Abstract
Cooperativity in transcription factor (TF) binding is essential in eukaryotic gene regulation, and arises through diverse mechanisms. Here we focus on one mechanism, collaborative competition, which is of interest because it arises both automatically, with no requirement for TF co-evolution, and spontaneously, with no requirement for ATP-dependent nucleosome remodeling factors. Previous experimental studies of collaborative competition analyzed cases in which target sites for pairs of cooperating TFs were contained within the same side of the nucleosome. Here we utilize new assays to measure cooperativity in protein binding to pairs of nucleosomal DNA target sites. We focus on the cases that are of greatest in vivo relevance, in which one binding site is located close to the end of a nucleosome, and the other binding site is located at diverse positions throughout the nucleosome. Our results reveal energetically significant positive (favorable) cooperativity for pairs of sites on the same side of the nucleosome, but, for the cases examined, energetically insignificant cooperativity between sites on opposite sides of the nucleosome. These findings imply a special significance for TF binding sites that are spaced within one half nucleosome length (74 bp) or less along the genome, and may prove useful for prediction of cooperatively acting TFs genome-wide.
Keywords: chromatin, gene regulation, cooperativity, transcription factor, DNA
Introduction
Cooperative action of multiple DNA binding transcription factors (TFs) is a hallmark of eukaryotic promoters and enhancers.1–3 Cooperative binding is required for most eukaryotic TFs to identify functional DNA target sites, because of the factors’ short degenerate target sites (~15 bits of information) compared to eubacterial TF’s (~25 bits). 4 Such information-poor target sites occur by happenstance every few thousand basepairs, raising the question of how the small fraction of bona-fide functional sites are specified. Nucleosomal organization of eukaryotic DNA does not eliminate this specificity problem, as only 75–90% of the genomic DNA is wrapped in nucleosomes 5. The smaller genomes of eubacteria are similarly occluded by DNA binding proteins,6 yet eubacterial TF’s have much greater specificity. Cooperative interactions between eukaryotic TF’s allow TF’s to collectively develop the sequence specificity that they lack individually.4 Here we extend our earlier analysis of a novel mechanism for cooperativity in TF binding, in which cooperativity arises spontaneously, through synergistic action of two or more TF’s against a nucleosome.7; 8
Cooperativity between eukaryotic TF’s can arise by conventional mechanisms. Two TFs can favorably bind each other while simultaneously binding their DNA targets, allowing the two TFs to effectively bind as a single unit with increased specificity. Alternatively, another protein can bridge two non-interacting TFs, again allowing the two TF’s to bind DNA as a single unit. Conventional TF cooperativity can be augmented by a DNA bending protein that brings two separately bound TFs into proximity, facilitating their interaction.
The above mechanisms require coevolution of different proteins, or of different binding surfaces of one protein, because the cooperativity derives ultimately from favorable specific protein-protein interactions.
In contrast, the nucleosomal organization of DNA makes possible a novel cooperativity between eukaryotic TF’s, which can occur with no requirement for coevolution. This cooperativity arises automatically, when two or more arbitrary TFs each seek individually to bind to target sites contained within one nucleosome.7–14 This cooperativity can arise with or without the assistance of an ATP-dependent nucleosome remodeling factor. In the former case, an initially bound TF can recruit a remodeler, which then disassembles or moves a nucleosome that covers a second TF’s binding site, thereby facilitating binding by the second TF.15; 16 However, nucleosome-dependent cooperativity can also occur spontaneously, without participation of remodelers. TFs can spontaneously bind to DNA target sites that, in the time-average, are buried inside nucleosomes, through frequent but transient “site exposure” conformational fluctuations that are inherent to nucleosomes;17–20 and binding of a first TF can enhance the accessibility of a second TF’s binding site, facilitating spontaneous binding by the second TF.7 The resulting “collaborative competition” of TF’s against a nucleosome occurs both in vitro7; 9 and in vivo8; 10; 21 without requirement for remodeler action,7; 11; 22 and may contribute to transcriptional regulation genome-wide.23; 24 We seek to understand the structural constraints governing cooperative TF binding via this remodeler-independent collaborative competition.
Existing studies suggest that all that is required to evolve cooperatively acting pairs of TFs by remodeler-independent collaborative competition is for binding sites for the two TFs to be juxtaposed close together along the DNA, within the same nucleosome.7; 12 However, previous in vitro analyses of collaborative competition only tested for cooperativity between sites on the same side of the nucleosome,7; 9 raising the question of whether comparable cooperativity also occurs between sites on opposite sides of the nucleosome. Indeed, binding to sites on opposite sides might occur with negative (unfavorable) cooperativity instead of positive (favorable) cooperativity.25; 26 Here we use restriction enzyme digestion and fluorescence resonance energy transfer (FRET) assays to further characterize cooperative interactions in a nucleosome. As we discuss below, for reasons both of plausible biological relevance and of technical feasibility, our analysis is restricted to cases in which at least one of the two potentially cooperating target sites is located not far from an end of the nucleosomal DNA. Our previous demonstration of reciprocity in cooperativity,7 together with our earlier and new findings of cooperativity between many different pairs of sites, make our findings more general. Our results again reveal energetically significant positive (favorable) cooperativity between pairs of sites on one side of the nucleosome, but, for the cases examined cooperativity between sites on opposite sides of the nucleosome is energetically insignificant. Our results imply specific lengthscales at which clustering of TF binding sites may be functionally relevant, and may prove helpful for improving genome-wide prediction of cooperatively acting TFs.
Results
Our earlier theoretical analysis of collaborative competition7 was based on data obtained by others,9 in which the binding sites investigated were located on the same side of the nucleosome. Those studies utilized a coupled equilibrium binding assay. Cooperative binding was manifested as an influence of the binding of one DNA binding protein on the binding of another DNA binding protein to the same nucleosome, with double-binding events detected using a gel mobility supershift assay.
If our understanding is correct, such nucleosome-dependent cooperative binding should be detectable using quite different assays as well. We therefore sought first to extend the previous work with two new assays for cooperative binding to nucleosomal target sites. One assay uses a restriction enzyme in place of one of the two DNA binding proteins which were used in the coupled binding assay described above. We then measure how a protein-DNA binding event at one location in the nucleosome influences the ease with which the restriction enzyme can cleave its target site, which is sterically occluded by the nucleosomal wrapping. This assay is attractive because it yields a positive signal (specific DNA cleavage) on a near-zero background while being carried out fully in solution, thereby avoiding some problems that can occur in gel mobility shift assays.27 Our other assay looks directly at the coupling between a protein-DNA binding event at one location in the nucleosome and the ease (energetic cost, or equivalently, the equilibrium probability) of making a second site more-available for binding of another protein, using a fluorescence resonance energy transfer (FRET) readout.28 This assay is attractive because it provides both evidence of cooperative coupling, and also insight into the underlying molecular mechanism of this coupling.
We began by using both assays to extend the previous findings of cooperativity between sites on the same side of the nucleosome.
Nucleosome binding cooperativity measured with restriction enzymes
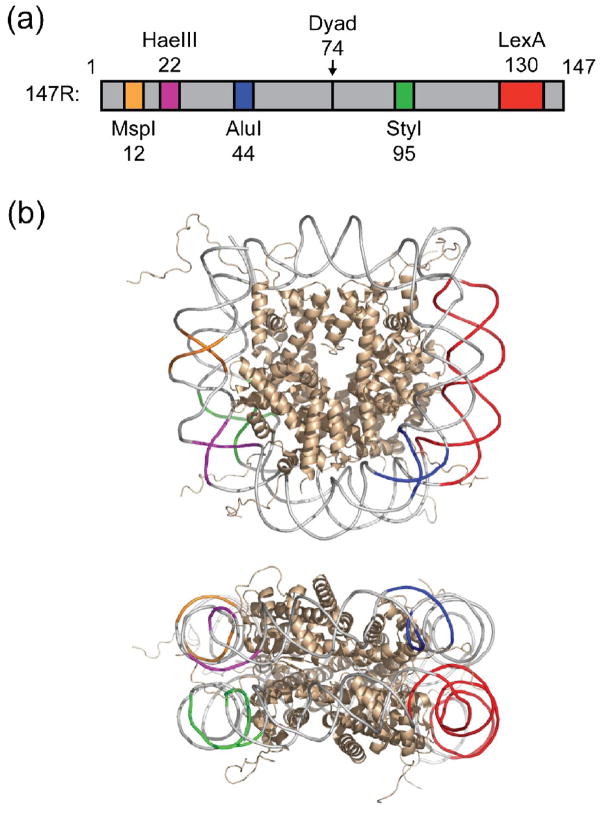
For the restriction enzyme digestion assay we utilized a nucleosomal DNA construct (“147R”) that we had extensively characterized and used in analyses of nucleosome conformational dynamics.19; 20 This DNA occupies a unique translational setting on the nucleosome, and contains within it a binding site for a site-specific DNA binding protein (the Escherichia coli LexA repressor protein) near one end of the DNA, and unique sites for several restriction enzymes at various other locations (Figure 1a, b). The LexA site is oriented so as to face “inward” toward the histone surface, while restriction enzymes generally embrace nearly the entire circumference of their DNA target sites. Thus binding by both LexA protein and the various restriction enzymes is sterically occluded by the nucleosomal wrapping of their respective DNA target sites.17; 19; 20 Nevertheless, spontaneous “site exposure” nucleosome conformational changes, in which segments of DNA unwrap from the histone core make these sites transiently accessible, allow binding by LexA, and binding and cleavage by restriction enzymes, but with a cost in occupancy (for LexA) and cleavage rate (for restriction enzymes) compared to naked DNA. For detailed analyses and tests of the site exposure mechanism, see refs. 17–20
Figure 1. DNA template for restriction enzyme digestion kinetic assay.
