Abstract
Mutations in MYO15A are associated with deafness in humans, and shaker 2 mice also exhibit a hearing loss due to defects of unconventional myosin 15a. We ascertained a consanguineous Pakistani family with recessively inherited moderate to severe hearing loss, which putatively segregated with markers linked to the DFNB3 locus. Prioritized sequencing of the second exon of MYO15A from the DNA of all affected individuals of family revealed a duplication of Cytosine in a stretch of seven repetitive C nucleotides (c.1185dupC). This mutation results in a frameshift and incorporates a stop codon in the open reading frame of MYO15A (p.E396fsX431). The findings of less severe hearing loss in families with linkage to DFNB3 are only reported for some individuals with mutations in exon 2 of MYO15A, which are further supported by this study. Therefore, on basis of linkage data and the presence of a less severe hearing loss phenotype, sequencing of a single exon of MYO15A can efficiently identify the causative mutations in patients from these families.
Keywords: MYO15A, unconventional myosins, DFNB3, hearing loss, deafness, Pakistan
1. Introduction
MYO15A is an unconventional myosin and has a role in the formation of stereocilia [1]. Myosins are actin-activated ATPase that use the hydrolysis of ATP to move on actin filaments. Mutations in MYO15A result in profound deafness in humans [2,3] while the orthologous gene is found mutated in mice [4]. Affected shaker 2 mice have mutations in Myo15a and exhibit head tossing and circling behaviour with profound deafness. Electron microscopic and immunohistochemical studies of inner ear of mutant mice show that hair cell stereocilia are properly positioned on the apical surface of hair cells of the inner ear, but the lengths of stereocilia are shorter than those of wild-type mice [4]. Additionally, MYO15A is also expressed in the pituitary [3] although no phenotype in humans or mice have been associated with altered function of this gland.
MYO15A has 66 exons. The second exon encodes a large N-terminal extension in one of the isoforms of MYO15A. A great number of missense and nonsense mutations have been identified in exons encoding motor, FERM and MyTH4 domain of MYO15A which cause profound deafness in humans [3,5–10]. Interestingly, however, three mutations were recently identified in the alternatively spliced exon 2, which may cause a less severe phenotype in some of the affected members in these families [6,7]. This suggests that mutations in exon 2 of MYO15A may be involved in genetics of less severe hearing loss.
2. Family and methods
Family HLRB3 with three affected children in a single consanguineous union was ascertained from Lahore, Pakistan (Fig. 1A). Institutional review board (IRB) approval was obtained at School of Biological Sciences, University of the Punjab, Lahore, Pakistan. Blood samples were collected for DNA analysis after written informed consent from each participant of family or from parents in case of minor children. DNA was extracted with a non-organic procedure [11,12]. Audiometry was performed with DANPLEX audiometer model DA65 (Denmark). Hearing of affected individuals of the family was evaluated at frequencies of 250Hz, 500Hz, 2000Hz, 4000Hz, and 8000Hz. A medical history was obtained from all participants of the family in order to minimize the presence of hearing loss due to medical and environmental causes. Vestibular system was evaluated by Romberg and tandem gait tests. Additionally, the heights of all affected individuals were also measured.
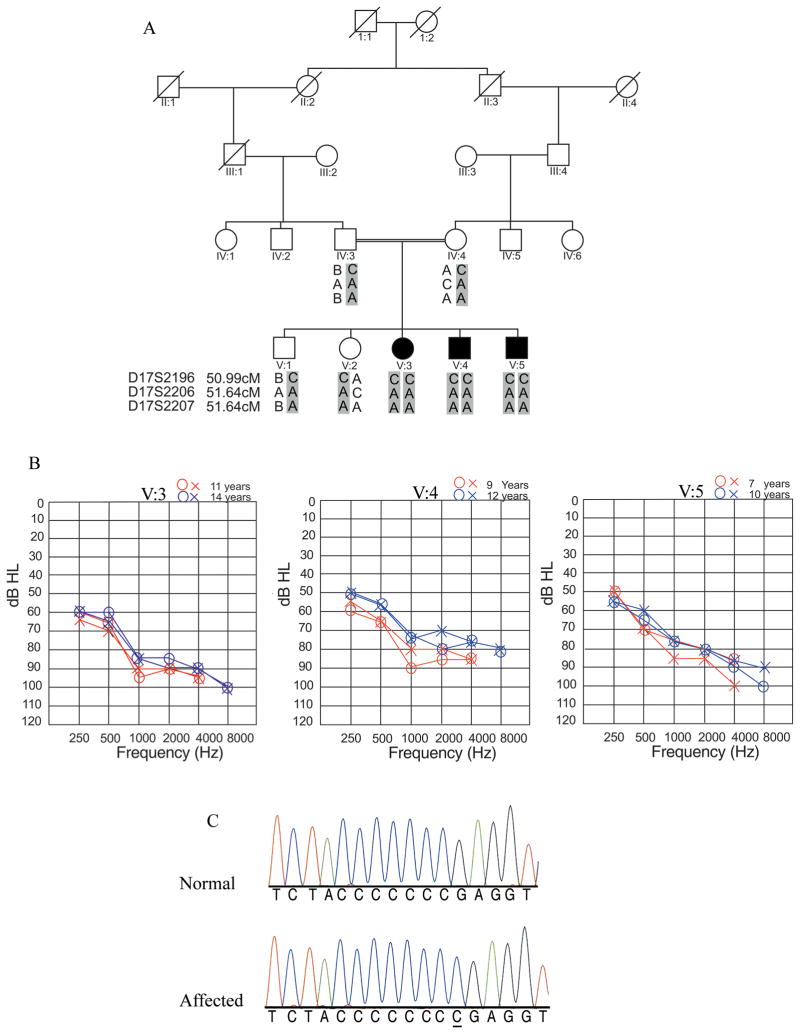
Figure 1. Phenotypic and genetic analysis for family HLRB3.
