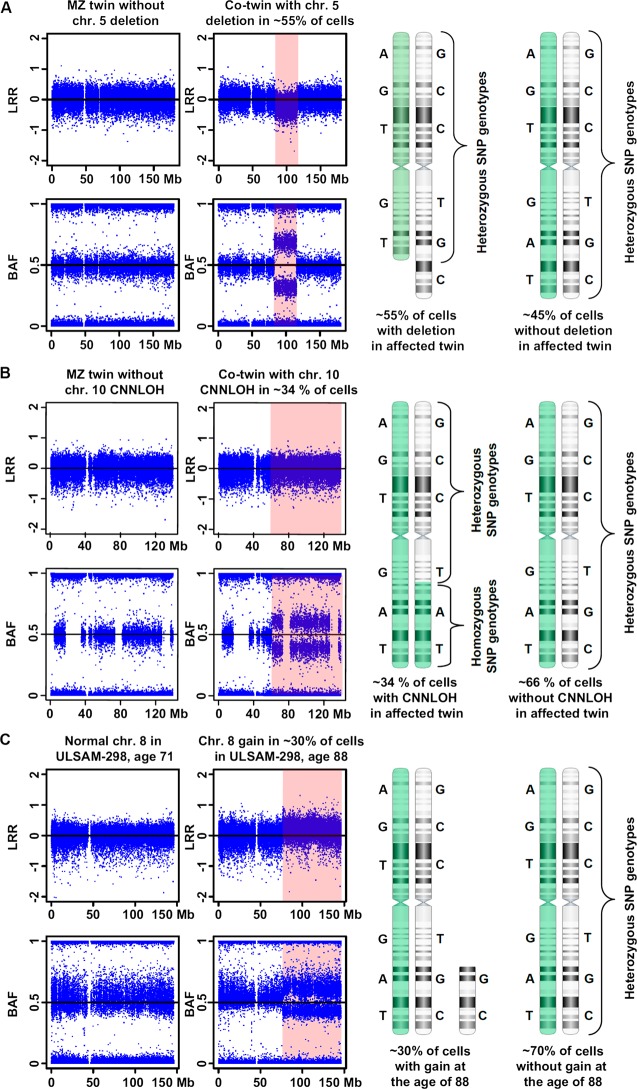
The papers that prompted this review1–3 are the latest contributions towards increasing awareness of post-zygotic variation as a widespread and easily detectable phenomenon with potentially important consequences for various human phenotypes.29–39 The three papers showed that normal cells accumulate structural aberrations with age, which are readily identified using genome scanning on SNP arrays. These structural changes fall into three major categories: deletions, gains, and copy number neutral loss of heterozygosity (CNNLOH, also called acquired uniparental disomy, aUPD) (figure 1). The size of these aberrations is highly variable, from a few kb to entire chromosomes. The relationship between age and mosaicism is strong and other tested co-variants, such as sex, ancestry, and smoking, did not have a significant effect on the mosaic status. A common thread in these reports is the detection of clonal expansions of blood cells that were affected with various aberrations, suggesting that these mutations convey a proliferative advantage for the cells carrying them. Forsberg et al1 showed the highest frequency of subjects affected with aberrations; that is, 3.4% of generally healthy people in the window of 55–90 years old show clones of nucleated cells containing megabase-range changes, which affect up to 60% of nucleated cells in blood. This number of ∼3% for mosaic mega-base range aberrations occurring among elderly/old subjects should be compared to ∼1% of mosaics for chromosomal aberrations described in a preselected cohort of children referred for clinical diagnostic testing.14 In addition, Forsberg et al1 showed, using a unique cohort of age stratified monozygotic twins sampled several times, that smaller structural aberrations (in the range a few kb) also accumulate with age, as they appear much more common in older subjects.
Figure 1.
An illustration of the three main types of post-zygotic structural genetic aberrations, selected from Forsberg et al.1 Panels A, B and C display a deletion, a copy number neutral loss of heterozygosity (CNNLOH, also called acquired uniparental disomy, aUPD), and a gain, respectively. Each panel is composed of images from Illumina single nucleotide polymorphism (SNP) beadchips showing a selected aberrant chromosome, with the affected regions highlighted in pink. The results from Illumina SNP arrays consist of two data tracks: log R ratio (LRR) values of fluorescent intensities from array probes (upper part), and B allele frequency (BAF) values representing the fraction of fluorescent intensity at each SNP accounted for by the B allele (lower part). Normally, BAF values cluster around 0 (AA genotype), 0.5 (AB) or 1 (BB). On the right hand side, a schematic explanatory figure displaying the mosaic mixture of cells with aberrant and wild-type chromosomes is shown. Two hypothetical homologous chromosomes (labelled in green and white) with heterozygous genotypes for six SNPs are shown. Panel A shows data for chromosome 5 in a monozygotic (MZ) twin pair sampled at the age of 77 years. MZ twin TP25-1 has a normal profile, while its co-twin TP25-2 has a 32.5 Mb deletion on 5q in approximately 55% of nucleated blood cells. This deletion is uncovered using both LRR (downward shift) and BAF (heterozygous SNPs cluster away from 0.5) data from the Illumina SNP array. The Illumina profile contains a mixture of genotypes from aberrant cells (approximately 55%) and wild-type cells representing approximately 45% of nucleated blood cells. Panel B shows data for chromosome 10 in MZ twin pair sampled at the age of 77 years. Twin TP12-1 shows a normal profile. Using BAF values a 76.5 Mb large CNNLOH/aUPD was identified on 10q in co-twin TP12-2. Quantification of cells containing the CNNLOH/aUPD suggests that 34% of cells are affected. As this aberration does not change the copy number of the aberrant segment, LRR values are normal. However, the genotypes of SNPs within this segment are all homozygous in aberrant cells. Panel C shows data for chromosome 8 from subject ULSAM-298 using two samples collected at the ages of 71 and 88 years. The sample collected at 71 years shows a normal profile, while the sample taken at the age of 88 years shows a 70 Mb gain of chromosome 8 in approximately 30% of cells, visible with both LRR and BAF data from the Illumina SNP array.
Comparison between frequencies of the three main classes of mega-base range structural mutations showed that deletions are far more common than gains. Another prominent finding is the high frequency of CNNLOH/aUPD. Forsberg et al,1 Laurie et al2 and Jacobs et al3 reported that CNNLOH/aUPD represent 22%, 34% and 48% of all mutations, respectively. Different scoring algorithms might explain differences between these three studies. It should also be pointed out that in cases where only a few percent of cells are affected, it might be difficult to discriminate CNNLOH/aUPD from a gain or a deletion event. Nevertheless, CNNLOH/aUPD appears to be a major class of somatic mutations, either the most common or second most common in frequency among the mega-base range aberrations. The simplest definition of CNNLOH/aUPD in the context of a single affected chromosome is the presence of both homologues of a pair of chromosomes from one parent only.40 CNNLOH/aUPD can affect the entire chromosome or smaller segments (segmental CNNLOH/aUPD, terminal or interstitial) with stretches of homozygosity. CNNLOH/aUPD should be considered a special case of structural variation since it does not change the copy number of the affected segment. It is, however, a result of a structural rearrangement, most commonly due to meiotic or mitotic nondisjunction/anaphase lag, alternatively mitotic recombination. From the disease point of view, CNNLOH/aUPD could result in: (1) an imprinting disorder, via loss or doubling of the expression of an imprinted gene; or (2) expression of a recessive trait (eg, a mutation in a tumour suppressor gene) in a non-Mendelian fashion. The latter is mediated by reduction to homozygosity causing a recessive phenotype to appear, which is inherited in an initially heterozygous state from the parents. The list of conditions associated with CNNLOH/aUPD is continuously growing40–43 and this trend is likely to continue due to an increasing awareness and application of SNP based arrays with ultra-high resolution in analyses of normal and disease related samples. CNNLOH/aUPD cannot be detected by cytogenetic analyses or by standard array-CGH. However, allelic ratio values from SNP based arrays, such as Illumina beadchips, are sensitive tools for the detection of constitutional (non-mosaic) and mosaic forms of CNNLOH/aUPD.14 The detection of CNNLOH/aUPD should be discussed in the context of next generation, highly parallel sequencing, gradually revolutionising the field. This approach is neither straightforward (from the data analysis point of view) nor inexpensive for detection of CNNLOH/aUPD, especially for samples affected with low level mosaicism. Therefore SNP microarrays should remain the preferred approach for such analyses.
