Abstract
Quantitative real-time reverse transcription polymerase chain reaction (qPCR), a sensitive technique for gene expression analysis, depends on the stability of the reference genes used for data normalization. Caragana intermedia, a native desert shrub with strong drought-resistance, sand-fixing capacity and high forage value that is widespread in the desert land of west and northwest China, has not been investigated regarding the identification of reference genes suitable for the normalization of qPCR data. In this study, 10 candidate reference genes were analyzed in C. intermedia subjected to different abiotic (osmotic, salt, cold and heat) stresses, in two distinct plant organs (roots and leaves). The expression stability of these genes was assessed using geNorm, NormFinder and BestKeeper algorithms. The best-ranked reference genes differed across the different sets of samples, but UNK2, PP2A and SAND were the most stable across all tested samples. UNK2 and SAND would be appropriate for normalizing gene expression data for salt-treated roots, whereas the combination of UNK2, SAND and EF-1α would be appropriate for salt-treated leaves. UNK1, UNK2 and PP2A would be appropriate for PEG-treated (osmotic) roots, whereas the combination of TIP41 and PP2A was the most suitable for PEG-treated leaves. SAND, PP2A and TIP41 exhibited the most stable expression in heat-treated leaves. In cold-treated leaves, SAND and EF-1α were the most stably expressed. To further validate the suitability of the reference genes identified in this study, the expression levels of DREB1 and DREB2 (homologs of AtDREB1 and AtDREB2) were studied in parallel. This study is the first systematic analysis for the selection of superior reference genes for qPCR in C. intermedia under different abiotic stress conditions, and will benefit future studies on gene expression in C. intermedia and other species of the leguminous genus Caragana.
Introduction
Quantitative real-time reverse transcription polymerase chain reaction (qPCR) is an efficient, specific, and reproducible method for quantifying transcript expression levels, and is widely used to analyze mRNA in different organisms [1], developmental stages [2], [3] and responses to abiotic and biotic stress [4]–[7]. However, the accuracy of qPCR is influenced by a number of variables, such as RNA stability, quantity, purity, enzymatic efficiency in cDNA synthesis and PCR amplification [8]. Thus, to avoid bias, a normalization step is an essential pre-requisite. The most accepted approach for normalization is to include one or a small number of reference genes (internal control genes), whose expression is presumed stable in control and experimental conditions [9], [10].
The traditional reference genes are mostly cellular maintenance genes, such as 18S ribosomal RNA (18S rRNA), actin (ACT), tubulin (TUB), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and elongation factor 1-α (EF1-α) [11], [12]. However, recent studies indicate that these genes are not always stably expressed when tested in other species or under a wider range of experimental treatments [13]–[15]. Recently, some new reference genes were identified by microarray analyses in Arabidopsis thaliana and soybean that show highly stable expression levels [16], [17]. These reference genes include SAND family protein (SAND), protein phosphatase 2A (PP2A), TIP41-like family protein (TIP41), F-box/kelch-repeat protein (F-box), phosphoenolpyruvate carboxylase-related kinase 1 (PEPKR1) and others. Many of these reference genes were found to outperform traditional reference genes, for example, PP2A in hybrid roses [18], SAND in buckwheat [19] and TIP41 in peanut [20] were the most stably expressed genes in those systems. Therefore, systematic validation of reference genes is essential for certain experimental conditions and in different species [10]. Statistical algorithms, such as geNorm [21], NormFinder [22], and BestKeeper [23], have been used to identify the best reference genes for qPCR data normalization under different experimental conditions.
To date, studies of reference gene expression in plants have mainly focused on model and important crop species, such as Arabidopsis [16], rice [24], poplar [25], soybean [15], [17], wheat [26], barley [27], tomato [13], Vitis [28], and Medicago truncatula [29]. However, no systematic analysis for the selection of reference genes for qPCR in Caragana intermedia has been found.
C. intermedia belongs to the family Fabaceae, and is a native desert shrub with strong drought-resistance, sand-fixing capacity and high forage value that is widespread in the desert land of west and northwest China [30]. From a scientific standpoint, it has proven an ideal material for studying the mechanisms of drought and salt tolerance of shrubs in China, because of its easy cultivation and strong abiotic resistance [30]–[32].
In this study, 10 candidate reference genes (ACT7, TUA5, EF-1α, PP2A, SAND, TIP41, F-box, PEPKR1, UNK1, UNK2) were selected because of their stable expression in microarray studies in A. thaliana and soybean [16], [17]. The stability of these genes was analyzed in C. intermedia subjected to different abiotic (osmotic, salt, cold and heat) stresses, in two distinct plant organs (roots and leaves). Furthermore, to validate the selection of candidate reference genes, the expression levels of DREB1 and DREB2 homologues were assessed using different reference genes. This work will benefit future studies on gene expression in C. intermedia and other species of the leguminous genus Caragana.
Results
Expression Profiling of Candidate Reference Genes
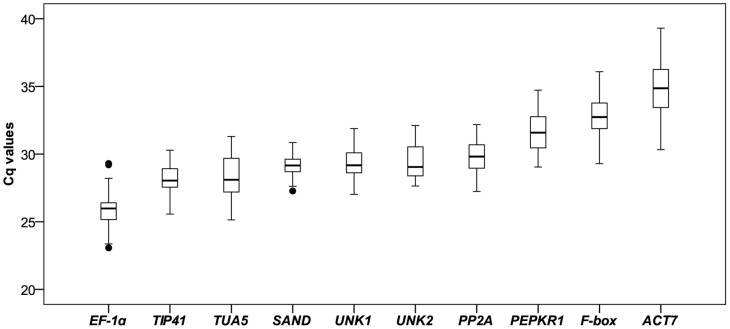
A total of 10 candidate reference genes were assessed using qPCR to quantify their mRNA levels (Table 1). The expression levels of the candidate reference genes were determined as quantification cycle (Cq) values, and the transcripts of these genes showed different levels of abundance (Figure 1). The mean Cq values of the genes ranged from 26–35, with most lying between 28 and 30 across all tested samples. EF-1α had the lowest Cq (mean Cq of 25.8), indicating the highest level of expression, SAND, PP2A, TIP41, TUA5, UNK1 and UNK2 were moderately expressed, F-box and ACT7 were expressed at low levels (mean Cq of 32.9 and 34.9). SAND showed the least gene expression variation (coefficient of variation, CV, of 3.05%), while ACT7 (5.78%) and TUA5 (5.74%) were the most variable across all samples.