(a) Schematic illustration of 147R DNA19; 20 indicating locations of recognition sites for the four restriction enzymes and the DNA binding protein LexA. (b) Crystal structure of the nucleosome55 depicting the binding sites for LexA (red) and the restriction enzymes MspI (orange), HaeIII (magenta), AluI (blue) and StyI (green). Histone proteins are shown in ribbon representation (colored wheat).
The idea of the cooperativity assay is that, if binding at the LexA site is energetically coupled to ease of binding at a restriction site, then by first adding sufficient LexA protein to drive its occupancy to near saturation, the ease of binding and concomitant rate of DNA cleavage by a restriction enzyme should be enhanced commensurately;7 and the quantitative magnitude of the digestion rate enhancement (after correction with a parallel control digestion on naked DNA) is a measure of the free energy of coupling between the LexA site and the restriction site – i.e., an apparent free energy of cooperativity.
We focus on experiments in which the LexA binding site is located near one end of the DNA because equilibrium binding in that region is orders of magnitude more favorable than at internal sites7; 29 and access occurs orders of magnitude more quickly, 19; Tims, 2011, p02085} and therefore is more likely to be relevant to stepwise invasion of nucleosomes in vivo (see Discussion). We synthesized 147R DNA by preparative scale PCR and purified it by HPLC. The purified DNA was then mixed with purified histone octamer in 2M NaCl and reconstituted into nucleosomes by salt gradient dialysis. The reconstituted nucleosomes were purified away from any free DNA and non-nucleosomal complexes by sucrose gradient ultracentrifugation (Supplemental Figure 1a). Native gel electrophoresis of the purified nucleosomes confirms that they are greater than 97% free of contaminating naked DNA (Supplemental Figure 1b). As expected, binding of LexA protein was greatly reduced relative to binding to naked DNA, yet binding could still be driven to near-saturation (Supplemental Figure 1c, d).
The StyI site (Figure 1a) tests our analysis and serves as an additional positive control to complement our earlier analysis of coupled equilibrium binding data:7 if cooperativity occurs between two DNA sites on the same side of the nucleosome, then binding of LexA at its target site should enhance the equilibrium accessibility of the StyI site, and thus also the rate of StyI cleavage. A typical StyI digestion experiment and quantitative analysis for both naked DNA and nucleosomal DNA is shown in Figure 2a–e. The reactions reach apparent endpoints at less than complete extent of digestion, due to slow inactivation of the enzyme during the course of the reaction (see Methods). We are careful to use the same enzyme concentrations for a given sample (nucleosomes or naked DNA) in the presence or absence of LexA protein, so that any time dependent changes in enzyme activity will be identical between the plus and minus LexA samples. Our analysis ignores the first timepoint after enzyme addition, as this can have contributions from any nucleosomes that dissociated during preparation and mixing of the reaction; and we analyze the initial velocity only, using exponential or linear fits (see Methods).
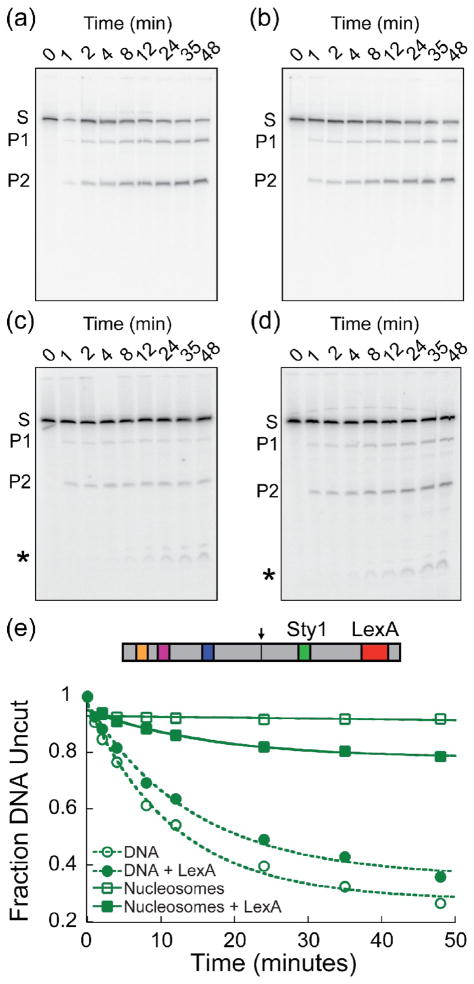
Figure 2. Nucleosome-dependent cooperativity between LexA and StyI sites.
Representative experiment probing accessibility at the StyI binding site in the absence and presence of saturating LexA. (a–d) Denaturing polyacrylamide gel analyses of StyI digestion reactions. Substrate (S) is converted to products (P1 and P2). Asterisk (*) indicates additional products resulting from StyI “star” activity. (a) < 0.5 nM naked DNA with 10 U/mL Sty1. (b) <0.5 nM naked DNA with 100 nM LexA and 10 U/mL StyI. (c) 5 nM nucleosomes with 5000 U/mL StyI. (d) 5 nM nucleosomes with 1 μM LexA and 5000 U/mL StyI. (e) Quantitative analysis of the digestion pattern resolved in the gels in panels a–d. Circles: naked DNA; squares: nucleosomes. Open datapoints: no LexA; filled datapoints: saturating LexA. The fraction of DNA uncut is plotted versus time. Data are fit to single exponential (a, b, d) or linear (c) functions. For nucleosomes, the zero time point is omitted from the analysis to exclude the digestion of any contaminating naked DNA, which is complete within the first minute of reaction (see Methods).
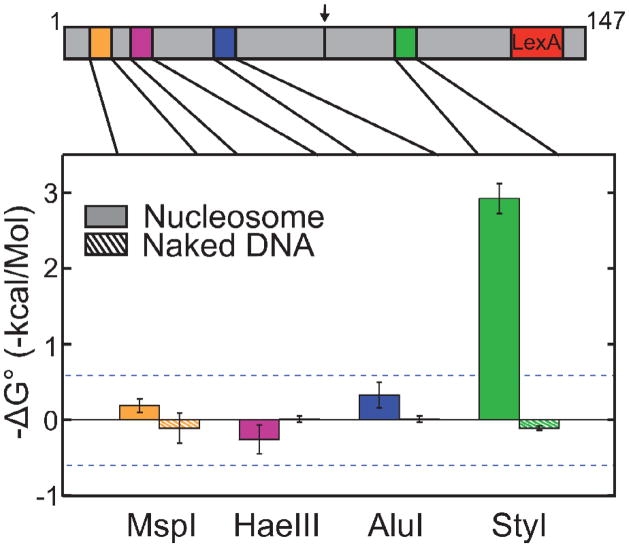
As expected, the presence of LexA bound at near saturation negligibly influenced the rate of StyI cleavage (Figure 2 a, b, e and Table 1). From many such experiments we calculate an apparent slight unfavorable (negative) cooperativity between LexA and StyI binding on naked DNA of 0.11 ± 0.03 kcal mol−1, a value that is small in comparison to the energies of inevitable thermal fluctuations (~0.6 kcal mo−1) and, by that metric, is insignificant.
