Abstract
Mutations in PJVK, encoding Pejvakin, cause autosomal recessive nonsyndromic hearing loss in humans at the DFNB59 locus on chromosome 2q31.2. Pejvakin is involved in generating auditory and neural signals in the inner ear. We have identified a consanguineous Pakistani family segregating sensorineural progressive hearing loss as a recessive trait, consistent with linkage to DFNB59. We sequenced PJVK and identified a novel missense mutation, c.1028 G>C in exon 7 (p.C343S) co-segregating with the phenotype in the family. The p.C343 residue is fully conserved among orthologs from different vertebrate species. We have also determined that mutations in PJVK are not a common cause of hearing loss in families with moderate to severe hearing loss in Pakistan. This is the first report of PJVK mutation in a Pakistani family and pinpoints an important residue for PJVK function.
Keywords: Pejvakin, Pakistan, Progressive hearing loss, DFNB59, Deafness
1. Introduction
Nonsyndromic recessively inherited hearing loss is the most common type of deafness. The majority of the affected individuals with recessively inherited hearing loss are described to suffer from prelingual profound deafness. This is perhaps due to the lower incidence of recessively inherited progressive hearing loss in contrast to non-progressive deafness. Therefore, progressive hearing loss has not been described as a frequent finding in recessively inherited hearing loss, although there are a few exceptions. For example mutations in MYO3A, PTPRQ and SERPINB6 cause progressive hearing loss in affected members in the reported families (Walsh et al., 2002; Sirmaci et al., 2010; Schraders et al., 2010b). Additionally, particular mutant alleles of TMC1, TMPRSS3, GRXCR1, PJVK, LOXHD1, GIPC3 and TPRN also lead to progressive hearing loss in affected individuals in a few families (Veske et al., 1996; Delmaghani et al., 2006; Grillet et al., 2009; Li et al., 2010; Schraders et al., 2010a; Charizopoulou et al., 2011; de Heer et al., 2011)
PJVK is located at chromosome 2q31.2, and encodes a protein, pejvakin which plays a role in neural transmission (Delmaghani et al., 2006). Mutations in PJVK may cause stable or progressive hearing loss with or without auditory neuropathy spectrum disorder (ANSD) (Delmaghani et al., 2006; Collin et al., 2007; Ebermann et al., 2007; Hashemzadeh Chaleshtori et al., 2007; Schwander et al., 2007; Shahin et al., 2010; Borck et al., 2011). An ANSD is diagnosed when auditory brain stem response (ABR) is absent, while otoacoustic emissions (OAE) are normal which is characteristic of normal outer hair cell function (Starr et al., 1996). A PJVK mutation has also been reported to cause vestibular dysfunction along with deafness in one family from Morocco (Ebermann et al., 2007). These studies have shown that frameshift mutations in PJVK usually result in cochlear deafness while missense mutations may result in deafness with auditory neuropathy spectrum disorder (Hashemzadeh Chaleshtori et al., 2007). However, it is interesting to note that the mutation, p.R183W, can cause either deafness with or without ANSD in different individuals (Delmaghani et al., 2006; Collin et al., 2007). This may be due to the involvement of a modifier gene which affects the outer hair cell function.
Pejvakin is expressed in the cell bodies of neurons, hair cells, supporting cells and spiral ganglion cells in the inner ear (Delmaghani et al., 2006; Collin et al., 2007). PJVK is comprised of 352 amino acids. It has a predicted nuclear localization signal (amino acids 249–258) and also contains a zinc finger motif (amino acids 305–331) [7]. Knock-in mice with a point mutation identical to that identified in humans with auditory neuropathy spectrum disorder and those with an ENU induced nonsense mutation in Pjvk have revealed that PJVK is involved in neural signal transmission though the mechanism is unknown. It has also demonstrated that PJVK is involved in generating an auditory signal but not in the development and maintenance of hair cells or spiral ganglion neurons (Delmaghani et al., 2006; Schwander et al., 2007).