Table 1. Caragana intermedia candidate reference genes descriptions and comparison with Arabidopsis orthologs.
| Gene symbol | Gene name | GenBankAccession | Arabidopsishomolog locus | Primer sequence (5′–3′) | Ampliconlength (bp) | Tm(°C) | PCRefficiency (%) |
| ACT7 | Actin7 | JX272637 | AT5G09810 | CCGAAGAGCATCCAGTTTTG | 58 | 78.5 | 100.67 |
| TCACGGTTAGCCTTGGGGTT | |||||||
| EF-1α | Elongation factor -1α | JX272638 | AT5G60390 | AGATGGTTCCCACTAAGCCTATG | 115 | 82.6 | 93.13 |
| ACACTCTTGATGACTCCAACTGC | |||||||
| TUA5 | Alpha Tubulin | JX272639 | AT5G19780 | CTGATGTGGTTGTGCTTTTGGAC | 183 | 80.8 | 93.11 |
| GGTTTGTCTGGAACTCGGTAATG | |||||||
| F-box | F-box/kelch-repeat protein | JX272640 | AT5G15710 | AATGGGTCGTGGAGGGTCTA | 71 | 82.3 | 94.80 |
| AACCCCTTGGCTTGGTCTTA | |||||||
| PEPKR1 | Phosphoenolpyruvate carboxylase-related Kinase 1 | JX272641 | AT1G12580 | GAACAGTTGGGTTGGGGACA | 181 | 82.4 | 94.05 |
| GATCCACAACATTCGGGTGC | |||||||
| PP2A | Protein phosphatase 2A | JX272642 | AT1G10430 | TTTCGGATAGGAGGAAATGCAC | 106 | 80.3 | 94.18 |
| TCAAGGCCACCAAAAGCGTA | |||||||
| SAND | SAND family protein | JX272643 | AT2G28390 | ATACTCGTCAACAGCAGAAA | 133 | 79.8 | 91.46 |
| GTCACCCAACATAAAAGAAC | |||||||
| TIP41 | TIP41-like family protein | JX272644 | AT4G34270 | CGTCCAAGAGTGGGAACAGA | 167 | 81.7 | 90.22 |
| GAACTTCAACAGGCGGCAAG | |||||||
| UNK1 | Hypothetical protein | JX272645 | AT1G31300 | CAATGTTGAGTGGGGAGGGA | 162 | 81.2 | 97.41 |
| CAACCAGCCAAGCAAGGAAT | |||||||
| UNK2 | Hypothetical protein | JX272646 | AT4G33380 | CAAAGATAGTGCTGCTGATTGC | 147 | 81.6 | 92.64 |
| TCCTGGTGTTTGTGCTGATAGA | |||||||
| DREB1* | Dehydration responsive element binding protein 1 | KC123242 | AT4G25480 | TTCTGACCCACAACCTTACTC | 167 | 86.5 | 96.40 |
| TCTTCTTGTTTGGTTCCCTTA | |||||||
| DREB2* | Dehydration responsive element binding protein 2 | JX272647 | AT2G40340 | GAAAGGGTGTATGAAAGGTA | 228 | 83.4 | 92.56 |
| GTTATGTGAGGGAAGTTGAG |
Notes: * used for normalization validation under cold or salt stress conditions.
Figure 1. Expression levels of candidate reference genes across all samples.
Lines across the boxes depict the medians. Boxes indicate the interquartile range. Whiskers represent 95% confidence intervals, black dots represent outliers.
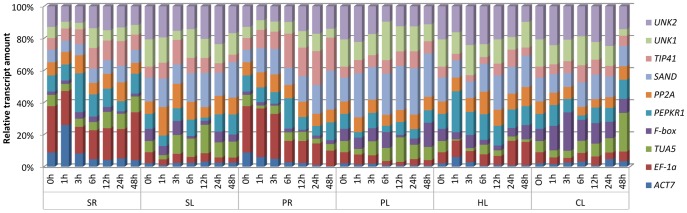
The variation in relative transcript amount of the reference genes across all tested samples is shown in Figure 2. Transcript amounts are represented as percentages, relative to the aggregated reference transcript pool for each sample. The proportions of PP2A, SAND and UNK2 transcript remained relatively constant, while those of ACT7, TUA5 and F-box were more variable across all samples. The transcript levels of UNK1 remained relatively constant in PEG-treated roots (PR) or leaves (PL), and those of TIP41 were also relatively constant in the PL and cold-treated leaves (CL). Although the expression level of EF-1α was more variable across all samples and particularly high in the PR and salt-treated roots (SR), its expression was relatively constant in the CL and salt-treated leaves (SL). These results clearly suggested that the expression level of none of the reference genes is truly constant, and varies in different spatial and temporal patterns and environmental conditions.
Figure 2. Distribution of relative transcript amount of the reference genes across all samples.
Transcript amount are represented as percentages of the aggregated 10-transcript pool for each sample. SR (0–48 h), roots exposed to high-salt; SL (0–48 h), leaves exposed to high-salt treatment; PR (0–48 h), roots exposed to PEG treatment; PL (0–48 h), leaves exposed to PEG treatment; HL (0–48 h), leaves exposed to heat treatment; CL (0–48 h), leaves exposed to cold treatment.
Expression Stability of Candidate Reference Genes
a) geNorm analysis
The expression stability of the 10 reference genes was assessed using the geNorm software. The geNorm algorithm is based on the principle that the logarithmically transformed expression ratio between two genes should be constant if both genes are stably expressed in a given sample set. The candidate reference genes were ranked by geNorm based on the expression stability value M, which is calculated for all genes being investigated (the lower the M value, the higher the gene's expression stability) [21].
