Abstract
Objective
To develop and evaluate a text mining system for extracting key clinical features from vaccine adverse event reporting system (VAERS) narratives to aid in the automated review of adverse event reports.
Design
Based upon clinical significance to VAERS reviewing physicians, we defined the primary (diagnosis and cause of death) and secondary features (eg, symptoms) for extraction. We built a novel vaccine adverse event text mining (VaeTM) system based on a semantic text mining strategy. The performance of VaeTM was evaluated using a total of 300 VAERS reports in three sequential evaluations of 100 reports each. Moreover, we evaluated the VaeTM contribution to case classification; an information retrieval-based approach was used for the identification of anaphylaxis cases in a set of reports and was compared with two other methods: a dedicated text classifier and an online tool.
Measurements
The performance metrics of VaeTM were text mining metrics: recall, precision and F-measure. We also conducted a qualitative difference analysis and calculated sensitivity and specificity for classification of anaphylaxis cases based on the above three approaches.
Results
VaeTM performed best in extracting diagnosis, second level diagnosis, drug, vaccine, and lot number features (lenient F-measure in the third evaluation: 0.897, 0.817, 0.858, 0.874, and 0.914, respectively). In terms of case classification, high sensitivity was achieved (83.1%); this was equal and better compared to the text classifier (83.1%) and the online tool (40.7%), respectively.
Conclusion
Our VaeTM implementation of a semantic text mining strategy shows promise in providing accurate and efficient extraction of key features from VAERS narratives.
Keywords: Medical informatics
Introduction
Spontaneous reporting systems (SRS), such as the vaccine adverse event reporting system (VAERS) play an important role in providing early evidence of new, serious, and unexpected adverse events after the use of medical products. In SRS, safety signals are typically identified using both qualitative and quantitative methods requiring time-intensive manual report review by medical experts. The standard approach to organizing the clinical data is to derive a description of the key features, including the diagnosis, time to onset, and alternative explanations that can be summarized across multiple cases for a ‘case series’ evaluation. Further evaluation of the summary data seeks unusual patterns among the key features or might include analysis of disproportionate reporting of a diagnosis after some medical products compared with others.1 During the period 2006–11, the average number of reports per year more than doubled from the previous 5 years to 32 200; this trend of increasing numbers of reports to VAERS makes the development of automated tools essential.
To increase the efficiency of manual report review effectively, any automated tool must reliably: (1) recognize and differentiate between merely a reported symptom and an actual diagnosis in the narrative; (2) extract the key features that help determine whether a real association exists between a medical exposure and a reported outcome (eg, timing of both the exposure and outcome, alternative explanations for the event such as past medical history and co-administered medications; (3) reduce the amount of text required to be read and interpreted; and (4) tag and organize the key medical concepts after extraction from the raw report data.
A variety of methods has been employed to process medical text, extract facts, and/or recognize a specific range of adverse events in patient records, but each of these methods has certain limitations.2–4 For example, machine learning techniques offer a promising solution but require large pre-annotated corpora for training, consuming considerable human resources.5 6 Similarly, other approaches based on the construction of controlled dictionaries are considered to be laborious, demanding, and costly because they must be informed by specialist knowledge.7 A number of text mining systems perform a part-of-speech-based tagging and shallow parsing that are followed by the named entity recognition, such as the Genia,8 cTakes6 and MedTAS/P systems.9 MedLEE grammar combines semantic and syntactic co-occurrence patterns.10 These systems do not have the capability to extract the key features required for safety surveillance using the case series framework without modification. While modification of one of these systems might be possible, we considered it would be simpler to develop a self-contained system. In this paper, we describe the development and evaluation of a text mining system specifically designed for VAERS that combines semantic tagging with rule-based techniques, to identify key clinical features and facilitate adverse event review.
Background
VAERS collects reports of adverse events following immunization (AEFI) with any US-licensed vaccine.11 AEFI reports may be submitted by medical experts and other members of the public. Safety surveillance in VAERS has two main purposes: first, to identify new and unexpected AEFI; second, to characterize further the safety profile of a vaccine by reviewing previously described AEFI in the context of comorbidities and other medical products among the general population.
Methods
With the case series evaluation framework as an overarching guide, five US Food and Drug Administration (FDA) medical epidemiologists with experience reviewing VAERS reports identified, by consensus, the set of key features of VAERS reports that should be extracted: (1) primary diagnostic features: diagnosis and the cause of death; (2) secondary diagnostic features: second-level diagnosis (stated as ‘ssessment’, ‘impression’, or ‘possible’ diagnosis), rule-out diagnosis, other reported symptoms and treatment; (3) causal assessment features: time to the onset of the first symptoms following vaccination, family and medical history; and (4) quality assessment features: the vaccine exposures and lot numbers. In this paper we focus on the performance of the first three feature categories.
Vaccine adverse event text mining
The Vaccine adverse event Text Mining (VaeTM) system comprises multiple components (see table 1) that together process the free text of VAERS reports and extract 11 predefined features (see supplementary figure 1, available online only). MedLEE developers argued that it is the semantic and not the syntactic patterns in the clinical documents that determine the underlying interpretation.12 This observation is particularly valid for VAERS narratives and inspired us in following a purely semantic text mining strategy. For example, ‘epinephrine’, ‘pain’ and ‘stomach’ were not considered as nouns but as ‘Drug’, ‘Symptom’ and ‘Anatomy’, respectively. Therefore, ‘pain’ could be either combined with ‘stomach’ in ‘stomach pain’ or used as a single term. Our semantic types corresponded to:
Table 1.
