SUMMARY
Cancer cells must regulate plasticity and invasion to survive and metastasize. However, the identification of targetable mechanisms to inhibit metastasis has been slow. Signaling programs that drive stem and progenitor cells during normal development offer an inroad to discover mechanisms common to metastasis. Using a chick embryo transplant model, we have compared molecular signaling programs of melanoma and their embryonic progenitors, the neural crest. We report that malignant melanoma cells hijack portions of the embryonic neural crest invasion program. Genes associated with neural crest induction, delamination, and migration are dynamically regulated by melanoma cells exposed to an embryonic neural crest microenvironment. Specifically, we demonstrate that metastatic melanoma cells exploit neural crest-related receptor tyrosine kinases to increase plasticity and facilitate invasion while primary melanocytes may actively suppress these responses under the same microenvironmental conditions. We conclude that aberrant regulation of neural crest developmental genes promotes plasticity and invasiveness in malignant melanoma.
Keywords: neural crest, melanoma, metastasis, cell migration, in vivo model
INTRODUCTION
The incidence of melanoma has drastically risen in the United States, up more than 1800% over the past 100 years (Rigel et al., 1996). Because mortality among melanoma cancer victims is directly associated with the metastatic properties of the tumor, early detection and accurate diagnosis of malignant melanoma would significantly improve patient treatment and disease outcome. However, multiple challenges, including a high rate of clinical misdiagnosis, remain before this goal can be realized in the clinic (Marghoob et al., 2009; Morton and Mackie, 1998).
The high rate of misdiagnosis of metastatic melanoma is in part due to a lack of definitive molecular biomarkers. Genes commonly associated with stemness and tumorigenicity are dynamically regulated and fail to distinguish between tumorigenic and non-tumorigenic melanoma cells, highlighting a critical need for molecular biomarkers of melanoma disease progression (Quintana et al., 2008; Roesch et al., 2010).
Melanoma is initiated by the neoplastic transformation of melanocytes. Melanocytes derive from a highly invasive, multi-potent embryonic cell population termed the neural crest. The neural crest gives rise to many cell types, including both mesenchymal and neural derivatives (Le Douarin et al., 2004). The embryonic neural crest program utilizes several cellular processes commonly associated with cancer metastasis, including EMT and cell invasion. This has led to the postulate that transformed melanocytes might be inherently predisposed to having invasive and metastatic traits as a result of their normal developmental differentiation program (Gupta et al., 2005; Nesbit et al., 1998; Strizzi et al., 2011). Thus, we hypothesize that the high degree of plasticity and the aggressive nature of malignant melanoma derive from the aberrant re-activation of the embryonic neural crest program, typically silenced through the process of normal melanocyte differentiation.
Much is known about the genesis and advancement of the neural crest (Gammill and Bronner-Fraser, 2002; Meulemans and Bronner-Fraser, 2004; Steventon et al., 2005). As such, the progressive steps requisite for the colonization of melanocytes in the skin are well established (Sommer, 2011). Neural crest induction occurs at the presumptive dorsal neural tube between the neural plate and the non-neural ectoderm. Neural crest induction is regulated by the potent embryonic morphogens Bmp, Wnt, and Fgf (Mayor et al., 1997; Taneyhill and Bronner-Fraser, 2005). Following induction, neural crest cells are further specified by signaling from Myc, Pax3, Snail, and Sox9 (Steventon et al., 2005). Neural crest cells must then undergo an epithelial-to-mesenchymal transition (EMT) in order to dissociate from the neural tube and begin the migratory phase. Neural crest cell migration relies on both attractive and inhibitory cell guidance mechanisms. These include positive chemoattractive signaling mediated through neuropilin-VEGF interactions (McLennan et al., 2010) and both attractive and repulsive cues through Eph receptor tyrosine kinase signaling (Harris et al., 2008; Santiago and Erickson, 2002; Smith et al., 1997). The understanding of the embryonic neural crest program has led to its use as a developmental model to study a wide array of cellular processes, including EMT, migration, pluripotency, and cell fate determination (Duband et al., 1995; Kulesa et al., 2004; Le Lievre and Le Douarin, 1975; Motohashi et al., 2011; Taneyhill et al., 2007).