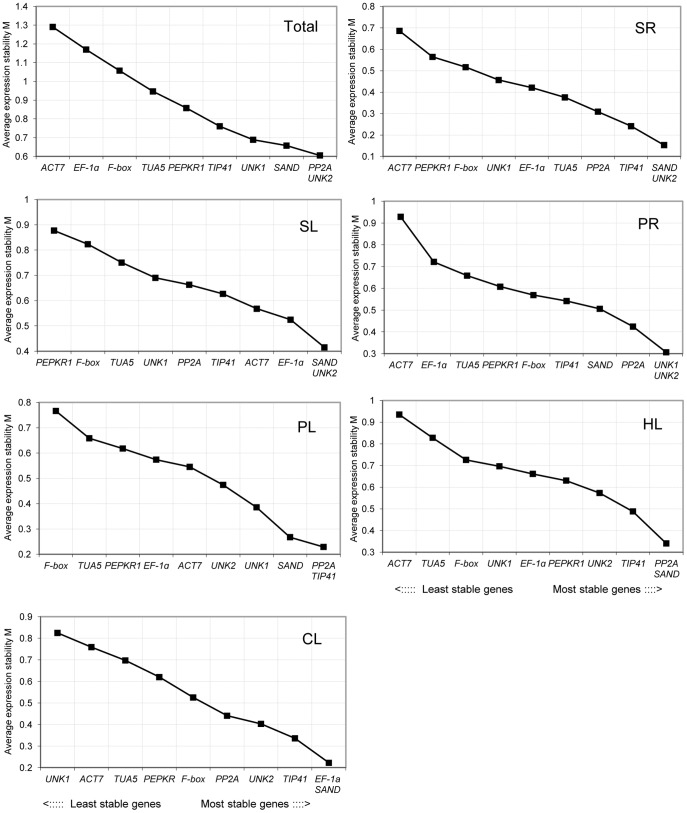
Figure 3 shows the ranking of the tested genes according to their expression stability in the C. intermedia samples, using data from six sets of treatment. When all 38 samples were analyzed together, UNK2, PP2A, and SAND were the most stable genes, while ACT7 and EF-1α were the least stable (in order). In salt stress treatments, UNK2 and SAND were the most stable genes, while ACT7 and PEPKR1 were the least stable in the SR and SL treatments, respectively. In the PR treatment, UNK2 and UNK1 were the most stable genes and ACT7 the least; In the PL treatment, TIP41 and PP2A were the most stable and F-box the least. In heat-treated leaves (HL), SAND and PP2A were the most stable genes and ACT7 was the least stably expressed. In the CL treatment, the expression levels of SAND and EF-1α were the most stable and UNK1 was the least stable. In addition, all of the tested reference genes showed relatively high stability with M values of less than 1.5, which is below the default limit of M ≤1.5.
Figure 3. Gene expression stability and ranking of 10 reference genes as calculated by geNorm.
Mean expression stability (M) was calculated following stepwise exclusion of the least stable gene across all treatment groups. The least stable genes are on the left, and the most stable on the right.
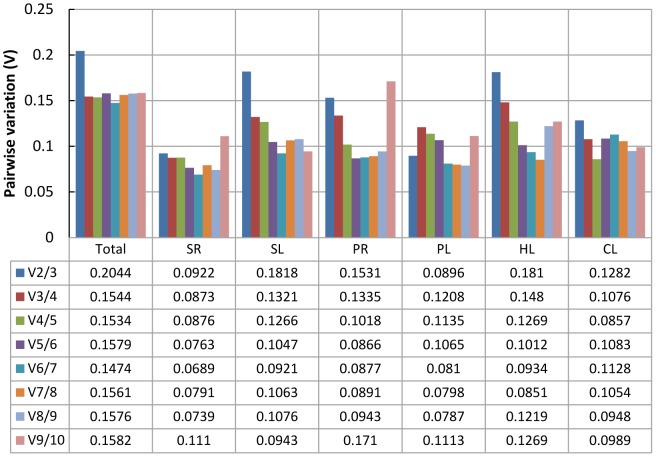
geNorm performs a stepwise calculation of the pairwise variation (Vn/Vn+1) between sequential normalization factors (NFn and NFn+1) to determine the optimal number of reference genes required for accurate normalization. A large variation means that the added gene had a significant effect and should preferably be included for calculation of a reliable normalization factor [21]. As shown in Figure 4, the inclusion of a fourth gene had no significant effect (that is, low V3/4 value) for all pooled samples or for the SL treatment, so three reference genes would be optimal for normalizing gene expression under those conditions. Similarly, two reference genes would be sufficient for the SR and PL treatments, four for the PR and CL treatments, and five for the HL treatment. In the practical application, three reference genes for the PR (V3/4 = 0.1335) and HL (V3/4 = 0.1480) treatments, and two for the CL treatment (V2/3 = 0.1282) could also be accepted using a threshold value of 0.15 [14], [21], [33], [34].
Figure 4. Determination of the optimal number of reference genes required for effective normalization.
Pairwise variation (Vn/Vn +1) analysis between the normalization factors (NFn and NFn +1) was performed by the geNorm program to determine the optimal number of reference genes, and carried out for qPCR data normalization in various sample pools.
b) NormFinder analysis
NormFinder program is a Visual Basic application tool for Microsoft Excel used to determine the expression stabilities of reference genes that ranks all reference gene candidates based on intra- and inter-group variations and combines both results into a stability value for each candidate reference gene [35]. The results of NormFinder analysis were slightly different from those of geNorm (Table 2). Both methods of analysis ranked as most stable UNK2 and SAND in the SR treatment; UNK2, SAND and EF-1α in the SL treatment; PP2A and TIP41 in the PL treatment; PP2A, UNK1 and UNK2 in the PR treatment. However, in the CL treatment, PP2A and UNK2 emerged as the most stably expressed, whereas they were ranked fifth and fourth, respectively, by geNorm. In the HL treatment, PP2A, UNK2 and SAND were in the top positions, while geNorm ranked UNK2 in the fourth position. When evaluated across all experimental samples, UNK2, PP2A and TIP41 were in the top positions, whereas TIP41 was ranked fifth by geNorm.