2. Patients and Methods
We recruited 50 consanguineous families with three or more individuals affected with nonsyndromic recessively inherited hearing loss and 57 sporadic individuals manifesting moderate to severe hearing loss from Pakistan after Institutional Review Board approval and informed written consent of the participants. Audiometry was performed with DANPLEX audiometer model DA65 (Denmark). Hearing of affected individuals was evaluated at frequencies of 250Hz, 500Hz, 2000Hz, 4000Hz, and 8000Hz in ambient noise conditions and ranged between 50–90 dB HL. All 50 families with a history of hearing loss and multiple affected individuals were identified with the help of social workers and audiologists from the Punjab province. Individuals with hearing loss were also questioned about problems related to their balance. Romberg and Tandem Gait tests were used to investigate the involvement of any vestibular dysfunction.
DNA was extracted from blood samples. GJB2 and all genes known to cause moderate to severe hearing loss or progressive deafness were excluded by sequencing, linkage analysis or homozygosity mapping with microsatellite markers. Linkage analysis and homozygosity mapping were performed with fluorescently labeled microsatellite markers which were located in close proximity to each of the screened deafness gene. Likelihood of odds (LOD) scores were calculated by using FASTLINK with a disease allele frequency of 0.001. The phenotype was coded as a fully penetrant autosomal recessive disorder. Screening of GJB2 and all 7 exons and flanking intronic regions of PJVK was performed by treatment of the PCR amplified products with Shrimp Alkaline Phosphatase and Exonuclease I (Fermentas, Glen Burnie, MD) followed by sequencing with Big Dye terminator, V.3.1 (Applied Biosystems, Foster City, CA).
3. Results
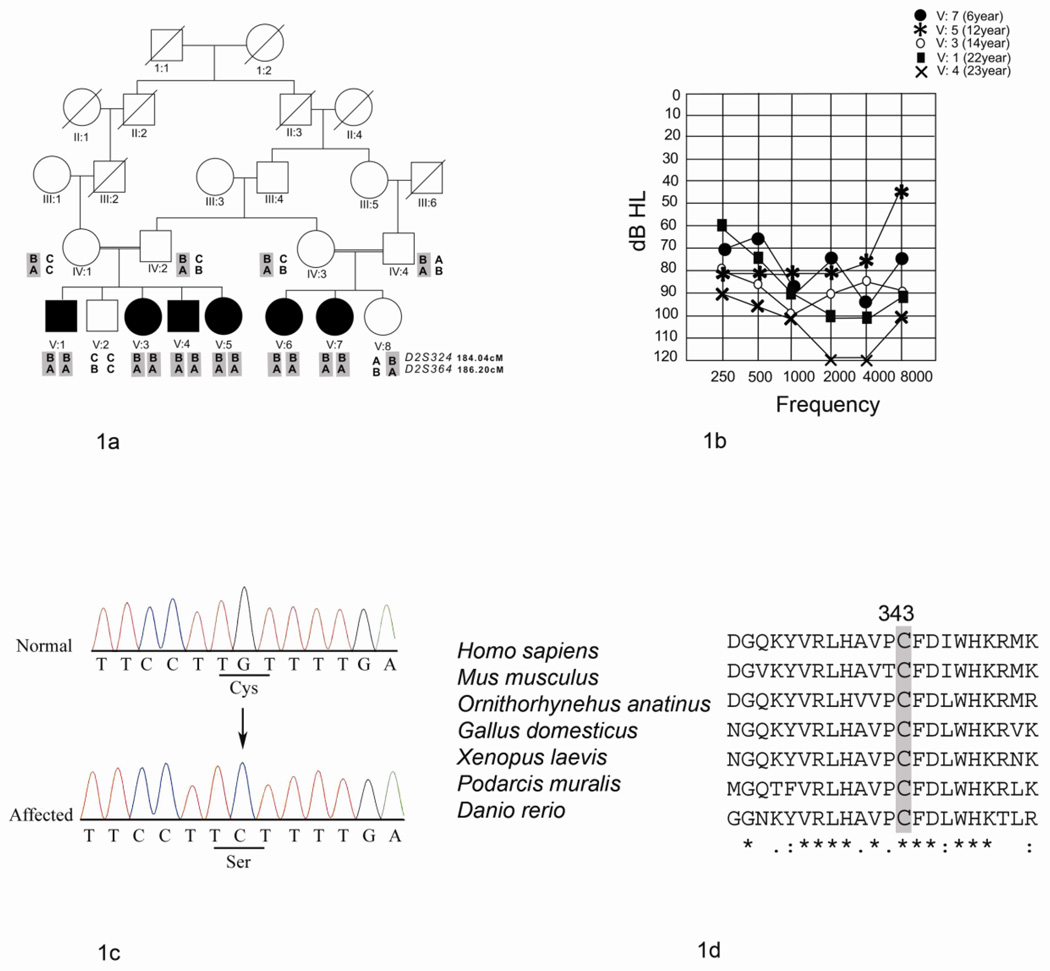
Family HLGM15 with six affected individuals was ascertained from Okara, Pakistan (Fig. 1A). The onset of hearing loss in this family was reported to be in early childhood. Audiometric assessment of the affected individuals in family revealed the presence of different degrees of deafness (Fig. 1B). Individual V: 4 was profoundly deaf while all others had a severe degree of hearing loss. The difference in hearing thresholds among the affected individuals was more noticeable at frequencies of 2, 4 and 8 KHz at which the younger individuals exhibited a less severe hearing loss than the older ones. The hearing loss of the affected individuals in family HLGM15 was self reported to gradually worsen with