Table 1.
Summary of observed rate constants (kobs) for nucleosome and naked DNA digestions
| Enzyme | LexA | Nucleosomal DNA (n) | Naked DNA (n) |
|---|---|---|---|
| StyI | + | 5.6 × 10−2 ± 8.0 × 10−3 (3) | 6.7 × 10−2 ± 9.0 × 10−3 (3) |
| − | 4.9 × 10−4 ± 1.1 × 10−4 (3) | 8.0 × 10−2 ± 1.4 × 10−2 (3) | |
| HaeIII | + | 7.9 × 10−4 ± 1.0 × 10−4 (7) | 3.4 × 10−1 ± 1.5 × 10−1 (7)* |
| − | 1.3 × 10−3 ± 2.7 × 10−4 (7) | 3.2 × 10−1 ± 1.4 × 10−1 (7)* | |
| AluI | + | 7.4 × 10−4 ± 3.4 × 10−4 (5) | 3.2 × 10−2 ± 3.1 × 10−3 (5) |
| − | 3.9 × 10−4 ± 1.9 × 10−4 (5) | 3.2 × 10−2 ± 4.9 × 10−3 (5) | |
| MspI | + | 2.6 × 10−1 ± 2.4 × 10−2 (2) | 3.0 × 10−1 ± 1.6 × 10−1 (2) |
| − | 1.9 × 10−1 ± 9.9 × 10−3 (2) | 3.4 × 10−1 ± 8.4 × 10−2 (2) |
Values are shown as the mean rate constants ± standard error (except for MspI, which shows standard deviation), where (n) is the number of independent experiments.
HaeIII digestions on naked DNA utilized different enzyme concentrations on different days, and were normalized for fixed enzyme concentration prior to averaging.
In contrast, the presence of LexA bound at near saturation greatly enhanced the rate of StyI cleavage of nucleosomal DNA compared to the same nucleosomes and conditions without bound LexA (Figure 2 c–e and Table 1), implying strong positive nucleosome-dependent cooperativity between the binding of LexA and StyI proteins to their respective nucleosomal DNA target sites. From many such experiments, we calculate an apparent positive cooperativity between LexA and StyI binding to the nucleosomal target sites of 2.94 ± 0.20 kcal mo−1. Correcting this value by the result from the naked DNA control yields a nucleosome-dependent cooperativity free energy between LexA and StyI binding of 3.05 ± 0.20 kcal mo−1, in good agreement with cooperativity free energies between analogous nucleosomal locations that we had deduced in our earlier analysis of coupled equilibrium binding data7 (see Discussion).
Note that 3 kcal mo−1 is an extremely large cooperativity, corresponding to a ~160-fold increase in probability of the doubly-bound state. For comparison, conventional protein-protein cooperativities such as with lambda repressor are typically only ~1–2 kcal mo−1, corresponding to ~5- to ~30-fold increases in probability of the doubly bound state; yet even these much smaller conventional cooperativities are of great biological significance.30
We conclude that the restriction enzyme digestion assay properly and sensitively monitors cooperativity between nucleosomal sites, and that this cooperativity can be highly significant.
Nucleosome binding cooperativity monitored by FRET
If our reasoning is correct – if binding of one protein makes it easier for another to bind – then we should not need to have the second protein present in the reaction at all: binding of LexA protein should increase the probability that sites further inside the nucleosome (for example, around the StyI site) should spend more time displaced from the surface of the histone octamer. We and others have shown previously that equilibrium measurement of FRET between two points, one on the DNA and one on the protein, can detect changes in the fraction of time that DNA spends partially unwrapped off the surface of the histone octamer, making binding sites in the unwrapped DNA more accessible.31–35
We created two new FRET systems that together investigate locations flanking the StyI site analyzed above. One of these FRET constructs places a fluorescence donor dye (Cy3) at basepair 35, the other at basepair 57, relative to a LexA binding site at basepairs 8–27 (Figure 3b, c, respectively). For comparison, the StyI site is 52 basepairs inward from the LexA site at basepairs 8–27 (measured from the nearest nucleosome end, i.e., basepairs 121–140 of the DNA as illustrated in Figure 1). Acceptor dyes were placed at engineered unique cysteines on the surface of the histone core nearby these DNA locations (H2B T112C and H4 L22C, respectively). Both of these engineered histones were utilized together with engineered H3 C110A, so that the other cysteines engineered at the desired locations would be unique within the histone octamer. Histone octamers were reconstituted from individually expressed and purified histones (see Methods).
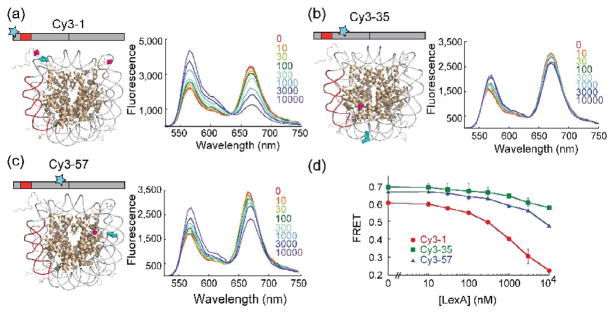
Figure 3. FRET assay for nucleosome-dependent cooperativity.
Assay probes cooperativity between the LexA site and additional locations on the same side of the nucleosome. Shown are schematic illustrations of DNA templates and labeled nucleosome structures, indicating locations of the LexA binding site (red), fluorescence donor dye (Cy3, Cyan) and fluorescence acceptor dyes (Cy5, Magenta), and normalized emission spectra of the labeled nucleosomes titrated with increasing concentrations of LexA (with excitation of the Cy3 donor). All histone octamers had the engineered mutation H3C110A together with an additional unique cysteine at a different location on one or another of the four histones. (a) Cy3-1 nucleosomes: Cy3 attached at bp 1, Cy5 attached at H3V35C. (b) Cy3-35 nucleosomes: Cy3 attached at bp 35, Cy5 attached at H2BT112C. (c) Cy3-57 nucleosome: Cy3 attached at bp 57, Cy5 attached at H4L22C. (d) Quantitative analysis of FRET efficiency changes during LexA titrations for Cy3-1 (red circles), Cy3-35 (green squares), and Cy3-57 (blue triangles).
For the Cy3-57, Cy5-H4L22C system, we estimate the expected donor/acceptor distance at ~2 nm, in comparison to a ~6 nm characteristic energy transfer distance for this dye pair;36 thus, we expect to, and indeed do (see below), observe a high FRET efficiency when the DNA is fully wrapped. A second copy of the acceptor, present in the other symmetry-related copy of H4 in the nucleosome, is much farther away (~8 nm) and thus contributes little to the FRET signal (Table 2). For the Cy3-35, Cy5-H2BT112C system, the closer acceptor is ~3 nm away, again much closer than the ~6 nm characteristic distance for energy transfer, so we again expect, and do (see below) observe high FRET efficiency for this dye pair as well. In this case, the second acceptor dye is also within transfer distance (5 nm), and thus also contributes to a high FRET efficiency for fully wrapped nucleosomes. For both systems, unwrapping the DNA to an extent sufficient to allow binding by LexA protein (i.e., at least several bp beyond the inner end of the LexA target site, in order to make room for the ~3 nm diameter LexA protein37) is expected to greatly increase the distances from the Cy3 donors to both Cy5 acceptors, leading to a decrease in FRET signal, as is indeed observed (see below).
Table 2.