Table 2. Expression stability of the reference genes calculated by NormFinder software.
| Rank | Total | SR | SL | PR | PL | HL | CL | |||||||
| Gene | Stability | Gene | Stability | Gene | Stability | Gene | Stability | Gene | Stability | Gene | Stability | Gene | Stability | |
| 1 | PP2A | 0.188 | UNK2 | 0.060 | UNK2 | 0.163 | PP2A | 0.157 | PP2A | 0.079 | PP2A | 0.238 | PP2A | 0.142 |
| 2 | UNK2 | 0.268 | SAND | 0.066 | SAND | 0.268 | UNK1 | 0.234 | TIP41 | 0.200 | UNK2 | 0.303 | UNK2 | 0.154 |
| 3 | TIP41 | 0.462 | PP2A | 0.186 | EF-1α | 0.283 | UNK2 | 0.301 | UNK2 | 0.268 | SAND | 0.306 | SAND | 0.160 |
| 4 | SAND | 0.465 | TIP41 | 0.255 | ACT7 | 0.356 | F-box | 0.304 | UNK1 | 0.316 | UNK1 | 0.326 | TIP41 | 0.197 |
| 5 | UNK1 | 0.514 | EF-1α | 0.262 | PP2A | 0.368 | PEPKR1 | 0.358 | SAND | 0.332 | PEPKR1 | 0.360 | EF-1α | 0.261 |
| 6 | PEPKR1 | 0.531 | F-box | 0.348 | TIP41 | 0.415 | EF-1α | 0.434 | ACT7 | 0.353 | TIP41 | 0.369 | F-box | 0.439 |
| 7 | TUA5 | 0.700 | TUA5 | 0.376 | UNK1 | 0.454 | TUA5 | 0.435 | EF-1α | 0.386 | EF-1α | 0.422 | PEPKR1 | 0.568 |
| 8 | F-box | 0.898 | UNK1 | 0.387 | TUA5 | 0.625 | SAND | 0.473 | PEPKR1 | 0.407 | F-box | 0.435 | ACT7 | 0.580 |
| 9 | EF-1α | 0.930 | PEPKR1 | 0.427 | F-box | 0.628 | TIP41 | 0.505 | TUA5 | 0.519 | TUA5 | 0.797 | TUA5 | 0.597 |
| 10 | ACT7 | 1.078 | ACT7 | 0.763 | PEPKR1 | 0.639 | ACT7 | 1.179 | F-box | 0.765 | ACT7 | 0.868 | UNK1 | 0.673 |
Notes: SR, roots exposed to high-salt treatment; SL, leaves exposed to high-salt treatment; PR, roots exposed to PEG treatment; PL, leaves exposed to PEG treatment; HL, leaves exposed to heat treatment; CL, leaves exposed to cold treatment.
c) BestKeeper analysis
BestKeeper determines the most stably expressed genes based on the coefficient of correlation to the BestKeeper Index, which is the geometric mean of the candidate reference gene Cq values. BestKeeper also calculates the standard deviation (SD) and the coefficient of variation (CV) based on the Cq values of all candidate reference genes [23]. Genes with SD greater than 1 are considered unacceptable [36]. Reference genes are identified as the most stable genes, i.e. those that exhibit the lowest coefficient of variance and standard deviation (CV ± SD) [37]. The results of BestKeeper analysis are shown in Table 3. In the PL and CL treatments, the same four genes were identified by both the BestKeeper and geNorm programs, although their rank order was slightly altered. In the SL treatment, UNK1 emerged as the most stably expressed (ranked seventh by geNorm and NormFinder). In the SR treatment, the same three genes, UNK2, SAND and TIP41, were identified by both the BestKeeper and geNorm programs, although their rank order was slightly altered. In the HL treatment, EF-1α emerged as the most stably expressed (ranked sixth by geNorm and seventh by NormFinder). When evaluated across all experimental samples, SAND, TIP41 and PP2A were in the top positions, whereas TIP41 was ranked fifth by geNorm and third by NormFinder.
Table 3. Expression stability of the reference genes calculated by BestKeeper software.
| Rank | Total | SR | SL | PR | PL | HL | CL |
| 1 | SAND | SAND | UNK1 | SAND | SAND | EF-1α | SAND |
| CV±SD | 2.22±0.65 | 0.68±0.19 | 1.48±0.43 | 1.32±0.37 | 2.01±0.57 | 1.25±0.30 | 0.97±0.27 |
| 2 | TIP41 | TIP41 | UNK2 | TIP41 | TIP41 | TIP41 | TIP41 |
| CV±SD | 2.89±0.81 | 0.74±0.20 | 1.57±0.40 | 1.54±0.45 | 2.23±0.62 | 1.54±0.39 | 1.22±0.34 |
| 3 | PP2A | UNK2 | SAND | UNK1 | UNK1 | F-box | EF-1α |
| CV±SD | 3.22±0.96 | 0.90±0.26 | 2.02±0.56 | 1.97±0.60 | 2.35±0.68 | 1.66±0.56 | 1.40±0.34 |
| 4 | UNK1 | UNK1 | EF-1α | UNK2 | PP2A | SAND | UNK2 |
| CV±SD | 3.23±0.95 | 0.95±0.27 | 2.14±0.51 | 2.08±0.60 | 2.41±0.69 | 1.86±0.53 | 1.80±0.47 |
| 5 | UNK2 | TUA5 | F-box | PP2A | UNK2 | PP2A | UNK1 |
| CV±SD | 3.58±1.05 | 1.14±0.32 | 2.20±0.73 | 2.13±0.60 | 3.05±0.85 | 2.05±0.57 | 1.80±0.49 |
| 6 | PEPKR1 | PP2A | PP2A | F-box | EF-1α | UNK1 | F-box |
| CV±SD | 3.74±1.18 | 1.34±0.38 | 2.50±0.67 | 2.16±0.73 | 3.11±0.86 | 2.57±0.74 | 2.11±0.71 |
| 7 | F-box | EF-1α | ACT7 | PEPKR1 | ACT7 | UNK2 | PP2A |
| CV±SD | 3.91±1.28 | 1.60±0.39 | 2.52±0.82 | 3.08±0.89 | 3.14±1.02 | 2.71±0.79 | 2.40±0.66 |
| 8 | EF-1α | PEPKR1 | TIP41 | TUA5 | F-box | PEPKR1 | ACT7 |
| CV±SD | 4.20±1.08 | 1.61±0.48 | 2.81±0.75 | 3.70±0.97 | 3.16±1.09 | 2.78±0.84 | 2.44±0.82 |
| 9 | TUA5 | F-box | PEPKR1 | EF-1α | TUA5 | ACT7 | TUA5 |
| CV±SD | 4.63±1.31 | 1.66±0.55 | 3.34±1.02 | 4.49±1.08 | 3.46±1.00 | 3.65±1.23 | 2.53±0.67 |
| 10 | ACT7 | ACT7 | TUA5 | ACT7 | PEPKR1 | TUA5 | PEPKR1 |
| CV±SD | 4.64±1.62 | 3.32±1.03 | 4.07±1.06 | 5.35±1.73 | 4.12±1.22 | 4.39±1.16 | 2.68±0.83 |
Notes: SR, roots exposed to high-salt treatment; SL, leaves exposed to high-salt treatment; PR, roots exposed to PEG treatment; PL, leaves exposed to PEG treatment; HL, leaves exposed to heat treatment; CL, leaves exposed to cold treatment. Descriptive statistics of 10 candidate genes based on the coefficient of variance (CV) and standard deviation (SD) of their Cq values were determined using the whole data set. Reference genes were identified as the most stable genes, i.e. those with the lowest coefficient of variance and standard deviation (CV ± SD).
