Abstract
Most of colorectal adenocarcinomas are believed to arise from adenomas, which are premalignant lesions. Sequencing the whole exome of the adenoma will help identifying molecular biomarkers that can predict the occurrence of adenocarcinoma more precisely and help understanding the molecular pathways underlying the initial stage of colorectal tumorigenesis. We performed the exome capture sequencing of the normal mucosa, adenoma and adenocarcinoma tissues from the same patient and sequenced the identified mutations in additional 73 adenomas and 288 adenocarcinomas. Somatic single nucleotide variations (SNVs) were identified in both the adenoma and adenocarcinoma by comparing with the normal control from the same patient. We identified 12 nonsynonymous somatic SNVs in the adenoma and 42 nonsynonymous somatic SNVs in the adenocarcinoma. Most of these mutations including OR6X1, SLC15A3, KRTHB4, RBFOX1, LAMA3, CDH20, BIRC6, NMBR, GLCCI1, EFR3A, and FTHL17 were newly reported in colorectal adenomas. Functional annotation of these mutated genes showed that multiple cellular pathways including Wnt, cell adhesion and ubiquitin mediated proteolysis pathways were altered genetically in the adenoma and that the genetic alterations in the same pathways persist in the adenocarcinoma. CDH20 and LAMA3 were mutated in the adenoma while NRXN3 and COL4A6 were mutated in the adenocarcinoma from the same patient, suggesting for the first time that genetic alterations in the cell adhesion pathway occur as early as in the adenoma. Thus, the comparison of genomic mutations between adenoma and adenocarcinoma provides us a new insight into the molecular events governing the early step of colorectal tumorigenesis.
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer in males and the second in females in the world [1]. In China, the incidence of CRC has been rising in the most recent years [2]. New insights into the pathogenesis of this lethal disease are needed. The tumorigenesis of CRC is characterized by a multi-stage pathological evolution process. Most of colorectal adenocarcinomas are believed to arise from adenomas, which are premalignant lesions. The CRC development involves multiple genetic alterations including both oncogenic mutations and loss of tumor suppressor genes [3], [4]. Among these genetic alterations, inactivation of the APC gene was often detected in small adenomas, the early stage of CRC development. KRAS mutations were detected when a small adenoma grows into a large adenoma (>1 cm diameter) [5]; and alterations in PIK3CA and TP53 or other genes [6]–[9] occurred during the development of invasive adenocarcinoma. With advances in sequencing technology, the whole exomes and even the whole genomes of individual colorectal adenocarcinomas have been sequenced and a comprehensive landscape of genetic alterations was delineated [10]–[12].
Patients with colorectal adenomas are at increased risk of developing colorectal cancer. The risk of metachronous neoplasm including adenocarcinoma can be predicted by the size and the pathology of initial adenomas after their initial polypectomy [13]. Adenomas have been frequently identified since colonoscopy was used for routine colorectal cancer screening. Nonetheless, the pathologic features of initial adenoma cannot adequately predict the occurrence of adenocarcinoma. Sequencing the whole exome of the adenoma will help understand the genetic characteristics of the adenoma and potentially identify molecular biomarkers that can predict the occurrence of adenocarcinoma more precisely. It will also help understand the molecular mechanisms underlying the initial stage of colorectal tumorigenesis and may potentially uncover the pathways that may be targeted to stop the tumorigenesis process at its early stage.
In this study, we performed the exome capture sequencing of normal mucosa, adenoma and adenocarcinoma tissues from the same CRC patient. Somatic single nucleotide variations (SNVs) were identified in both the adenoma and the adenocarcinoma by comparing with the normal control from the same patient. Similar enrichment in nucleotide transitions was found in the adenoma and the adenocarcinoma while the mutation rate in the adenoma was lower than that in the adenocarcinoma. Functional annotation of the mutated genes showed several pathways including Wnt pathway, cell adhesion pathway and ubiquitin mediated proteolysis pathway were altered genetically in the early stage of colorectal tumorigenesis. The comparison between adenoma and carcinoma provides us a new insight into the molecular events governing the early step of CRC tumorigenesis.
Materials and Methods
This study was proved by the Ethical Committee of the 2nd Affiliated Hospital Zhejiang University School of Medicine and Zhejiang University Cancer Institute. For human tissue samples, written informed consent was obtained in compliance with the consent procedure approved by the Ethical Committee.
Patient Samples
Initial tumor specimens were obtained from a 66-year-old Chinese man with no family history of CRC who underwent a resection of colon cancer. Two polyps locating about 4 cm near the primary tumor in the resected colon were also resected. Part of the primary tumor and one of the polyps were dissected and snap frozen in liquid nitrogen. The resected adenocarcinoma was well to moderately differentiated and invaded into sub-mucosa without regional lymph node metastasis. The resected polyp was a tubular adenoma. Portions of the patient’s adenocarcinoma, adenoma, normal mucosa and peripheral blood were banked for exome capture sequencing and digital gene expression profiling. Additional 215 cases of fresh frozen tissues of adenocarcinoma were obtained from the tissue bank of Zhejiang University Cancer Institute. Seventy-three pairs of formaldehyde fixed-paraffin embedded (FFPE) tissues of matched adenoma and adenocarcinoma from the same patients were obtained from the pathology department of the 2nd affiliated hospital of medical school of Zhejiang University. These additional specimens were used as a validation sample set. All the tissue samples were reviewed under microscope by 2 pathologists and the tumor contents of the adenoma and adenocarcinoma samples were more than 70%.