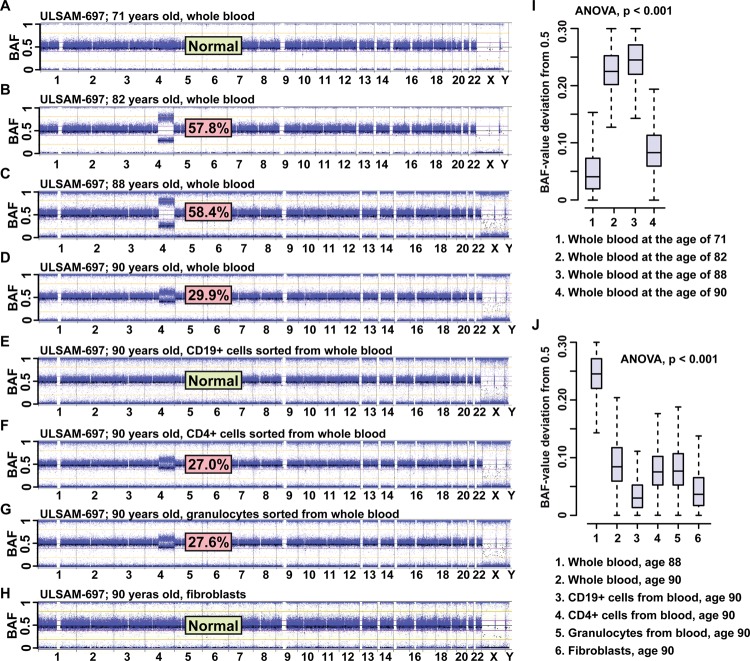
In addition to showing a high frequency of post-zygotic structural aberrations in normal cells, Forsberg et al1 also showed variable dynamics of cell clones affected with aberrations in different individuals, by studying 2–4 longitudinal samples collected many years apart from the same subject (figure 2). A more or less rapid relative increase in frequency of cells affected by a certain abnormality was observed in many cases and the rate of this increase varied between different subjects and different aberrations. Interestingly, in multiple subjects that were studied in longitudinal fashion, a decrease in the number of affected cells in the oldest samples was observed, which suggest a self-correcting process in the haematopoietic system. This decrease suggests that the initially expanding cell clones, possessing a higher proliferative potential, are not immortalised and follow the normal apoptotic programme. Furthermore, new blood samples from subjects that were studied longitudinally provided an opportunity for sorting blood cells into several sub-compartments, such as CD4 T cells, CD19 B cells, and granulocytes. In one illustrative subject (ULSAM-697), who is generally healthy, we described a >100 Mb CNN-LOH/aUPD of chromosome 4 using four time points: 71, 82, 88, and 90 years (figure 2). This aberration was not detectable at the age of 71 years, reached ∼58% at the ages of 82 and 88 years, and decreased radically to ∼30% of cells at the age of 90 years. Sorting of cells at the age of 90 years showed that CD4 T cells and granulocytes were affected to a similar degree, as identified in DNA from unsorted blood at the same age. However, CD19 B cells were unexpectedly free from this aberration. Thus, both myeloid and lymphoid lineages were affected to a similar degree, with the notable exception of B lymphocytes. It should be stressed that aberrations of 4q are typical for myelodysplastic syndrome (MDS), but this individual does not have any symptoms of the disorder. In addition, considering the rapid decrease of the cell clone carrying the aberrant 4q between samplings at 88 and 90 years, it is likely that this subject should soon be free from aberrant cells, which emphasises the self-eliminating property of the system. Moreover, all three reports1–3 observed a frequent coexistence of two (or more) aberrations in the blood of a single person. Longitudinal analyses of subjects showing multiple aberrations revealed variable dynamics of changes for different aberrations over time, pointing to the coexistence of different cell clones in blood, each affected with a distinct aberration.1
Figure 2.
The whole genome profiles in longitudinal analysis of 4 peripheral blood samples collected from subject ULSAM-697 at the ages of 71, 82, 88, and 90 years (panels A, B, C and D, respectively). This figure illustrates a clonal cell expansion containing a terminal CNNLOH/aUPD encompassing 103 Mb of the long arm of chromosome 4, with an increase and a decrease in the number of cells at different ages (data from ref. [1]). Each panel is composed of images from Illumina SNP beadchips showing the BAF-values, as CNNLOH/aUPD is not detectable using LRR data (see Fig. 1). The estimated percentage of cells displaying CNNLOH/aUPD on chromosome 4 is shown for each studied sample. This aberration was not detectable at the age of 71, reached approximately 58% at the ages of 82 and 88 years and decreased radically to approximately 30% of cells at the age of 90 years. This figure also displays the BAF-profiles for the whole genome from genotyping of sorted blood cells (CD19+ B lymphocytes, CD4+ T lymphocytes, and granulocytes) as well as skin fibroblasts collected at the age of 90 years (panels E, F, G and H, respectively). Sorting of blood cells at the age of 90 years showed that CD4+ T-cells and granulocytes were affected to a similar degree, as identified in DNA from unsorted blood at the same age. However, CD19+ B-cells were unexpectedly free from this aberration. Thus, both myeloid and lymphoid lineages were affected to a similar degree, with the notable exception of B-lymphocytes. Panels I and J show statistical analysis of data. Panel I shows comparisons of “BAF-value deviation from 0.5” for heterozygous probes only and within the aberrant region of 4q, derived from analysis displayed in panels A through D. Similar analysis is shown in panel J for data derived from panels E through H. The proportion of cells with the 4q aberration changes with time and between different types of cells. These changes are significantly different between all samplings (ANOVA p<0.001; Tukey's test for multiple comparisons).
The results on expanding-contracting, potentially pre-cancerous clones, which are subject to auto-correction,1 are in good agreement with data showing expansions of pre-leukaemic clones containing gene fusions specific to acute leukaemia described in newborns.44 Thus, throughout the lifetime, peripheral blood likely contains multiple aberrant expanding and contracting cell clones and these can persist in circulation for many years, if not decades. This issue requires further studies and one intriguing question in this context is: which are the cells that are giving rise to these clonal expansions? We can only speculate that these might be very early progenitors for multiple lineages of haematopoiesis or perhaps even haematopoietic stem cells (HSC). Other interesting and related questions are: why do humans in the age window of 55–90 years develop so frequently post-zygotic aberrant cell clones, present at the high frequency (5–95% of all nucleated cells) in peripheral blood? In other words, why are such clonal expansions present in blood at much lower frequencies below the age of 55 years? One plausible explanation is related to immuno-senescence and accumulation of random mutations with age. Immuno-senescence involves loss of cell diversity in elderly/old subjects, preferentially in B and T cell lineages.45–48 This loss of diversity of clones might be caused by depletion of the complexity in the pool of HSC, due to detrimental mutations forcing the affected cells into apoptosis/growth arrest. The stem cells remaining in the pool also accumulate mutations with age, but these mutations might, on the contrary, be promoting their proliferation. As such a process gradually progresses with age, a threshold effect is reached and the frequency of aberrant clones rise above the detection limit of array based analyses, which is ∼5% of all nucleated blood cells.14 49
The results presented by Forsberg et al,1 Laurie et al2 and Jacobs et al3 likely represent only ‘the tip of an iceberg’ and there are many arguments supporting this assumption. Perhaps the strongest argument is derived from the above discussed predictions of the number and consequences of mutations that we can expect to develop within a single human soma. The largest category of post-zygotic mutations is likely never detected, if they are detrimental and lead to apoptosis/growth arrest of the affected cell(s). This category of mutations is probably largely responsible for the development of age related loss of diversity of cells in the human immune system, characteristic for the immuno-senescence.45–48 Another category of undetected mutations is phenotypically neutral, not leading to a sufficient proliferative advantage of affected cells, over all the other nucleated cells in the peripheral blood. Genetic events in this category are beyond the reach of array based analyses, but could be studied using the next generation sequencing with a deep coverage. Furthermore, another argument is related to the fact that we have so far only studied blood, which is quite special, compared to solid tissues. Extrapolation on the level of post-zygotic mosaicism beyond blood DNA using similar resolution of analysis is currently difficult. In addition, blood is composed of numerous cell types with discrepancies in their longevity and their natural rate of replenishment, but blood DNA is routinely studied without cell sorting. Much lower levels of mosaicism could be detected by analysing well defined subsets of blood cells, especially for cell clones representing a minority of circulating cells. This is essential for analyses of human disorders where a certain subset of cells (from blood or elsewhere) can be suspected as being important for the development of particular phenotypes. Moreover, the SNP arrays used1–3 interrogated only in the order of 0.4-1 million nucleotides with an uneven distribution of data points. This has important implications for a likely high false-negative rate of mutation discovery, especially for structural rearrangements below 50 kb in size. Finally, balanced inversions and translocations would have escaped detection by our method. Thus, future studies should be directed towards better defined subpopulations of cells using a considerably higher resolution approach. Whole genome sequencing would definitely suffice with regard to the resolution of analysis. However, this method is still expensive and is not established to analyse all types of mutations, especially when structural variation is considered. A recent comparative study using different sequencing platforms of a single genome at high coverage illustrated this notion.50 The concordance rate between two platforms was low; 88% for calling of single nucleotide variants and only 26% for indels. In summary, in order to see more of the iceberg, we should address a number of points discussed above.