VaeTM components and their functions
| Components and subcomponents | Functions |
| 1. Pre-processor | Prepares the text for the main processing. |
| 1.1 Sentence tokenizer | Splits text into sentences using a period. |
| 1.2 Word tokenizer | Splits each sentence into tokens. |
| 1.3 Normalizer | Removes punctuation marks and converts text to lowercase (1st normalization step). |
| Removes the tokens tagged as ‘Unimportant’ after their tagging by the semantic tagger (2nd normalization step). | |
| Removes an irrelevant tagged token that disrupts the contiguous tokens of a feature (3rd normalization step). | |
| 2. VAERS dictionary | Includes 55 000 entries (each entry includes to a term and its tag that corresponds to a semantic type). |
| 3. Semantic tagger | Tags the tokens based on the dictionary entries. |
| 4. Grammar rules | Define the relationships between tags (ie, the semantic types). |
| 5. Rule-based parser | Parses the text by executing the grammar rules after: (1) the 2nd normalization step, and (2) the 3rd normalization step. |
| 6. Features extractor | Extracts the predefined features. |
VAERS, vaccine adverse event reporting system; VaeTM, vaccine adverse event text mining.
symptoms, diagnoses, drugs, and anatomical terms;
acronyms, modifiers and general terms;
elements that composed the information related to vaccination, for example, time modifiers and time units;
terms denoting negation, probability, impression, assessment and certainty.
The ‘Modifier’ type included two main groups of terms: (1) syndrome/disease names (terms such as ‘Sjogren’), and (2) other attributive modifiers (adjectives such as ‘cardiovascular’, participles such as ‘persisting’ and adverbs such as ‘continuously’). Furthermore, the ‘generalTerm’ type included more general medical terms (nouns such as ‘syndrome’ and ‘injury’). Any term that did not fit into any of the above distinct types was not included in any of the above semantic types and was considered as ‘Unimportant’.
Our semantic text mining strategy was supported by two of the VaeTM components: the VAERS dictionary and the grammar rules.
VAERS dictionary
The existing (vaccine) adverse event ontologies and terminologies have been either inadequately described13 or developed for different purposes.14 Therefore, a dictionary of approximately 55 000 entries made out of other dictionaries and resources was created with each entry including the term and the appropriate tag corresponding to the semantic type, for example, ‘benzphetamine-Drug’. The multi-word phrases (eg, ‘acute abdomen’) representing symptoms or diagnoses were not added as phrase entries to the dictionary but were composed from the single-word terms with the help of grammar rules (described below). In accordance with the principles of the ontology-driven information extraction,15 this technique allowed the development and the active use of a flexible semantic dictionary.
Various resources were used to create the VAERS dictionary: SNOMED CT, Medical Dictionary for Regulatory Activities (MedDRA), the NIH SPECIALIST lexical tools, two medical textbooks,16 17 the Drugs@FDA database,18 and the list of vaccines published by the US Centers for Disease Control and Prevention.8 The extraction of temporal information related to vaccination was supported by the inclusion of terms tagged as: (1) time modifiers and units (see supplementary table 1, available online only), and (2) numerical words. Supplementary table 1 (available online only) also includes abbreviations and the terms denoting negation, probability, impression, assessment, and certainty.
As in MedEx,19 the VaeTM semantic tagger combined look-up and regular expression methods to map the tokens of the free text to the entries of the VAERS dictionary and tag them accordingly. In addition, the VaeTM parser and the normalizer allowed the composition of the multi-word phrases no matter whether the individual words were contiguous or not, a problem that has been identified but treated differently before.20
Grammar rules
The grammar rules supported the extraction of the 11 predefined features; each rule incorporated certain tags and, in some cases, other nested rules. Recognizing the difficulty to differentiate consistently between symptoms and diagnoses (eg, ‘diarrhea’ is reported either as a diagnosis or as a symptom in VAERS), we created two sets of rules. First, to support the extraction of medical terms that were described as symptoms and diagnoses in the sources we used for the development of our dictionary, we created two basic rules (namely ‘MAIN_SYMPTOM’ and ‘MAIN_DIAGNOSIS’, respectively). The basic rules combined a number of tags (‘Symptom’, ‘Diagnosis’, ‘Acronym’, ‘Modifier’, ‘Anatomy’, and ‘generalConcept’) and were treated as semantically equal, ie, symptoms were not distinguished from diagnoses; this was accomplished in the next step with the main rules.
Second, 11 main rules were formed to support the extraction of the predefined features. Some of them incorporated tags for certain keywords that: (1) acted as ‘triggers’ for certain features, for example, ‘dx’ for ‘DIAGNOSIS’, and (2) represented the contextual information of negation, possibility, impression, assessment and certainty. Previous natural language processing studies have reported the use of triggers to extract contextual features, such as negation (NegEx algorithm),21 temporality (ConText system),22 smoking status (HITEx system)23 and uncertainty (MedLEE system).10 Certain separators (commas, semicolons, the words ‘and/or’ and the symbol ‘&’) were incorporated into the grammar rules to separate contiguous tokens appropriately (eg, to split the sequence ‘dx anaphylaxis and urticarial rash’ into two diagnoses ‘anaphylaxis’ and ‘urticarial rash’). Also, some secondary rules were created better to support the extraction of contextual features and along with the basic rules were nested within some of the main rules. Supplementary appendix 1 (available online only) includes the details about the structure of the main rules.