DNA and RNA Extraction
DNA was extracted from fresh frozen tissues using a DNeasy Blood & Tissue Kit (Qiagen, Germany) and from FFPE tissues using a QIAamp DNA FFPE Tissue Kit (Qiagen, Germany) according to the manufacturer’s instructions. RNA was extracted from fresh frozen tissues using TRIzol (Invitrogen, CA, USA) according to the manufacturer’s instructions. DNA without whole genomic amplification was directly used for exome capture.
Public Genome Data
The human reference genome sequences (NCBI build 36.1/hg18) were downloaded from the UCSC (University of California, Santa Cruz) database (http://genome.ucsc.edu). Human reference mRNA sequences (Refseqs of coding mRNA) were downloaded from NCBI (http://www.ncbi.nlm.nih.gov/RefSeq). Known SNPs (single nucleotide polymorphism) were downloaded from the UCSC database (single nucleotide polymorphism database, dbSNP version 132).
Exome Capture, Sequencing and Mapping
Exome capture was performed using the NimbleGen 2.1 M Human Exome Array (Roche NimbleGen, WI, USA) which includes 180,000 coding exon (28.4 Mb) and adjacent regions (5.7 M) of one human genome (http://www.nimblegen.com/products/seqcap/arrays/exome/). Genomic DNA was randomly fragmented by nebulization to an average size of 500 bp. A pair of linkers was ligated to each end of DNA fragments. The linker-ligated DNA fragments were then hybridized to the NimbleGen 2.1 M Human Exome Array. Unbound fragments were washed away; and the target-enriched DNA was kept and then eluted. Enriched samples were amplified by ligation-mediated PCR (LM-PCR).
The captured DNA fragments were randomly ligated by DNA ligase. The ligated long exon-enriched DNA was sheared to small pieces in about 200 bp on average, and then ligated with Illumina compatible adapters. The resulted DNA fragments were subjected to the standard Solexa library preparation. The exome-enriched shotgun library was sequenced at the Illumina GAII platform (Illumina, CA, USA). Raw data from GA sequencing were processed by the Illumina Pipeline v1.3.1 software. The low quality reads were discarded (fractions of N bases > = 0.1 and fractions of bases with quality less than 5> = 0.5). The clean reads were aligned against the human reference genome (NCBI build 36.1/hg18) using the Burrows-Wheeler Alignment tool (BWA) [14]. The alignment results were further processed sequentially with local realignment, duplicate read marking, and base quality recalibration by using the Picard (developed at the Broad Institute [15]) and GATK [16], [17] pipeline software.
The chi-square test was used for comparison of the enrichment in nucleotide transitions and mutation rate between the adenoma and the adenocarcinoma.
Data Deposition and Availability
Raw data (Fastaq sequence) has been uploaded to the Short Read Archive database and the accession number is SRA052805.
Somatic Single Nucleotide Variation Analysis
Somatic single nucleotide variations of the adenoma and the adenocarcinoma were called using SAMTools (The Sequence Alignment/Map Tools) [18]. The following filtering criteria were applied: 1) the CLR score of each mutation must be more than 15; 2) only 1 alternative allele supporting read is allowed in normal control; 3) there should be at least 4 alternative allele supporting reads in tumor (adenoma or adenocarcinoma); 4) the total number of MQ0 reads in normal and tumor should be no more than 1. Finally, the mapped reads of all the SNVs were manually reviewed by using the Integrative Genomics Viewer (IGV) [19].
Digital Gene Expression Profiling
Digital gene expression profiling was applied using deep sequencing of cDNA tags [20], [21]. The libraries for sequencing tags were prepared using Illumina Gene Expression Sample Prep Kit (Illumina, CA, USA) according to manufacturer’s instruction. Briefly, Messenger RNA was purified using Oligo(dT) magnetic beads (Illumina, CA, USA) from total RNA. The first and second-strand cDNA were synthesized using Oligo(dT) as the primer. The bead-bound cDNA was digested with restriction enzyme NlaIII (Illumina, CA, USA), which recognized and cut the CATG sites. Then, the Illumina adaptor 1 was ligated to the sticky 5' end of the digested fragments. MmeI (an endonuclease provided by Illumina with a different recognition site and a digestion site) cut at 17 bp downstream of the CATG site and subsequently produced tags with adaptor 1. After removing 3' fragments, Illumina adaptor 2 was ligated to the 3' ends of tags, thereby acquiring tags with different adaptors at both ends to form a tag library. Finally, the tag libraries were sequenced on an Illumina GA II platform.
Raw sequencing data were processed by removing adapters and low quality reads. All the clean tags were mapped to the reference sequences (NCBI build 36.1/hg18). The number of unambiguous clean tags was calculated for each gene and normalized to TPM (number of transcripts per million clean tags) [20], [21].
Sanger Sequencing and Sequenom Validation
All the somatic single nucleotide mutations were validated using both Sanger sequencing and Sequenom iPLEX genotyping system (Sequenom Inc., CA, USA).
Primers for amplification and sanger sequencing validation were designed by using an online tool, Primer3 (http://frodo.wi.mit.ed/primer3/), to target regions flanking point mutations and were listed in Table S5. PCR amplifications were performed using HotStarTaq Plus DNA Polymerase (Qiagen, Germany) following manufacturer’s instruction. Sanger sequencing was performed on an ABI 3730 Capillary DNA Analyzer. Sequence trace files were manually analyzed for point mutations.