Reference Gene Validation
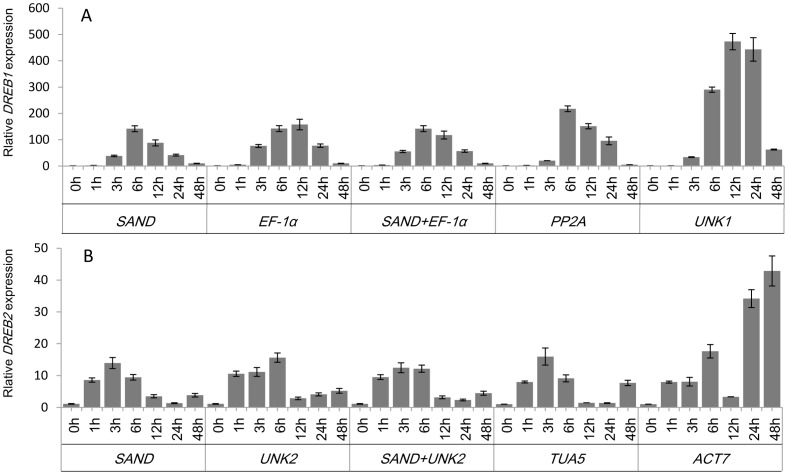
To validate the selection of candidate reference genes, the expression pattern of DREB1 and DREB2 were analyzed using the selected reference genes (Figure 5). In A. thaliana, expression of the DREB1 was induced by low-temperature stress, whereas expression of the DREB2 was induced by drought and high salinity [38]. In this study, expression of DREB1 in cold-stressed leaves and expression of DREB2 in salt-stressed roots were assessed.
Figure 5. Relative quantification of DREB1 and DREB2 expression using validated reference genes for normalization.
The results are represented as mean fold changes in relative expression compared to the first sampling stage (0 h). cDNA samples were taken from the same set used for gene expression stability analysis. (A), leaves were collected from three-week-old seedlings subjected to cold stress after 0, 1, 3, 6, 12, 24 and 48 h treatment. (B), roots were collected from three-week-old seedlings subjected to salt stress after 0, 1, 3, 6, 12, 24 and 48 h treatment.
When the two most stable reference genes, SAND and EF-1α were used for normalization, the expression levels of DREB1 increased sharply after 3 h of treatment, peaked at 6 h, and thereafter decreased (Figure 5A). When the least stable gene UNK1 was used for normalization, the expression patterns and transcript levels were very different. For DREB2, when the two most stable reference genes, UNK2 and SAND were used for normalization, the transcript levels of DREB2 increased rapidly from 1 h, peaked at 3–6 h, and thereafter decreased. The expression level of DREB2 peaked at 3 h using just SAND and at 6 h using just UNK2. Similar expression patterns were generated when the less stable reference gene TUA5 was employed (Figure 5B). Normalization based on the least stable reference gene ACT7 found that the transcript level increased rapidly and peaked at 6 h, decreased between 6 and 24 h, and thereafter increased again up to 48 h, which obviously differed from normalization against SAND and UNK2.
Discussion
In plant molecular biological research, qPCR has become an important tool for understanding gene expression in different experimental conditions. For accurate qPCR measurements, endogenous reference genes are used as internal controls. An ideal reference gene should be representative of the overall expression across all possible tissues (cells) and experimental conditions [10]. However, such a perfect reference gene is impossible and nothing close has yet been reported. This means that reference genes need to be validated under certain experimental conditions and among various species.
C. intermedia is a native desert shrub that is widespread in the desert land of west and northwest China, but the application of qPCR in this species has been limited by a lack of information on reference gene stability in a variety of experimental contexts. Here, we describe the analysis of 10 candidate reference genes to improve relative quantification by qPCR for gene expression analysis in C. intermedia. Using three algorithms (geNorm, NormFinder and BestKeeper), we evaluated the expression stability of these ten genes under different abiotic (osmotic, salt, cold and heat) stress conditions in roots and leaves. As far as can be ascertained, this is the first systematic study of the expression stability of reference genes for qPCR in C. intermedia under different abiotic stress conditions.
geNorm, NormFinder and BestKeeper are often used to select reference genes. Because they employ different strategies, they can give different results [39], [40]. For example, in the SL treatment, UNK1 emerged as the most stably expressed using BestKeeper, while it was ranked seventh by geNorm and NormFinder. In the HL treatment, BestKeeper selected EF-1α as the most stably expressed gene, but it was ranked sixth by geNorm and seventh by NormFinder. Finally, in the CL treatment, SAND and EF-1α were identified as the most stably expressed by geNorm, but were ranked third and fifth, respectively, by NormFinder. We considered the results of the three algorithms together when determining suitable reference genes for qPCR normalization (Table S1) [18].
In the SL treatment, the pairwise variation V3/4 value (0.1321), calculated by geNorm, suggested the use of UNK2, SAND and EF-1α for normalization. These three genes were also identified by NormFinder. BestKeeper, unlike the other two programs, ranked UNK1 as most stable (ranked seventh by geNorm and NormFinder), UNK2 as second, SAND as third and EF-1α as fourth. Based on these results, we recommended UNK2 combined with SAND and EF-1α as the best combination of stable reference genes for qPCR in the SL treatment.