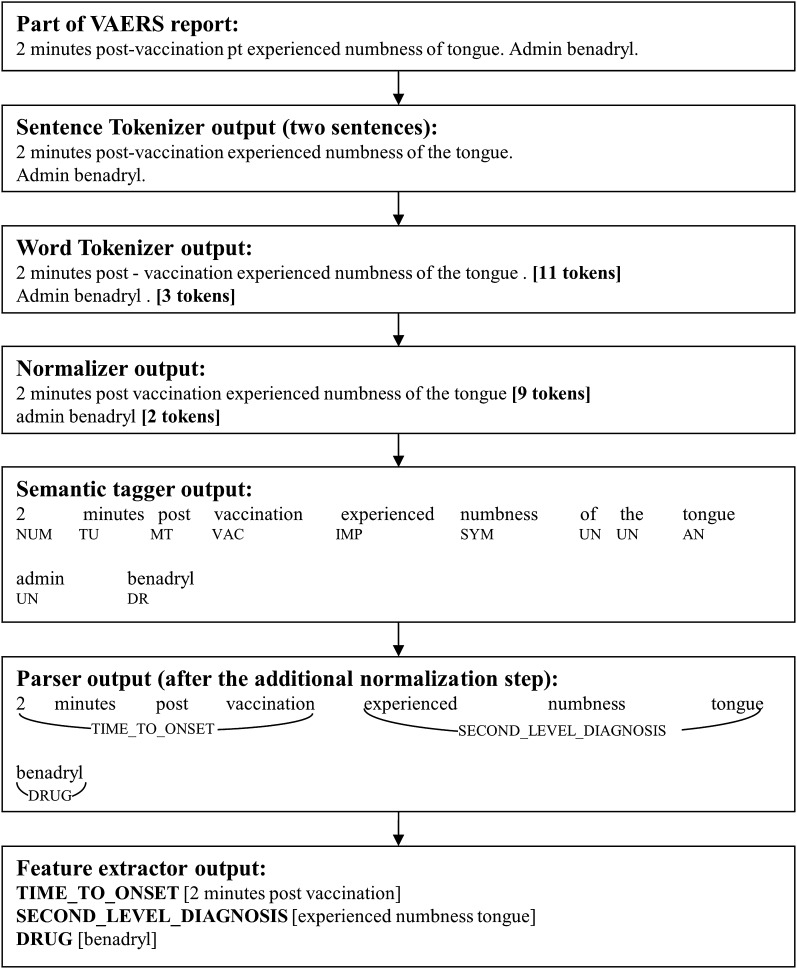
A fully worked example of a sentence processed through VaeTM components is illustrated in figure 1. Python (version 2.6.4) was used to develop the VaeTM.
Figure 1.
Example of a sentence processed through VaeTM components. AN, Anatomy; DR, Drug; IMP, Impression; MT, ModifierTime; NUM, Number; SYM, Symptom; TU, TimeUnit; UN, Unimportant; VAC, Vaccination; VAERS, vaccine adverse event reporting system.
Evaluation
There were two purposes of the evaluation process. First, to assess the completeness of the VaeTM automated feature extraction compared to the manual feature extraction by physicians applying the same extraction rules. Second, to explore the feasibility of VaeTM output for an information retrieval (IR)-based approach at the first major step in the case series evaluation, namely case classification.
Completeness of feature extraction
In the first part, three evaluation rounds were performed. In the first evaluation, a corpus of 100 text files was created from VAERS reports received from 3 February 2011 to 28 February 2011. The second evaluation consisted of a corpus of equal size from a random selection of reports submitted to VAERS in 2010. A third evaluation of a subset of reports with high frequency of TIME_TO_ONSET and MEDICAL_HISTORY features was also conducted. We applied a query to all VAERS reports that were received before 1 January 2010 using specific keywords and key phrases (such as ‘history’ and ‘after vaccination’). From the resulting subset (N=1265) 100 reports were randomly selected.
All corpora were split into two sets of 50 text files each and each set was independently reviewed by experienced VAERS reviewers. The descriptive statistics for the three corpora are presented in table 2.
Table 2.
Brief statistics for the three corpora used in the evaluations
| No of reports | Total no of sentences (mean±SD)* | Total no of tokens (mean±SD)† | Total no of unique tokens‡ | |
| Corpus 1 | 100 | 2206 (22±14) | 22 076 (10±8) | 3013 |
| Corpus 2 | 100 | 1770 (18±12) | 17 589 (10±9) | 2650 |
| Corpus 3 | 100 | 1600 (16±7) | 22 422 (14±11) | 2786 |
Mean±SD over the reports.
Mean±SD over the sentences.
After removing stop-words and conjunctions, numbers, duplicates, dates (full dates, months and weekdays) and misspelled words.
Three medical reviewers participated in the three evaluations; two as primary manual extractors and one as the consensus annotator. All medical reviewers attended a 1-h training session in which the schema for the annotation of reports was described. The consensus annotator reviewed the annotations of the primary extractors and resolved any deviations from the reference schema to develop the ‘reference standard’ that was used for comparison with the VaeTM output. Medical reviewers followed the same principles that the VaeTM algorithm was built on. The VaeTM was modified after the first evaluation to improve performance during the second evaluation.