Approximate distances between dye pairs
| FRET Construct (Acceptor Location) | Distance from Cy3 to closer Cy5, nm | Distance from Cy3 to further Cy5, nm |
|---|---|---|
| Cy3-1 (Cy5-H3V35C) | 2 | 7 |
| Cy3-35 (Cy5-H2BT112C) | 3 | 5 |
| Cy3-57 (Cy5-H4L22C) | 2 | 8 |
As a positive control, we also repeated an earlier experiment in which we placed a Cy3 donor at the end (basepair 1) of the nucleosomal DNA, beyond the LexA site, with the Cy5 acceptor placed on a nearby unique cysteine at H3 V35C (C110A) (Figure 3a). The two Cy5 acceptors are located ~2 and ~7 nm away from the Cy3 donor in this system (Table 2), again yielding high FRET efficiency for the fully wrapped nucleosome. We showed earlier that LexA binding on this construct is made possible by spontaneous nucleosomal site exposure: DNA undergoes frequent large scale partial unwrapping fluctuations off of the histone surface20 (whether LexA protein is present or not19), rendering the LexA target site freely accessible; and subsequent binding by LexA to the now-accessible target site traps the nucleosome in this open state. In this open state, the point on the DNA to which the Cy3 FRET donor dye is attached is much further away from both Cy5 acceptor dyes compared to the fully wrapped nucleosome, leading to the observed large reduction in FRET efficiency.20
As expected, titration of all three FRET constructs with increasing concentrations of LexA resulted in an increase in Cy3 emission and a decrease in Cy5 emission (Figure 3a–c), implying a decrease in apparent FRET efficiency (Figure 3d). Possible alternative interpretations of the apparent LexA-dependent FRET decreases were explicitly tested and excluded in an earlier analysis of the end-labeled construct (Figure 3a).20 That study showed that the apparent FRET change did not depend on the chemical identify of the dyes; moreover LexA titration of a related FRET system in which a LexA site was present, but was located on the opposite side of the nucleosome from the DNA-linked FRET donor left the FRET signal unchanged. These observations implied that the fluorescence changes are measuring real changes in FRET efficiency, not nonspecific effects on dye spectral properties. We subsequently analyzed several closely related FRET systems both with and without LexA sites, both for individual FRET-labeled nucleosomes and FRET-labeled nucleosomes in the middle of three-nucleosome long chains, and found that in all cases the FRET changes appropriately mirrored LexA binding.38 Most recently, for a series of FRET systems where the LexA site was moved progressively inward, the rate of LexA binding, calculated from the time-dependent FRET changes, decreased dramatically with each step in a manner that accorded with the decreasing position-dependent equilibrium constant for site exposure, providing further evidence that the FRET system was correctly monitoring LexA binding.39
Thus, the apparent decreased FRET efficiency measured in our new experiments implies an increase in the time-averaged separation between fluorescence donor and acceptor dyes. We refrain from making detailed claims about exact distance changes from these FRET experiments, because of the difficulty of exactly knowing all required parameters. We note that, based on our earlier analyses of nucleosome dynamics, which suggest a stepwise unwrapping of nucleosomal DNA from one end, we might have expected binding by LexA to increase the accessibility at basepair 35 more than at basepair 57, whereas our FRET results apparently show the opposite. This could mean that DNA does not necessarily unwrap only from an end,31; other possible explanations include environmental effects on the Cy3 or Cy5 emission unique to the Cy3-35 or Cy3-57 FRET pair; or possibly that the Cy3 and/or Cy5 dye locations used in this construct may interfere sterically with the nucleosome structure, even though our modeling had suggested that there should be no interference.
Taken together, these results mean that binding of LexA protein near the end of the nucleosomal DNA cooperatively influences the behavior of DNA stretches elsewhere in the nucleosome. These results also shed light on the nature of the cooperative coupling mechanism: the increase in time-averaged separation between inner-more stretches of the nucleosomal DNA and histone would presumably make it easier for something else to bind to those DNA stretches – precisely as found in the restriction enzyme digestion assay.
In summary, both the quantitative analysis of the StyI restriction enzyme digestion kinetics and the qualitative analysis of the FRET experiments demonstrate cooperative coupling between LexA binding near one DNA end and DNA wrapping elsewhere along that same side of the nucleosome DNA, confirming key predictions of our previous theoretical analysis.7 Thus, three independent kinds of assay, investigating many different pairs of proteins, locations of DNA binding sites, and DNA sequence contexts, all point to the same conclusion: cooperativity is ubiquitous for pairs of sites on the same side of the nucleosome.
Cooperativity between two sides of the nucleosome
We next sought to test for cooperative effects of protein binding on one side of the nucleosome with access to sites on the other side. Of the two new solution-based assays, the restriction enzyme digestion assay is both technically simpler and also more sensitive, since it looks for a positive signal (DNA cleavage) on top of a near zero background, rather than a (potentially small) decrease in an initially-large signal. We therefore used the restriction enzyme assay to address this question.
We utilized the DNA construct of Figure 1a for analysis of cooperative binding at three distinct locations on the side of the nucleosome opposite to the LexA binding site. These three sites have three key attributes. The HaeIII site (bp22) and AluI site (bp44) together flank a location (bp35) for which cooperativity to the LexA site was detected by FRET when the LexA site was on the same side of the nucleosome as the FRET donor/acceptor pair. The location of the AluI site relative to an end of the nucleosome is similar to that of the StyI site, for which cooperativity to the LexA site was detected by StyI restriction cleavage kinetics when the LexA site was on the same side of the nucleosome as the restriction site. Finally, like the LexA site itself, the location of the MspI site (bp12) is within a DNA region (relative to the nearest end of the nucleosome) that is most-easily occupiable by other DNA binding proteins see Discussion).
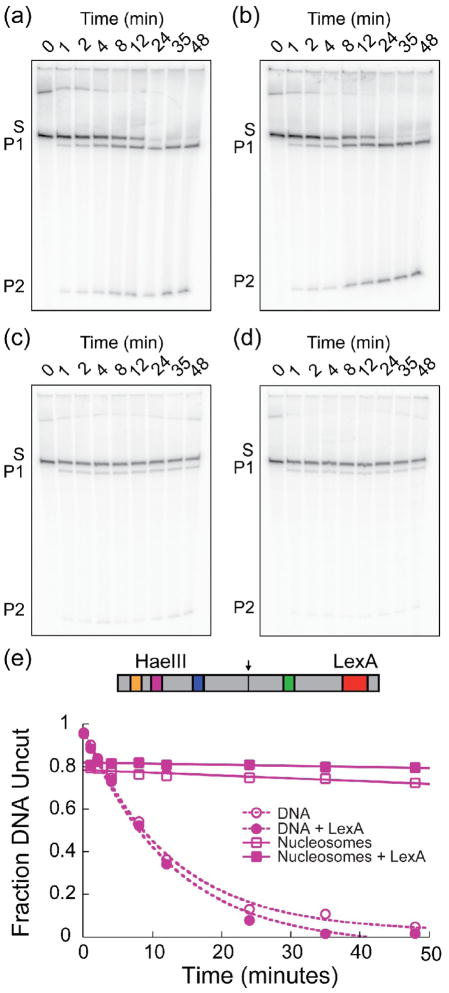
Results from the restriction enzyme assay probing cooperativity between LexA binding on one side of the nucleosome and these three sites on the opposite side of the nucleosome are presented in Figure 4 and Supplemental Figures 2, 3 for HaeIII, MspI, and AluI, respectively, along with their naked DNA controls. As expected, LexA binding negligibly influenced the digestions on the naked DNA. However, in contrast to the results from both the restriction enzyme and FRET assays probing cooperativity between sites on the same side of the nucleosome, LexA binding did not facilitate binding to nucleosomal DNA significantly, at any of these three new sites.
Figure 4. No nucleosome-dependent cooperativity between LexA and HaeIII sites.
Representative experiment probing accessibility at the HaeIII binding site in the absence and presence of saturating LexA. (a–d) Denaturing polyacrylamide gel analyses of HaeIII digestion reactions. Substrate (S) is converted to products (P1 and P2) over time. (a) <0.5 nM naked DNA with 0.625 U/mL HaeIII. (b) <0.5 nM naked DNA with 100 nM LexA and 0.625 U/mL HaeIII. (c) 5 nM nucleosomes with 5000 U/mL HaeIII. (d) 5 nM nucleosomes with 1 μM LexA and 5000 U/mL HaeIII. (e) Quantitative analysis of the gels in panels (A–D). The fraction of DNA uncut is plotted versus time. Data are fit to single exponential (naked DNA) or linear (nucleosomal DNA) functions. For nucleosomes, the zero time point is omitted from data analysis (see Methods).
Quantitative results from several such experiments at each site are presented in Figure 5. LexA binding marginally enhanced accessibility of nucleosomal DNA at the MspI and AluI sites (implying slight positive, i.e., favorable, cooperativity), and marginally decreased accessibility at the nucleosomal HaeIII site (implying slight negative cooperativity); but the quantitative magnitudes of cooperativity in each case are small in comparison to the energies of typical thermal fluctuations, and by that metric are insignificant. Similarly, as expected, none of the influences on the naked DNA controls are energetically significant.
Figure 5. Position dependent cooperativity across the nucleosome.