For Sequenom iPLEX genotyping, all the steps were performed following the manufacturer’s application guide (Sequenom Inc., CA, USA). Briefly, PCR amplification primers and extension probe were designed using the Sequenom MassARRAY Assay Design 3.0 software. PCR amplification and single-base extension were performed following manufacturer’s protocols (Sequenom Inc., CA, USA). After removing residual salt, the purified extension products were dispensed onto a 384-element SpectroCHIP bioarray (Sequenom Inc., CA, USA). SpectroCHIPs were analyzed by a matrix-assisted laser desorption/ionization–time of flight (MALDI-TOF) mass spectrometer. The data was processed and analyzed using MassARRAY Workstation (version 3.3).
Results
Sequence Coverage, Analysis of Mutations and Digital Gene Expression Profiling
Exome capture was performed on the normal mucosa, adenoma, and adenocarcinoma tissues from the same patient by using NimbleGen 2.1 M Human Exome Array. The target regions of exome capture include 180,000 coding exon (28.4 Mb) and adjacent regions (5.7 M). Combining parallel sequencing on an Illumina GAII platform, we generated approximately 4.6, 4.4 and 4.3 billion bases of effective sequence data with an average read length of 69 bases for normal mucosa, adenoma and carcinoma, respectively. After mapping to the human reference genome (NCBI36.1/HG18) using Burrows-Wheeler Alignment tool (BWA) [14], we obtained the average depth of each base in the target regions as 46.69×, 46.44× and 45.12× (Table 1) for each sample. The coverage of target regions was more than 98% in all the three samples including adenocarinoma, adenoma, and normal mucosa.
Table 1. Summary of sequencing coverage of the normal mucosa, adenoma and adenocarcinoma from the same patient.
| Category | Normal Mucosa | Adenoma | Adenocarcinoma |
| Total raw reads | 72586262 | 68993041 | 70103375 |
| Mapping rate | 91.70% | 90.70% | 92.70% |
| Total effective reads | 66535567 | 62548877 | 65016433 |
| Total effective yield(Mb) | 4603.18 | 4350.15 | 4456.06 |
| Average read length(bp) | 69.18 | 69.55 | 68.54 |
| Average sequencing depth on target | 46.69 | 46.44 | 45.12 |
| Base covered on target | 33627496 | 33597920 | 33559156 |
| Coverage of target regions | 98.60% | 98.50% | 98.40% |
All the mapped sequences (in the target regions and near the target regions) were used for the mutation analysis. Comparing with the sequences data of normal mucosa as the normal control, somatic single nucleotide variations (SNVs) in adenoma and adenocarcinoma were called by SAMtools [18]. We further selected the somatic SNVs in the coding sequence (CDS) of adenoma and adenocarcinoma by excluding those SNVs in flanking sequences like splicing sites, 5′-untranselated regions (UTR), 3′-UTR, introns and intergenic regions. After filtering against the single nucleotide polymorphism database (dbSNP 132) (Table S1), the numbers of the potential nonsynonymous somatic SNVs in the adenoma and adenocarcinoma prior to being validated by the Sanger sequencing and Sequenom genotyping were 16 and 44, respectively.
Simultaneously, digital gene expression profiling of the three samples including adenocarcinoma, adenoma, and normal mucosa were performed with tag based RNA sequencing. The number of clean tags obtained from these three samples was 7492596, 5291578 and 3318930, respectively. All the tags were mapped to the reference genome and only the unambiguous mapped tags were counted for each gene. The differentially expressed genes were selected through adenocaicinoma and adenoma compared with normal mucosa. The differential expressed genes in adenoma were enriched in 9 functional pathways including ribosome, cell adhesion molecules, and peroxisome proliferator-activated receptors (PPAR) signaling pathway (p<0.01) while the differential expressed genes in carcinoma were enriched in 24 pathways (p<0.01) (Table S3 and S4) by using the KEGG pathway analysis tool [22].
Mutation Spectrum of the Adenoma and the Adenocarcinoma from the same Patient
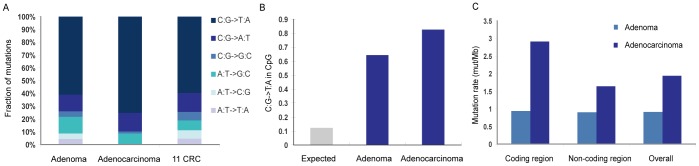
While investigating the mutation spectrum in adenoma and adenocarcinoma, we found that C: G->T: A transitions were the most significant changes in both adenoma and adenocarcinoma (Figure 1A). This feature of colorectal cancer was previously reported [10]. Our data show a significantly higher enrichment of C: G->T: A in 5′-CpG-3′ dinucleotides in both adenoma (p = 2.78E-09) and carcinoma (p = 3.14E-54) compared to that expected by chance, respectively (Figure 1B). Given the fact that DNA methylation occurs almost exclusively within the context of CpG dinucleotides [23], the enrichment of C:G->T:A in 5′-CpG-3′ dinucleotides may be associated with the extensive methylation of 5′-CpG-3′ dinucleotides in colon cancer [24], [25].
Figure 1. Somatic SNVs pattern in the adenoma and the adenocarcinoma.
(A) Somatic mutation spectrum in adenoma and adenocarcinoma, similar with 11 colorectal cancers previously reported [10], [11]. (B) Fractions of guanine mutations at 5′-CpG-3′ dinucleotides in the exome of adenoma and adenocarcinoma. (C) Prevalence of somatic SNVs in the coding region and non-coding region of the exome of the adenoma and the adenocarcinoma.