To evaluate the primary extractors' adherence to the reference schema, we calculated an adjusted version of the inter annotator agreement (IAA) per feature24:
where matched_annotations included the number of features that were correctly annotated by the primary annotators, and total_annotations the matched, the missed and the incorrectly annotated features.
VaeTM processed the text files of the corpora; the extracted features were compared with the ‘reference standard’. The consensus annotator identified the full, conceptual and non-matches per feature in each report. A match was considered to be ‘full’ when there was a complete textual agreement between the reference standard and the machine feature extraction (true positive (TP) cases); ‘conceptual’ when there was some textual agreement with minor contextual differences; ‘none’ when either there was no agreement or when VaeTM failed to extract any text for an existing feature (false negative (FN) cases). The ‘non-matches’ also included cases in which VaeTM extracted a non-existing feature (false positive cases). Based on the above counts the standard text extraction metrics of recall, precision and F-measure were calculated per feature:
We also made the assumption that ‘conceptual-matches’ could be treated either as TP or FN based on whether they weighed the same as a full or non-match, respectively. Based on this assumption, we calculated the lenient (conceptual-match as a TP case) and the strict version (conceptual-match as a FN case) of the above metrics.
We also conducted a qualitative error analysis to examine the differences between the ‘reference standard’ and the VaeTM output. Guided by the approach of Roberts et al,24 we identified cases in which VaeTM did not:
Extract a feature (occurrence).
Extract all the elements of a feature (textual extent).
Assign the same feature type for the same span of text (typing).
Separate a multi-element feature (term decomposition).
Select all the appropriate neighboring modifiers for a key term (granularity).
Assign a term to the correct feature due to its ambiguity (term ambiguity).
Capture the anatomic sub-locations (locus specification).
Furthermore, we examined the sensitivity and specificity of the ability of VaeTM to extract a diagnosis ‘fully’ or ‘conceptually’ from the free text. To measure this, we calculated the proportion of reports with at least one correct diagnosis (as there may be more than one diagnosis per report) extracted from the VAERS report in the first and second corpus.
Use case: IR for case classification
We recently developed a rule-based text mining algorithm to identify possible anaphylaxis reports over a large set of VAERS reports; the rules of the algorithm represented the criteria of the Brighton collaboration (BC) case definition for anaphylaxis.25 Although useful, the further development of rule-based systems is limited by the labor required to customize the rules for a broad range of individual conditions. To explore the potential for automated case classification, we used a previously developed training and testing set that were randomly formed from 6034 VAERS reports for H1N1 vaccine.25 These reports had been classified by the FDA medical experts as potentially positive or negative for anaphylaxis.
VaeTM processed the symptom text of each report and extracted the corresponding features. The medical terms included in the DIAGNOSIS, the CAUSE_OF_DEATH, the SECOND_LEVEL_DIAGNOSIS and the SYMPTOM features were mapped to the medical terms that appear in the major and minor criteria of the BC case definition for anaphylaxis.26
According to IR theory, it is possible to represent both the reports and the anaphylaxis queries as vectors whose components are the criteria of the BC case definition. Then a score can be assigned to each report by calculating the cosine similarity of the two vectors (see supplementary appendix 2, available online only).27 After calculating the scores for all reports in the training and testing set, we used the scores of the training set as the predictors and the medical experts' classification as the gold standard to build the receiver operating characteristic (ROC) curve; specificity and sensitivity were calculated for the best cut-off point. The scores of the testing set were used to evaluate the performance of this approach.
We compared the performance of this method with both a text classification algorithm for anaphylaxis (hereafter, ‘Text classifier’) developed in our previous study25 and an online classifier (‘ABC tool’), which is offered by the BC to its members.28 The ‘ABC tool’ allows confirmation of anaphylaxis based on user input for specific criteria of the corresponding case definitions. The same VaeTM terms that were mapped to the BC criteria were also used to feed the ‘ABC tool’. The ‘ABC tool’ will not perform any classification for anaphylaxis when both criteria are not satisfied: (1) sudden onset and (2) rapid progression of signs and symptoms. As this information is not always available in the narratives of spontaneous reports that later prove to meet the case definition, the use of these criteria for the initial screening of reports would be counterproductive, so we considered them as present to facilitate the process.