Shown are the coupling free energy between each restriction enzyme and LexA, for nucleosomes (solid bars) or naked DNA (hatched). Error bars: standard errors, for MspI (n=2), HaeIII (n=7), AluI (n=5) and Sty1 (n=3). Dashed horizontal lines represent the numerical value of kBT, the characteristic energy of thermal fluctuations, where KB is Boltzmann’s constant and T the absolute temperature.
In summary, our new assays confirm our previous theoretical analysis, revealing significant positive cooperativity between diverse sites on the same side of the nucleosome, but in addition show that this positive cooperativity does not extend to diverse sites on the other side of the nucleosome. Moreover, a possible negative cooperativity between the two sides of the nucleosome, which was anticipated theoretically, is similarly seen not to occur.
Discussion
In this study we examined how spontaneous binding of a protein to a nucleosomal DNA target site influences the spontaneous accessibility and concomitant protein binding at other DNA target sites contained inside the same nucleosome, a process that we refer to as collaborative competition. Previous in vitro studies provide a theoretical analysis and experimental demonstration of collaborative competition for TFs with binding sites on the same side of the nucleosome,7; 9; 12; 40 however, these previous studies have not tested whether cooperativity also occurs if the binding sites for the factors are positioned on opposite sides of the nucleosome. Our study extends the earlier analyses to ask if cooperativity occurs between sites near one end of the nucleosome and other sites distributed along the full nucleosome length. Interestingly, binding to sites on opposite sides has been proposed to occur, but to do so with negative (unfavorable) cooperativity instead of positive (favorable) cooperativity.25; 26. Our new study tests this question, and introduces two additional assays for collaborative competition.
For reasons both of plausible biological relevance and of technical feasibility, our analysis is restricted to cases in which at least one of the potentially cooperating target sites is located not far from an end of the nucleosomal DNA. This is because binding to sites far inside the nucleosome is suppressed (in both the equilibrium and kinetic senses) by many orders of magnitude relative to binding near the ends of the nucleosomal DNA.18; 20; 29; 39 For example, the LexA protein used in our present study binds to its site at basepairs 8–27, with 300 nM affinity, and on the sub-second timescale, while moving the site further inside the nucleosome by just 10 bp, to basepairs 18–37, reduces the accessibility of the LexA target site so greatly that we achieve only partial occupancy by LexA even at the exceedingly high concentration of 10 μM, the limit of what we can manage with the stocks that we are able to prepare, and the rate of binding is similarly reduced by orders of magnitude.39 Thus, given realistic in vivo TF concentrations, sites relatively near to an end of the nucleosomal DNA are the only ones on which typical TFs could achieve high occupancy if binding individually via spontaneous nucleosome site exposure;18; 41; 42, and therefore remodeler-independent collaborative competition of TFs against a common nucleosome most plausibly would occur with at least one of the two TFs binding to a site near a nucleosome end. Finally, as a technical matter for our in vitro studies, sites near the nucleosome ends are also the only ones for which we can drive occupancy to high levels in vitro, as required for sensitive tests of cooperativity.
For these reasons, we focused our analysis on systems in which the target sites for at least one (LexA), and sometimes both (LexA and MspI), of the DNA binding proteins were located within high-accessibility DNA stretches near the nucleosome ends. The MspI site is related by symmetry to the LexA site on the opposite side of the nucleosome, and thus is comparably close to the DNA end. As expected, this site proved highly accessible to MspI. The HaeIII site tests a location further inside the nucleosome, with reduced but still significant accessibility to HaeIII when binding individually. And both the HaeIII and AluI sites are symmetrically disposed relative to the StyI site (Figure 1) and basepairs 35 and 57 (Figure 3), for which positive cooperativity with bound LexA was observed. Thus, we consider that the sites we tested represent the most plausible ones at which to look for cooperative interactions with the LexA site. And for these sites on opposite sides of the nucleosome that we examined, our clear result is that neither positive nor negative cooperativity occurs to significant extent.
Our earlier analysis of coupled equilibrium binding experiments (for sites located on the same side of the nucleosome) showed that cooperativity in binding of pairs of proteins to nucleosomal DNA target sites occurs reciprocally: the amount by which the first protein’s binding enhances the subsequent binding of the second protein, regardless of which of the two proteins binds first.7 This reciprocity holds as long as the system is in equilibrium, which is the case in our studies of both restriction enzyme digestion and simple binding.18 Our new study extends our earlier work with two additional types of experiment: the restriction enzyme/LexA experiment and the LexA/FRET experiment. Thus three independent kinds of experimental test of collaborative competition all lead to the same conclusion: cooperativity between pairs of sites on the same side of the nucleosome occurs ubiquitously.
Our new results with the LexA/StyI system are also in good quantitative accord with the predictions of our theoretical model for cooperativity between pairs of sites on the same side of the nucleosome.7 In that theoretical analysis, the free energy of cooperativity is exactly equal to minus one times the free energy cost of DNA unwrapping sufficient to expose the outer-more site. This free energy cost in turn is simply the (log of the) position-dependent equilibrium constant for accessibility at that outer-more site, easily measured in experiment.18 We showed earlier that this model, with no adjustable parameters, provided a reasonably good quantitative explanation of several different coupled equilibrium binding experiments. Our new results provide an additional test of these ideas. The cooperativity measured here between LexA and Sty I is approximately −3 kcal mol−1 (Figure 5), which corresponds to ~160-fold enhancement of Sty I accessibility induced by LexA binding. According to our theoretical model, the expected cooperativity between the LexA and Sty I sites should be equal to −1 times the free energy of site exposure of the outer-more target site, which in this case is the LexA site. And according to our initial analysis of site exposure equilibria,18 the free energy of site exposure of the LexA site is to be calculated as the ratio of affinities of LexA protein for the target site in the nucleosome versus for the same site as naked DNA. We recently reported the measurement of these quantities:39 the affinity of LexA protein for this same target site as naked DNA was 1–3 nM, while LexA binding to the same site in the nucleosome had an apparent affinity of ~300 nM. Thus the nucleosomal organization of the LexA site decreases LexA binding affinity by ~150-fold, implying that the free energy of site exposure of the LexA site is ~3 kcal mol−1, in very good agreement with our current measurement.
Conclusions
Spatial clustering of TF binding sites is widely used as one of many important features in algorithms for genome-wide prediction of TF binding sites and TF cooperativity.23; 43–49 Our earlier studies showed that collaborative competition can occur when the two TF binding sites are contained within the same nucleosome, i.e., are separated by ~147bp or less, suggesting that this may be a natural lengthscale over which TF site clustering might usefully be defined. Our new results show further that, for remodeler-independent collaborative competition to occur at plausible in vivo TF concentrations, the two TF sites should be contained not just within the same nucleosome, but within the same side of the same nucleosome. This implies a potential special significance for TF sites that are spaced by distances of ~74bp or less, and, especially, for TF sites that are spaced by 74 bp or less and that are additionally known to be located inside the same competing nucleosome.
We suggest that, in addition to the diverse features that are already utilized in predictive algorithms, consideration of TF site spacings relative to the nucleosome lengthscale, and, ideally, relative to detailed nucleosome locations, may further improve understanding and genome-wide prediction of functionally cooperative transcription factors.
Materials and Methods
DNA preparation
The DNA construct, 147R, was amplified by PCR from the high-affinity nucleosome positioning sequence 601, modified to incorporate a LexA binding site centered at 130 bp as described previously.20 147R was PCR amplified, concentrated and buffer exchanged into H2O and purified by reverse-phase HPLC on a Zorbax column (Agilent). The scheme for HPLC purification was 10–15% (V/V) gradient of acetonitrile (ACN) in 0.1M triethanolamine acetate (TEAA), pH 7.0, at 1 mL/min flow rate over 20 minutes. The peak containing fractions (absorbance at 260 nm) were lyophilized to remove ACN and the DNA was resuspended, washed, and concentrated into storage buffer (1 mM Tris HCl, pH 8.0, 0.1 mM EDTA) using a YM30 centricon filter (Amicon). 147R was labeled with 32P at the 5′ ends by T4 polynucleotide kinase (New England Biolabs). Cy3-1 labeled DNAs for FRET analysis were prepared as described 20. Cy3-35 and Cy3-57 DNAs were prepared by the same procedure except that the second generation dye labeled PCR primer had the Cy3 attached off the DNA base at nucleotide position 35 or 57, respectively, instead of at the 5′-end of the primer. In this case, the primers for labeling were purchased with amino-dT residues at the position to be labeled; these were then subsequently derivatized with amine-reactive Cy3, and the resulting dye-labeled oligonucleotides purified by reverse phase HPLC. Subsequent steps were as for Cy3-1.