In the HL treatment, the pairwise variation V3/4 value (0.148) indicated that the three most stable genes (SAND, PP2A and TIP41) can be used for normalization. BestKeeper ranked EF-1α as most stable (ranked seventh by geNorm and NormFinder), TIP41 as second. NormFinder, ranked PP2A as the best reference gene and SAND and TIP41 as third and sixth, respectively. Based on these results, we inferred that SAND, PP2A and TIP41 would be appropriate for qPCR in the HL treatment.
In the CL treatment, the pairwise variation V2/3 value (0.1282) indicated that the two top ranked genes (SAND and EF-1α) can be used for normalization. NormFinder ranked SAND as third, and EF-1α as fifth, however, their stability values did not differ substantially from those of higher-ranked genes (e.g. 0.160 for SAND versus 0.142 for first-ranked PP2A). BestKeeper ranked SAND as most stable, and EF-1α as third, similar the results of geNorm. Altogether, we recommended SAND and EF-1α to be the suitable reference genes for qPCR in the CL treatment. Similarly, UNK2 and SAND would be sufficient for the SR treatment, PP2A, UNK2 and UNK1 for the PR treatments, and SAND, PP2A and TIP41 for the PL treatment.
In summary, UNK2, SAND and PP2A were the most stably expressed genes, while ACT7 was the most variable, over all samples. UNK2 has been noted as showing stable expression across tissues and developmental stages in tomato [13], soybean [41] and aspen [42]. SAND and PP2A have been noted as showing stable expression across tissues and different abiotic and biotic stress conditions in roses [18] and buckwheat [19]. The weakness of ACT7 was also seen in soybean, where its expression was found to be variable [41].
The transcript levels of DREB2 peaked at 3–6 h and then began to decline at 12 h in roots under salt stress conditions when UNK2 and SAND were used for normalization (Figure 5B). A similar expression pattern was described under salt stress conditions in Caragana korshinskii [43], indicating that the reference genes identified in this study are suitable under such conditions. In the expression profile or transcript abundance quantification produced from normalization using the least stable gene ACT7, the transcript level increased rapidly and peaked at 6 h, decreased between 6 and 24 h, and thereafter increased again up to 48 h, which obviously differed from normalization against SAND and UNK2. Obviously, ACT7 is not a suitable reference gene to normalize gene expression in C. intermedia under such conditions. These results indicate that the incorrect use of reference genes without validation may reduce precision or produce misleading results.
Conclusions
To our knowledge, this study is the first systematic analysis for the selection of superior reference genes for qPCR in C. intermedia under different abiotic (osmotic, salt, cold and heat) stress conditions. Analysis of expression stability using geNorm, NormFinder and BestKeeper revealed that UNK2, PP2A and SAND could be considered to be appropriate reference genes for gene expression analysis of different tissues under different abiotic stress conditions, whereas ACT7, PEPKR1 and F-box showed relatively low expression stability. This work will benefit future studies on gene expression under different abiotic stress conditions in C. intermedia and other species of the leguminous genus Caragana.
Materials and Methods
Plant Materials and Treatments
Seeds of C. intermedia were collected from the Experimental Base (Hohhot, Inner Mongolia, China), Research Institute of Forestry, Chinese Academy of Forestry. Seeds were washed three times with tap water, and then sown in plastic pots filled with peat soil in a growth chamber with a 16 h light/8 h dark photoperiod at 25/22°C day/night temperatures and relative humidity 80%. For salt and osmotic stress treatments, three-week-old seedlings were carefully removed from the soil to avoid injury, their roots were washed cleanly with tap water, and they were placed in NaCl (200 mM) or PEG6000 (15%) solutions, respectively, for 0, 1, 3, 6, 12, 24, or 48 h in the growth chamber. For the cold and heat stress treatments, the seedlings in pots were grown at 4°C or 42°C, respectively, for 0, 1, 3, 6, 12, 24, or 48 h.
Leaves were collected from the three-week-old seedlings subjected to all four treatments, and roots were collected from the seedlings subjected to salt and osmotic stress treatments. These were immediately frozen in liquid nitrogen and stored at −80°C. Samples above were collected from 3 seedlings to give 3 replicas.
Total RNA Isolation and cDNA Synthesis
Total RNA was extracted from treated tissues using Trizol reagent (Invitrogen, USA) according to the manufacturer's instructions. The remaining DNA was removed by RNase-free DNase according to the manufacturer’s instructions (Promega, USA). Total RNA concentration and purity was determined using a Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies, USA). RNA samples with an absorbance ratio at OD260/280 between 1.9 and 2.2 and OD260/230 ≈ 2.0 were used for further analysis. RNA integrity was verified by 1.5% agarose gel electrophoresis. Samples with 28S/18S ribosomal RNA between 1.5 and 2.0 and without smears on the agarose gel were used for subsequent experiments.
For each sample, 1 µg of total RNA was reverse transcribed using the RevertAidTM First Strand cDNA Synthesis Kit (Fermentas, Germany) in a 20 µl reaction using oligo dT primers according to manufacturer’s instructions. The cDNAs were diluted 1∶30 with nuclease-free water prior to the qPCR analyses.
Selection of Candidate Reference Genes
Potential homologues of the ten published reference genes were identified from the transcriptome data sequences of C. intermedia seedlings (unpublished data) (Table 1).
The candidate reference genes comprised ACT7 (actin 7), EF-1α (elongation factor -1α), TUA5 (alpha tubulin), PP2A (protein phosphatase2A), SAND (SAND-family protein), TIP41 (TIP41-like protein), F-box (F-box/kelch-repeat protein), PEPKR1 (phosphoenolpyruvate carboxylase-related kinase 1), UNK1 (hypothetical protein) and UNK2 (hypothetical protein), which were previously shown to have highly stable expression levels by microarray analysis in A. thaliana and soybean [16], [17].