The mutation rate in the adenoma is different from that in the adenocarcinoma of this case (Figure 1C). The adenoma has a much lower mutation rate in both the coding region (0.94 vs. 2.91 mutations/Mb, p = 1.98E-06) and the overall sequencing region (0.91 vs. 1.93 mutations/Mb, p = 1.75E-09) compared with the adenocarcinoma. The coding region in the adenocarcinoma has a significantly higher mutation rate than the overall sequencing region (2.91 vs. 1.93 mutations/Mb, p = 0.0045). However, the mutation rate of the coding region in the adenoma is similar to that of the overall sequencing region (0.94 vs. 0.91 mutations/MB, p = 0.995).
Validation of Mutations Identified in the Adenoma and the Adenocarcinoma from the same Patient
Nonsynonymous SNVs were validated by Sanger sequencing in combination with Sequenom genotyping. Taken together, 12 SNVs (75.0%) out of 16 nonsynonymous somatic SNVs in the adenoma were confirmed (Table 2); and 42 SNVs (95.4%) out of 44 nonsynonymous somatic SNVs in the adenocarcinoma were confirmed (Table 3). Our result is consistent with previous reports showing that the number of nonsynonymous SNVs in the coding region of CRC is between 7 and 113 [10]–[12]. Among these mutated genes, mutations in APC, FBXW7, KIAA1409, COL4A6, FAM181A, TFR2 and NRXN3 were previously reported in colon adenomas or adenocarcinomas [10]–[12]. In addition, 5 out of 12 mutated genes identified in our cases of adenoma and 15 of 42 mutated genes identified in our cases of adenocarcinomas were reported previously to have genetic mutations in non-colorectal cancers [26]. Genetic alterations in the remaining mutated genes have not been reported in the past in colorectal cancer or any other malignancies.
Table 2. Validated somatic mutation in the colorectal adenoma.
| Sample | Gene | Genomic Change | Amino acid | Reported[10]–[12], [26] |
| Adenoma | OR6X1 | g.chr11:123129881C->T | p.A186T | |
| Adenoma | SLC15A3 | g.chr11:60461415A->G | p.I533T | |
| Adenoma | KRTHB4 | g.chr12: 51061435C->T | p.A352T | |
| Adenoma | RBFOX1 | g.chr16: 7569894C->T | p.R149W | |
| Adenoma | LAMA3 | g.chr18: 19762624T->C | p.L2778S | * |
| Adenoma | CDH20 | g.chr18: 57354795G->A | p.R454Q | * |
| Adenoma | BIRC6 | g.chr2: 32494045G->T | p.E728X | * |
| Adenoma | APC | g.chr5: 112203247G->T | p.E1353X | * |
| Adenoma | NMBR | g.chr6: 142438569C->T | p.R361H | * |
| Adenoma | GLCCI1 | g.chr7: 8092565C->T | p.L506F | |
| Adenoma | EFR3A | g.chr8: 133057465G->A | p.G390E | |
| Adenoma | FTHL17 | g.chrX: 30999856C->T | p.D46N |
Table 3. Validated somatic mutation in the colorectal adenocarcinoma.
| Sample | Gene | Genomic Change | Amino acid | Reported[10]–[12], [26] |
| Adenocarcinoma | PGM1 | g.chr1: 63886845 G->A | p.R405H | * |
| Adenocarcinoma | DTL | g.chr1:210341029G->A | p.G692R | |
| Adenocarcinoma | PPP1R3C | g.chr10: 93380476 G->A | p.R48X | |
| Adenocarcinoma | OR51E2 | g.chr11: 4660017 G->T | p.F167L | |
| Adenocarcinoma | RRP8 | g.chr11:6579205A->G | p.S223P | |
| Adenocarcinoma | NARS2 | g.chr11:77963118G->A | p.H22Y | |
| Adenocarcinoma | FAM109A | g.chr12:110285370C->T | p.R95H | |
| Adenocarcinoma | FAM181A | g.chr14: 93461390 G->T | p.R7I | * |
| Adenocarcinoma | NRXN3 | g.chr14:78245443T->C | p.F78S | * |
| Adenocarcinoma | KIAA1409 | g.chr14:93242879C->T | p.P2418L | * |
| Adenocarcinoma | TGM7 | g.chr15: 41358655 C->T | p.M597I | |
| Adenocarcinoma | CORO1A | g.chr16: 30105713 C->T | p.R133C | |
| Adenocarcinoma | KRTHA1 | g.chr17: 36805323 G->A | p.R223W | |
| Adenocarcinoma | GNAL | g.chr18:11743892G->A | p.R191Q | |
| Adenocarcinoma | PPAP2C | g.chr19: 238556 C->A | p.D134Y | * |
| Adenocarcinoma | FLJ37549 | g.chr19: 42852592 G->A | p.R100W | |
| Adenocarcinoma | RYR1 | g.chr19: 43768631 G->A | p.E4977K | |
| Adenocarcinoma | GSK3A | g.chr19: 47429297 T->C | p.Q328R | |
| Adenocarcinoma | ATF2 | g.chr2: 175691277 G->A | p.A89V | * |
| Adenocarcinoma | ZFP36L2 | g.chr2:43305936C->A | p.G171C | |
| Adenocarcinoma | NFATC2 | g.chr20: 49525428 C->A | p.L503F | |
| Adenocarcinoma | ZFP64 | g.chr20: 50202574 C->T | p.V468I | |
| Adenocarcinoma | GRIK1 | g.chr21: 29945440 C->T | p.V275I | |