In an attempt to take advantage of established neural crest biology and the ancestral relationship between melanoma and the neural crest, our laboratory has developed a model using the chick embryo to study the metastatic behaviors of human melanoma cells. We have recently shown that human metastatic melanoma cells, when transplanted into the chick embryonic neural crest microenvironment, respond to cues from surrounding host tissues and emigrate along stereotypic migratory routes traveled by neural crest cells, while poorly invasive melanoma cells do not (Hendrix et al., 2007; Kulesa et al., 2006). We postulate that the observed melanoma cell migration in the chick embryo mimics many aspects of melanoma tumor dissemination and metastasis. This provides a model in which to study the profile of metastatic melanoma cells in vivo. Using a combination of laser capture microdissection (LCM) and gene profiling of small cell populations (≤10 cells), we compared genes and signaling pathways exhibited by highly invasive versus poorly invasive human melanoma cells and primary melanocytes. We focused on genes critically involved in the neural crest-to-melanocyte differentiation program. Herein we demonstrate that invasive cells display an enhanced ability to respond to signals within the chick embryonic neural crest microenvironment. This is evident by the dynamic regulation of genes commonly associated with neural crest induction, specification, EMT, and migration. Most notable are the dramatic upregulation of BMP4, and the dynamic regulation of the Eph/ephrin gene families observed in transplanted melanoma cells. Our results indicate that malignant melanoma cells exploit portions of the embryonic neural crest program to increase plasticity and facilitate metastasis.
RESULTS
Molecular Analysis of Transplanted Cells
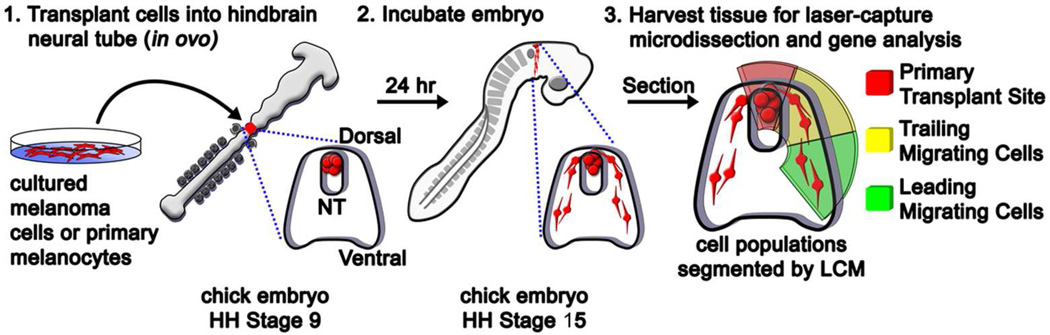
Our primary aim was to provide an in vivo comparison of genes typically associated with the embryonic neural crest developmental program between malignant melanoma and primary melanocytes. We analyzed three different human cell lines: c8161 highly aggressive human melanoma cells, c81-61 poorly aggressive human melanoma cells, and primary human melanocytes (Welch et al., 1991). c8161 and c81-61 are isogenic cell lines, making them ideal for this study. Small clusters (containing roughly 300 cells) of c8161 cells, c81-61 cells, or primary human melanocytes were transplanted into the dorsal neural tube at the axial level of the hindbrain (rhombomere 4) of stage 9–10 [Hamburger and Hamilton staging, (Hamburger and Hamilton, 1951)] chick embryos in ovo. Following re-incubation for 24 hours, embryos were harvested and prepared for laser capture microdissection (LCM). Following microdissection of 10 cells, 90 neural crest-related genes were subsequently analyzed by RT-qPCR. Analysis of cultured cells was performed in like manner following isolation of approximately 100 cells by serial dilution (see Figure 1 for a workflow of our experimental design).
Figure 1. A cartoon depicting the experimental methods.
Cells were transplanted into chick embryo neural tubes at HH Stage 9 and the egg was re-incubated for 24 hours. Following incubation, tissue was harvested and cells from different regions of the transplant were isolated by LCM: cells at the migratory front (leading), trailing cells, and cells from the primary transplant site, shown in green, yellow, and red respectively.