Results
Quantitative difference analysis (completeness of extraction)
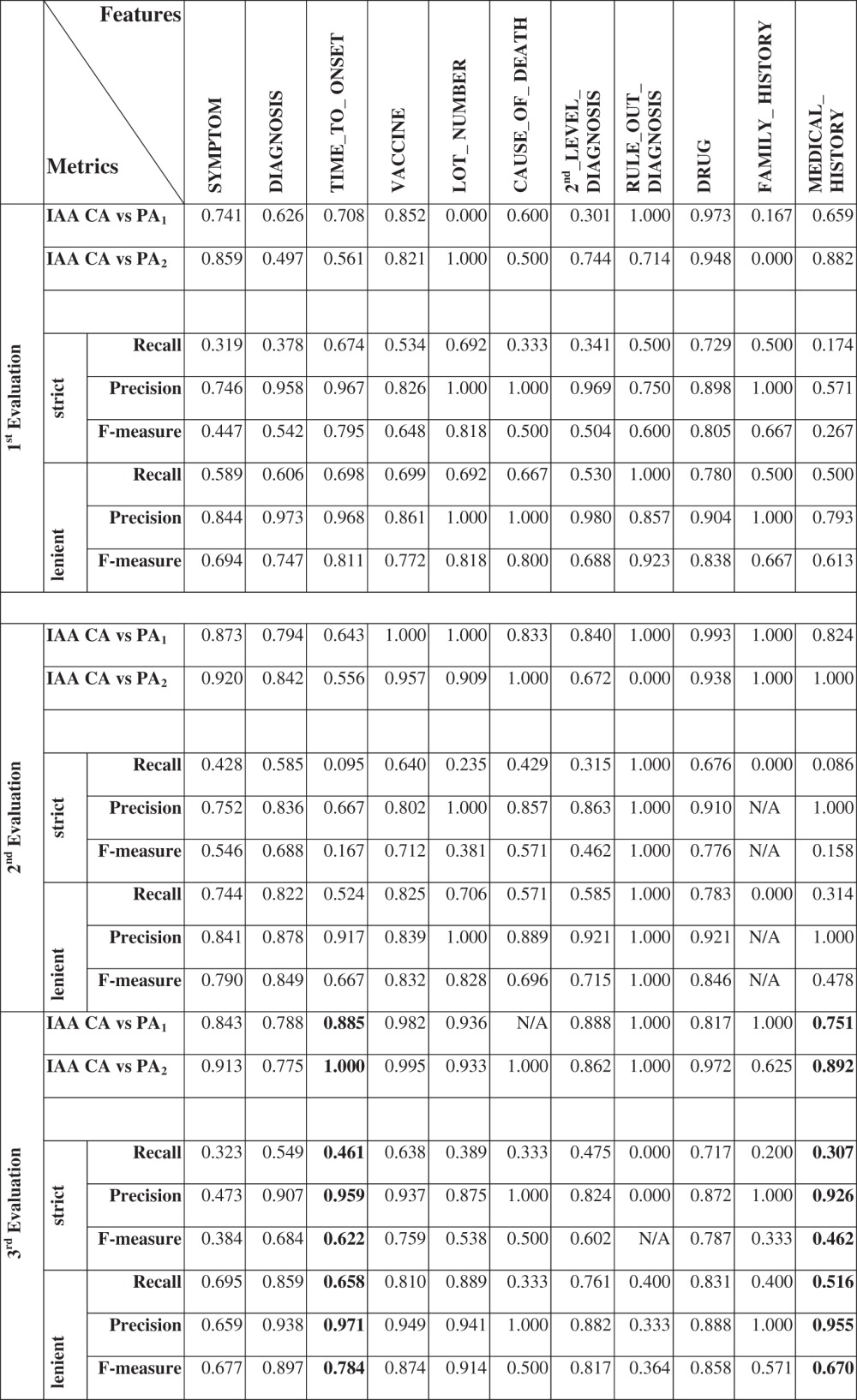
Certain features rarely (CAUSE_OF_DEATH, RULE_OUT_DIAGNOSIS and FAMILY_HISTORY) or less frequently (LOT_NUMBER, TIME_TO_ONSET and MEDICAL_HISTORY) appeared in the first two evaluations (see supplementary table 2, available online only). The metrics in table 3 were calculated based on the numbers included in supplementary tables 2 and 3 (available online only). The performance of VaeTM for the frequently appearing features was significantly improved in the second evaluation. DIAGNOSIS, DRUG, VACCINE and SYMPTOM features were reliably extracted (second evaluation lenient F-measure, 0.849, 0.846, 0.832, and 0.790, respectively). However, in terms of SECOND_LEVEL_DIAGNOSIS, the improvement was marginal (lenient F-measure increased from 0.688 to 0.715).
Table 3.
Features descriptive statistics and inter annotator agreement (strict and lenient) measures
|
CA, consensus annotator; IAA, inter annotator agreement; PA, primary annotator.
VaeTM's extraction of at least one DIAGNOSIS feature per report was highly specific (100%, 95% CI 82.20% to 100%), and sensitive (77.90%, 95% CI 66.75% to 86.26%) in the first evaluation; this was also shown by the area under the ROC curve (AUC 88.96%, 95% CI 84.30% to 93.62%). In the second evaluation specificity decreased but sensitivity increased (specificity 93.91%, 95% CI 78.38% to 98.94%; sensitivity 91.04%, 95% CI 80.88% to 96.31%; AUC 92.49%, 95% CI 87.11% to 97.87%).
The results for the set of 100 reports enriched for TIME_TO_ONSET and MEDICAL_HISTORY (table 3) showed that VaeTM performed well in terms of the selected as well as the remaining features. Again, it is not possible to draw any conclusions for the rarely appearing CAUSE_OF_DEATH (N=3), RULE_OUT_DIAGNOSIS (N=5) and FAMILY_HISTORY (N=10).
