Abstract
Background
HIV-1-infected individuals with higher viral set points progress to AIDS more rapidly than those with lower set points. Predicting viral set point early following infection can contribute to our understanding of early control of HIV-1 replication, to predicting long-term clinical outcomes, and to the choice of optimal therapeutic regimens.
Methods
In a longitudinal study of 10 untreated HIV-1-infected patients, we used gene expression profiling of peripheral blood mononuclear cells to identify transcriptional networks for viral set point prediction. At each sampling time, a statistical analysis inferred the optimal transcriptional network that best predicted viral set point. We then assessed the accuracy of this transcriptional model by predicting viral set point in an independent cohort of 10 untreated HIV-1-infected patients from Malawi.
Results
The gene network inferred at time of enrollment predicted viral set point 24 weeks later in the independent Malawian cohort with an accuracy of 87.5%. As expected, the predictive accuracy of the networks inferred at later time points was even greater, exceeding 90% after week 4. The composition of the inferred networks was largely conserved between time points. The 12 genes comprising this dynamic signature of viral set point implicated the involvement of two major canonical pathways: interferon signaling (p<0.0003) and membrane fraction (p<0.02). A silico knockout study showed that HLA-DRB1 and C4BPA may contribute to restricting HIV-1 replication.
Conclusions
Longitudinal gene expression profiling of peripheral blood mononuclear cells from patients with acute HIV-1 infection can be used to create transcriptional network models to early predict viral set point with a high degree of accuracy.
Keywords: Bioinformatics, HIV, systems biology, immunology, microbiology, HIV, microarray
Introduction
The best established parameter for predicting the rate of clinical progression in HIV-1-infected individuals is the set point plasma virus RNA level,1 usually called viral set point. By a few weeks after infection, peak virus replication is reached in most infected patients. As immune responses are mobilized, partial containment of HIV-1 occurs and a steady-state level of virus replication is reached. This viral set point is associated with the rate of disease progression.1 A high viral set point is associated with rapid disease progression, while a lower viral set point is associated with slower disease progression.
A delineation of the mechanisms that contribute to establishing the set point virus load in HIV-1-infected individuals would add important information to our understanding of how HIV-1 replication is contained. Studies in HIV-1-infected humans and simian immunodeficiency virus (SIV)-infected rhesus monkeys have implicated virus-specific CD8+ cytotoxic T lymphocytes in the early control of AIDS virus replication.2–4 Genome-wide association studies have underscored the importance of this cellular immune response in early HIV-1 control through demonstrating the contribution of MHC genes in determining viral set point.5 6 However, this genome-wide association studies work also suggests that other as yet undefined factors also contribute to early HIV-1 control.
The set point plasma HIV-1 RNA level can provide a useful clinical tool for determining the timing for initiating anti-retroviral therapy for infected individuals. For example, patients having high set point values can be started on aggressive anti-retroviral therapy and patients having low set point values can be monitored without initiating therapy. However, if an acutely-infected patient presents to a physician before set point virus replication is reached, the appropriate therapy for an infected individual can be difficult to determine. Therefore, a means of predicting set point plasma virus RNA levels in a recently infected patient would provide a useful tool for establishing a treatment strategy for that individual.
RNA microarray technology provides both a powerful tool for exploring mechanisms underlying biologic phenomena and a means of categorizing those phenomena into groups with differing clinical outcomes.7 Therefore, the whole blood RNA transcriptional profile of individuals during the period of acute HIV-1 infection may provide a gene expression signature that may be associated with particular clinical sequelae. The present study was done to determine computationally whether the expression of a limited network of genes by peripheral blood mononuclear cells sampled from HIV-1-infected individuals early after exposure to the virus can predict viral set point.