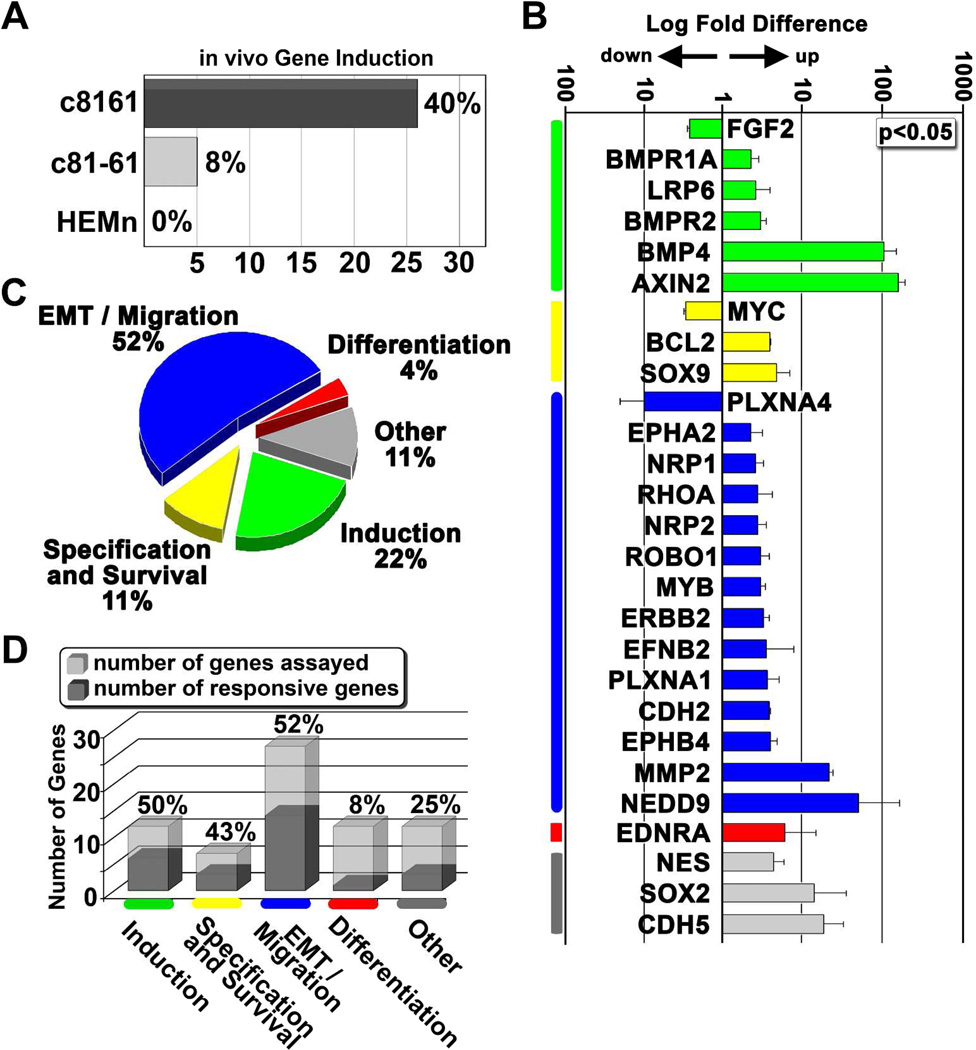
In order to evaluate transcriptional changes resulting from tumor cell transplantation into the chick embryo microenvironment, we first examined gene expression changes in transplanted melanoma cells and melanocytes harvested from the dorsal neural tube versus cultured cells. We observed that the aggressive melanoma cells exhibited a much higher capacity to respond to signals associated with the embryonic neural crest microenvironment (Figure 2). That is, 40% of the analyzed genes showed a significant induction in c8161 cells (Figure 2A). In contrast, only 8% of the genes were induced in c81-61 cells and no gene induction was observed in melanocytes (Figure 2A). Strikingly, 85% of responsive genes function in the early stages of the neural crest program, with over 50% of the genes associated with EMT and cell migration (Figure 2B,C-genes were grouped and colored based on their predominant role in the avian neural crest program; this color scheme is maintained throughout the text). Genes associated with growth, pluripotency, and survival of the neural crest were also represented (Figure 2B). However, genes involved in neural crest differentiation remained predominantly unchanged (Figure 2D). Thus, invasive c8161 melanoma cells possess the ability to sense and respond to the embryonic neural crest microenvironment.
Figure 2. Differential expression of neural crest-related genes from human melanoma and melanocyte cells following exposure to the chick embryonic microenvironment.