Phenotypic relevance of post-zygotic mosaicism
Reports on mosaic mutations causing Mendelian and non-Mendelian conditions are continuously accumulating. A few recent examples of conditions associated with post-zygotic variation are: Proteus syndrome,51 different vascular anomalies,52 Ollier disease/Mafucci syndrome/metaphyseal chondromatosis,53 54 CLOVES syndrome (Congenital, Lipomatous, Overgrowth, Vascular malformations, Epidermal nevi and Spinal/Skeletal anomalies and/or Scoliosis),55 and congenital dyskeratosis.56 Post-zygotic mosaicism can result in a milder phenotype, can cause reversion of disease phenotype, or can unmask an expression of a mutation that would otherwise be lethal to the embryo. It is likely that many instances of post-zygotic mosaicism are not clinically recognised since the patient may show a borderline, mild clinical phenotype due to a low proportion of cells carrying a mutation. Another reason underlying the ascertainment bias is that post-zygotic variation is primarily relevant for sporadic cases (de novo mutations) with no previous family history of a disease. The steadily growing body of data indicates that somatic mosaicism for pathogenic mutations affecting known disease genes should be seen as a rule applicable to the vast majority of disease related genes, rather than as an exception. As comprehensive reviews on this subject are published,29–34 37–39 57–62 we will only discuss two well studied genes providing insights into the role of somatic mosaicism on the phenotype. Duchenne muscular dystrophy (DMD) is an X chromosome linked, lethal neuromuscular disorder, affecting one in 3500 liveborn males. The DMD gene shows interesting findings with regard to somatic mosaicism.63–65 Its mutation spectrum is atypical as up to 75% of DMD cases are due to structural rearrangements; that is, a deletion or duplication of one or more exons. This gene contains two mutational hot spots involving distal (exons 45–52) and proximal (exons 2–7) regions.66 There is a difference in the distribution of rearrangements within the gene in patients showing mosaicism versus non-mosaic cases. Deletions in patients showing somatic mosaicism are preferentially clustered around exon 2.67 68 This suggests that the mechanism behind generation of these structural rearrangements is different in mitosis versus meiosis. The third interesting aspect of the DMD gene is a reversion of disease phenotype in muscle fibres of DMD patients, via mitotic rearrangements restoring the reading frame and allowing some dystrophin expression to occur. In several cases, the reverting mutation appeared to be in the distal deletion hotspot, supporting the suggestion that this region is unstable. Somatic reversions have also been described for other diseases.31–33 37 56 69–71
Neurofibromatosis type 1 (NF1) is an inherited tumour syndrome caused by mutations in the NF1 gene on 17q.72–74 Approximately 5% of patients are affected by large (1.2–1.4 Mb) deletions removing NF1, along with other genes.75 76 Most of these large deletions are the result of non-allelic homologous recombination between segmental duplications, flanking the NF1 gene. In the important study by Kehrer-Sawatzki et al,75 mosaicism for the NF1 gene deletions was detected in up to 40% of cases, when sporadic NF1 patients were specifically targeted for analysis of deletions using DNA from several tissues. Mosaic patients also lacked the cognitive defects and facial dysmorphology typically associated with NF1 microdeletions, suggesting a genotype–phenotype correlation. In patients with mosaicism, the proportion of cells with the deletion was 91–100% in peripheral leucocytes, but was much lower (51–80%) in buccal smears or peripheral skin fibroblasts. Detailed analysis of the deletion breakpoints revealed additional surprising results. In contrast to the typical NF1 deletion of 1.4 Mb (occurring between the major segmental duplications flanking the gene, also known as type 1 deletions), seven of the eight mosaic deletions were 1.2 Mb in size (known as type 2 deletions) and were the product of recombination between the SUZ12 gene and a highly similar pseudogene.75 77 Thus, type I NF1 microdeletions occur by intra-chromosomal recombination during meiosis, while the type II deletions are mediated by intra-chromosomal recombination during mitosis. This scenario is reminiscent of the above described findings for the DMD gene, pointing again to a different mechanism behind the generation of some structural rearrangements in meiosis and mitosis. The NF1 gene can also be somatically mutated in human glioblastoma multiforme and leukaemia.78 79
The three papers1–3 have pointed out the cancer related aspect of clonal cell expansions in the blood of elderly/old individuals. Laurie et al2 and Jacobs et al3 showed that individuals affected by post-zygotic aberrations have a considerably increased risk of hematological malignancies/cancers, with the relative risk increasing 10- and 35-fold, respectively. These numbers are higher by at least an order of magnitude, compared to the risk estimates from GWAS.4 The report by Jacobs et al3 (see figures 2 and 3 in their article3) compared cohorts of cancer-affected and cancer-free subjects. The vast majority, if not all, of aberrations that were observed in the cancer-affected cohort were also seen in cancer-free subjects, although at lower frequency. A detailed inspection of the regions with aberrations is interesting when viewed in the context of the two most common hematological malignancies of the elderly, namely chronic lymphocytic leukaemia (CLL) and MDS. Numerous uncovered chromosomal aberrations in blood have previously been described in patients affected with these disorders, which suggests that these mutations are not cancer specific. They represent rather an early pre-cancerous change, possibly predisposing to the development of malignancy/cancer later in life, presumably after acquisition of additional mutations and further in vivo selection for clones with the highest proliferative potential.
It should be stressed that, considering the frequency of CLL and MDS in the general population, the majority of these discovered post-zygotic aberrations will not lead to a clinically manifested disease, reinforcing the issue of the self correcting haematopoietic system. A comparison of the total number of subjects affected with post-zygotic aberrations1–3 and the literature for CLL80–87 and MDS88–94 suggests that the number of mutations related to MDS is higher when compared with those relevant for CLL. The most commonly observed and MDS related changes are: 4q CNNLOH/aUPD (targeting the TET2 tumour suppressor gene)95; deletions of 5q and 5q-CNNLOH/aUPD; monosomy 7 and deletions of 7q (targeting the EZH2 gene); trisomy 8; deletions of 11q and 11q-CNNLOH/aUPD (targeting the CBL gene); monosomy 17, deletions of 17p and 17p-CNNLOH/aUPD; deletions of 20q; as well as trisomy 21. The corresponding list of aberrations related to CLL is: 11q deletions and 11q-CNNLOH/aUPD; trisomy 12; 13q deletions and 13q-CNNLOH/aUPD; monosomy 17, deletions of 17p and 17p-CNNLOH/aUPD as well as 22q deletions and 22q-CNNLOH/aUPD (possibly targeting the PRAME gene). This overrepresentation of MDS related aberrations may seem surprising since CLL is usually considered to be the more common malignancy of the elderly. However, this MDS biased portrait of post-zygotic aberrations is in agreement with studies showing that the aging of the human immune system is connected with the relative depletion of lymphoid precursors and an increase of the myeloid counterparts.
The human haematopoietic system undergoes a dramatic shift with age. This includes a reduced cellularity of the bone marrow,96 reduced lymphopoiesis,45 and a decreased complexity of T cells46 and B cells.47 Nevertheless, the frequency of HSC appear to be high in the elderly, although their developmental trajectories are changing from a lymphoid dominated developmental pattern in the young to a more myeloid dominated developmental pattern in the elderly.48 97 98 HSC from both the young and elderly had the potential to generate lymphoid and myeloid lineages in culture. However, HSC from the elderly individuals have a more myeloid biased differentiation potential as compared to HSC from young subjects.48 In line with this, mutations in the TET2 gene, which are frequently found in patients with MDS, were observed in the blood of phenotypically normal humans with clonal haematopoiesis.95 Thus, considering the above literature, we would argue that the age dependent shift between lymphoid and myeloid lineages mirrors well the picture of MDS and CLL related aberrations in the peripheral blood of elderly/old humans.