Results
Transcriptional network predictors of viral set point
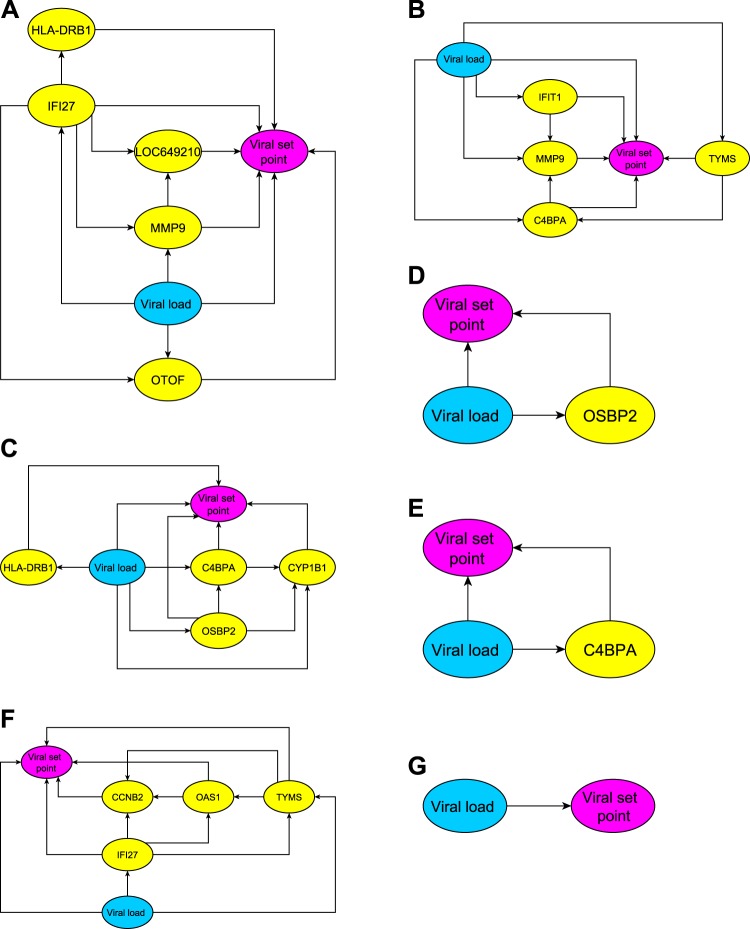
Viral set point predictors were developed from the longitudinal gene expression data from the first cohort of untreated, acutely HIV-1-infected individuals. We collected the peripheral blood mononuclear cells of the patients during a course of 24 weeks: enrollment, weeks 1, 2, 4, 12, and 24. At each time point, we used a multivariate statistical analysis, known as Bayesian networks, to infer a transcriptional network comprised of viral load and transcripts, and then identified the dependency pattern between the network and viral set point. Figure 1 shows the inferred networks at each time, where an arrow represents the target node's dependency on its source node. The network models were used to predict viral set point by substituting the viral load and the expression levels of the genes in the networks. Table 1 reports the predicted viral set points of individual patients obtained from leave-one-out cross-validation calculations. Table 2 summarizes the predictive accuracy, and shows that the transcriptional network models can achieve at least 90% accuracy.
Figure 1.
The transcriptional networks inferred from the mRNA or viral load data for predicting viral set point. An arrow indicates that the source node regulates its target node. The transcriptional networks suggest how gene products and viral replication may interact to modulate viral set point. Each network can further serve as a predictive model for determining viral set point: the viral set point can be determined by substituting the expression levels of the signature genes and viral load into the network model. The panels present the networks at different times: (A) enrollment, (B) week 1, (C) week 2, (D) week 4, (E) week 12, (F) week 24, and (G) using only viral load measurements.
Table 1.