A) The percentage of total analyzed genes that showed a significant induction following transplantation. c8161 is an invasive human melanoma cell line. c81-61 is a poorly invasive isogenic counterpart to c8161. HEMn are primary human melanocytes. B) A graph showing log fold changes in gene expression following transplantation of c8161 melanoma into the embryo. All genes are normalized to gene expression values acquired from cultured cells, represented by the baseline expression level of 1. All changes reported were statistically significant with a p-value < 0.05 based on the parametric Limma statistical analysis (Jeanmougin et al., 2010) and adjusted using the Benjamini-Hochberg method. Error bars represent the standard deviation of three biological replicates. Genes are grouped based on their involvement with the neural crest program: induction (green), specification and survival (yellow), EMT and migration (blue), differentiation (red). Genes not specifically associated with any of these steps were grouped under the category “other” (gray). C) A pie chart depicting the percentage of induced genes from Figure 2C involved in aspects of the neural crest program: induction, specification, EMT and migration, and differentiation. D) A bar graph illustrating the percent induction of genes involved in the neural crest program, separated by the phases of the neural crest program.
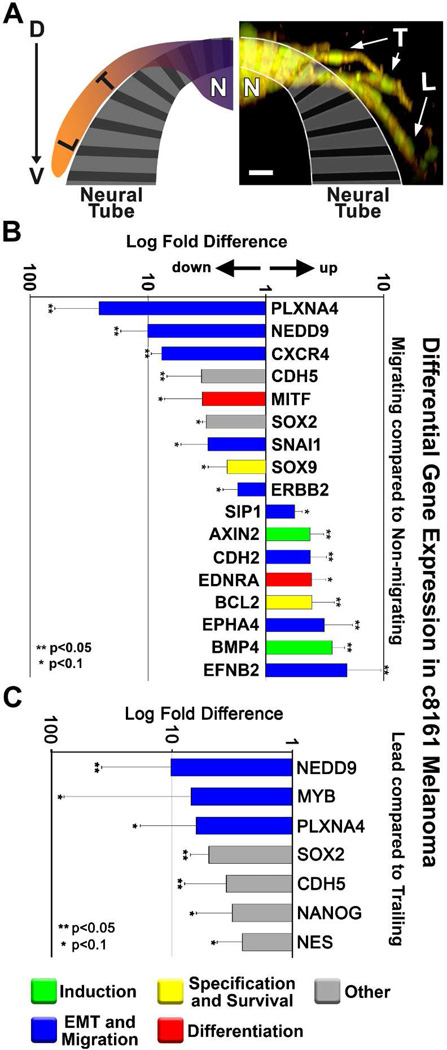
We observed that c81-61 cells and primary melanocytes did not emigrate from the dorsal neural tube, but remained clumped at the transplant site. Moreover, not all c8161 cells migrated from the graft site within the dorsal neural tube. In an effort to identify genes important to cell invasion, we used LCM to extract and compare invading c8161 cells with cells that remained in the neural tube (Figure 3A). RT-qPCR analysis revealed dynamic changes in a broad spectrum of genes, including genes involved in receptor-mediated or chemokine-based cell guidance (PLXNA4, CXCR4, EPHA4, EFNB2), suggesting that c8161 cells quickly adapt to their local microenvironment (Figure 3B).
Figure 3. Differential gene expression of migrating compared to non-migrating melanoma cells.
A) An immunofluorescent image and accompanying cartoon depicting the biological domains from which melanoma cells were harvested for gene analysis. Domains include “L” for leading cells, “T” for trailing cells, and “N” for non-migrating cells. c8161 melanoma cells are dual-labeled with an H2B:cerulean nuclear marker (pseudo-colored green) and a Gap43:YFP membrane marker. The image is displayed as an XZ projection of an XY z-stack. The image was acquired with a Zeiss 20X NA 1.0 objective. The image is viewed dorsal (D) to ventral (V) along the y-axis. The scale bar represents 30 microns. B) A graph depicting relative fold-changes in gene expression observed in migrating c8161 melanoma cells. Values are displayed as log fold difference. C) A graph depicting relative log-fold changes in gene expression observed in leading c8161 melanoma cells. Error bars represent the standard deviation of three biological replicates. ** depicts changes with p<0.05, * depicts changes with p<0.1 based on Limma statistical analysis.
We then asked whether leading migratory cells were different from the trailing cell populations. Interestingly, this analysis revealed seven genes that were differentially regulated among lead and trailing cell populations (Figure 3C). Four of the down-regulated genes (NEDD9, PLXNA4, SOX2 and CDH5) were also identified in the previous comparison of migrating versus non-migrating cells, suggesting a continued downward trend displayed by migrating cells. However, MYB, NANOG, and NES show exclusive down-regulation in lead cells compared to trailing cells. These observations highlight the dynamic temporal and/or spatial gene regulation exhibited by migratory cells in our in vivo model system and suggest either a response to a distinct microenvironment or complex communication dynamics between the migratory lead and trailing populations.