One of the intriguing questions raised in the recent papers1–3 is: which other phenotypes (other than hematological cancers and non-cancer related) can be linked to clonal cell expansions in blood harbouring different aberrations? Our results provide one illustrative example, regarding a non-cancer related hematological phenotype. One subject displayed a 20q deletion, which was barely detectable at the age of 71 years. The number of cells containing the 20q deletion was estimated to be ∼50% when he was 75 years old and he had ∼36% aberrant cells at the age of 88 years. In between the samplings at 75 and 88 years, he was diagnosed with idiopathic thrombocytopenic purpura, which might be due to clonal expansion of 20q deletion cells and suppression of normal thrombocyte production. In line with the above example, future studies aiming at correlations of phenotype with a better defined post-zygotic mutation profile should be informative.
Conclusions, open questions, challenges and opportunities
The three papers1–3 have raised a number of questions and challenges, but also point to opportunities in connection with future investigations of post-zygotic mutations. These studies suggest a likely and largely unexplored impact of post-zygotic variation on common human phenotypes, not necessarily restricted to cancer. Sporadic disorders, defined as a lack of similar cases among the closest relatives of an affected patient, are common in medicine. We therefore argue that studies of differences in the post-zygotic mutational profile of appropriate target cells, in comparison with other normal cells of the same patient, will be highly informative. The non-heritable causes of human disease have traditionally been ascribed to environmental factors. With few exceptions, however, such as smoking for lung cancer or alcohol for liver cirrhosis, specific identification of most of these factors has proven elusive for common multifactorial diseases and methodological breakthroughs likely to change this are nowhere in sight. Post-zygotic mutations are clearly not heritable, and cannot therefore explain the ‘missing heritability’. However, they might be a part of the non-heritable disease causality, which has, until now, been underestimated in importance and routinely ascribed to the environment. The new evidence discussed here strongly suggests that a sizeable part of the non-heritable causes of human disease can be ascribed to stochastic molecular events that are readily amenable to well established paradigms of analysis.
These recent results1–3 should also be discussed in the general context of aging, longevity and age associated diseases. Aging has been defined as a complex process of cellular senescence of adult tissues that results in compromised stress response, homeostatic imbalance, and elevated risk of disease.99 100 The dramatic rise of the human lifespan (by 20 years during the second half of the 20th century) is calling for more research focused on healthy aging and age associated conditions. This life extending trend is expected to continue worldwide, with an average human lifespan rising another 10 years by the year 2050.101 By itself, aging is the largest risk factor for the majority of common human disorders.102 Studies of aging human cohorts collected in the longitudinal fashion and using the approach described recently1–3 (ie, analysis of post-zygotic structural aberrations that are accumulating during lifetime) may be fruitful for uncovering mutations that are causative for many of common human disorders. It should be stressed that the result of Laurie et al2 and Jacobs et al3 indicate that CNV analysis of post-zygotic changes yields considerably stronger predictions of disease risk, when compared with typical results from germline variants discovered in GWAS.4 This is a strong argument in favour of the extension of analyses targeting post-zygotic variation. Finally, a possible consequence of the accumulation of post-zygotic aberrations is that some of the clonal cell expansions might actually entail an increased lifespan for people affected with them, via enhanced function of the immune system, which is possibly stretching over many years of life. This issue should also be investigated in further detail.
The recent literature provides a rough ‘post-zygotic variation baseline’,1–3 defining what can be expected when the bulk genome derived from all cells present in the peripheral blood is scanned in young/middle aged and elderly/old subjects. However, this portrait of post-zygotic variation is not necessarily representative for all cell clones in circulation (see above, discussion about subject ULSAM-697) (figure 2). We should gain more insight into post-zygotic variation across various ages, when the blood is sorted into at least a few cellular sub-compartments. We would argue that such analyses will yield important information with regard to another hidden layer of post-zygotic variation, which might be useful for genotype–phenotype correlations in conditions related to dysfunctions of the haematopoietic system; for example, autoimmune or other chronic inflammatory conditions. Furthermore, it is equally important to assess the level of post-zygotic variation in at least a few other human tissues across different age groups. These should preferably represent at least one non-mesodermal lineage of embryonic development, as the most popular sources of DNA from different human tissues (blood and fibroblasts) are both of mesodermal origin. In conclusion, a major consequence of the recent results is that a profile of variation in a single human tissue collected at one time point cannot be used as a surrogate representing a faithful portrait of variation present in other tissues nor in the same tissue throughout lifetime. In line with this, future studies of genetic but not inherited mechanisms behind sporadic complex diseases should be directed towards an analysis of the cells, which are presumed to cause the phenotype under investigation. Such an approach should maximise the success rate for uncovering a truly pathogenic variation.
One of the strengths of the recent analyses1–3 is that the studied cells had not been manipulated in vitro, providing a representative snapshot picture of a dynamic system taken at a certain age. In this context, a concern should be raised regarding the use of lymphoblastoid cell lines (LCLs) as a source of DNA for similar studies. LCLs are Epstein–Barr virus transformed B lymphocytes and are usually cultured in vitro for a prolonged time. LCLs are polyclonal in the beginning, and then become gradually oligoclonal and monoclonal after prolonged culturing.103 104 Thus, these cultured cells might acquire a new genotype, which was not present in the original B lymphocytes that gave rise to the LCL. Indeed, a recent analysis of one parent–offspring trio performed in the context of the 1000 Genomes Project showed that the majority of de novo mutations present in the LCL of the offspring was neither present in parents nor was it detectable in DNA derived from total peripheral blood DNA of the offspring.105 Another independent study has recently confirmed this conclusion.106 Accordingly, these de novo mutations were likely artefacts induced by in vitro culturing. An alternative unfavourable scenario is that cultured LCLs may conceal post-zygotic mutations. This is because the variation studied via LCLs is representative for only a fraction of B lymphocytes and the latter are a minority of all circulating cells in peripheral blood. Furthermore, it has been shown that cells affected by some chromosomal rearrangements are less efficiently cultured in vitro, when compared to normal euploid cells,107 108 which might lead to a selective removal of cells with a variant genotype. Thus, LCLs should be restricted for studies of genetic variation.
Forsberg et al1 showed that the post-zygotic genome of normal blood is dynamic. Peripheral blood likely contains throughout lifetime multiple aberrant expanding–contracting cell clones. The available data are still limited but suggest that such clones can persist in circulation of elderly/old people for a decade or more. The currently available results provide a clear link between these aberrant expanding–contracting clones and hematological malignancies/cancers. However, the frequency of subjects affected with aberrant clones typical for MDS or CLL, for example, is considerably higher than the frequency of these diseases in the general population. Thus, not all subjects containing the pre-cancerous clones will develop malignancy/cancer and it is important to follow up this topic with description of causative factors promoting the development of these diseases. Furthermore, we envisage that the genotype–phenotype relationships based on the presence of specific aberrant cell clones (in blood and in other tissues) will be expanded to non-cancer related phenotypes. The medical literature provides many examples of diseases related to the haematopoietic system with fluctuating disease course, with relapses or even self healing; for example, asthma, multiple sclerosis, Crohn's disease, and inflammatory bowel disease, to mention a few. It might be relevant to search for expanding–contracting cell clones with post-zygotic mutations in different cellular sub-compartments of blood in such patients. Furthermore, in order to exploit this line of research maximally, the human post-zygotic genomes of several tissues should be monitored in a longitudinal fashion, using samples collected at multiple time points throughout life. Such analyses will require modifications to the currently applied bio-banking procedures for sample collection from large population based cohorts and ethical approvals that justify such collections.
Acknowledgments
We thank Nick CP Cross, Maj Hulten, Richard Rosenquist Brandell, Eva Tiensuu Janson, Chiara Rasi, and Constantin Polychronakos for critical review of the manuscript and constructive comments. This work was supported by the Ellison Medical Foundation, Swedish Cancer Society, Swedish Research Council, the Science for Life Laboratory-Uppsala and Uppsala University.
Footnotes
Contributors: All authors (LF, DA and JD) contributed to the conception and design, analysis and interpretation of data, review of the literature, and revising it critically for important intellectual content as well as the final approval of the version to be published. LF and JD wrote the manuscript.
Funding: None.
Competing interests: None.
Provenance and peer review: Commissioned; internally peer reviewed.