Predicted viral set points at individual times of the leave-one-out cross-validation and the independent test
| Leave-one-out cross-validation | Measured viral set point | ||||||
| Enrol. | Week 1 | Week 2 | Week 4 | Week 12 | Week 24 | ||
| Subject A | NA | 4.35 | 4.92 | 4.78 | 4.76 | 5.01 | 4.92 |
| Subject B | 4.38 | NA | NA | 4.47 | 4.23 | 4.31 | 4.12 |
| Subject C | 3.76 | 3.97 | NA | NA | 3.44 | 3.62 | 3.56 |
| Subject D | 4.94 | 4.88 | 4.93 | 4.99 | 4.83 | 5.13 | 5.12 |
| Subject E | 4.82 | 4.68 | NA | NA | 4.94 | NA | 4.66 |
| Subject F | 5.06 | 5.16 | 4.82 | 5.10 | 5.04 | 4.77 | 5.20 |
| Subject G | 4.05 | 4.54 | 4.72 | 4.13 | 4.40 | 3.99 | 3.96 |
| Subject H | 5.01 | 5.15 | 5.04 | 4.97 | 4.92 | 5.35 | 5.19 |
| Subject I | 4.89 | 5.06 | 5.22 | 5.03 | 5.17 | 5.11 | 5.34 |
| Subject J | 4.01 | NA | NA | 4.26 | 4.14 | NA | 3.82 |
| Independent validation on Malawian data | Measured viral set point | ||||||
| Enrol. | Week 1 | Week 2 | Week 4 | Week 12 | Week 24 | ||
| Subject 1 | 3.06 | 3.70 | NA | NA | NA | 4.28 | 3.79 |
| Subject 2 | 3.69 | NA | NA | 3.49 | 3.47 | 4.00 | 3.51 |
| Subject 3 | 3.92 | NA | 4.14 | 4.06 | 3.67 | 4.53 | 4.51 |
| Subject 4 | 4.30 | 4.51 | 4.36 | 3.77 | 4.73 | 4.89 | 4.97 |
| Subject 5 | 4.82 | 4.93 | 4.70 | 5.08 | NA | 4.71 | 5.43 |
| Subject 6 | 4.12 | 4.13 | 3.95 | 4.02 | 3.62 | 4.32 | 3.67 |
| Subject 7 | 5.06 | 5.50 | 4.97 | 5.78 | 5.15 | 5.50 | 5.76 |
| Subject 8 | NA | NA | NA | NA | 3.92 | NA | 4.30 |
| Subject 9 | NA | 3.79 | NA | 3.83 | 3.84 | NA | 3.42 |
| Subject 10 | 2.29 | 3.43 | NA | NA | NA | NA | 2.19 |
Note: The not available entries are due to lack of quality RNA samples for gene expression assessment.
Table 2.
Accuracy of viral set point prediction in terms of normalized root-mean-square-error
| Leave-one-out cross-validation | ||||||
| Enrol. | Week 1 | Week 2 | Week 4 | Week 12 | Week 24 | |
| Transcriptional network model | 95.54% | 94.01% | 93.96% | 94.91% | 94.97% | 96.84% |
| Viral load alone prediction | 88.43% | 89.45% | 87.14% | 93.73% | 93.62% | 94.37% |
| Independent validation on Malawian data | ||||||
| Enrol. | Week 1 | Week 2 | Week 4 | Week 12 | Week 24 | |
| Transcriptional network model | 88.56% | 84.86% | 88.92% | 90.99% | 91.75% | 90.71% |
| Viral load alone prediction | 83.63% | 78.26% | 84.09% | 86.79% | 89.97% | 91.58% |
In addition to this leave-one-out cross-validation, we applied the network models to predict viral set points of an independent Malawian cohort. Table 1 reports the predicted viral set points of individual subjects, and table 2 presents the predictive accuracy at specified times. The independent validation achieves 87.5% predictive accuracy in the early weeks and over 90% accuracy in the later weeks following infection.
Signature genes of transcriptional network predictor
Table 3 lists the signature genes associated with the set point viral load prediction at specified times. There were only a small number of genes that, in association with viral load, predicted the set point viral load. There are more signature genes at early times following the infection, (from enrollment to week 2) than from week 4 to week 24. Therefore, in the early weeks following infection, when viral load was a less dominant predictor of set point, we need to take into account more transcripts interacting with virus to achieve an accurate prediction of set point viral load.
Table 3.
The signature genes in the transcriptional network models
| Enrol. | Week 1 | Week 2 | Week 4 | Week 12 | Week 24 |
| HLA-DRB1 | C4BPA | C4BPA | OSBP2 | C4BPA | CCNB2 |
| IFI27 | IFIT1 | CYP1B1 | IFI27 | ||
| LOC649210 | MMP9 | HLA-DRB1 | OAS1 | ||
| MMP9 | TYMS | OSBP2 | TYMS | ||
| OTOF |
There are 19 represented genes, 12 of which are unique. Six of the 12 genes are repeated (shown in boldface).