age. The affected individuals had some degree of oral speech. They faced difficulties in understanding conversations, especially in a noisy environment. The data and self reports from the patients suggest that the hearing loss in members of family HLGM15 is progressive in nature. However, audiometry was only performed at the time of enrollment in study when the patients were 6, 8, 14, 12, 22 and 23 years old, respectively.
Fig. 1. Phenotype and genotype analysis for family HLGM15.
A) Pedigree and haplotypes of family HLGM15. Genetic distances of markers in cM are taken from the Rutgers’ genetic map. Affected individuals are homozygous for alleles of markers D2S324 and D2S364, which flank PJVK. The deafness associated haplotype is shaded in gray.
B) Pure Tone air conduction results for affected individuals (V:1, V:3, V:4, V:5 and V:7) of family HLGM15. The results are shown for the better hearing ear for each individual. The younger affected individuals (V:7 and V:5) have a severe degree of hearing loss. The oldest affected individual V:4 manifests profound deafness.
C) Partial sequence electropherograms of PJVK for the normal individual V: 2 and the affected individual V:5 showing the missense mutation c.1028 G>C. The arrow indicates the location of the transversion. The affected codon is underlined in both the wild type and the affected individual’s sequence trace. Colour figure can be viewed in the online issue at the journal website.
D) ClustalW multiple alignment of PJVK sequences from different vertebrate species showing conservation of p.C343. The identical amino acid residues to p.C343 are shaded in gray. Amino acid residues with complete identity in all the species are indicated by an asterisk below the alignment. The colon and the period indicate amino acid substitutions with highly conserved and less conserved residues, respectively.
The patients did not report any problems with their balance. Moreover, the Romberg and Tandem Gait tests were also normal suggesting that the vestibular system was not overtly affected. However, we could not rule out subtle equilibrium defects and the presence of ANSD due to the unavailability of ENG and OAE testing facilities.
Hearing loss in family HLGM15 showed linkage to the DFNB59 (Fig. 1A) with LOD scores of 4.1 with markers D2S324 and D2S364 at θ = 0. Out of the 57 sporadic individuals with moderate to severe hearing loss who participated in this study, only two patients showed homzygosity for marker alleles of D2S324 and D2S364. This could be consistent with the presence of homozygous mutations in PJVK.