In the first evaluation, the low IAA (table 3) could be explained by difficulties associated with distinguishing the DIAGNOSIS from SECOND_LEVEL_DIAGNOSIS, as well as SYMPTOM from both types of diagnoses among the primary annotators. Even though the IAA for DIAGNOSIS and SECOND_LEVEL_DIAGNOSIS was improved in the second evaluation, the primary annotators were still confused with the triggers that differentiate the two types of diagnoses. These issues were resolved in the third evaluation in which IAA was significantly improved. In the end, the low IAA in the first two evaluations did not impact the adequacy of the reference standard as a true comparator to assess the performance of the text miner because all the identified differences were reconciled appropriately using the same rules of the text mining algorithm.
Qualitative difference error analysis
The qualitative differences between VaeTM and the reference standard in the first evaluation are summarized in supplementary table 4 (available online only). We determined that VaeTM could not extract:
Non-medical periphrases, for example, ‘patient was able to answer yes or no’.
General statements, for example, ‘no further seizures’.
Periphrastic description of treatment, for example, ‘anti-seizure medication’.
Features without a trigger, for example, ‘The patient does not drink alcohol’ as MEDICAL_HISTORY.
The initiation of a condition (eg, ‘started to blister’), the alteration of a status (eg, ‘adenoma increased in size’), and the discontinuation of a medication (eg, ‘stopped treatment with Lamictal’).
A feature acting as a modifier within another feature, for example, in ‘penicillin allergy’ in which penicillin modified allergy.
Standalone adjectives denoting a condition, for example, ‘patient was hypoxic’.
Several additional sources of errors were attributed to missing inflections of existing terms in the VAERS dictionary (eg, past participles and adverbs). VaeTM was updated to address these weaknesses, however some occurrence, textual extent, granularity and term ambiguity issues persisted in the second evaluation (table 4).
Table 4.
Examples of differences between VaeTM and reference standard in the second evaluation
| Sentence* | Reference standard* | VaeTM† | Type of difference |
| With dx: Post-viral syndrome | Post-viral syndrome (DIAGNOSIS) | viral syndrome (SYMPTOM) | Occurrence |
| pins and needles in hands, which moved to elbows | pins and needles in hands, which moved to elbows (SYMPTOM) | Not extracted | Occurrence |
| Left arm pain since seasonal flu shot | seasonal flu shot (VACCINE) | flu shot (VACCINE) | Textual extent |
| Assessment: Guillain–Barre syndrome | Guillain-Barré syndrome (SECOND_LEVEL_DIAGNOSIS) | assessment guillain barre (SECOND_LEVEL_DIAGNOSIS) | Textual extent |
| Following administration of hepatitis B vaccine | Hepatitis B (VACCINE) | hepatitis vaccine (VACCINE) | Granularity |
| Approximately a year later the patient had tested positive for high-risk HPV | high-risk HPV (SYMPTOM) | hpv (VACCINE) | Term ambiguity |
The extracted span of text is followed by the feature name in brackets; the type of difference is also included per example, while ‘Not extracted’ means that VaeTM did not extract anything.
Span of text as in the original report without any normalization.
Span of text as normalized by VaeTM.
HPV, human papillomavirus; VaeTM, vaccine adverse event text mining.
Use case
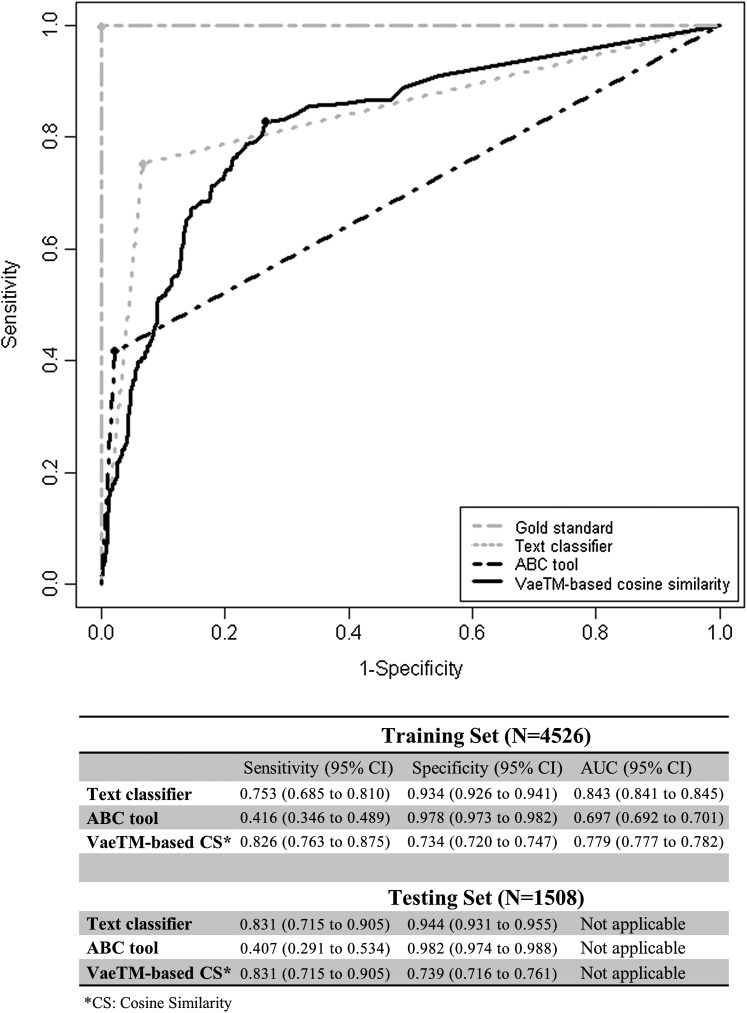
The ROC in figure 2 illustrates the classification of anaphylaxis reports in the training set using the ‘Text classifier’, the ‘ABC tool’ and the VaeTM-based score. At the best cut-off point of the curves (marked with the dots) sensitivity and specificity were calculated (figure 2). The ‘Text classifier’ and the ‘ABC tool’ also classified the reports of the testing set, while the cosine similarity threshold that resulted in the best cut-off point in the ROC curve was used accordingly. In terms of sensitivity, our approach performed equally well (83.1%) and significantly better compared to the ‘Text classifier’ (83.1%) and the ‘ABC tool’ (40.7%), respectively. However, specificity was lower (73.9%).