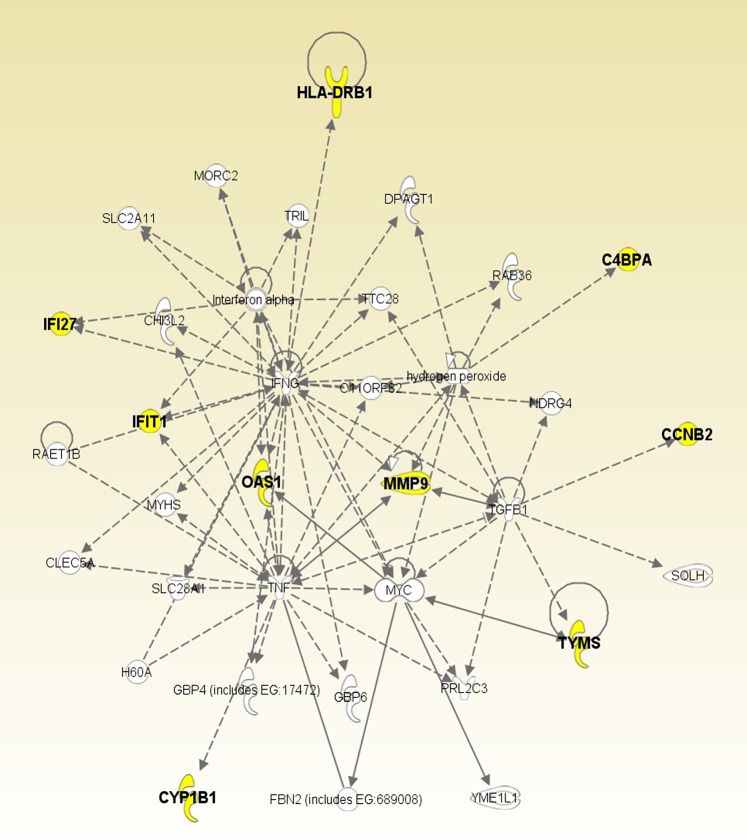
There are 12 genes in these networks (C4BPA, CCNB2, CYP1B1, HLA-DRB1, IFI27, IFIT1, LOC649210, MMP9, OAS1, OSBP2, OTOF, TYMS), 6 of which (C4BPA, HLA-DRB1, IFI27, MMP9, OSBP2, TYMS) are repeated. For example, C4BPA appears 3 times (weeks 1, 2, and 12). The pathway analyses performed using DAVID Bioinformatics Resources8 and Ingenuity Pathways Analysis (Ingenuity® Systems, http://www.ingenuity.com) shows that the association with interferon signaling (p<0.0003) and the membrane fraction (p<0.02) is over-represented in this collection of signature genes. Furthermore, 9 of the 12 signature genes (C4BPA, CCNB2, CYP1B1, HLA-DRB1, IFI27, IFIT1, MMP9, OAS1, TYMS) are related to each other through 4 hubs: hydrogen peroxide, IFNG, TGFB1, and TNF (figure 2). The crucial mechanisms associated with these genes are cell-cell signaling and interaction (p<0.001), cell cycle (p<0.001), molecular transport (p<0.001), immune cell trafficking (p<0.001), and hematological system development and function (p<0.01).
Figure 2.
Regulatory relationships among the signature genes. The names of the nine signature genes are shown in bold. The signature genes are compactly related to each other through 4 hubs: hydrogen peroxide, IFNG, TGFB1 and TNF.