Sequencing analysis revealed the presence of a homozygous transversion mutation, c.1028 G>C, in exon 7 (p.C343S) of PJVK in affected individuals in family HLGM15 (Fig. 1C) and absence of mutations in samples of the two sporadically affected individuals. The parents and all except one member of family HLGM15 were heterozygous for this mutation. One individual, V: 2, did not inherit the deafness linked haplotype and also lacked the mutation. The absence of c.1028 G>C in 400 chromosomes from ethnically matched controls further supported the correlation of the mutation with the phenotype.
4. Discussion
The mutation p.C343S identified in family HLGM15 results in a missense mutation and causes the replacement of cysteine to serine. Cysteine (p.C343) in PJVK is conserved in all vertebrate species including mouse, platypus, hen, lizard, frog and zebra fish (Fig. 1D). Cysteine residues usually have important structural roles due to their capability of making disulphide bridges which stabilize the three dimensional structures of proteins. The disulphide bridges can be intra- or inter-molecular. These bonds may be involved in stabilizing the folding of polypeptide chains and help in prevention of degradation of proteins (Branden and Tooze, 1991). The missense mutation p.C343S may cause structural changes in PJVK affecting its function, and ultimately cause the observed phenotype in the affected individuals.
PJVK mutations seem to be important cause of hearing loss in Iranians and Arabs, (Delmaghani et al., 2006; Hashemzadeh Chaleshtori et al., 2007; Shahin et al., 2010). The hearing loss is usually stable and severe to profound in the affected individuals of these populations. Currently, there are only two reports of PJVK mutations with progressive hearing loss (Table 1). A mutation reported in a Moroccan family causes moderate to profound hearing loss which is progressive in nature (Ebermann et al., 2007). Hearing loss associated with another mutation in PJVK reported in affected individuals from Iran is also progressive and moderate to profound in degree (Schwander et al., 2007). However, PJVK has not been previously reported as a cause of either stable or progressive deafness in Pakistan.
Table 1.
Mutations in PJVK and associated phenotypes
| Mutation | Amino Acid change | Mutation | Population | Phenotype |
|
|---|---|---|---|---|---|
| Reference | |||||
| ANSD | Hearing loss | ||||
| c.113_114insT | p.V38VfsX10* | Frameshift | Morocco | Absent | Progressive, |
| (Ebermann et al. |
Moderate to |
||||
| profound 2007) | |||||
| with vestibular dysfunction |
|||||
| c.122delA | p.K41SfsX17 | Frameshift | Iran | status | Progressive, |
| (Schwander et al. | |||||
| unknown 2007) |
Moderate to | ||||
| profound | |||||
| c.1028 G>C | p.C343S | Missense | Pakistan | Status | Progressive, |
| This study | |||||
| unknown | Moderate to profound |
||||
Mutations in PJVK associated with progressive hearing loss. All mutations are numbered with respect to PJVK open reading frame (GenBank, NM_001042702) with the “A” of the initiation codon numbered as “1”. ANSD, Auditory neuropathy spectrum disorder
The number has been corrected from that in the original publication.
The two mutations previously implicated in causing progressive hearing loss are frameshift mutations and are predicted to result in formation of truncated proteins or in their complete absence due to the nonsense mediate decay of the mRNA. It can be speculated that the missense mutation affecting PJVK in deaf individuals in HLGM15 may also ultimately result in loss of function of the protein and lead to progressive hearing loss. There is no correlation between ages of affected individuals and severity of hearing loss in the patients in the Moroccan and Iranian families. In contrast, the older affected individuals of family HLGM15 exhibit a more severe hearing loss than the younger patients.