| Adenocarcinoma | KRTAP19-7 | g.chr21:30855383G->A | p.R33C | |
| Adenocarcinoma | CESK1 | g.chr22: 15453028 G->A | p.T138M | |
| Adenocarcinoma | KLHL22 | g.chr22: 19126407 T->C | p.M620V | |
| Adenocarcinoma | ALPK1 | g.chr4: 113573037 C->T | p.P962L | * |
| Adenocarcinoma | FBXW7 | g.chr4: 153471357 G->A | p.R367X | * |
| Adenocarcinoma | APC | g.chr5: 112179160 C->T | p.R302X | * |
| Adenocarcinoma | APC | g.chr5: 112203106 G->T | p.E1306X | * |
| Adenocarcinoma | POU4F3 | g.chr5:145700003C->T | p.R274W | |
| Adenocarcinoma | FLT4 | g.chr5:179989704G->A | p.S174L | * |
| Adenocarcinoma | FAM54A | g.chr6: 136602344 C->A | p.L274F | |
| Adenocarcinoma | KIF25 | g.chr6: 168185641 C->T | p.R264X | |
| Adenocarcinoma | SDK1 | g.chr7: 4085690 G->A | p.R1091Q | |
| Adenocarcinoma | TFR2 | g.chr7:100063331A->G | p.F552L | |
| Adenocarcinoma | CUX1 | g.chr7:101631839G->A | p.G848S | |
| Adenocarcinoma | NRG1 | g.chr8:32733489G->A | p.R123H | |
| Adenocarcinoma | OR13J1 | g.chr9:35860034C->T | p.R122H | * |
| Adenocarcinoma | COL4A6 | g.chrX:107305660C->T | p.V904I | * |
| Adenocarcinoma | WDR44 | g.chrX:117411223G->A | p.E263K | * |
Mutation Analysis in Additional Cases of Adenoma and Adenocarcinoma
In order to know the prevalence of the above-identified mutations in CRC patients, we analyzed all the 54 SNVs in 73 samples of matched adenoma and adenocarcinoma from the same patients and additional 215 samples of adenocarcinomas by sanger sequencing in combination with sequenome genotyping. We found that the same SNVs of APC and FBXW7 repeatedly occurred in these tumors (Table 4). The E1353X mutation of APC was detected in one additional case of adenoma, and was also reported in colorectal adenocarcinomas [27]. The R302X and E1306X mutations of APC were detected in several other adenocarcinomas. The R367X mutation of FBXW7 was detected in several cases of both adenomas and adenomcarcinomas (Table 4). It should be noted that the remaining cases of adenomas and adenocarcinomas may have mutations in other residues of APC and FBXW7. Similarly, although the remaining SNVs were not detected in any of the 73 cases of adenomas and 288 cases of adenocarcinomas, we cannot exclude the possibility that mutations occur in other regions of these 54 genes.
Table 4. Recurrent mutations in 288 additional cases of colorectal tumors.
| Gene | Mutation | Frequency | SampleID | Tumor |
| APC | g.chr5: 112203247 G->T | 1/73 | 10-027 | Adenoma |
| FBXW7 | g.chr4: 153471357 G->A | 2/73 | 10-005 | Adenoma |
| 10-006 | Adenoma | |||
| APC | g.chr5: 112179160 C->T | 3/288 | 10-058 | Adenoarcinoma |
| 10-040 | Adenoarcinoma | |||
| 252T | Adenoarcinoma | |||
| APC | g.chr5: 112203106 G->T | 2/288 | B7C8 | Adenoarcinoma |
| K3A5 | Adenoarcinoma | |||
| FBXW7 | g.chr4: 153471357 G->A | 1/288 | 187T | Adenoarcinoma |
Pathway Analysis of Mutated Genes in the Adenoma and the Adenocarcinoma
All the validated, mutated genes were annotated through the KEGG pathway analysis [22], the Gene Ontology analysis [28], and literature review (Table S2). We found that several cellular pathways that shared between adenoma and adenocarcinoma (Table 5). First, APC was mutated in both adenoma and adenocarcinoma; thus this study confirmed that the Wnt pathway was activated in the early stage of colorectal tumor development. Second, our study supported the importance of the cell adhesion pathway in the initial process of colorectal tumor development. We found that CDH20 and LAMA3 were mutated in the adenoma while NRXN3 and COL4A6 were mutated in the adenocarcinoma. The products of all these four genes are involved in cell adhesion. Third, the ubiquitination process was also found to be altered in both adenoma and adenocarcinoma. BIRC6, which was found to be mutated in the adenoma, is an ubiquitin-conjugating enzyme (E2) and involved in the apoptosis process. FBXW7, which was found to be mutated in the adenocarcinoma, is a subunit of ubiquitin ligase (E3) and is involved in the cell cycle regulation. Finally, genes involved in olfactory transduction and neuroactive ligand-receptor interaction pathway were also mutated in both the adenoma and the adenocarcinoma.