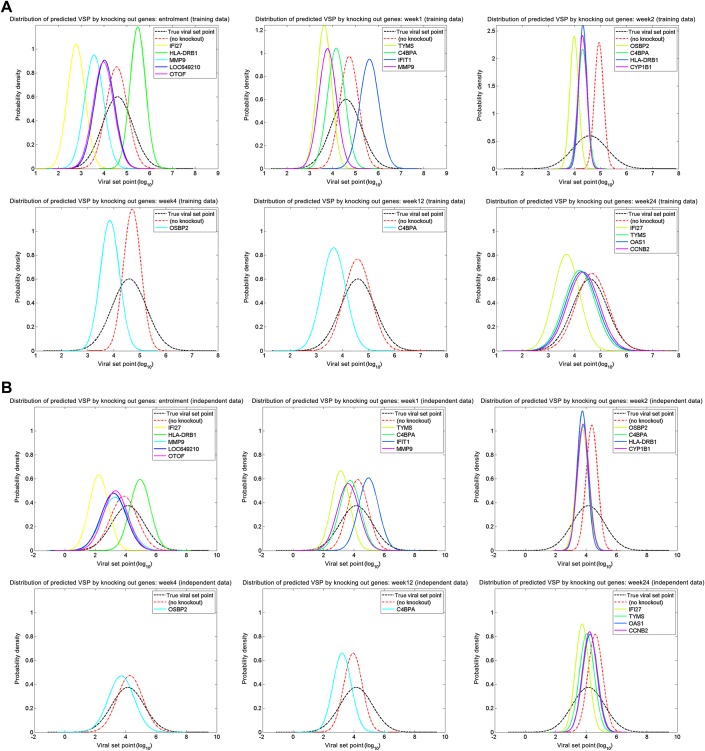
In silico knockout study
The viral set point predictors allowed us to conduct a computational knockout study. Knocking out a gene was computationally equivalent to setting the expression level of the gene to zero in the model. We knocked out a signature gene at a time, and then predicted viral set point. Figure 3 illustrates that the results, where each curve depicts the distribution of predicted viral set points when the corresponding gene was eliminated. When the predicted viral set point is greater than the actual value, it implies that the gene knockout inhibits HIV-1 replication; similarly, the predicted viral set point smaller than the actual value indicates that the gene knockout promotes HIV-1 replication. The results at early times (ie, enrollment and week 1) show that HLA-DRB1 and C4BPA are able to decrease HIV viremia; this finding is consistent with the ability of HLA genes to restrict AIDS progression.9 10
Figure 3.
In Silico Knockout Study. Each curve depicts the distribution of predicted viral set points when the corresponding gene was eliminated. When the predicted viral set point becomes greater (smaller) than the actual value, it implies that the gene knock-out inhibits (promotes) HIV replication. (A) Training data; (B) Independent test data.
Comparison between transcriptional network and viral load alone models
A viral load measurement correlated with viral set point with increasing accuracy, as the time of that measurement began to approximate the time of the steady-state level of viral replication in an individual.11 We created a univariate regression model that uses viral load alone to predict set point. A graphical representation of the regression model in which viral load is the single variable determining viral set point is shown in figure 1G. Table 2 also reports the predictive accuracy for viral set point generated using the regression models. At early times following infection (ie, enrollment, and weeks 1 and 2), the transcriptional network models predicted set point with 5% greater accuracy than the viral load alone model (p<0.05). As expected, when the time following infection more closely approximated chronic infection (weeks 4, 12, and 24), the predictive accuracy of the two types of models were comparable.
Comparison between transcriptional network and correlating predictors
We further contrast our transcriptional network with correlating predictors. Correlating predictors, or correlation-based predictors, are conventional techniques to search for the best transcripts whose expression levels statistically significantly correlate with continuous outcomes and are able to predict the outcomes. We used a statistical package in Matlab (MathWorks, Natick, Massachusetts, USA) to find the correlating transcripts of viral set point at each time. The results are summarized in table 4. We noted that only three transcripts (IFI27, HLA-DRB5, MMP9) were selected for viral set point prediction. However, the correlating predictors have worse predictive accuracy than transcriptional network model (p<0.005).
Table 4.
Correlating predictors and their predictive accuracy
| Enrol. | Week 1 | Week 2 | Week 4 | Week 12 | Week 24 | |
| Best predictive transcript | IFI27 | HLA-DRB5 | MMP9 | HLA-DRB5 | HLA-DRB5 | IFI27 |
| Leave-one-out cross-validation | 85.69% | 52.28% | 53.59% | 65.26% | 49.88% | 70.76% |
| Independent validation | 79.80% | 41.12% | 51.45% | 35.80% | 40.31% | 59.61% |
Discussion
Various strategies for predicting viral set point in HIV-1-infected humans and SIV-infected- or simian HIV (SHIV)-infected-rhesus monkeys have been explored. The peak plasma SIV RNA levels during primary infection are associated with set point levels in rhesus monkeys, with high peak values predicting high set point values.12 The magnitude of a vaccine-elicited CD8+ cytotoxic T lymphocyte response has also been shown to predict the post-challenge set point plasma virus RNA level in rhesus monkeys following a pathogenic CXCR4-tropic SHIV challenge.13 The utility of these predictive values in a clinical setting is, however, limited because of the difficulty of ascertaining when an individual became infected with HIV-1 and determining the magnitude of a cytotoxic T lymphocyte population.