We were unable to determine whether auditory neuropathy spectrum disorder was present in affected individuals of HLGM15 with the PJVK mutation. So far, only missense mutations affecting PJVK, (p.T54I and p.R183W), have been shown to cause auditory neuropathy spectrum disorder (Delmaghani et al., 2006). Since p.C343S is also a missense mutation, it is tempting to speculate that the affected individuals in these families may also suffer from ANSD. However, in the absence of otoacoustic emissions data, a definite conclusion cannot be reached since it has been shown that one missense mutation (p.R183W) can cause either auditory neuropathy spectrum disorder or cochlear deafness (Delmaghani et al., 2006; Collin et al., 2007).
The presence of PJVK mutations in only one out of 50 families who participated in this study, and the additional absence of mutations in 57 sporadic individuals with hearing loss suggests that PJVK is not a significant contributor to etiology of moderate to severe hearing loss in Pakistan. However, the contribution of PJVK to progressive deafness remains to be determined in a large subset of individuals with a similar phenotype.
In this study we have investigated one of the few families from Pakistan in which affected individuals suffer from a gradual loss in hearing. So far, only one mutation in TMPRSS3 has been reported to cause postlingual, progressive hearing loss in patients in a family from Pakistan (Veske et al., 1996; Scott et al., 2001). A few individuals with mutations in BSND and GIPC3 may also have a progressive hearing loss since some younger affected patients have better hearing thresholds as compared to the older individuals in their families (Riazuddin et al., 2009; Rehman et al., 2011). Our results further confirm that PJVK is one of the genes to be considered in the etiology of progressive hearing loss in different world populations including Pakistan.
Highlights.
The phenotype in a family with hearing loss was linked to DFNB59.
A novel missense mutation p.C343S was identified in PJVK.
p.C343S is only the third PJVK mutation associated with progressive hearing loss.
ACKNOWLEDGEMENTS
We thank the participants and Mr. M. Saddique & Dr. Nasir Mahmood for their cooperation. We are grateful to Dr. Thomas Friedman for review of the manuscript and Rasheeda Bashir for assistance. Supported by R01TW007608 from the Fogarty International Center and National Institute of Deafness and other Communication Disorders, National Institutes of Health, USA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: The authors declare that they have no conflict of interest
Contributor Information
Ghulam Mujtaba, Email: gmuj2006@yahoo.com.
Ihtisham Bukhari, Email: bukhari5408@gmail.com.
Amara Fatima, Email: amara_lhr@yahoo.com.
Sadaf Naz, Email: naz.sbs@pu.edu.pk.
REFERENCES
- Borck G, et al. High frequency of autosomal-recessive DFNB59 hearing loss in an isolated Arab population in Israel. Clin. Genet. 2011 doi: 10.1111/j.1399-0004.2011.01741.x. PMID 21696384. [DOI] [PubMed] [Google Scholar]
- Branden C, Tooze J. Introduction to protein structure. New York: Garland Publishing inc; 1991. p. 5. [Google Scholar]
- Charizopoulou N, et al. Gipc3 mutations associated with audiogenic seizures and sensorineural hearing loss in mouse and human. Nat. Commun. 2011;2:201–212. doi: 10.1038/ncomms1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin RW, et al. Involvement of DFNB59 mutations in autosomal recessive nonsyndromic hearing impairment. Hum. Mutat. 2007;28:718–723. doi: 10.1002/humu.20510. [DOI] [PubMed] [Google Scholar]
- de Heer AM, et al. Progressive sensorineural hearing loss and normal vestibular function in a Dutch DFNB7/11 family with a novel mutation in TMC1. Audiol. Neurootol. 2011;16:93–105. doi: 10.1159/000313282. [DOI] [PubMed] [Google Scholar]