Table 5. Mutated pathways in adenoma and adenocarcinoma.
| Pathways involved in carcinogenesis | Mutated genes in Adenoma | Mutated genes in Adenocarcinoma |
| Cell adhesion pathway | CDH20; LAMA3 | NRXN3;COL4A6 |
| Wnt signaling pathway | APC | APC |
| Ubiquitin mediated proteolysis pathway | BIRC6 | FBXW7 |
| Olfactory transduction pathway | OR6X1 | OR13J1; OR51E2; GNAL |
| Neuroactive ligand-receptor interaction | NMBR | GRIK1 |
| Glycerolipid metabolism | PPAP2C | |
| MAPK signaling pathway | ATF2 | |
| Aminoacyl-tRNA biosynthesis | NARS2 | |
| Cytokine-cytokine receptor interaction | FLT4 | |
| Chemokine signaling pathway | GSK3A | |
| Calcium signaling pathway | GNAL | |
| ErbB signaling pathway | NRG1 | |
| Glycolysis/Gluconeogenesis | PGM1 | |
| Insulin signaling pathway | PPP1R3C |
To study how gene mutations affect the expression of genes belonging to the pathways that were found to be altered genetically in both adenoma and adenocarcinoma, we performed the real time PCR analysis of 12 representative mutated genes among those listed in Table 5. As shown in Figure S1, all 12 genes are consistently downregulated in both the adenoma and the adenocarcinoma comparing to the normal colorectal tissue from the same index patient. This has suggested that the mutations identified in these genes may have led to the downregulation of their gene expression in either adenoma or adenocarcinoma of this patient. Interestingly, even though these genes are not mutated in both adenoma and adenocarcinoma, their expression levels are consistently downregulated in both tissues, suggesting that other mechanisms may have led to the downregulation of gene expression.
Discussion
Our study, for the first time, described the somatic mutation in the whole exome of a colorectal adenoma. The adenoma had a smaller number of somatic mutations but demonstrating a similar pattern of enrichment in nucleotide transitions and involving similar functional pathways as the adenocarcinoma from the same patient. Identification of these gene alterations and their associated pathways in the adenoma has provided new sights into the molecular process of developmental course from colorectal adenoma to colorectal adenocarcinoma.
We identified 12 somatic nonsynonymous SNVs in the adenoma. The APC gene was the only gene reportedly associated with colon cancer. It was found to be mutated in both adenoma and adenocarcinoma. Four other genes were reported as mutated genes in other types of cancer. First, a missense mutation in LAMA3 in adenoma changed from leucine to serine. LAMA3 is involved in cell adhesion, signal transduction and differentiation and its mutations were previously reported in brain cancer [29], ovary cancer [15] and melanoma [30]. Second, a missense SNV in CDH20 changed from arginine to glutamine. CDH20 encodes a type II classical cadherin which is involved in cell to cell adhesion and its mutation was previously reported in breast cancer [10]. Third, a nonsense mutation was found in BIRC6 and would result in a truncated protein. The mutations in BIRC6 were previously reported in several kinds of cancer including melanoma [31], glioma [29], breast cancer [11] and pancreatic cancer [32]. Its gene product, also known as APOLLON, acts as an ubiquitin-conjugating enzyme (E2) and inhibits apoptosis by facilitating the degradation of apoptotic proteins by ubiquitination. Fourth, a missense SNV was found in NMBR. NMBR encodes a Neuromedin B receptor, and its mutations were previously reported in gliomas [29], [33].
There were 42 somatic nonsynonymous SNVs in the adenocarcinoma. Among them, APC and FBXW7 were reported as two mutated gene ‘mountains’ in colorectal carcinomas [11]. In addition, 5 other mutated genes were previously reported in colon cancers including FAM181A, NRXN3, KIAA1409, TFR2, and COL4A6. The mutation of FAM181A was reported in colon cancers [12]. But its function is not clear, so the further funcational study will be needed. NRXN3 functions in the vertebrate nervous system as cell adhesion molecules and receptors and was previously found to be mutated in colon cancer [12], breast cancer [10], glioma [33], squamous cell carcinoma of the head and neck [34], [35] and ovary cancer [15]. The mutation of KIAA1409, which is an unknown protein, was previously reported in colon cancers [11], ovary cancer [15], and melanoma [30]. TFR2 encodes a single-pass type II membrane protein involved in iron uptake and was previously found to be mutated in colon cancer [12] and liver cancer [36]. COL4A6 encodes one of the six subunits of type IV collagen likely involved in cell to cell adhesion and was found to be mutated in ovary cancer [15] and colon cancer [11]. We observed mutations in other 8 genes in the adenocarcinoma. Their mutations were only reported in non-CRC malignancies. Thus, they are newly discovered gene mutations in CRCs.