The early containment of an AIDS virus infection is mediated to a significant extent by CD8+ cytotoxic T lymphocytes. This has been shown most dramatically in SIV-infected rhesus monkeys, when depletion of CD8+ lymphocytes by infusion of a monoclonal anti-CD8 antibody eliminates early virus containment, and animals die before they reach a set point level of virus replication.14 It is therefore likely that many of the signature gene transcripts are expressed in CD8+ T lymphocytes or cells that regulate these effector lymphocytes.
A number of the 12 transcripts associated with viral control in the present study have been implicated in HIV-1 human interactions. Some (IFI27, IFIT1, MMP9, and OAS1) have been shown to be induced by HIV-1 infection of cell lines in vitro.15–17 Some have been reported to interact directly with HIV-1 proteins: CCNB2 interacts with HIV-1 Vpr in the induction of cell cycle arrest; the expression of IFI27, MMP9 and OAS1 are upregulated by Tat.16 18–20 The group of transcripts also includes interferon inducible genes with well documented antiviral activity: IFI27, IFIT1 and OAS1.15–17 Interestingly, HLA-DRB1**1303 has been associated with decreased virus load as well as strong, polyfunctional mucosal CD4+ T cell responses in HIV-1-infected individuals.21 22 Other of these transcripts have not previously been associated with HIV-1 biology or infections, and two have no previously reported functions.
A microarray assay focused on the signature transcripts defined in this study might be used in a clinical setting. Rather than employing an array that monitors 44 000 gene transcripts like the one used in this study, a microarray assay focused on the limited number of signature genes defined in the present study could be devised. Such an assay might then be used in association with other clinical data to determine whether a newly diagnosed HIV-1-infected patient should receive immediate treatment or monitored until the progression of disease warrants instituting treatment.
Materials and methods
Patient information
A cohort of 10 acute HIV-1-infected patients was enrolled from 2 USA and 6 African sites. In this cohort, there are 2 Caucasians, 1 African American, 2 African Blacks, 2 African Chewas, 1 African Ngoni, 1 African Lomwe, and 1 African Tumbuka. At enrollment they were verified as acute Fiebig stages 4 to 6 (plasma RNA+, third generation EIA+, Western blot indeterminant or +).23 Follow-up samples were collected from patients at 1, 2, 4, 12, and 24 weeks post-enrollment. All patients were untreated throughout the 24-week period of study.
A second cohort of 10 untreated Malawians with acute HIV-1 infection in Fiebig stages 4–6 was enrolled in the study as an independent validation set. Blood samples from these patients were collected at enrollment and at the same intervals as the first cohort (at 1, 2, 4, 12, and 24 weeks after enrollment).
Whole blood sample collection and microarray hybridization
Whole blood was collected using standardized conditions into Tempus vacutainer tubes. Total RNA was isolated from lysates of the whole blood.24 All samples passing quality control were then amplified and labeled using the Illumina TotalPrep-96 RNA amplification kit. Amplified RNA was then hybridized to Illumina HT-12 V3 beadchips and scanned on an Illumina Beadstation 500 according to the protocol detailed by the manufacturer (http://www.illumina.com). The gene expression of the first cohort was assessed at Baylor Institute for Immunology Research, and the second cohort was assessed at Duke University.
Viral set point measurement
Viral set point, the steady-state viral load after acute infection, was determined for all subjects by an experienced infectious-disease clinician following the standard protocol.5
Statistical analysis of transcriptional network
To model the interactions among transcripts, viral load, and viral set point, we carried out a multivariate dependency analysis using dynamic Bayesian networks.25 26 Such an analysis has already been applied to several types of genomic data including gene regulation,27 protein-protein interactions,28 SNPs29 30 and pedigrees.31 A Bayesian network is a directed acyclic graph in which nodes represent random variables and arcs define directed dependencies quantified by conditional probability distributions. Besides inferring transcriptional interactions, Bayesian networks can be exploited for prediction. Thus, we reasoned that a network capturing the relationship between transcripts and viral set point might be used to compute the most probable quantity for viral set point when given the levels of an individual's expression of these transcripts.