- Delmaghani S, et al. Mutations in the gene encoding pejvakin, a newly identified protein of the afferent auditory pathway, cause DFNB59 auditory neuropathy. Nat. Genet. 2006;38:770–778. doi: 10.1038/ng1829. [DOI] [PubMed] [Google Scholar]
- Ebermann I, et al. Truncating mutation of the DFNB59 gene causes cochlear hearing impairment and central vestibular dysfunction. Hum. Mutat. 2007;28:571–577. doi: 10.1002/humu.20478. [DOI] [PubMed] [Google Scholar]
- Grillet N, et al. Mutations in LOXHD1, an evolutionarily conserved stereociliary protein, disrupt hair cell function in mice and cause progressive hearing loss in humans. Am. J. Hum. Genet. 2009;85:328–337. doi: 10.1016/j.ajhg.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashemzadeh Chaleshtori M, et al. Novel mutations in the pejvakin gene are associated with autosomal recessive non-syndromic hearing loss in Iranian families. Clin. Genet. 2007;72:261–263. doi: 10.1111/j.1399-0004.2007.00852.x. [DOI] [PubMed] [Google Scholar]
- Li Y, et al. Mutations in TPRN cause a progressive form of autosomal-recessive nonsyndromic hearing loss. Am. J. Hum. Genet. 2010;86:479–484. doi: 10.1016/j.ajhg.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman AU, et al. Mutations of GIPC3 cause nonsyndromic hearing loss DFNB72 but not DFNB81 that also maps to chromosome 19p. Hum. Genet. 2011;130:759–765. doi: 10.1007/s00439-011-1018-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riazuddin S, et al. Molecular basis of DFNB73: mutations of BSND can cause nonsyndromic deafness or Bartter syndrome. Am. J. Hum. Genet. 2009;85:273–280. doi: 10.1016/j.ajhg.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schraders M, et al. Homozygosity mapping reveals mutations of GRXCR1 as a cause of autosomal-recessive nonsyndromic hearing impairment. Am. J. Hum. Genet. 2010a;86:138–147. doi: 10.1016/j.ajhg.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schraders M, et al. Mutations in PTPRQ are a cause of autosomal-recessive nonsyndromic hearing impairment DFNB84 and associated with vestibular dysfunction. Am. J. Hum. Genet. 2010b;86:604–610. doi: 10.1016/j.ajhg.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwander M, et al. A forward genetics screen in mice identifies recessive deafness traits and reveals that pejvakin is essential for outer hair cell function. J. Neurosci. 2007;27:2163–2175. doi: 10.1523/JNEUROSCI.4975-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott HS, et al. Insertion of beta-satellite repeats identifies a transmembrane protease causing both congenital and childhood onset autosomal recessive deafness. Nat. Genet. 2001;27:59–63. doi: 10.1038/83768. [DOI] [PubMed] [Google Scholar]
- Shahin H, et al. Five novel loci for inherited hearing loss mapped by SNP-based homozygosity profiles in Palestinian families. Eur. J. Hum. Genet. 2010;18:407–413. doi: 10.1038/ejhg.2009.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirmaci A, et al. A truncating mutation in SERPINB6 is associated with autosomal-recessive nonsyndromic sensorineural hearing loss. Am. J. Hum. Genet. 2010;86:797–804. doi: 10.1016/j.ajhg.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr A, Picton TW, Sininger Y, Hood LJ, Berlin CI. Auditory neuropathy. Brain. 1996;119(Pt 3):741–753. doi: 10.1093/brain/119.3.741. [DOI] [PubMed] [Google Scholar]
- Veske A, et al. Autosomal recessive non-syndromic deafness locus (DFNB8) maps on chromosome 21q22 in a large consanguineous kindred from Pakistan. Hum. Mol. Genet. 1996;5:165–168. doi: 10.1093/hmg/5.1.165. [DOI] [PubMed] [Google Scholar]
- Walsh T, et al. From flies' eyes to our ears: mutations in a human class III myosin cause progressive nonsyndromic hearing loss DFNB30. Proc. Natl. Acad. Sci. U S A. 2002;99:7518–7523. doi: 10.1073/pnas.102091699. [DOI] [PMC free article] [PubMed] [Google Scholar]