Open Access: This is an Open Access article distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 3.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/3.0/
References
- 1.Forsberg L, Rasi C, Razzaghian H, Pakalapati G, Waite L, Thilbeault K Stanton, Ronowicz A, Wineinger N, Tiwari H, Boomsma D, Westerman M, Harris J, Lyle R, Essand M, Eriksson F, Strachan E, O'Hanlon T, Rider L, Miller F, Giedraitis V, Lannfelt L, Ingelsson M, Piotrowski A, Pedersen N, Absher D, Dumanski J. Age-related somatic structural changes in the nuclear genome of human blood cells. Am J Hum Genet 2012;90:217–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laurie CC, Laurie CA, Rice K, Doheny KF, Zelnick LR, McHugh CP, Ling H, Hetrick KN, Pugh EW, Amos C, Wei Q, Wang LE, Lee JE, Barnes KC, Hansel NN, Mathias R, Daley D, Beaty TH, Scott AF, Ruczinski I, Scharpf RB, Bierut LJ, Hartz SM, Landi MT, Freedman ND, Goldin LR, Ginsburg D, Li J, Desch KC, Strom SS, Blot WJ, Signorello LB, Ingles SA, Chanock SJ, Berndt SI, Le Marchand L, Henderson BE, Monroe KR, Heit JA, de Andrade M, Armasu SM, Regnier C, Lowe WL, Hayes MG, Marazita ML, Feingold E, Murray JC, Melbye M, Feenstra B, Kang JH, Wiggs JL, Jarvik GP, McDavid AN, Seshan VE, Mirel DB, Crenshaw A, Sharopova N, Wise A, Shen J, Crosslin DR, Levine DM, Zheng X, Udren JI, Bennett S, Nelson SC, Gogarten SM, Conomos MP, Heagerty P, Manolio T, Pasquale LR, Haiman CA, Caporaso N, Weir BS. Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nat Genet 2012;44:642–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jacobs KB, Yeager M, Zhou W, Wacholder S, Wang Z, Rodriguez-Santiago B, Hutchinson A, Deng X, Liu C, Horner MJ, Cullen M, Epstein CG, Burdett L, Dean MC, Chatterjee N, Sampson J, Chung CC, Kovaks J, Gapstur SM, Stevens VL, Teras LT, Gaudet MM, Albanes D, Weinstein SJ, Virtamo J, Taylor PR, Freedman ND, Abnet CC, Goldstein AM, Hu N, Yu K, Yuan JM, Liao L, Ding T, Qiao YL, Gao YT, Koh WP, Xiang YB, Tang ZZ, Fan JH, Aldrich MC, Amos C, Blot WJ, Bock CH, Gillanders EM, Harris CC, Haiman CA, Henderson BE, Kolonel LN, Le Marchand L, McNeill LH, Rybicki BA, Schwartz AG, Signorello LB, Spitz MR, Wiencke JK, Wrensch M, Wu X, Zanetti KA, Ziegler RG, Figueroa JD, Garcia-Closas M, Malats N, Marenne G, Prokunina-Olsson L, Baris D, Schwenn M, Johnson A, Landi MT, Goldin L, Consonni D, Bertazzi PA, Rotunno M, Rajaraman P, Andersson U, Freeman LE Beane, Berg CD, Buring JE, Butler MA, Carreon T, Feychting M, Ahlbom A, Gaziano JM, Giles GG, Hallmans G, Hankinson SE, Hartge P, Henriksson R, Inskip PD, Johansen C, Landgren A, McKean-Cowdin R, Michaud DS, Melin BS, Peters U, Ruder AM, Sesso HD, Severi G, Shu XO, Visvanathan K, White E, Wolk A, Zeleniuch-Jacquotte A, Zheng W, Silverman DT, Kogevinas M, Gonzalez JR, Villa O, Li D, Duell EJ, Risch HA, Olson SH, Kooperberg C, Wolpin BM, Jiao L, Hassan M, Wheeler W, Arslan AA, Bueno-de-Mesquita HB, Fuchs CS, Gallinger S, Gross MD, Holly EA, Klein AP, LaCroix A, Mandelson MT, Petersen G, Boutron-Ruault MC, Bracci PM, Canzian F, Chang K, Cotterchio M, Giovannucci EL, Goggins M, Bolton JA Hoffman, Jenab M, Khaw KT, Krogh V, Kurtz RC, McWilliams RR, Mendelsohn JB, Rabe KG, Riboli E, Tjonneland A, Tobias GS, Trichopoulos D, Elena JW, Yu H, Amundadottir L, Stolzenberg-Solomon RZ, Kraft P, Schumacher F, Stram D, Savage SA, Mirabello L, Andrulis IL, Wunder JS, Garcia A Patino, Sierrasesumaga L, Barkauskas DA, Gorlick RG, Purdue M, Chow WH, Moore LE, Schwartz KL, Davis FG, Hsing AW, Berndt SI, Black A, Wentzensen N, Brinton LA, Lissowska J, Peplonska B, McGlynn KA, Cook MB, Graubard BI, Kratz CP, Greene MH, Erickson RL, Hunter DJ, Thomas G, Hoover RN, Real FX, Fraumeni JF, Jr, Caporaso NE, Tucker M, Rothman N, Perez-Jurado LA, Chanock SJ. Detectable clonal mosaicism and its relationship to aging and cancer. Nat Genet 2012;44:651–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Hum Genet 2012;90:7–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maher B. Personal genomes: The case of the missing heritability. Nature 2008;456:18–21 [DOI] [PubMed] [Google Scholar]
- 6.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature 2009;461:747–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eichler EE, Flint J, Gibson G, Kong A, Leal SM, Moore JH, Nadeau JH. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet 2010;11:446–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zuk O, Hechter E, Sunyaev SR, Lander ES. The mystery of missing heritability: Genetic interactions create phantom heritability. Proc Natl Acad Sci U S A 2012;109:1193–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson E. The cell in heredity and development. 3rd edn New York: The Macmillian Company, 1925 [Google Scholar]
- 10.Cotterman CW. Somatic mosaicism for antigen A2. Acta genetica et statistica medica 1956;6:520–1 [DOI] [PubMed] [Google Scholar]
- 11.Delhanty JD. Mechanisms of aneuploidy induction in human oogenesis and early embryogenesis. Cytogenet Genome Res 2005;111:237–44 [DOI] [PubMed] [Google Scholar]
- 12.Vanneste E, Voet T, Le Caignec C, Ampe M, Konings P, Melotte C, Debrock S, Amyere M, Vikkula M, Schuit F, Fryns JP, Verbeke G, D'Hooghe T, Moreau Y, Vermeesch JR. Chromosome instability is common in human cleavage-stage embryos. Nat Med 2009;15:577–83 [DOI] [PubMed] [Google Scholar]
- 13.Hassold T. Mosaic trisomies in human spontaneous abortions. Hum genet 1982;61:31–5 [DOI] [PubMed] [Google Scholar]
- 14.Conlin LK, Thiel BD, Bonnemann CG, Medne L, Ernst LM, Zackai EH, Deardorff MA, Krantz ID, Hakonarson H, Spinner NB. Mechanisms of mosaicism, chimerism and uniparental disomy identified by SNP array analysis. Hum Mol Genet 2010;19:1263–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ballif BC, Rorem EA, Sundin K, Lincicum M, Gaskin S, Coppinger J, Kashork CD, Shaffer LG, Bejjani BA. Detection of low-level mosaicism by array CGH in routine diagnostic specimens. Am J Med Genet A 2006;140:2757–67 [DOI] [PubMed] [Google Scholar]
- 16.Veltman JA, Brunner HG. De novo mutations in human genetic disease. Nat Rev Genet 2012;13:565–75 [DOI] [PubMed] [Google Scholar]
- 17.Goriely A, Wilkie AO. Paternal age effect mutations and selfish spermatogonial selection: causes and consequences for human disease. Am J Hum Genet 2012;90:175–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strachan T, Read A. Human Molecular Genetics 3. 3rd edn New York: Garland Publishing, 2004 [Google Scholar]
- 19.Rubin H. The disparity between human cell senescence in vitro and lifelong replication in vivo. Nat Biotechnol 2002;20:675–81 [DOI] [PubMed] [Google Scholar]
- 20.Takubo K, Izumiyama-Shimomura N, Honma N, Sawabe M, Arai T, Kato M, Oshimura M, Nakamura K. Telomere lengths are characteristic in each human individual. Exp Gerontol 2002;37:523–31 [DOI] [PubMed] [Google Scholar]
- 21.Nakamura K, Izumiyama-Shimomura N, Sawabe M, Arai T, Aoyagi Y, Fujiwara M, Tsuchiya E, Kobayashi Y, Kato M, Oshimura M, Sasajima K, Nakachi K, Takubo K. Comparative analysis of telomere lengths and erosion with age in human epidermis and lingual epithelium. J Invest Dermatol 2002;119:1014–19 [DOI] [PubMed] [Google Scholar]
- 22.Baird DM. Telomere dynamics in human cells. Biochimie 2008;90:116–21 [DOI] [PubMed] [Google Scholar]