Our analysis began by selecting gene transcripts with at least a twofold change in expression with respect to enrollment over the entire 24-weeks of the study. We then log10-transformed all data, including gene expression, viral loads and viral set points. Finally, we analyzed the data using a Bayesian network method. Our aim was to search for the most probable network of gene-gene dependency for each time point. To find such a network, the analysis explored a space of different network models, scored each model by its posterior probability conditional on the available data, and returned the model with maximum posterior probability. This probability was computed by Bayes theorem as , where p(D|M) is the probability that the observed data are generated from the network model M and p(M) is the prior probability encoding knowledge about the model M before seeing any data. We assumed that all models were equally likely a priori, so p(M) is uniform and p(M|D) becomes proportional to p(D|M), a quantity known as marginal likelihood. The marginal likelihood averages the likelihood functions for different parameter values and is calculated as where p(D|θ) is the traditional likelihood function and p(θ) is the prior probability density of parameters. When we assumed the log-transformed data to be Gaussian, has a closed-form solution.25 We used a greedy search algorithm25 with order permutation to identify the most probable network model with highest marginal likelihood .
Evaluation of predictive accuracy
Viral set point is a continuous quantity that we want to predict. The popular method to evaluate the prediction of continuous variables is based on root-mean-square-error (RMSE):
where N is the number of subjects, and and are the predicted and actual values, respectively. However, this method does not explain the degree of error/accuracy. When two distinct cohorts share the same predictive RMSE, one cohort with higher actual values has greater predictive accuracy than the other. To better evaluate the predictive accuracy, we in this paper define the accuracy A as follows:
where NRMSE is the normalized RMSE. Unlike RMSE, the normalized RMSE encodes the error rate, and helps us better quantify the predictive accuracy.
Predictive validation
We used the first data set to infer a transcriptional network model. To assess the robustness of the transcriptional network for sampling variability, we used a leave-one-out cross-validation strategy: a single observation from the data was treated as the validation sample, the remaining observations were used to re-estimate the parameters of the network model, and the newly parameterized model was used to predict the viral set point of the validation sample. This process was repeated until each observation in the data set was used once as the validation sample. We then evaluated the predicted set points by mean error rates deviating from the true values.
To confirm the results, we used the transcriptional network models learned from the first data set to predict the viral set points of 10 Malawian patients who were not included in the network learning process. The validation was performed by quantile-normalizing the Malawian data to the first data, followed by log10-transformation and viral load prediction. The measure of predictive accuracy was the same as the leave-one-out cross-validation.
Footnotes
Contributors: HHC designed the method, conducted the analysis, and prepared the manuscript; KAS collected Malawian samples; JAS, JB, and DC collected samples for building up transcriptional network models; BFH, MFR, and NLL directed the study and prepared the manuscript.
Funding: This work was supported by NIH grant 5U19A1067854-05.
Competing interests: None.
Patient consent: Obtained.
Ethics approval: Ethics approval was provided by Duke Human Vaccine Institute, Duke University.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1. Mellors JW, Rinaldo CR, Jr, Gupta P, et al. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 1996;272:1167–70 [DOI] [PubMed] [Google Scholar]
- 2. Koup RA, Safrit JT, Cao Y, et al. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J Virol 1994;68:4650–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kuroda MJ, Schmitz JE, Charini WA, et al. Emergence of CTL coincides with clearance of virus during primary simian immunodeficiency virus infection in rhesus monkeys. J Immunol 1999;162:5127–33 [PubMed] [Google Scholar]
- 4. Schmitz JE, Kuroda MJ, Santra S, et al. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 1999;283:857–60 [DOI] [PubMed] [Google Scholar]
- 5. Fellay J, Shianna KV, Ge D, et al. A whole-genome association study of major determinants for host control of HIV-1. Science 2007;317:944–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pereyra F, Jia X, McLaren PJ, et al. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 2010;330:1551–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Golub TR, Slonim DK, Tamayo P, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 1999;286:531–7 [DOI] [PubMed] [Google Scholar]