Histone octamer and LexA preparation
Purified chicken erythrocyte core histones and core particle DNA were prepared from chicken blood as described.50 Recombinant wild type (H2A, H2B, and H4) and engineered H3 C110A and H3 C110A V35C Xenopus histones were expressed in E. coli, purified, reconstituted into histone octamers, and labeled with Cy5 as described.20 Two additional engineered histone mutants, H2B T112C and H4 L22C were prepared and purified by the same methods. LexA protein was prepared from the expression plasmid pJWL228 (gift of J. Little, University of Arizona, Tucson) and purified to near-homogeneity as described.51
Nucleosome reconstitution
Trace quantities of γ-32P-ATP labeled 147R plus bulk nucleosome core particle DNA were reconstituted with chicken core histone octamer at a 0.37:1 (HO: DNA) by salt gradient dialysis and purified by 5–30% sucrose gradient ultracentrifugation as described elsewhere (Supplemental Figure 1a).18; 52 Purified nucleosomes were stored at 4 °C in storage buffer (5 mM Tris-HCl, 0.5 mM EDTA, 5 mM NaCl). A native gel of the purified nucleosomes reveals less than 3% contaminating naked DNA (Supplemental Figure 1b). The small amount of contaminating naked DNA is due to unstable nucleosomes in the digestion buffer as well as nucleosomes that fall apart during gel electrophoresis.18; 53 The small fraction of nucleosomes that are unstable in our digestion assays are cut immediately by the restriction enzymes and are excluded in the subsequent analysis.18 Nucleosomes for FRET analyses were reconstituted using tracer concentrations of Cy3-labeled DNA plus bulk nucleosome core particle DNA together with Cy5-labeled histone octamers, and were purified by sucrose gradient ultracentrifugation as described.19; 20
LexA gel mobility shift assay
147R as naked DNA (1 nM) or incorporated into nucleosomes (5 nM) were titrated with increasing concentration of purified LexA in restriction enzyme digestion buffer. Samples were equilibrated for 2 minutes and loaded onto a running 5% (W/V) polyacrylamide gel at 300 volts (Supplemental Figure 1c, d). Gels were dried and analyzed by Image Quant software using a Storm Phosphorimager (GE Healthcare).
Restriction enzyme digestions
All restriction enzymes were purchased from New England Biolabs. Enzyme concentrations varied from 0.1 U/mL to 5000U/mL, when necessary, enzymes were diluted in NEB Dilution Buffer A before addition to the reactions. All digests were carried out at 30 °C in reaction buffer (50 mM NaCl, 10.25 mM Tris-HCl (pH 7.9), 5 mM MgCl2, 1mM DTT, 0.25 mM EDTA, 5% glycerol, 1mg/mL BSA). LexA or an equivalent volume of LexA storage buffer (10 mM Pipes pH 7.0, 0.1mM EDTA, 200mM NaCl, 10% glycerol) was added at either 100 nM (for naked DNA) or 1 μM (for nucleosomes) and allowed 5 minutes to bind to <0.1 nM naked DNA or 5 nM nucleosomes prior to the start of the reaction. For a given set of experiments (naked DNA or nucleosome DNA +/− LexA) all reactions were conducted at the same time. Reactions were initiated by addition of enzyme and time point aliquots were quenched immediately with addition of equal volume quench buffer (20 mM EDTA, 0.1 % SDS, 1 mg/mL proteinase K). For loading samples into gels, formamide loading dye was added to each time point before incubation for 5 minutes at 94 °C. Digestions were resolved on 8% (StyI, AluI) or 12% (HaeIII, MspI) denaturing urea polyacrylamide gels in 2X TBE (90mM Tris-Borate, 2mM EDTA).
Analysis of digestion experiments
The results of the digestion experiments were resolved by gel electrophoresis, dried and imaged as described above. The gels were analyzed by measuring the intensity of the products and substrates using the program Image Quant (Molecular Dynamics). The intensities of the substrate and product bands were measured by drawing individual boxes around each band. The local background intensity was subtracted from each band from a box of the same size close to the actual band, declared to be background. To determine the amount of DNA that is uncut, counts in the substrate band were divided by the sum of counts in the substrate plus product bands (S/(S+P1+P2)).
We noticed that in many cases the digestions do not proceed to completion, yielding different extents of digestion for different enzymes. We believe that the major effect responsible for the different amplitudes of the exponential decays (different final endpoints) is inactivation of the restriction enzymes over time. For example, in the Sty I digest of naked DNA (Figure 2e), even the naked DNA is not digested to completion. However control experiments with higher enzyme concentrations allow digestion in much shorter time, and these reactions on the same DNAs do go to completion, proving that all of the DNA is indeed digestible. To address the problem of the enzyme activity changing over time, we use the same enzyme concentrations for a given nucleosome (or naked DNA) sample in the absence and presence of LexA protein; we carry out the reactions for both nucleosomes and naked DNA over the same period of time, so that the decrease in restriction enzyme activity over the nearly hour long sampling will be eliminated from our analysis during calculation of the coupling free energy.
In fitting the data points we restrict the analysis to the initial velocity. Additionally, the first time point following addition of the restriction enzyme can reflect a contribution from nucleosomes that had dissociated before the reaction was started; therefore the initial time point for each experiment was excluded from the analysis. The data were fit to a first order exponential decay curve (or, in a cases of very slow/little digestion, the data were fit to a straight line, representing the leading term in an exponential decay).
An additional complication came from Sty1, which has published “star” activity at high enzyme and glycerol concentrations (New England Biolabs), which we observed in the nucleosome digestions. In our quantitative analyses for this enzyme, we only included the bona fide specific product bands in the calculation of total band intensity in each lane.
Calculation of coupling free energy
The kinetic analysis of the site exposure mechanism and collaborative competition are described in detail elsewhere.7; 18 For this study, the cutting rates determined in the restriction digestion assays (in the presence and absence of LexA) can be used to determine coupling free energy between the two proteins by:
| (1) |
Where R is the Boltzmann constant, T is the temperature and is the position dependent equilibrium constant for site exposure. is experimentally determined by dividing the observed digestion rate on nucleosomal DNA by the observed digestion rate on naked DNA while taking into account the difference in enzyme concentration for nucleosomes vs. naked DNA:
| (2) |
If LexA is assumed to have no effect on accessibility of the binding sites on naked DNA, the expression for determining the coupling free energy is simplified to:
| (3) |
In reality, LexA might affect the rate of restriction enzyme cutting on naked DNA, therefore the apparent coupling free energy was also measured on naked DNA and subtracted from the result on nucleosomes. The coupling free energy was calculated separately for each individual set of experiments.
Fluorescence measurements
Fluorescence measurements were carried out as described.54 For all experiments, nucleosome solutions were diluted to 5 nM final concentration in 0.5 x TE, pH 8.0 (TE is 10 mM Tris, 1 mM Na3EDTA). Samples were excited at 515 nm. We calculated the proximity ratio (A/(D+A)), where A is the (replicate-averaged) Cy5 emission intensity (670 nm), and D the (replicate-averaged) Cy3 emission intensity (570 nm), as a proxy for the FRET efficiency.
Supplementary Material
(a) Sucrose gradient purification of nucleosomes (blue) and naked DNA (pink). Nucleosomes are reconstituted by gradual salt dialysis and separated from naked DNA and nonnucleosomal contaminants on a 5–30% (wt/vol) sucrose gradient. Naked DNA is separated from unincorporated 32P also on a 5–30% (wt/vol) sucrose gradient. (b) Native gel electrophoresis of purified naked DNA (Lane 1) and nucleosomes (Lane 2). W: loading well of the gel; N and D, nucleosomes and naked DNA, respectively. L: 50 bp ladder. Phosphorimager analysis reveals less than 3% contaminating naked DNA in nucleosome sample. (c) Native gel mobility shift assay for LexA binding to naked DNA; same experimental conditions as for digestion assay. 100 bp ladder is included as size standards (not shown). D, mobility of naked DNA; D-LexA, mobility of DNA-LexA complex. W indicates loading well. (d) Native gel mobility shift assay for LexA binding to nucleosomes. D, mobility of contaminating naked DNA (or nucleosomes that did not survive electrophoresis); N, nucleosomes; N-LexA, nucleosomes stably bound by LexA.