- 23.Baird DM, Britt-Compton B, Rowson J, Amso NN, Gregory L, Kipling D. Telomere instability in the male germline. Hum Mol Genet 2006;15:45–51 [DOI] [PubMed] [Google Scholar]
- 24.Frank SA. Somatic evolutionary genomics: mutations during development cause highly variable genetic mosaicism with risk of cancer and neurodegeneration. Proc Natl Acad Sci U S A 2010;107(Suppl 1):1725–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lynch M. Rate, molecular spectrum, and consequences of human mutation. Proc Natl Acad Sci U S A 2010;107:961–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lynch M. Evolution of the mutation rate. Trends Genet 2010;26:345–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lupski JR. Genomic rearrangements and sporadic disease. Nat Genet 2007; 39(7 Suppl):S43–7 [DOI] [PubMed] [Google Scholar]
- 28.Itsara A, Wu H, Smith JD, Nickerson DA, Romieu I, London SJ, Eichler EE. De novo rates and selection of large copy number variation. Genome Res 2010;20:1469–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall JG. Review and hypotheses: somatic mosaicism: observations related to clinical genetics. Am J Hum Genet 1988;43:355–63 [PMC free article] [PubMed] [Google Scholar]
- 30.Gottlieb B, Beitel LK, Trifiro MA. Somatic mosaicism and variable expressivity. Trends Genet 2001;17:79–82 [DOI] [PubMed] [Google Scholar]
- 31.Youssoufian H, Pyeritz RE. Mechanisms and consequences of somatic mosaicism in humans. Nat Rev Genet 2002;3:748–58 [DOI] [PubMed] [Google Scholar]
- 32.Erickson RP. Somatic gene mutation and human disease other than cancer. Mutat Res 2003;543:125–36 [DOI] [PubMed] [Google Scholar]
- 33.Hirschhorn R. In vivo reversion to normal of inherited mutations in humans. J Med Genet 2003;40:721–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Notini AJ, Craig JM, White SJ. Copy number variation and mosaicism. Cytogenet Genome Res 2008;123:270–7 [DOI] [PubMed] [Google Scholar]
- 35.Piotrowski A, Bruder C, Andersson R, Diaz de Ståhl T, Menzel U, Poplawski A, von Tell D, Crasto C, Bogdan A, Bartoszewski R, Bebok Z, Krzyżanowski M, Jankowski Z, Partridge C, Komorowski J, Dumanski J. Somatic mosaicism for copy number variation in differentiated human tissues. Hum Mutat 2008;29:1118–24 [DOI] [PubMed] [Google Scholar]
- 36.Bruder C, Piotrowski A, Gijsbers A, Andersson R, Erickson S, Diaz de Ståhl T, Menzel U, Sandgren J, von Tell D, Poplawski A, Crowley M, Crasto C, Partridge E, Tiwari H, Allison D, Komorowski J, van Ommen G-J, Boomsma D, Pedersen N, den Dunnen J, Wirdefeldt K, Dumanski J. Phenotypically concordant and discordant monozygotic twins display different DNA copy-number-variation profiles. Am J Hum Genet 2008;82:763–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Erickson RP. Somatic gene mutation and human disease other than cancer: an update. Mutat Res 2010;705:96–106 [DOI] [PubMed] [Google Scholar]
- 38.De S. Somatic mosaicism in healthy human tissues. Trends Genet 2011;27:217–23 [DOI] [PubMed] [Google Scholar]
- 39.Dumanski JP, Piotrowski A. Structural genetic variation in the context of somatic mosaicism. In: Feuk L, ed. Genomic Structural Variation. Vol 838 New York: Humana Press, 2012:249–72 [DOI] [PubMed] [Google Scholar]
- 40.Kotzot D. Complex and segmental uniparental disomy updated. J Med Genet 2008;45:545–56 [DOI] [PubMed] [Google Scholar]
- 41.Feinberg AP. Genomic imprinting and cancer. In: Vogelstein B, Kinzler KW, eds. The genetic basis of human cancer. 2nd edn New York: McGraw-Hill, 2002 [Google Scholar]
- 42.Engel E. A fascination with chromosome rescue in uniparental disomy: Mendelian recessive outlaws and imprinting copyrights infringements. Eur J Hum Genet 2006;14:1158–69 [DOI] [PubMed] [Google Scholar]
- 43.Kotzot D. Prenatal testing for uniparental disomy: indications and clinical relevance. Ultrasound Obstet Gynecol 2008;31:100–5 [DOI] [PubMed] [Google Scholar]
- 44.Mori H, Colman SM, Xiao Z, Ford AM, Healy LE, Donaldson C, Hows JM, Navarrete C, Greaves M. Chromosome translocations and covert leukemic clones are generated during normal fetal development. Proc Natl Acad Sci U S A 2002;99:8242–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Linton PJ, Dorshkind K. Age-related changes in lymphocyte development and function. Nat Immunol 2004;5:133–9 [DOI] [PubMed] [Google Scholar]
- 46.Naylor K, Li G, Vallejo AN, Lee WW, Koetz K, Bryl E, Witkowski J, Fulbright J, Weyand CM, Goronzy JJ. The influence of age on T cell generation and TCR diversity. J Immunol 2005;174:7446–52 [DOI] [PubMed] [Google Scholar]
- 47.Gibson KL, Wu YC, Barnett Y, Duggan O, Vaughan R, Kondeatis E, Nilsson BO, Wikby A, Kipling D, Dunn-Walters DK. B-cell diversity decreases in old age and is correlated with poor health status. Aging cell 2009;8:18–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pang WW, Price EA, Sahoo D, Beerman I, Maloney WJ, Rossi DJ, Schrier SL, Weissman IL. Human bone marrow hematopoietic stem cells are increased in frequency and myeloid-biased with age. Proc Natl Acad Sci U S A 2011;108:20012–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Razzaghian HR, Shahi MH, Forsberg LA, de Stahl TD, Absher D, Dahl N, Westerman MP, Dumanski JP. Somatic mosaicism for chromosome X and Y aneuploidies in monozygotic twins heterozygous for sickle cell disease mutation. Am J Med Genet A 2010;152A:2595–8 [DOI] [PubMed] [Google Scholar]
- 50.Lam HY, Clark MJ, Chen R, Natsoulis G, O'Huallachain M, Dewey FE, Habegger L, Ashley EA, Gerstein MB, Butte AJ, Ji HP, Snyder M. Performance comparison of whole-genome sequencing platforms. Nat Biotechnol 2012;30:78–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lindhurst MJ, Sapp JC, Teer JK, Johnston JJ, Finn EM, Peters K, Turner J, Cannons JL, Bick D, Blakemore L, Blumhorst C, Brockmann K, Calder P, Cherman N, Deardorff MA, Everman DB, Golas G, Greenstein RM, Kato BM, Keppler-Noreuil KM, Kuznetsov SA, Miyamoto RT, Newman K, Ng D, O'Brien K, Rothenberg S, Schwartzentruber DJ, Singhal V, Tirabosco R, Upton J, Wientroub S, Zackai EH, Hoag K, Whitewood-Neal T, Robey PG, Schwartzberg PL, Darling TN, Tosi LL, Mullikin JC, Biesecker LG. A mosaic activating mutation in AKT1 associated with the Proteus syndrome. N Engl J Med 2011;365:611–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Limaye N, Boon LM, Vikkula M. From germline towards somatic mutations in the pathophysiology of vascular anomalies. Hum Mol Genet 2009;18:R65–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pansuriya TC, van Eijk R, d'Adamo P, van Ruler MA, Kuijjer ML, Oosting J, Cleton-Jansen AM, van Oosterwijk JG, Verbeke SL, Meijer D, van Wezel T, Nord KH, Sangiorgi L, Toker B, Liegl-Atzwanger B, San-Julian M, Sciot R, Limaye N, Kindblom LG, Daugaard S, Godfraind C, Boon LM, Vikkula M, Kurek KC, Szuhai K, French PJ, Bovee JV. Somatic mosaic IDH1 and IDH2 mutations are associated with enchondroma and spindle cell hemangioma in Ollier disease and Maffucci syndrome. Nat Genet 2011;43:1256–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vissers LE, Fano V, Martinelli D, Campos-Xavier B, Barbuti D, Cho TJ, Dursun A, Kim OH, Lee SH, Timpani G, Nishimura G, Unger S, Sass JO, Veltman JA, Brunner HG, Bonafe L, Dionisi-Vici C, Superti-Furga A. Whole-exome sequencing detects somatic mutations of IDH1 in metaphyseal chondromatosis with D-2-hydroxyglutaric aciduria (MC-HGA). Am J Med Genet A 2011;155A:2609–16 [DOI] [PubMed] [Google Scholar]