Figure 2.
Receiver operating characteristic curve illustrating the classification of anaphylaxis reports based on the three approaches. Sensitivity and specificity have been calculated for the best cut-off point; the area under the curve (AUC) has also been calculated in the training set. VaeTM, vaccine adverse event text mining.
Discussion
To our knowledge, this is the first attempt to create a fully automated feature extraction tool for the VAERS spontaneous reporting system. We have demonstrated that it is possible to extract clinically significant medical concepts efficiently from VAERS narratives using a novel text miner that combines a small lexicon with a flexible set of grammar rules and use it to classify a set of reports accurately following an IR approach.
The principal finding is that VaeTM successfully extracts information despite the known variability and idiosyncrasies of VAERS reports (lenient F-measure in the second evaluation ≥79% for the six key high frequency features and lenient F-measures of 78.4% and 67% for TIME_TO_ONSET and MEDICAL_HISTORY in the enriched dataset). Our analyses of the sensitivity (91%) and specificity (94%) of VaeTM in detecting at least one correct diagnosis per report in the second evaluation, coupled with the low estimated false positive rate of diagnoses (lenient precision in the second evaluation 0.878; table 3), suggests that the VaeTM system might be sufficiently reliable for routine surveillance.
While the ability to summarize the most salient clinical information in a report is immediately helpful to the FDA, more formal classification of the extracted features would provide even greater gains in reducing the labor required for adverse event review. We demonstrated that the combination of a general purpose feature extractor (VaeTM) and a general IR algorithm (cosine similarity) can provide results as good as a specific rule-based classification algorithm. In particular, sensitivity was higher (83.1% vs 40.7) and equal (83.1% vs 83.1%) when compared to the ‘ABC tool’ and the ‘Text classifier’ in the testing set, respectively. Although the cost of decreased specificity should not be ignored, high sensitivity is often a higher priority in safety surveillance, as the only cost is the manual review of some extra false positive reports. While the rule-based ‘Text classifier’ performed best, it is labor intensive to pursue this direction for a broader range of conditions. The combination of VaeTM with a general IR algorithm, such as cosine similarity, offers potential for a generalizable approach to adverse event classification without the requirement for customized rules.
The question remains as to how such a tool might be integrated into current pharmacovigilance practice. Anaphylaxis is an AEFI of sufficient interest for vaccine safety surveillance that VaeTM could be used to identify such reports in routine surveillance at the FDA. Also, the tool offers the possibility of distilling a long report into a short informative statement combining the extracted features with the structured information in VAERS, eg, ‘nausea, vomiting and diarrhea occurred 2 days after receipt of trivalent influenza vaccine in a 63 year old woman with a prior history of diabetes and taking insulin’. Such statements might be used by pharmacovigilance reviewers in daily surveillance of serious reports as an initial screen instead of the entire report. Further development of these applications is currently underway. Tools such as VaeTM should improve the aggregation of data, making more time available for critical assessment and decision making by pharmacovigilance experts.
Limitations
Our study has three primary limitations. First, the small corpora may limit the extraction efficacy of VaeTM in terms of the variety of identifiable medical concepts, abbreviations and acronyms. However, human annotation requires considerable effort, and further investigation may find that certain features that are present rarely (family history, and rule out diagnosis) or infrequently (past medical history, vaccine lot, cause of death and onset interval) in VAERS reports may not significantly impact routine safety surveillance because of the overall extent of true missing information in the remainder of the reports. Second, it is important to note that VaeTM cannot overcome the traditional limitations of passive surveillance: variable data quality, completeness, susceptibilities to reporting bias, and underreporting. Finally, it is not known whether the performance of VaeTM is generalizable to other SRS containing different medical products, different and unique terminology, and other key features.
Conclusion
This work has examined and demonstrated the feasibility of efficiently extracting key descriptors from the narratives of a safety surveillance system using a general purpose tool. We have also demonstrated the feasibility of applying VaeTM output to case classification for possible anaphylaxis. Semantic text mining might offer significant gains in efficiency and reproducibility for safety surveillance.
Supplementary Material
Footnotes
Contributors: TB developed the VaeTM tool, analyzed the data, drafted and revised the paper; ThB acted as the consensus annotator, collected the evaluation data and revised the paper; SW and EJW acted as the primary annotators and revised the paper; MN revised the draft paper; RB revised the draft paper, created the mapping table for the BC criteria and supervised the study. All authors participated in the design of the evaluation plan, the monitoring of the process and the VaeTM updates.