A) Pedigree of HLRB3 along with haplotypes for markers on chromosome 17p11.2. Genetic distances of markers in cM are taken from Rutgers’ human genetic map. Affected individuals are homozygous for alleles of markers D17S2196, D17S2206 and D17S2207. The gray bar highlights the haplotype carrying the disease allele. Different sizes of alleles are denoted by letters (A, B and C).
B) Audiograms for all affected family members of family HLRB3. Audiometry was conducted at two different age groups (11, 9, 7 and 14, 12, 10) for the affected children V: 3, V: 4 and V: 5 respectively. “O” indicates air conduction for right ear, while “X” indicates air conduction for left ear. Audiograms of all affected individuals in family HLRB3 revealed moderate to severe hearing loss at two different time intervals.
C) A single base pair homozygous duplication of cytosine (c.1185dupC) in a stretch of seven “C” nucleotides was identified in DNA of affected individuals of family HLRB3 segregating with the phenotype. The duplication is underlined in the trace from the affected individual.
Eight DNA samples were available for genotyping from family HLRB3. The family was screened with fluorescently labeled markers for loci known to cause less severe hearing loss phenotypes. These included DFNB2, DFNB3, DFNB4, DFNB8, DFNB16, DFNB21, DFNB22, DFNB25, DFNB29, DFNB30, DFNB32, DFNB33, DFNB49, DFNB59, DFNB72 and DFNB79. DFNB1 was excluded by direct sequencing of GJB2. Two point LOD scores were calculated by using FASTLINK with a disease allele frequency of 0.001. The phenotype was coded as a fully penetrant autosomal recessive disorder. Primers were designed to amplify MYO15A exon 2 in 7 overlapping fragments (sequences for oligonucleotides are available on request). The PCR products were treated with Shrimp Alkaline Phosphatase and Exonuclease I (Fermentas, Glen Burnie, MD) and sequenced with Big Dye Terminators V. 3.1 (Applied Biosystems, Foster City, CA).
3. Results
Audiometric assessments revealed moderate to severe hearing loss in all affected individuals in family HLRB3 at all frequencies (Fig. 1B), classified with respect to the ear with the better hearing. The loss of hearing was profound in degree at the frequency of 8000 Hz in the oldest affected individual (V:3). Audiometric assessments held three years apart did not reveal progression of hearing loss in any of the affected individuals. Clinical and physical examination suggested that deafness was not due to syndromes and other environmental factors. Physical measurements of all three family HLRB3 affected individuals (ages 14, 12, and 10 years) were normal and within the expected range for their age groups [13]. The patients reported no problem with balance and tandem gait tests and Romberg tests were normal indicating that vestibular system was not affected. However, ENG could not be performed to conclusively rule out this possibility. The presence of problems related to balance is possible in the affected individuals since there is loss of vestibular function in deaf individuals from Bengkala when the mutation disrupts MYO15A isoform 2 [14].
The affected children V:3, V:4 and V:5 were homozygous for alleles of three microsatellite markers at the DFNB3 locus, D17S2196, D17S2206 and D17S2207 spanning and within MYO15A while the parents were heterozygous for alleles at these markers (Fig. 1A). A maximum two point LOD score of 2.3 was obtained at recombination fraction θ = 0 with the marker D17S2196.
A less severe hearing loss due to mutations in MYO15A has only been reported if mutations lie in the second exon of this gene, which has a total of 66 exons. We therefore prioritized the second exon of MYO15A for sequencing analysis. Sequencing of the amplicons from exon 2 of MYO15A revealed a duplication of a Cytosine in a stretch of 7 repetitive C nucleotides, (c.1185dupC) in the DNA of all affected individuals in family HLRB3 (Fig. 1C). Other unaffected individuals were carriers for the mutation.