In addition to the APC/Wnt pathway known to be involved in the development of the colorectal adenoma, this study has identified a few new cellular pathways that are altered in the adenoma at the genetic level. These pathways are also altered at the genetic level in the adenocarcinoma, suggesting that they may functionally drive colorectal tumorigenesis. First, CDH20 and LAMA3, which are both involved in cell adhesion, were found to be mutated in the colorectal adenoma. This is quite interesting as the cell adhesion pathway is thought to be important for cancer cell invasion and metastasis, which are the characteristics of the later stage of cancer development. Our finding thus provides direct evidence supporting a recently proposed hypothesis that the capacity of metastasis may have been acquired by the cancer cells at the early stage of their development [37]. Second, our result also showed that ubiquitin mediated proteolysis pathway is altered at the genetic level in the early stage of tumorigenesis. BIRC6 which encodes E2 ubiquitin-conjugating enzyme is involved in ubiquitin mediated proteolysis. Although we found that FBXW7 which encodes E3 ubiquitin-ligase enzyme is mutated only in colorectal adenocarcinomas [38], previous report has identified somatic mutations in FBXW7 in adenomas [39]. Recently, whole-exome sequencing of premalignant lesions of pancreatic adenocarcinoma also revealed recurrent mutations in components of ubiquitin-dependent pathways [40], [41]. Our study thus suggested that the ubiquitin mediated proteolysis may play a role in the early stage of tumorigenesis in the CRC. Third, we found OR6X1 and NMBR, which are involved in olfactory transduction and neuroactive ligand-receptor interaction, respectively, are mutated in the adenoma. Many proteins involved in neuronal transduction are also involved in cell-cell communication, which again is important for cancer cell invasion and metastasis. The functions of the mutations of OR6X1 and NMBR in the early stage of colorectal tumorigenesis remain to be explored. Multiple genes involved in olfactory transduction and neuroactive ligand-receptor interaction including OR13J1, OR51E2, GNAL, and GRIK1 were also found by this study to be mutated in the adenocarcinoma, suggesting these two classes of genes warrant further investigation. Finally, although neither the remaining mutations found in the adenoma nor the genetic alterations in their involved pathways are observed in the adenocarcinoma, their importance in the development of colorectal adenoma cannot be excluded.
Although the host environmental factors were largely eliminated by analyzing the mutation profiles of the adenoma and the adenocarcinoma from the same patient, it is not surprising that mutations in the adenoma are not present in the adenocarcinoma from the same patient because they may have arisen from two independent tumorigenesis processes. Nevertheless, we found that mutations affecting the same cellular pathways were found in both the adenoma and the adenocarcinoma from the same patient. It is also not surprising that identical mutations identified in the adenoma from one patient were not found in adenomas from additional patients tested in this study. We anticipate that different adenomas harbor mutations in different regions on the same genes that were found to be mutated in one adenoma. If these genes are important for the development of adenoma, we may see them repeatedly altered in multiple adenomas by sequencing their entire exome. However, none of them except APC are anticipated to be frequently mutated in the adenoma. The same cellular pathways such as the cell adhesion and ubiquitin-dependent pathways may be altered in other components at the genetic level more frequently in the adenomas and therefore warrant further investigation.
Supporting Information
mRNA expression of genes whose mutations were identified in either adenoma or adenocarcinoma by the exome capture sequencing. Gene expression levels were measured by the quantitative real-time PCR analysis on RNA purified from the normal mucosa, adenoma and adenocarcinoma tissue from the same patient, respectively. Exome capture sequencing was performed on these same tissues. Each real-time PRC experiment was repeated for at least three times. The GAPDH expression level was used as an internal control. Expression levels relative to those in normal mucosa after being normalized with the GAPDH expression level are shown in histogram.
(TIF)
The process of selecting somatic nonsynonymous mutations in adenoma and adenocarcinoma.
(XLS)
Functional annotation and Pathway analysis of somatic mutaiton in adenoma and adenocarcinoma.
(XLS)
Pathway analysis of differential expressed genes in adenoma.
(XLS)
Pathway analysis of differential expressed genes in adenocarcinoma.
(XLS)
Primers for validation of somatic SNVs.
(XLS)
Funding Statement
This project is supported by the National Natural Science of China Foundation 30973382 and the National High Technology Research and Development Program of China (2006AA02A302,2012AA02A204,2012AA02A506). Lei Zheng is supported by an NIH K23 CA148964-01 and a Johns Hopkins University School of Medicine Clinician Scientist Award. Lei Zheng is an adjunct "Guangji Scholar" at the Second Affiliated Hospital of Zhejiang University School of medicine. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jemal A, Bray F (2011) Center MM, Ferlay J, Ward E, et al (2011) Global cancer statistics. CA Cancer J Clin 61: 69–90. [DOI] [PubMed] [Google Scholar]
- 2.Jie He PZ, Wanqing Chen (2011) CHINESE CANCER REGISTRY ANNUAL REPORT: Military Medical Science Press (People's Republic of China). 50 p.