- 8. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44–57 [DOI] [PubMed] [Google Scholar]
- 9. Carrington M, Nelson GW, Martin MP, et al. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science 1999;283:1748–52 [DOI] [PubMed] [Google Scholar]
- 10. Kiepiela P, Leslie AJ, Honeyborne I, et al. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 2004;432:769–75 [DOI] [PubMed] [Google Scholar]
- 11. Kelley CF, Barbour JD, Hecht FM. The relation between symptoms, viral load, and viral load set point in primary HIV infection. J Acquir Immune Defic Syndr 2007;45:445–8 [DOI] [PubMed] [Google Scholar]
- 12. Whitney JB, Luedemann C, Bao S, et al. Monitoring HIV vaccine trial participants for primary infection: studies in the SIV/macaque model. AIDS 2009;23:1453–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tsukamoto T, Yuasa M, Yamamoto H, et al. Induction of CD8+ cells able to suppress CCR5-tropic simian immunodeficiency virus SIVmac239 replication by controlled infection of CXCR4-tropic simian-human immunodeficiency virus in vaccinated rhesus macaques. J Virol 2007;81:11640–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Matano T, Shibata R, Siemon C, et al. Administration of an anti-CD8 monoclonal antibody interferes with the clearance of chimeric simian/human immunodeficiency virus during primary infections of rhesus macaques. J Virol 1998;72:164–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cheon H, Stark GR. Unphosphorylated STAT1 prolongs the expression of interferon-induced immune regulatory genes. Proc Natl Acad Sci U S A 2009;106:9373–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim SY, Li J, Bentsman G, et al. Microarray analysis of changes in cellular gene expression induced by productive infection of primary human astrocytes: implications for HAD. J Neuroimmunol 2004;157:17–26 [DOI] [PubMed] [Google Scholar]
- 17. Reynolds JL, Mahajan SD, Aalinkeel R, et al. Methamphetamine and HIV-1 gp120 effects on lipopolysaccharide stimulated matrix metalloproteinase-9 production by human monocyte-derived macrophages. Immunol Invest 2011;40:481–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mouland AJ, Coady M, Yao XJ, et al. Hypophosphorylation of poly(A) polymerase and increased polyadenylation activity are associated with human immunodeficiency virus type 1 Vpr expression. Virology 2002;292:321–30 [DOI] [PubMed] [Google Scholar]
- 19. Izmailova E, Bertley FM, Huang Q, et al. HIV-1 Tat reprograms immature dendritic cells to express chemoattractants for activated T cells and macrophages. Nat Med 2003;9:191–7 [DOI] [PubMed] [Google Scholar]
- 20. Ju SM, Song HY, Lee JA, et al. Extracellular HIV-1 Tat up-regulates expression of matrix metalloproteinase-9 via a MAPK-NF-kappaB dependent pathway in human astrocytes. Exp Mol Med 2009;41:86–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Julg B, Moodley ES, Qi Y, et al. Possession of HLA class II DRB1*1303 associates with reduced viral loads in chronic HIV-1 clade C and B infection. J Infect Dis 2011;203:803–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ferre AL, Hunt PW, McConnell DH, et al. HIV controllers with HLA-DRB1*13 and HLA-DQB1*06 alleles have strong, polyfunctional mucosal CD4+ T-cell responses. J Virol 2010;84:11020–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fiebig EW, Wright DJ, Rawal BD, et al. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS 2003;17:1871–9 [DOI] [PubMed] [Google Scholar]
- 24. Ovcharenko D, Jarvis R, Hunicke-Smith S, et al. High-throughput RNAi screening in vitro: from cell lines to primary cells. RNA 2005;11:985–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chang HH, Ramoni MF. Transcriptional network classifiers. BMC Bioinformatics 2009;10(Suppl 9):S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chang HH, Ramoni MF. Inferring nonstationary gene networks from longitudinal gene expression microarrays. J Signal Processing Systems 2011;65:261–73 [Google Scholar]
- 27. Friedman N. Inferring cellular networks using probabilistic graphical models. Science 2004;303:799–805 [DOI] [PubMed] [Google Scholar]
- 28. Jansen R, Yu H, Greenbaum D, et al. A Bayesian networks approach for predicting protein-protein interactions from genomic data. Science 2003;302:449–53 [DOI] [PubMed] [Google Scholar]
- 29. Sebastiani P, Ramoni MF, Nolan V, et al. Genetic dissection and prognostic modeling of overt stroke in sickle cell anemia. Nat Genet 2005;37:435–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chang HH, McGeachie M, Alterovitz G, et al. Mapping transcription mechanisms from multimodal genomic data. BMC Bioinformatics 2010;11(Suppl 9):S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lauritzen SL, Sheehan NA. Graphical models for genetic analysis. Statist Sci 2004;18:489–514 [Google Scholar]