4. Discussion
DFNB3 is the third common locus for recessively inherited profound deafness in the Pakistani population and mutations in MYO15A account for 5% of recessively inherited severe to profound deafness [5]. Mutations of MYO15A are also a significant contributor to profound deafness in many other world populations [3,6–10]. However, a less severe hearing loss due to mutations of MYO15A has been reported only in a few members of two Pakistani and a single Turkish family [6,7] and all of these mutations are in exon 2 of MYO15A (summarized in Table 1). The hearing loss in family HLRB3 was not found to be progressive and was of similar severity in all three affected individuals. In the two previously reported families from Pakistan with mutations in exon 2 of MYO15A [6], affected individuals had slight retention of hearing at low frequencies while the loss of hearing was profound at high frequencies in all except one individual. One affected individual in the Pakistani family, with p.G1112fsX1124 mutation, suffered from moderate to severe hearing loss involving all frequencies. Interestingly, in the family from Turkey [7], three affected individuals had similar hearing loss as in those in the Pakistani families (Table 1). However, their affected mother had moderate to severe hearing loss at 47 years of age and her hearing was considerably better than the affected children, one of whom was 24 years old. Additionally, she reported that her hearing had gotten worse in the past eight years, suggesting that hearing loss was progressive in nature. The degree of hearing loss of her 28 year old affected offspring was not determined by audiometry but was reported to be residual due to his good oral skills.
Table 1.
List of all mutations in exon 2 encoding N-terminal domain of MYO15A.
| Country | Exon | Amino acid change | Consequence | Phenotype | Reference |
|---|---|---|---|---|---|
| Pakistan | 2 | p.E1105X | Nonsense | Residual hearing at low frequencies | [6] |
| Pakistan | 2 | p.G1112fsX1124 | Frameshift | Residual hearing at low frequencies and moderate to severe HL in one individual | [6] |
| Turkey | 2 | p.Y289X | Nonsense | Moderate to severe HL in one individual (47 yrs). Residual hearing in 28 yr old individual. Profound deafness in three individuals (~24 yrs) with residual hearing at low frequencies. | [7] |
| Pakistan | 2 | p.E395fsX431 | Frameshift | Moderate to severe HL (10–14 yrs). | This report |
All four mutations are predicted to result in production of either mutant shortened proteins for MYO15A long isoform 1 or its complete absence due to nonsense mediated decay of the prematurely truncated transcript. HL; Hearing loss.
The mutation c.1185dupC identified in family HLRB3 results in a frameshift and introduces an eventual stop codon in the MYO15A open reading frame (p.E396fsX431). It is possible that this mutant transcript will be degraded by the mRNA surveillance system [15] due to presence of a premature stop codon in the transcript. However, even if the transcript is translated it is predicted to produce a short 430 amino acid protein with 35 wrongly incorporated amino acid residues and will lack most of the domains of MYO15A, which have been shown to be necessary for its activity and function
Genotype-phenotype correlations of MYO15A mutations and degree of hearing loss suggest that mutations in all 66 exons cause profound deafness while those in exon 2 can also result in some residual hearing at low frequencies or moderate to severe hearing loss. A role of modifier genes in determining the level of deafness is also suggested since individuals with identical mutations in exon 2 of MYO15A may exhibit different degrees of hearing loss (Table 1). A mouse model with targeted disruption of long isoform of Myo15a will be helpful in understanding the role of this isoform in the inner ear.
In Pakistan, seventeen missense, nonsense, and splice site mutations (p.Q1229X, c.8486G >A, IVS4+1G>T, p.T1253fsX1277, p.Y1392X, p.K1557E, p.G1706V, p.L1730P, p.G2018R, p.Q2021X, p. T22051, p.G2244E, p.V2266M, p.2720H, p.V2940fsX3034, p.L3160F and p.Q3492X) in exons encoding motor and first FERM domains of MYO15A have been reported which cause profound deafness [5,6]. Identification of additional mutant alleles and the prevalence of MYO15A mutations in less severe hearing loss remain to be determined in a large subset of families with a similar phenotype from Pakistan.
The identification of a new mutation in exon 2 of MYO15A in family HLRB3 together with less severe hearing loss of the affected individuals supports the hypothesis that mutations in exon 2 may have less debilitating effects on hearing, in at least some affected individuals. It also facilitates rapid identification of mutations in MYO15A when linkage of less severe hearing loss to DFNB3 is detected.
Acknowledgments
We thank the family members for participating in this study. We appreciate the help of Mr. Haider Ali for audiometric examinations and help in sample collections from the participants. We are grateful to Tom Friedman for providing some of the primers for STR analysis and critical review of the manuscript. This research was supported by Grant number R01TW007608 from the Fogarty International Center and National Institute of Deafness and other Communication Disorders, National Institutes of Health, USA.