PCR Primer Design and Test of Amplification Efficiency
Primers were designed using the Primer Premier 5 software (http://www.PremierBiosoft.com/primerdesign/primerdesign.html) with melting temperatures 58–62°C, primer lengths 20–24 bp, GC content 45–55% and amplicon lengths 50–230 bp (Table 1, Table S2 and Figure S1). For each primer pair, amplification efficiency estimates were derived from a standard curve generated from a serial dilution of pooled cDNA (1, 10, 102, 103, 104, 105 × dilutions; each gene in triplicate) (Figure S2). Mean quantification cycle (Cq) values of each ten-fold dilution were plotted against the logarithm of the pooled cDNA dilution factor. Efficiency (E) for each gene was determined with the slope of a linear regression model [44] using the Cq values and the following equation was used:
Quantitative Real-time RT-PCR
qPCR reactions were carried out with an ABI Prism 7700 Sequence Detection System (Applied Biosystems, USA), using SYBR® Premix Ex Taq™ (Takara, Japan) in a 20 µl reaction volume (containing 2 µl diluted cDNA, 10 µl 2 × SYBR Premix Ex Taq™, 0.4 µl ROX Reference Dye, and 0.4 µl each primer). The reaction conditions were: an initial denaturation step of 95°C/30 s, followed by 40 cycles of 95°C/5 s and 60°C/30 s. The dissociation curve was obtained by heating the amplicon from 60 to 95°C (Figure S3). All qPCR reactions were carried out in biological and technical triplicate. A non-template control was also included in each run for each gene. The final quantification cycle (Cq) values were the means of nine values (biological triplicate, each in technical triplicate).
Statistical Analysis
Three different types of Microsoft Excel-based software, geNorm [45], NormFinder [46] and BestKeeper [47], were used to rank the expression stability of reference genes across all of the experimental sets. Following qPCR data collection, Cq values were converted to relative quantities using the formula: 2−ΔCq, in which ΔCq = each corresponding Cq value − minimum Cq value. The sample with the maximum expression level (the minimum Cq value) was used as a calibrator and was set to a value of 1. Relative quantities were used for geNorm and NormFinder, while BestKeeper analyses were based on untransformed Cq values. All three software packages were used according to the manufacturer’s instructions. All other multiple comparisons were performed with SPSS17.0.
Supporting Information
qPCR amplification specificity of the 10 reference genes, and DREB1 and DREB2 . Amplification fragments were separated by 2% agarose gel electrophoresis.
(PDF)
Amplification efficiencies of the 10 reference genes, and DREB1 and DREB2 .
(PDF)
Melting curves of the 10 reference genes, and DERB1 and DREB2 .
(PDF)
The ranking of 10 reference genes by geNorm, NormFinder, and Bestkeeper.
(DOC)
List of amplified sequences of the 10 reference genes, and DREB1 and DREB2.
(DOC)
Acknowledgments
We are very grateful to Prof. Qing-Yin Zeng (State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, Beijing, China) for good suggestions and careful proofreading of the manuscript, and Prof. Xinmin Li (Department of Pathology and Laboratory Medicine, University of California at Los Angeles, USA) for thoughtful suggestions and critical comments, and Dr. Tao Wu (Yunnan Academy of Forestry, Kunming, Yunnan, China) for suggestions and help with statistical analysis. We are also grateful to the Inner Mongolia Academy of Forestry Science for the help in collecting experimental materials.
Funding Statement
This work was supported by the National Basic Research Program of China (2009CB119106) and the National High Technology Research and Development Program of China (2011AA100203). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Ohdan T, Francisco PB Jr, Sawada T, Hirose T, Terao T, et al. (2005) Expression profiling of genes involved in starch synthesis in sink and source organs of rice. J Exp Bot 56: 3229–3244. [DOI] [PubMed] [Google Scholar]
- 2. Vaucheret H, Vazquez F, Crete P, Bartel DP (2004) The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev 18: 1187–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Koo SC, Bracko O, Park MS, Schwab R, Chun HJ, et al. (2010) Control of lateral organ development and flowering time by the Arabidopsis thaliana MADS-box Gene AGAMOUS-LIKE6 . Plant J 62: 807–816. [DOI] [PubMed] [Google Scholar]
- 4. Qiu JL, Fiil BK, Petersen K, Nielsen HB, Botanga CJ, et al. (2008) Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J 27: 2214–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huang J, Gu M, Lai Z, Fan B, Shi K, et al. (2010) Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiology 153: 1526–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Portieles R, Ayra C, Gonzalez E, Gallo A, Rodriguez R, et al. (2010) NmDef02, a novel antimicrobial gene isolated from Nicotiana megalosiphon confers high-level pathogen resistance under greenhouse and field conditions. Plant Biotechnol J 8: 678–690. [DOI] [PubMed] [Google Scholar]
- 7. Ren X, Chen Z, Liu Y, Zhang H, Zhang M, et al. (2010) ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis . Plant J 63: 417–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mahoney DJ, Carey K, Fu MH, Snow R, Cameron-Smith D, et al. (2004) Real-time RT-PCR analysis of housekeeping genes in human skeletal muscle following acute exercise. Physiol Genomics 18: 226–231. [DOI] [PubMed] [Google Scholar]
- 9. Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, et al. (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55: 611–622. [DOI] [PubMed] [Google Scholar]
- 10. Guenin S, Mauriat M, Pelloux J, Van Wuytswinkel O, Bellini C, et al. (2009) Normalization of qRT-PCR data: the necessity of adopting a systematic, experimental conditions-specific, validation of references. J Exp Bot 60: 487–493. [DOI] [PubMed] [Google Scholar]
- 11. Radonic A, Thulke S, Mackay IM, Landt O, Siegert W, et al. (2004) Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun 313: 856–862. [DOI] [PubMed] [Google Scholar]
- 12. Nicot N, Hausman JF, Hoffmann L, Evers D (2005) Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J Exp Bot 56: 2907–2914. [DOI] [PubMed] [Google Scholar]
- 13. Exposito-Rodriguez M, Borges AA, Borges-Perez A, Perez JA (2008) Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol 8: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jain M, Nijhawan A, Tyagi AK, Khurana JP (2006) Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biophys Res Commun 345: 646–651. [DOI] [PubMed] [Google Scholar]
- 15. Jian B, Liu B, Bi Y, Hou W, Wu C, et al. (2008) Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Mol Biol 9: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis . Plant Physiol 139: 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Libault M, Thibivilliers S, Bilgin D, Radwan O, Benitez M, et al. (2008) Identification of four soybean reference genes for gene expression normalization. Plant Genome 1: 44–54. [Google Scholar]