Representative experiment probing accessibility at the MspI site in the absence and presence of saturating LexA. (a–d) Denaturing polyacrylamide gel analyses of MspI digestion reactions. Substrate (S) is converted to products (P1 and P2). (a) <0.5 nM naked DNA with 0.83 U/mL MspI. (b) <0.5 nM naked DNA with 100 nM LexA and 0.8 U/mL MspI. (c) 5 nM nucleosomes with 10000 u/mL MspI. (d) 5 nM nucleosomes with 1 μM LexA and 10000 U/mL MspI. (e) Quantitative analysis of the gels in panels (a–d). The fraction of DNA uncut is plotted versus time; data are fit to single exponentials. For nucleosomes, the zero time point is omitted from the analysis.
Representative experiment probing accessibility at the AluI site in the absence and presence of saturating LexA. (a–d) Denaturing polyacrylamide gel analyses of AluI digestion reactions. Substrate (S) is converted to products (P1 and P2). (a) <0.5 nM naked DNA with 4 U/mL AluI. (b) <0.5 nM naked DNA with 100 nM LexA and 4 U/mL AluI. (c) 5 nM nucleosomes with 4000 u/mL AluI. (d) 5 nM nucleosomes with 1 μM LexA and 4000 U/mL AluI. (e) Quantitative analysis of the digestion pattern resolved in the gels of panels (a–d). The fraction of DNA uncut is plotted versus time. Data are fit to single exponential (naked DNA) or linear (nucleosomal DNA) functions. For nucleosomes, the zero time point is omitted from the analysis.
Acknowledgments
We thank members of the Widom lab for discussion, and the Keck Biophysics Facility at Northwestern University for the use of instruments. J.W. acknowledges research support from the NIH and a Morris Belkin Visiting Professorship at the Weizmann Institute of Science. H.S.T. acknowledges support from an NIH Cell and Molecular Basis of Disease Training Grant.
Abbreviations used
- TF
transcription factor
- FRET
fluorescence resonance energy transfer
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Carey M. The enhanceosome and transcriptional synergy. Cell. 1998;92:5–8. doi: 10.1016/s0092-8674(00)80893-4. [DOI] [PubMed] [Google Scholar]
- 2.Levine M, Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–51. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- 3.Hare EE, Peterson BK, Iyer VN, Meier R, Eisen MB. Sepsid even-skipped enhancers are functionally conserved in Drosophila despite lack of sequence conservation. PLoS Genetics. 2008;4:e1000106. doi: 10.1371/journal.pgen.1000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wunderlich Z, Mirny LA. Different gene regulation strategies revealed by analysis of binding motifs. Trends Genet. 2009;25:434–40. doi: 10.1016/j.tig.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Holde KE. Chromatin. 1989. [Google Scholar]
- 6.Kao-Huang Y, Revzin A, Butler AP, O’Conner P, Noble DW, von Hippel PH. Nonspecific DNA binding of genome-regulating proteins as a biological control mechanism: measurement of DNA-bound Escherichia coli lac repressor in vivo. Proc Natl Acad Sci USA. 1977;74:4228–32. doi: 10.1073/pnas.74.10.4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Polach KJ, Widom J. A model for the cooperative binding of eukaryotic regulatory proteins to nucleosomal target sites. Journal of Molecular Biology. 1996;258:800–12. doi: 10.1006/jmbi.1996.0288. [DOI] [PubMed] [Google Scholar]
- 8.Miller JA, Widom J. Collaborative competition mechanism for gene activation in vivo. Molecular and Cellular Biology. 2003;23:1623–32. doi: 10.1128/MCB.23.5.1623-1632.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adams CC, Workman JL. Binding of disparate transcriptional activators to nucleosomal DNA is inherently cooperative. Molecular and Cellular Biology. 1995;15:1405–21. doi: 10.1128/mcb.15.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vashee S, Willie J, Kodadek T. Synergistic activation of transcription by physiologically unrelated transcription factors through cooperative DNA-binding. Biochem Biophys Res Commun. 1998;247:530–5. doi: 10.1006/bbrc.1998.8820. [DOI] [PubMed] [Google Scholar]
- 11.Vashee S, Melcher K, Ding WV, Johnston SA, Kodadek T. Evidence for two modes of cooperative DNA binding in vivo that do not involve direct protein-protein interactions. Curr Biol. 1998;8:452–8. doi: 10.1016/s0960-9822(98)70179-4. [DOI] [PubMed] [Google Scholar]
- 12.Mirny LA. Nucleosome-mediated cooperativity between transcription factors. Proc Natl Acad Sci USA. 2010;107:22534–9. doi: 10.1073/pnas.0913805107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raveh-Sadka T, Levo M, Segal E. Incorporating nucleosomes into thermodynamic models of transcription regulation. Genome Research. 2009;19:1480–96. doi: 10.1101/gr.088260.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teif VB, Rippe K. Nucleosome mediated crosstalk between transcription factors at eukaryotic enhancers. Phys Biol. 2011;8:044001. doi: 10.1088/1478-3975/8/4/044001. [DOI] [PubMed] [Google Scholar]
- 15.Chávez S, Beato M. Nucleosome-mediated synergism between transcription factors on the mouse mammary tumor virus promoter. Proc Natl Acad Sci USA. 1997;94:2885–90. doi: 10.1073/pnas.94.7.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular Cell. 2010;38:576–89. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Polach KJ, Widom J. Restriction enzymes as probes of nucleosome stability and dynamics. Meth Enzymol. 1999;304:278–98. doi: 10.1016/s0076-6879(99)04017-3. [DOI] [PubMed] [Google Scholar]
- 18.Polach KJ, Widom J. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. Journal of Molecular Biology. 1995;254:130–49. doi: 10.1006/jmbi.1995.0606. [DOI] [PubMed] [Google Scholar]
- 19.Li G, Levitus M, Bustamante C, Widom J. Rapid spontaneous accessibility of nucleosomal DNA. Nat Struct Mol Biol. 2005;12:46–53. doi: 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
- 20.Li G, Widom J. Nucleosomes facilitate their own invasion. Nat Struct Mol Biol. 2004;11:763–9. doi: 10.1038/nsmb801. [DOI] [PubMed] [Google Scholar]
- 21.Yu L, Morse RH. Chromatin opening and transactivator potentiation by RAP1 in Saccharomyces cerevisiae. Molecular and Cellular Biology. 1999;19:5279–88. doi: 10.1128/mcb.19.8.5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanaka M. Modulation of promoter occupancy by cooperative DNA binding and activation-domain function is a major determinant of transcriptional regulation by activators in vivo. Proc Natl Acad Sci USA. 1996;93:4311–5. doi: 10.1073/pnas.93.9.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernstein BE, Liu CL, Humphrey EL, Perlstein EO, Schreiber SL. Global nucleosome occupancy in yeast. Genome Biol. 2004;5:R62. doi: 10.1186/gb-2004-5-9-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tirosh I, Barkai N. Two strategies for gene regulation by promoter nucleosomes. Genome Research. 2008;18:1084–91. doi: 10.1101/gr.076059.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prinsen P, Schiessel H. Nucleosome stability and accessibility of its DNA to proteins. Biochimie. 2010;92:1722–8. doi: 10.1016/j.biochi.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 26.Möbius W, Neher RA, Gerland U. Kinetic accessibility of buried DNA sites in nucleosomes. Phys Rev Lett. 2006;97:208102. doi: 10.1103/PhysRevLett.97.208102. [DOI] [PubMed] [Google Scholar]
- 27.Fried M, Crothers DM. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Research. 1981;9:6505–25. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clegg RM. Fluorescence resonance energy transfer and nucleic acids. Meth Enzymol. 1992;211:353–88. doi: 10.1016/0076-6879(92)11020-j. [DOI] [PubMed] [Google Scholar]
- 29.Anderson JD, Widom J. Sequence and position-dependence of the equilibrium accessibility of nucleosomal DNA target sites. Journal of Molecular Biology. 2000;296:979–87. doi: 10.1006/jmbi.2000.3531. [DOI] [PubMed] [Google Scholar]
- 30.Ptashne M. A Genetic Switch: Phage Lambda Revisited. Cold Spring Harbor Laboratory Press; 2004. [Google Scholar]