Footnotes
Conflict of Interest: The authors declare that they have no conflict of interest
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Rasheeda Bashir, Email: rzssff_phd@yahoo.com.
Amara Fatima, Email: amara_lhr@yahoo.com.
Sadaf Naz, Email: naz.sbs@pu.edu.pk.
References
- 1.Anderson DW, Probst FJ, Belyantseva IA, et al. The motor and tail regions of myosin XV are critical for normal structure and function of auditory and vestibular hair cells. Hum Mol Genet. 2000;9:1729–38. doi: 10.1093/hmg/9.12.1729. [DOI] [PubMed] [Google Scholar]
- 2.Friedman TB, Liang Y, Weber JL, et al. A gene for congenital, recessive deafness DFNB3 maps to the pericentromeric region of chromosome 17. Nat Genet. 1995;9:86–91. doi: 10.1038/ng0195-86. [DOI] [PubMed] [Google Scholar]
- 3.Wang A, Liang Y, Fridell RA, et al. Association of unconventional myosin MYO15 mutations with human nonsyndromic deafness DFNB3. Science. 1998;280:1447–1451. doi: 10.1126/science.280.5368.1447. [DOI] [PubMed] [Google Scholar]
- 4.Probst FJ, Fridell RA, Raphael Y, et al. Correction of deafness in shaker-2 mice by an unconventional myosin in a BAC transgene. Science. 1998;280:1444–1447. doi: 10.1126/science.280.5368.1444. [DOI] [PubMed] [Google Scholar]
- 5.Liburd N, Ghosh M, Riazuddin S, et al. Novel mutations of MYO15A associated with profound deafness in consanguineous families and moderately severe hearing loss in a patient with Smith-Magenis syndrome. Hum Genet. 2001;109:535–541. doi: 10.1007/s004390100604. [DOI] [PubMed] [Google Scholar]
- 6.Nal N, Ahmed ZM, Erkal E, et al. Mutational spectrum of MYO15A: the large N-terminal extension of myosin XVA is required for hearing. Hum Mutat. 2007;28:1014–1019. doi: 10.1002/humu.20556. [DOI] [PubMed] [Google Scholar]
- 7.Cengiz FB, Duman D, Sirmaci A, et al. Recurrent and private MYO15A mutations are associated with deafness in the Turkish population. Genet Test Mol Biomarkers. 2010;14:543–550. doi: 10.1089/gtmb.2010.0039. [DOI] [PubMed] [Google Scholar]
- 8.Kalay E, Uzumcu A, Krieger E, et al. MYO15A (DFNB3) mutations in Turkish hearing loss families and functional modeling of a novel motor domain mutation. Am J Med Genet A. 2007;143A:2382–2389. doi: 10.1002/ajmg.a.31937. [DOI] [PubMed] [Google Scholar]
- 9.Shearer AE, Hildebrand MS, Webster JA, et al. Mutations in the first MyTH4 domain of MYO15A are a common cause of DFNB3 hearing loss. Laryngoscope. 2009;119:727–733. doi: 10.1002/lary.20116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belguith H, Aifa-Hmani M, Dhouib H, et al. Screening of the DFNB3 locus: identification of three novel mutations of MYO15A associated with hearing loss and further suggestion for two distinctive genes on this locus. Genet Test Mol Biomarkers. 2009;13:147–151. doi: 10.1089/gtmb.2008.0077. [DOI] [PubMed] [Google Scholar]
- 11.Grimberg J, Nawoschik S, Belluscio L, McKee R, Turck A, Eisenberg A. A simple and efficient non-organic procedure for the isolation of genomic DNA from blood. Nucleic Acids Res. 1989;17:8390. doi: 10.1093/nar/17.20.8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1998;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kelly AM, Shaw NJ, Thomas AMC, Pynset BP, Baker DJ. Growth of Pakistani children in relation to the 1990 growth standards. Archives of Disease in Childhood. 1997;77:401–405. doi: 10.1136/adc.77.5.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedman TB, Hinnant JT, Ghosh M, Boger ET, et al. DFNB3, spectrum of MYO15A recessive mutant alleles and an emerging genotype-phenotype correlation. In: Cremers CWRJ, Smith RJH, editors. Genetic Hearing Impairment Adv Otorhinolaryngol. Vol. 61. Basel: Karger; 2002. pp. 124–130. [DOI] [PubMed] [Google Scholar]
- 15.Hentze MW, Kulozik AE. A perfect message: RNA surveillance and nonsense- mediated decay. Cell. 1999;96:307–310. doi: 10.1016/s0092-8674(00)80542-5. [DOI] [PubMed] [Google Scholar]