- 3. Fearon ER, Vogelstein B (1990) A genetic model for colorectal tumorigenesis. Cell 61: 759–767. [DOI] [PubMed] [Google Scholar]
- 4. Jones S, Chen WD, Parmigiani G, Diehl F, Beerenwinkel N, et al. (2008) Comparative lesion sequencing provides insights into tumor evolution. Proc Natl Acad Sci U S A 105: 4283–4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, et al. (1988) Genetic alterations during colorectal-tumor development. N Engl J Med 319: 525–532. [DOI] [PubMed] [Google Scholar]
- 6. Baker SJ, Fearon ER, Nigro JM, Hamilton SR, Preisinger AC, et al. (1989) Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science 244: 217–221. [DOI] [PubMed] [Google Scholar]
- 7. Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, et al. (1995) Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science 268: 1336–1338. [DOI] [PubMed] [Google Scholar]
- 8. Thiagalingam S, Lengauer C, Leach FS, Schutte M, Hahn SA, et al. (1996) Evaluation of candidate tumour suppressor genes on chromosome 18 in colorectal cancers. Nat Genet 13: 343–346. [DOI] [PubMed] [Google Scholar]
- 9. Samuels Y, Velculescu VE (2004) Oncogenic mutations of PIK3CA in human cancers. Cell Cycle 3: 1221–1224. [DOI] [PubMed] [Google Scholar]
- 10. Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, et al. (2006) The consensus coding sequences of human breast and colorectal cancers. Science 314: 268–274. [DOI] [PubMed] [Google Scholar]
- 11. Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, et al. (2007) The genomic landscapes of human breast and colorectal cancers. Science 318: 1108–1113. [DOI] [PubMed] [Google Scholar]
- 12. Bass AJ, Lawrence MS, Brace LE, Ramos AH, Drier Y, et al. (2011) Genomic sequencing of colorectal adenocarcinomas identifies a recurrent VTI1A-TCF7L2 fusion. Nat Genet 43: 964–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang G, Zheng W, Sun QR, Shu XO, Li WD, et al. (1998) Pathologic features of initial adenomas as predictors for metachronous adenomas of the rectum. J Natl Cancer Inst 90: 1661–1665. [DOI] [PubMed] [Google Scholar]
- 14. Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cancer Genoma Atlas Research Network (2011) Integrated genomic analyses of ovarian carcinoma. Nature 474: 609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, et al. (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, et al. (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43: 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, et al. (2011) Integrative genomics viewer. Nat Biotechnol 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. t Hoen PA, Ariyurek Y, Thygesen HH, Vreugdenhil E, Vossen RH, et al. (2008) Deep sequencing-based expression analysis shows major advances in robustness, resolution and inter-lab portability over five microarray platforms. Nucleic Acids Res 36: e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Morrissy AS, Morin RD, Delaney A, Zeng T, McDonald H, et al. (2009) Next-generation tag sequencing for cancer gene expression profiling. Genome Res 19: 1825–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M (2012) KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res 40: D109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, et al. (2006) DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet 38: 1378–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olivier M, Hussain SP, Caron de Fromentel C, Hainaut P, Harris CC (2004) TP53 mutation spectra and load: a tool for generating hypotheses on the etiology of cancer. IARC Sci Publ: 247–270. [PubMed]
- 25. Costello JF, Fruhwald MC, Smiraglia DJ, Rush LJ, Robertson GP, et al. (2000) Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat Genet 24: 132–138. [DOI] [PubMed] [Google Scholar]
- 26. Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, et al. (2007) Patterns of somatic mutation in human cancer genomes. Nature 446: 153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lovig T, Meling GI, Diep CB, Thorstensen L, Norheim Andersen S, et al. (2002) APC and CTNNB1 mutations in a large series of sporadic colorectal carcinomas stratified by the microsatellite instability status. Scand J Gastroenterol 37: 1184–1193. [DOI] [PubMed] [Google Scholar]
- 28. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, et al. (2008) An integrated genomic analysis of human glioblastoma multiforme. Science 321: 1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wei X, Walia V, Lin JC, Teer JK, Prickett TD, et al. (2011) Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat Genet 43: 442–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, et al. (2010) A comprehensive catalogue of somatic mutations from a human cancer genome. Nature 463: 191–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, et al. (2008) Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321: 1801–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cancer Genoma Atlas Research Network (2008) Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455: 1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, et al. (2011) Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science 333: 1154–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, et al. (2011) The mutational landscape of head and neck squamous cell carcinoma. Science 333: 1157–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li M, Zhao H, Zhang X, Wood LD, Anders RA, et al. (2011) Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nat Genet 43: 828–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scheel C, Onder T, Karnoub A, Weinberg RA (2007) Adaptation versus selection: the origins of metastatic behavior. Cancer Res 67: 11476–11479; discussion 11479–11480. [DOI] [PubMed]
- 38. Sancho R, Jandke A, Davis H, Diefenbacher ME, Tomlinson I, et al. (2010) F-box and WD repeat domain-containing 7 regulates intestinal cell lineage commitment and is a haploinsufficient tumor suppressor. Gastroenterology 139: 929–941. [DOI] [PubMed] [Google Scholar]
- 39. Kemp Z, Rowan A, Chambers W, Wortham N, Halford S, et al. (2005) CDC4 mutations occur in a subset of colorectal cancers but are not predicted to cause loss of function and are not associated with chromosomal instability. Cancer Res 65: 11361–11366. [DOI] [PubMed] [Google Scholar]
- 40. Wu J, Jiao Y, Dal Molin M, Maitra A, de Wilde RF, et al. (2011) Whole-exome sequencing of neoplastic cysts of the pancreas reveals recurrent mutations in components of ubiquitin-dependent pathways. Proc Natl Acad Sci U S A 108: 21188–21193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanda M, Matthaei H, Wu J, Hong SM, Yu J, et al.. (2012) Presence of somatic mutations in most early-stage pancreatic intraepithelial neoplasia. Gastroenterology 142: 730–733 e739. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
mRNA expression of genes whose mutations were identified in either adenoma or adenocarcinoma by the exome capture sequencing. Gene expression levels were measured by the quantitative real-time PCR analysis on RNA purified from the normal mucosa, adenoma and adenocarcinoma tissue from the same patient, respectively. Exome capture sequencing was performed on these same tissues. Each real-time PRC experiment was repeated for at least three times. The GAPDH expression level was used as an internal control. Expression levels relative to those in normal mucosa after being normalized with the GAPDH expression level are shown in histogram.
(TIF)
The process of selecting somatic nonsynonymous mutations in adenoma and adenocarcinoma.
(XLS)
Functional annotation and Pathway analysis of somatic mutaiton in adenoma and adenocarcinoma.
(XLS)
Pathway analysis of differential expressed genes in adenoma.
(XLS)
Pathway analysis of differential expressed genes in adenocarcinoma.
(XLS)
Primers for validation of somatic SNVs.
(XLS)