- 55.Kurek KC, Luks VL, Ayturk UM, Alomari AI, Fishman SJ, Spencer SA, Mulliken JB, Bowen ME, Yamamoto GL, Kozakewich HP, Warman ML. Somatic mosaic activating mutations in PIK3CA cause CLOVES syndrome. Am J Hum Genet 2012;90:1108–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jongmans MC, Verwiel ET, Heijdra Y, Vulliamy T, Kamping EJ, Hehir-Kwa JY, Bongers EM, Pfundt R, van Emst L, van Leeuwen FN, van Gassen KL, Geurts van Kessel A, Dokal I, Hoogerbrugge N, Ligtenberg MJ, Kuiper RP. Revertant somatic mosaicism by mitotic recombination in dyskeratosis congenita. Am J Hum Genet 2012;90:426–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alvarado C, Beitel LK, Sircar K, Aprikian A, Trifiro M, Gottlieb B. Somatic mosaicism and cancer: a micro-genetic examination into the role of the androgen receptor gene in prostate cancer. Cancer Res 2005;65:8514–18 [DOI] [PubMed] [Google Scholar]
- 58.Gottlieb B, Chalifour LE, Mitmaker B, Sheiner N, Obrand D, Abraham C, Meilleur M, Sugahara T, Bkaily G, Schweitzer M. BAK1 gene variation and abdominal aortic aneurysms. Hum Mutat 2009;30:1043–7 [DOI] [PubMed] [Google Scholar]
- 59.Lutskiy MI, Park JY, Remold SK, Remold-O'Donnell E. Evolution of highly polymorphic T cell populations in siblings with the Wiskott-Aldrich Syndrome. PLoS One 2008;3:e3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Iourov IY, Vorsanova SG, Yurov YB. Somatic genome variations in health and disease. Current genomics 2010;11:387–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hulten MA, Jonasson J, Nordgren A, Iwarsson E. Germinal and Somatic Trisomy 21 Mosaicism: how common is it, what are the implications for individual carriers and how does it come about? Curr Genomics 2010;11:409–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hultén M, Jonasson J, Iwarsson E, Uppal P, Vorsanova S, Yurov Y, Iourov I. Trisomy 21 mosaicism: we may all have a touch of Down syndrome. Cytogenet Genome Res 2012; in press [DOI] [PubMed] [Google Scholar]
- 63.Mandel JL. Dystrophin. The gene and its product. Nature 1989;339:584–6 [DOI] [PubMed] [Google Scholar]
- 64.Den Dunnen JT, Grootscholten PM, Dauwerse JG, Walker AP, Monaco AP, Butler R, Anand R, Coffey AJ, Bentley DR, Steensma HY, Van ommen GJ. Reconstruction of the 2.4 Mb human DMD-gene by homologous YAC recombination. Hum Mol Genet 1992;1:19–28 [DOI] [PubMed] [Google Scholar]
- 65.Roberts RG, Coffey AJ, Bobrow M, Bentley DR. Exon structure of the human dystrophin gene. Genomics 1993;16:536–8 [DOI] [PubMed] [Google Scholar]
- 66.White SJ, den Dunnen JT. Copy number variation in the genome; the human DMD gene as an example. Cytogenet Genome Res 2006;115:240–6 [DOI] [PubMed] [Google Scholar]
- 67.Passos-Bueno MR, Bakker E, Kneppers AL, Takata RI, Rapaport D, den Dunnen JT, Zatz M, van Ommen GJ. Different mosaicism frequencies for proximal and distal Duchenne muscular dystrophy (DMD) mutations indicate difference in etiology and recurrence risk. Am J Hum Genet 1992;51:1150–5 [PMC free article] [PubMed] [Google Scholar]
- 68.White SJ, Aartsma-Rus A, Flanigan KM, Weiss RB, Kneppers AL, Lalic T, Janson AA, Ginjaar HB, Breuning MH, den Dunnen JT. Duplications in the DMD gene. Hum Mutat 2006;27:938–45 [DOI] [PubMed] [Google Scholar]
- 69.Kvittingen EA, Rootwelt H, Berger R, Brandtzaeg P. Self-induced correction of the genetic defect in tyrosinemia type I. J Clin Invest 1994;94:1657–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ellis NA, Lennon DJ, Proytcheva M, Alhadeff B, Henderson EE, German J. Somatic intragenic recombination within the mutated locus BLM can correct the high sister-chromatid exchange phenotype of Bloom syndrome cells. Am J Hum Genet 1995;57:1019–27 [PMC free article] [PubMed] [Google Scholar]
- 71.Gregory JJ, Jr, Wagner JE, Verlander PC, Levran O, Batish SD, Eide CR, Steffenhagen A, Hirsch B, Auerbach AD. Somatic mosaicism in Fanconi anemia: evidence of genotypic reversion in lymphohematopoietic stem cells. Proc Natl Acad Sci U S A 2001;98:2532–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cawthon RM, Weiss R, Xu GF, Viskochil D, Culver M, Stevens J, Robertson M, Dunn D, Gesteland R, O'Connell P, et al. A major segment of the neurofibromatosis type 1 gene: cDNA sequence, genomic structure, and point mutations. Cell 1990;62:193–201 [DOI] [PubMed] [Google Scholar]
- 73.Viskochil D, Buchberg AM, Xu G, Cawthon RM, Stevens J, Wolff RK, Culver M, Carey JC, Copeland NG, Jenkins NA, White R, O'Connell P. Deletions and translocation interrupt a cloned gene at the neurofibromatosis type 1 locus. Cell 1990;62:187–92 [DOI] [PubMed] [Google Scholar]
- 74.Wallace MR, Marchuk DA, Andersen LB, Letcher R, Odeh HM, Saulino AN, Fountain JW, Brereton A, Nicholson J, Mitchell AL, Brownstein BH, Collins FS. Type 1 neurofibromatosis gene: Identification of a large transcript disrupted in three NF1 patients. Science 1990;249:181–6 [DOI] [PubMed] [Google Scholar]
- 75.Kehrer-Sawatzki H, Kluwe L, Sandig C, Kohn M, Wimmer K, Krammer U, Peyrl A, Jenne DE, Hansmann I, Mautner VF. High frequency of mosaicism among patients with neurofibromatosis type 1 (NF1) with microdeletions caused by somatic recombination of the JJAZ1 gene. Am J Hum Genet 2004;75:410–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mantripragada KK, Thuresson AC, Piotrowski A, Diaz de Stahl T, Menzel U, Grigelionis G, Ferner RE, Griffiths S, Bolund L, Mautner V, Nordling M, Legius E, Vetrie D, Dahl N, Messiaen L, Upadhyaya M, Bruder CE, Dumanski JP. Identification of novel deletion breakpoints bordered by segmental duplications in the NF1 locus using high resolution array-CGH. J Med Genet 2006;43:28–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Petek E, Jenne DE, Smolle J, Binder B, Lasinger W, Windpassinger C, Wagner K, Kroisel PM, Kehrer-Sawatzki H. Mitotic recombination mediated by the JJAZF1 (KIAA0160) gene causing somatic mosaicism and a new type of constitutional NF1 microdeletion in two children of a mosaic female with only few manifestations. J Med Genet 2003;40:520–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O'Kelly M, Tamayo P, Weir BA, Gabriel S, Winckler W, Gupta S, Jakkula L, Feiler HS, Hodgson JG, James CD, Sarkaria JN, Brennan C, Kahn A, Spellman PT, Wilson RK, Speed TP, Gray JW, Meyerson M, Getz G, Perou CM, Hayes DN. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer cell 2010;17:98–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Haferlach C, Grossmann V, Kohlmann A, Schindela S, Kern W, Schnittger S, Haferlach T. Deletion of the tumor-suppressor gene NF1 occurs in 5% of myeloid malignancies and is accompanied by a mutation in the remaining allele in half of the cases. Leukemia 2012;26:834–9 [DOI] [PubMed] [Google Scholar]
- 80.Dohner H, Stilgenbauer S, Benner A, Leupolt E, Krober A, Bullinger L, Dohner K, Bentz M, Lichter P. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med 2000;343:1910–16 [DOI] [PubMed] [Google Scholar]
- 81.Lehmann S, Ogawa S, Raynaud SD, Sanada M, Nannya Y, Ticchioni M, Bastard C, Kawamata N, Koeffler HP. Molecular allelokaryotyping of early-stage, untreated chronic lymphocytic leukemia. Cancer 2008;112:1296–305 [DOI] [PubMed] [Google Scholar]