Funding: This project was supported in part by the appointment of Taxiarchis Botsis to the Research Participation Program at the Center for Biologics Evaluation and Research administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the US Department of Energy and the US Food and Drug Administration.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: For access to this software for research use, please contact the FDA Technology Transfer Program at techtransfer@fda.hhs.gov. Access to this software for commercial use is available through the NIH Office of Technology Transfer at http://www.ott.nih.gov/licensing_royalties/licensing_overview.aspx.
References
- 1. Food and Drug Administration Guidance for Industry: Good Pharmacovigilance Practices and Pharmacoepidemiologic Assessment. Rockville: Food and Drug Administration, 2005 [Google Scholar]
- 2. Melton GB, Hripcsak G. Automated detection of adverse events using natural language processing of discharge summaries. J Am Med Inform Assoc 2005;12:448–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Penz JF, Wilcox AB, Hurdle JF. Automated identification of adverse events related to central venous catheters. J Biomed Inform 2007;40:174–82 [DOI] [PubMed] [Google Scholar]
- 4. Mykowiecka A, Marciniak M, Kupsc A. Rule-based information extraction from patients' clinical data. J Biomed Inform 2009;42:923–36 [DOI] [PubMed] [Google Scholar]
- 5. Meystre SM, Savova GK, Kipper-Schuler KC, et al. Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inform 2008:128–44 [PubMed] [Google Scholar]
- 6. Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010;17:507–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sebastiani F. Machine learning in automated text categorization. ACM computing surveys (CSUR) 2002;34:47 [Google Scholar]
- 8. Kim JD, Ohta T, Tsujii J. Corpus annotation for mining biomedical events from literature. BMC Bioinformatics 2008;9:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Coden A, Savova G, Sominsky I, et al. Automatically extracting cancer disease characteristics from pathology reports into a disease knowledge representation model. J Biomed Inform 2009;42:937–49 [DOI] [PubMed] [Google Scholar]
- 10. Friedman C. A broad-coverage natural language processing system. Proc AMIA Symp 2000:270–4 [PMC free article] [PubMed] [Google Scholar]
- 11. Varricchio F, Iskander J, Destefano F, et al. Understanding vaccine safety information from the Vaccine Adverse Event Reporting System. Pediatr Infect Dis J 2004;23:287–94 [DOI] [PubMed] [Google Scholar]
- 12. Friedman C, Alderson PO, Austin JH, et al. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc 1994;1:161–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Herman TD, Liu F, Sagaram D, et al. Creating a vaccine adverse event ontology for public health. AMIA Annu Symp Proc 2005:978. [PMC free article] [PubMed] [Google Scholar]
- 14. Chang A, Schyve PM, Croteau RJ, et al. The JCAHO patient safety event taxonomy: a standardized terminology and classification schema for near misses and adverse events. Int J Qual Health Care 2005;17:95–105 [DOI] [PubMed] [Google Scholar]
- 15. Spasic I, Ananiadou S, McNaught J, et al. Text mining and ontologies in biomedicine: making sense of raw text. Brief Bioinform 2005;6:239–51 [DOI] [PubMed] [Google Scholar]
- 16. Domino FJ, Griffith HW. The 5-minute Clinical Consult. Philadelphia, PA: Lippincott Williams & Wilkins, 2010 [Google Scholar]
- 17. LeBlond RF, DeGowin RL, Brown DD. DeGowin's Diagnostic Examination. Columbus, OH: McGraw-Hill Professional, 2008 [Google Scholar]
- 18. Drugs@FDA Data Files. http://www.fda.gov/Drugs/InformationOnDrugs/ucm079750.htm (accessed 1 Oct 2010).
- 19. Xu H, Stenner SP, Doan S, et al. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc 2010;17:19–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Friedman C, Shagina L, Lussier Y, et al. Automated encoding of clinical documents based on natural language processing. J Am Med Inform Assoc 2004;11:392–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chapman WW, Bridewell W, Hanbury P, et al. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform 2001;34:301–10 [DOI] [PubMed] [Google Scholar]
- 22. Harkema H, Dowling JN, Thornblade T, et al. ConText: an algorithm for determining negation, experiencer, and temporal status from clinical reports. J Biomed Inform 2009;42:839–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zeng QT, Goryachev S, Weiss S, et al. Extracting principal diagnosis, co-morbidity and smoking status for asthma research: evaluation of a natural language processing system. BMC Med Inform Decis Mak 2006;6:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Roberts A, Gaizauskas R, Hepple M, et al. Building a semantically annotated corpus of clinical texts. J Biomed Inform 2009;42:950–66 [DOI] [PubMed] [Google Scholar]
- 25. Botsis T, Nguyen MD, Woo EJ, et al. Text mining for the Vaccine Adverse Event Reporting System: medical text classification using informative feature selection. J Am Med Inform Assoc 2011;18:631–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ruggeberg JU, Gold MS, Bayas JM, et al. Anaphylaxis: case definition and guidelines for data collection, analysis, and presentation of immunization safety data. Vaccine 2007;25:5675–84 [DOI] [PubMed] [Google Scholar]
- 27. Manning CD, Raghavan P, Schutze H. Introduction to Information Retrieval, Vol. 1 New York, NY: Cambridge University Press, 2008 [Google Scholar]
- 28. ABC tool: Electronic Concultant. http://brightoncollaboration.org/public/what-we-do/capacity/abc.html (accessed 14 Jun 2012).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.