- 18. Klie M, Debener T (2011) Identification of superior reference genes for data normalisation of expression studies via quantitative PCR in hybrid roses (Rosa hybrida). BMC Res Notes 4: 518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Demidenko NV, Logacheva MD, Penin AA (2011) Selection and validation of reference genes for quantitative real-time PCR in buckwheat (Fagopyrum esculentum) based on transcriptome sequence data. PLoS One 6: e19434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chi X, Hu R, Yang Q, Zhang X, Pan L, et al. (2012) Validation of reference genes for gene expression studies in peanut by quantitative real-time RT-PCR. Mol Genet Genomics 287: 167–176. [DOI] [PubMed] [Google Scholar]
- 21. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, et al. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Andersen CL, Jensen JL, Orntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64: 5245–5250. [DOI] [PubMed] [Google Scholar]
- 23. Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol Lett 26: 509–515. [DOI] [PubMed] [Google Scholar]
- 24. Kim BR, Nam HY, Kim SU, Kim SI, Chang YJ (2003) Normalization of reverse transcription quantitative-PCR with housekeeping genes in rice. Biotechnol Lett 25: 1869–1872. [DOI] [PubMed] [Google Scholar]
- 25. Brunner AM, Yakovlev IA, Strauss SH (2004) Validating internal controls for quantitative plant gene expression studies. BMC Plant Biol 4: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Paolacci AR, Tanzarella OA, Porceddu E, Ciaffi M (2009) Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Mol Biol 10: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Faccioli P, Ciceri GP, Provero P, Stanca AM, Morcia C, et al. (2007) A combined strategy of “in silico” transcriptome analysis and web search engine optimization allows an agile identification of reference genes suitable for normalization in gene expression studies. Plant Mol Biol 63: 679–688. [DOI] [PubMed] [Google Scholar]
- 28. Coito JL, Rocheta M, Carvalho L, Amâncio S (2012) Microarray-based uncovering reference genes for quantitative real time PCR in grapevine under abiotic stress. BMC Res Notes 5: 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kakar K, Wandrey M, Czechowski T, Gaertner T, Scheible WR, et al. (2008) A community resource for high-throughput quantitative RT-PCR analysis of transcription factor gene expression in Medicago truncatula . Plant Methods 4: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xu ZZ, Zhou GS, Wang YH (2007) Combined effects of elevated CO2 and soil drought on carbon and nitrogen allocation of the desert shrub Caragana intermedia . Plant and Soil 301: 87–97. [Google Scholar]
- 31. Guo WH, Li B, Zhang XS, Wang RQ (2010) Effects of water stress on water use efficiency and water balance components of Hippophae rhamnoides and Caragana intermedia in the soil-plant-atmosphere continuum. Agroforest Syst 80: 423–435. [Google Scholar]
- 32. Shi SQ, Shi Z, Jiang ZP, Qi LW, Sun XM, et al. (2010) Effects of exogenous GABA on gene expression of Caragana intermedia roots under NaCl stress: regulatory roles for H2O2 and ethylene production. Plant Cell Environ 33: 149–162. [DOI] [PubMed] [Google Scholar]
- 33. Condori J, Nopo-Olazabal C, Medrano G, Medina-Bolivar F (2011) Selection of reference genes for qPCR in hairy root cultures of peanut. BMC Res Notes 4: 392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Perez R, Tupac-Yupanqui I, Dunner S (2008) Evaluation of suitable reference genes for gene expression studies in bovine muscular tissue. BMC Mol Biol 9: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhong HY, Chen JW, Li CQ, Chen L, Wu JY, et al. (2011) Selection of reliable reference genes for expression studies by reverse transcription quantitative real-time PCR in litchi under different experimental conditions. Plant Cell Rep 30: 641–653. [DOI] [PubMed] [Google Scholar]
- 36. Migocka M, Papierniak A (2010) Identification of suitable reference genes for studying gene expression in cucumber plants subjected to abiotic stress and growth regulators. Mol Breeding 28: 343–357. [Google Scholar]
- 37. Chang E, Shi S, Liu J, Cheng T, Xue L, et al. (2012) Selection of reference genes for quantitative gene expression studies in Platycladus orientalis (Cupressaceae) Using real-time PCR. PLoS One 7: e33278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, et al. (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis . Plant Cell 10: 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mallona I, Lischewski S, Weiss J, Hause B, Egea-Cortines M (2010) Validation of reference genes for quantitative real-time PCR during leaf and flower development in Petunia hybrida . BMC Plant Biol 10: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mafra V, Kubo KS, Alves-Ferreira M, Ribeiro-Alves M, Stuart RM, et al. (2012) Reference genes for accurate transcript normalization in citrus genotypes under different experimental conditions. PLoS One 7: e31263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hu R, Fan C, Li H, Zhang Q, Fu YF (2009) Evaluation of putative reference genes for gene expression normalization in soybean by quantitative real-time RT-PCR. BMC Mol Biol 10: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gutierrez L, Mauriat M, Guenin S, Pelloux J, Lefebvre JF, et al. (2008) The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol J 6: 609–618. [DOI] [PubMed] [Google Scholar]
- 43. Wang X, Chen X, Liu Y, Gao H, Wang Z, et al. (2011) CkDREB gene in Caragana korshinskii is involved in the regulation of stress response to multiple abiotic stresses as an AP2/EREBP transcription factor. Mol Biol Rep 38: 2801–2811. [DOI] [PubMed] [Google Scholar]
- 44. Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.geNorm software website. Available: http://medgen.ugent.be/jvdesomp/geNorm. Accessed 2011 Dec, 13.
- 46.NormFinder software website. Available: http://www.mdl.dk/publicationsnormfinder.htm. Accessed 2011 Dec, 13.
- 47.BestKeeper software website. Available: http://www.gene-quantification.de/bestkeeper.html. Accessed 2011 Dec, 13.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
qPCR amplification specificity of the 10 reference genes, and DREB1 and DREB2 . Amplification fragments were separated by 2% agarose gel electrophoresis.
(PDF)
Amplification efficiencies of the 10 reference genes, and DREB1 and DREB2 .
(PDF)
Melting curves of the 10 reference genes, and DERB1 and DREB2 .
(PDF)
The ranking of 10 reference genes by geNorm, NormFinder, and Bestkeeper.
(DOC)
List of amplified sequences of the 10 reference genes, and DREB1 and DREB2.
(DOC)