- 82.Kujawski L, Ouillette P, Erba H, Saddler C, Jakubowiak A, Kaminski M, Shedden K, Malek SN. Genomic complexity identifies patients with aggressive chronic lymphocytic leukemia. Blood 2008;112:1993–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gunn SR, Bolla AR, Barron LL, Gorre ME, Mohammed MS, Bahler DW, Mellink CH, van Oers MH, Keating MJ, Ferrajoli A, Coombes KR, Abruzzo LV, Robetorye RS. Array CGH analysis of chronic lymphocytic leukemia reveals frequent cryptic monoallelic and biallelic deletions of chromosome 22q11 that include the PRAME gene. Leuk Res 2009;33:1276–81 [DOI] [PubMed] [Google Scholar]
- 84.Pfeifer D, Pantic M, Skatulla I, Rawluk J, Kreutz C, Martens UM, Fisch P, Timmer J, Veelken H. Genome-wide analysis of DNA copy number changes and LOH in CLL using high-density SNP arrays. Blood 2007;109:1202–10 [DOI] [PubMed] [Google Scholar]
- 85.Gunnarsson R, Staaf J, Jansson M, Ottesen AM, Goransson H, Liljedahl U, Ralfkiaer U, Mansouri M, Buhl AM, Smedby KE, Hjalgrim H, Syvanen AC, Borg A, Isaksson A, Jurlander J, Juliusson G, Rosenquist R. Screening for copy-number alterations and loss of heterozygosity in chronic lymphocytic leukemia—a comparative study of four differently designed, high resolution microarray platforms. Genes Chromosomes Cancer 2008;47:697–711 [DOI] [PubMed] [Google Scholar]
- 86.Grubor V, Krasnitz A, Troge JE, Meth JL, Lakshmi B, Kendall JT, Yamrom B, Alex G, Pai D, Navin N, Hufnagel LA, Lee YH, Cook K, Allen SL, Rai KR, Damle RN, Calissano C, Chiorazzi N, Wigler M, Esposito D. Novel genomic alterations and clonal evolution in chronic lymphocytic leukemia revealed by representational oligonucleotide microarray analysis (ROMA). Blood 2009;113:1294–303 [DOI] [PubMed] [Google Scholar]
- 87.Gunnarsson R, Isaksson A, Mansouri M, Goransson H, Jansson M, Cahill N, Rasmussen M, Staaf J, Lundin J, Norin S, Buhl AM, Smedby KE, Hjalgrim H, Karlsson K, Jurlander J, Juliusson G, Rosenquist R. Large but not small copy-number alterations correlate to high-risk genomic aberrations and survival in chronic lymphocytic leukemia: a high-resolution genomic screening of newly diagnosed patients. Leukemia 2010;24:211–15 [DOI] [PubMed] [Google Scholar]
- 88.Bernasconi P, Boni M, Cavigliano PM, Calatroni S, Giardini I, Rocca B, Zappatore R, Dambruoso I, Caresana M. Clinical relevance of cytogenetics in myelodysplastic syndromes. Ann N Y Acad Sci 2006;1089:395–410 [DOI] [PubMed] [Google Scholar]
- 89.Haase D. Cytogenetic features in myelodysplastic syndromes. Ann Hematol 2008;87:515–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Grand FH, Hidalgo-Curtis CE, Ernst T, Zoi K, Zoi C, McGuire C, Kreil S, Jones A, Score J, Metzgeroth G, Oscier D, Hall A, Brandts C, Serve H, Reiter A, Chase AJ, Cross NC. Frequent CBL mutations associated with 11q acquired uniparental disomy in myeloproliferative neoplasms. Blood 2009;113:6182–92 [DOI] [PubMed] [Google Scholar]
- 91.Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, Waghorn K, Zoi K, Ross FM, Reiter A, Hochhaus A, Drexler HG, Duncombe A, Cervantes F, Oscier D, Boultwood J, Grand FH, Cross NC. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet 2010;42:722–6 [DOI] [PubMed] [Google Scholar]
- 92.Boultwood J, Pellagatti A, McKenzie AN, Wainscoat JS. Advances in the 5q- syndrome. Blood 2010;116:5803–11 [DOI] [PubMed] [Google Scholar]
- 93.Bejar R, Levine R, Ebert BL. Unraveling the molecular pathophysiology of myelodysplastic syndromes. J Clin Oncol 2011;29:504–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tiu RV, Gondek LP, O'Keefe CL, Elson P, Huh J, Mohamedali A, Kulasekararaj A, Advani AS, Paquette R, List AF, Sekeres MA, McDevitt MA, Mufti GJ, Maciejewski JP. Prognostic impact of SNP array karyotyping in myelodysplastic syndromes and related myeloid malignancies. Blood 2011;117:4552–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Busque L, Patel JP, Figueroa ME, Vasanthakumar A, Provost S, Hamilou Z, Mollica L, Li J, Viale A, Heguy A, Hassimi M, Socci N, Bhatt PK, Gonen M, Mason CE, Melnick A, Godley LA, Brennan CW, Abdel-Wahab O, Levine RL. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat Genet 2012;44:1179–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ogawa T, Kitagawa M, Hirokawa K. Age-related changes of human bone marrow: a histometric estimation of proliferative cells, apoptotic cells, T cells, B cells and macrophages. Mech Ageing Dev 2000;117:57–68 [DOI] [PubMed] [Google Scholar]
- 97.Beerman I, Maloney WJ, Weissmann IL, Rossi DJ. Stem cells and the aging hematopoietic system. Curr Opin Immunol 2010;22:500–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Beerman I, Bhattacharya D, Zandi S, Sigvardsson M, Weissman IL, Bryder D, Rossi DJ. Functionally distinct hematopoietic stem cells modulate hematopoietic lineage potential during aging by a mechanism of clonal expansion. Proc Natl Acad Sci U S A 2010;107:5465–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rakyan VK, Down TA, Maslau S, Andrew T, Yang TP, Beyan H, Whittaker P, McCann OT, Finer S, Valdes AM, Leslie RD, Deloukas P, Spector TD. Human aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domains. Genome res 2010;20:434–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Berdasco M, Esteller M. Hot topics in epigenetic mechanisms of aging: 2011. Aging cell 2012;11:181–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cruzen C, Colman RJ. Effects of caloric restriction on cardiovascular aging in non-human primates and humans. Clin Geriatr Med 2009;25:733–43, ix–x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Harman D. The aging process: major risk factor for disease and death. Proc Natl Acad Sci U S A 1991;88:5360–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nilsson K, Ponten J. Classification and biological nature of established human hematopoietic cell lines. Int J Cancer 1975;15:321–41 [DOI] [PubMed] [Google Scholar]
- 104.Giovanella B, Nilsson K, Zech L, Yim O, Klein G, Stehlin JS. Growth of diploid, Epstein-Barr virus-carrying human lymphoblastoid cell lines heterotransplanted into nude mice under immunologically privileged conditions. Int J Cancer 1979;24:103–13 [DOI] [PubMed] [Google Scholar]
- 105.Conrad DF, Keebler JE, DePristo MA, Lindsay SJ, Zhang Y, Casals F, Idaghdour Y, Hartl CL, Torroja C, Garimella KV, Zilversmit M, Cartwright R, Rouleau GA, Daly M, Stone EA, Hurles ME, Awadalla P. Variation in genome-wide mutation rates within and between human families. Nat Genet 2011;43:712–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Londin ER, Keller MA, D'Andrea MR, Delgrosso K, Ertel A, Surrey S, Fortina P. Whole-exome sequencing of DNA from peripheral blood mononuclear cells (PBMC) and EBV-transformed lymphocytes from the same donor. BMC Genomics 2011;12:464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Reeser SL, Wenger SL. Failure of PHA-stimulated i(12p) lymphocytes to divide in Pallister-Killian syndrome. Am J Med Genet 1992;42:815–19 [DOI] [PubMed] [Google Scholar]
- 108.Priest JH, Rust JM, Fernhoff PM. Tissue specificity and stability of mosaicism in Pallister-Killian +i(12p) syndrome: relevance for prenatal diagnosis. Am J Med Genet 1992;42:820–4 [DOI] [PubMed] [Google Scholar]