- 31.Böhm V, Hieb AR, Andrews AJ, Gansen A, Rocker A, Tóth K, Luger K, Langowski J. Nucleic Acids Research. 2010. Nucleosome accessibility governed by the dimer/tetramer interface. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gansen A, Valeri A, Hauger F, Felekyan S, Kalinin S, Tóth K, Langowski J, Seidel C. Nucleosome disassembly intermediates characterized by single-molecule FRET. Proc Natl Acad Sci USA. 2009 doi: 10.1073/pnas.0903005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang JG, Narlikar GJ. FRET-based methods to study ATP-dependent changes in chromatin structure. Methods. 2007;41:291–5. doi: 10.1016/j.ymeth.2006.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koopmans WJA, Buning R, Schmidt T, van Noort J. spFRET using alternating excitation and FCS reveals progressive DNA unwrapping in nucleosomes. Biophysical Journal. 2009;97:195–204. doi: 10.1016/j.bpj.2009.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelbauskas L, Sun J, Woodbury N, Lohr D. Nucleosomal stability and dynamics vary significantly when viewed by internal versus terminal labels. Biochemistry. 2008;47:9627–35. doi: 10.1021/bi8000775. [DOI] [PubMed] [Google Scholar]
- 36.Iqbal A, Arslan S, Okumus B, Wilson TJ, Giraud G, Norman DG, Ha T, Lilley DMJ. Orientation dependence in fluorescent energy transfer between Cy3 and Cy5 terminally attached to double-stranded nucleic acids. Proc Natl Acad Sci USA. 2008;105:11176–81. doi: 10.1073/pnas.0801707105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knegtel RM, Fogh RH, Ottleben G, Rüterjans H, Dumoulin P, Schnarr M, Boelens R, Kaptein R. A model for the LexA repressor DNA complex. Proteins. 1995;21:226–36. doi: 10.1002/prot.340210305. [DOI] [PubMed] [Google Scholar]
- 38.Poirier M, Oh E, Tims H, Widom J. Dynamics and function of compact nucleosome arrays. Nat Struct Mol Biol. 2009 doi: 10.1038/nsmb.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tims HS, Gurunathan K, Levitus M, Widom J. Dynamics of Nucleosome Invasion by DNA Binding Proteins. Journal of Molecular Biology. 2011 doi: 10.1016/j.jmb.2011.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Teif VB, Ettig R, Rippe K. A lattice model for transcription factor access to nucleosomal DNA. Biophysical Journal. 2010;99:2597–607. doi: 10.1016/j.bpj.2010.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Widom J. Structure, dynamics, and function of chromatin in vitro. Annu Rev Biophys Biomol Struct. 1998;27:285–327. doi: 10.1146/annurev.biophys.27.1.285. [DOI] [PubMed] [Google Scholar]
- 42.Anderson JD, Thåström A, Widom J. Spontaneous access of proteins to buried nucleosomal DNA target sites occurs via a mechanism that is distinct from nucleosome translocation. Molecular and Cellular Biology. 2002;22:7147–57. doi: 10.1128/MCB.22.20.7147-7157.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y, Zhang XS, Xia Y. Predicting eukaryotic transcriptional cooperativity by Bayesian network integration of genome-wide data. Nucleic Acids Research. 2009;37:5943–58. doi: 10.1093/nar/gkp625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu X, Lin J, Masuda T, Esumi N, Zack DJ, Qian J. Genome-wide prediction and characterization of interactions between transcription factors in Saccharomyces cerevisiae. Nucleic Acids Research. 2006;34:917–27. doi: 10.1093/nar/gkj487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pilpel Y, Sudarsanam P, Church GM. Identifying regulatory networks by combinatorial analysis of promoter elements. Nat Genet. 2001;29:153–9. doi: 10.1038/ng724. [DOI] [PubMed] [Google Scholar]
- 46.Balaji S, Babu MM, Iyer LM, Luscombe NM, Aravind L. Comprehensive analysis of combinatorial regulation using the transcriptional regulatory network of yeast. Journal of Molecular Biology. 2006;360:213–27. doi: 10.1016/j.jmb.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 47.Das D, Banerjee N, Zhang MQ. Interacting models of cooperative gene regulation. Proc Natl Acad Sci USA. 2004;101:16234–9. doi: 10.1073/pnas.0407365101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berman BP, Pfeiffer BD, Laverty TR, Salzberg SL, Rubin GM, Eisen MB, Celniker SE. Computational identification of developmental enhancers: conservation and function of transcription factor binding-site clusters in Drosophila melanogaster and Drosophila pseudoobscura. Genome Biol. 2004;5:R61. doi: 10.1186/gb-2004-5-9-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morozov A, Fortney K, Gaykalova D, Studitsky V, Widom J, Siggia E. Using DNA mechanics to predict in vitro nucleosome positions and formation energies. Nucleic Acids Research. 2009 doi: 10.1093/nar/gkp475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feng HP, Scherl DS, Widom J. Lifetime of the histone octamer studied by continuous-flow quasielastic light scattering: test of a model for nucleosome transcription. Biochemistry. 1993;32:7824–31. doi: 10.1021/bi00081a030. [DOI] [PubMed] [Google Scholar]
- 51.Little JW, Kim B, Roland KL, Smith MH, Lin LL, Slilaty SN. Cleavage of LexA repressor. Meth Enzymol. 1994;244:266–84. doi: 10.1016/0076-6879(94)44022-0. [DOI] [PubMed] [Google Scholar]
- 52.Thåström A, Lowary PT, Widom J. Measurement of histone-DNA interaction free energy in nucleosomes. Methods. 2004;33:33–44. doi: 10.1016/j.ymeth.2003.10.018. [DOI] [PubMed] [Google Scholar]
- 53.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. Journal of Molecular Biology. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 54.Tims HS, Widom J. Stopped-flow fluorescence resonance energy transfer for analysis of nucleosome dynamics. Methods. 2007;41:296–303. doi: 10.1016/j.ymeth.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423:145–50. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(a) Sucrose gradient purification of nucleosomes (blue) and naked DNA (pink). Nucleosomes are reconstituted by gradual salt dialysis and separated from naked DNA and nonnucleosomal contaminants on a 5–30% (wt/vol) sucrose gradient. Naked DNA is separated from unincorporated 32P also on a 5–30% (wt/vol) sucrose gradient. (b) Native gel electrophoresis of purified naked DNA (Lane 1) and nucleosomes (Lane 2). W: loading well of the gel; N and D, nucleosomes and naked DNA, respectively. L: 50 bp ladder. Phosphorimager analysis reveals less than 3% contaminating naked DNA in nucleosome sample. (c) Native gel mobility shift assay for LexA binding to naked DNA; same experimental conditions as for digestion assay. 100 bp ladder is included as size standards (not shown). D, mobility of naked DNA; D-LexA, mobility of DNA-LexA complex. W indicates loading well. (d) Native gel mobility shift assay for LexA binding to nucleosomes. D, mobility of contaminating naked DNA (or nucleosomes that did not survive electrophoresis); N, nucleosomes; N-LexA, nucleosomes stably bound by LexA.
Representative experiment probing accessibility at the MspI site in the absence and presence of saturating LexA. (a–d) Denaturing polyacrylamide gel analyses of MspI digestion reactions. Substrate (S) is converted to products (P1 and P2). (a) <0.5 nM naked DNA with 0.83 U/mL MspI. (b) <0.5 nM naked DNA with 100 nM LexA and 0.8 U/mL MspI. (c) 5 nM nucleosomes with 10000 u/mL MspI. (d) 5 nM nucleosomes with 1 μM LexA and 10000 U/mL MspI. (e) Quantitative analysis of the gels in panels (a–d). The fraction of DNA uncut is plotted versus time; data are fit to single exponentials. For nucleosomes, the zero time point is omitted from the analysis.
Representative experiment probing accessibility at the AluI site in the absence and presence of saturating LexA. (a–d) Denaturing polyacrylamide gel analyses of AluI digestion reactions. Substrate (S) is converted to products (P1 and P2). (a) <0.5 nM naked DNA with 4 U/mL AluI. (b) <0.5 nM naked DNA with 100 nM LexA and 4 U/mL AluI. (c) 5 nM nucleosomes with 4000 u/mL AluI. (d) 5 nM nucleosomes with 1 μM LexA and 4000 U/mL AluI. (e) Quantitative analysis of the digestion pattern resolved in the gels of panels (a–d). The fraction of DNA uncut is plotted versus time. Data are fit to single exponential (naked DNA) or linear (nucleosomal DNA) functions. For nucleosomes, the zero time point is omitted from the analysis.