Comparison of Transplanted c8161 Cells to the Host Neural Crest
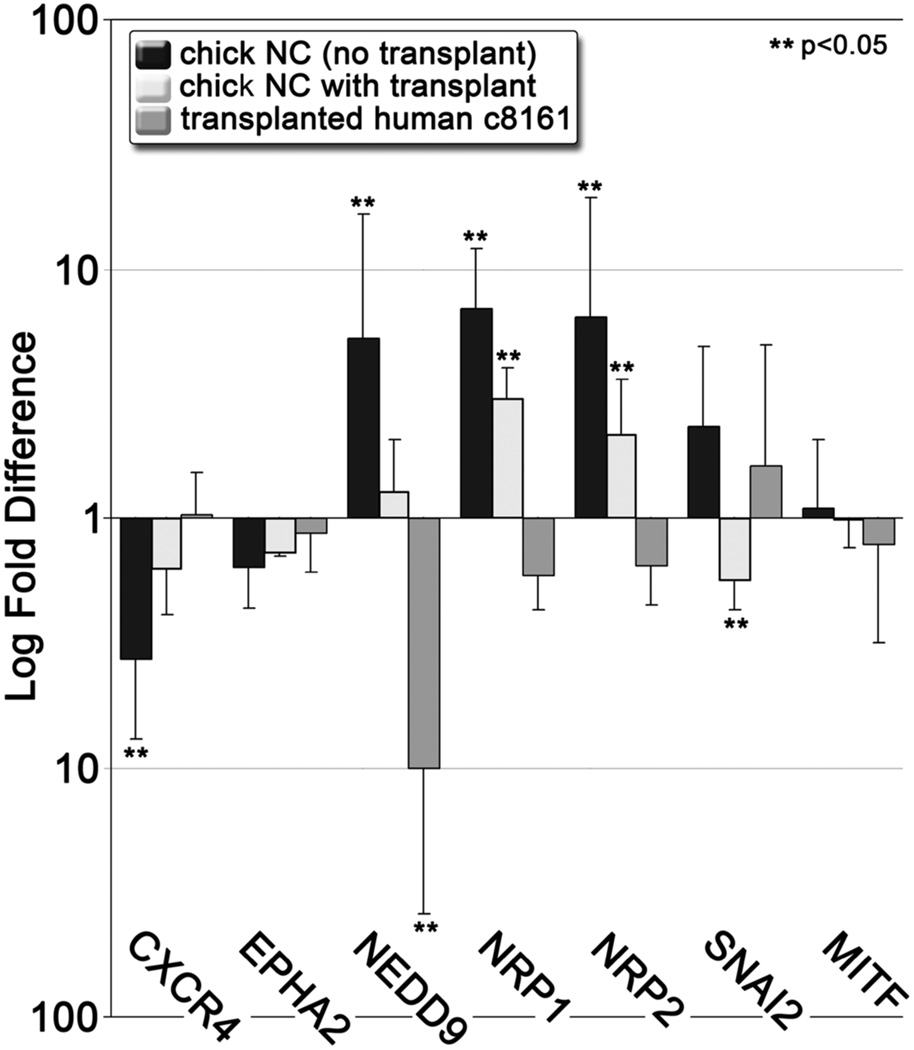
To determine whether gene expression patterns observed in migrating c8161 cells mimicked the expression patterns displayed by the host neural crest, we isolated chick neural crest cells from age-matched embryos in the presence or absence of transplanted c8161 cells. We compared gene expression levels of several chick genes known to be regulated during the migratory phase of the neural crest developmental program, including EMT and differentiation factors (SNAI2 and MITF), and factors involved in migration, including chemoattraction and pathway guidance (CXCR4, EPHA2, NEDD9, NRP1, NRP2). We observed that the lead migratory population of wild-type chick neural crest displayed significant changes in gene expression in CXCR4, NEDD9, NRP1, and NRP2 (Figure 4). However, c8161 gene expression directly opposed that observed in neural crest cells for these genes (Figure 4). The discrepant gene expression patterns we observed between c8161 and chick neural crest cells imply that c8161 cells are not constrained to the precise recapitulation of the neural crest developmental program.
Figure 4. A comparison of the differential expression of neural crest-related genes from the host chick neural crest and transplanted c8161 melanoma cells.
A graph depicting expression in lead cell populations relative to trailing cell populations. Error bars represent the standard deviation of three biological replicates. ** show statistically significant changes (p<0.05).
Comparison of Transplanted c8161 Melanoma Cells to Primary Melanocytes
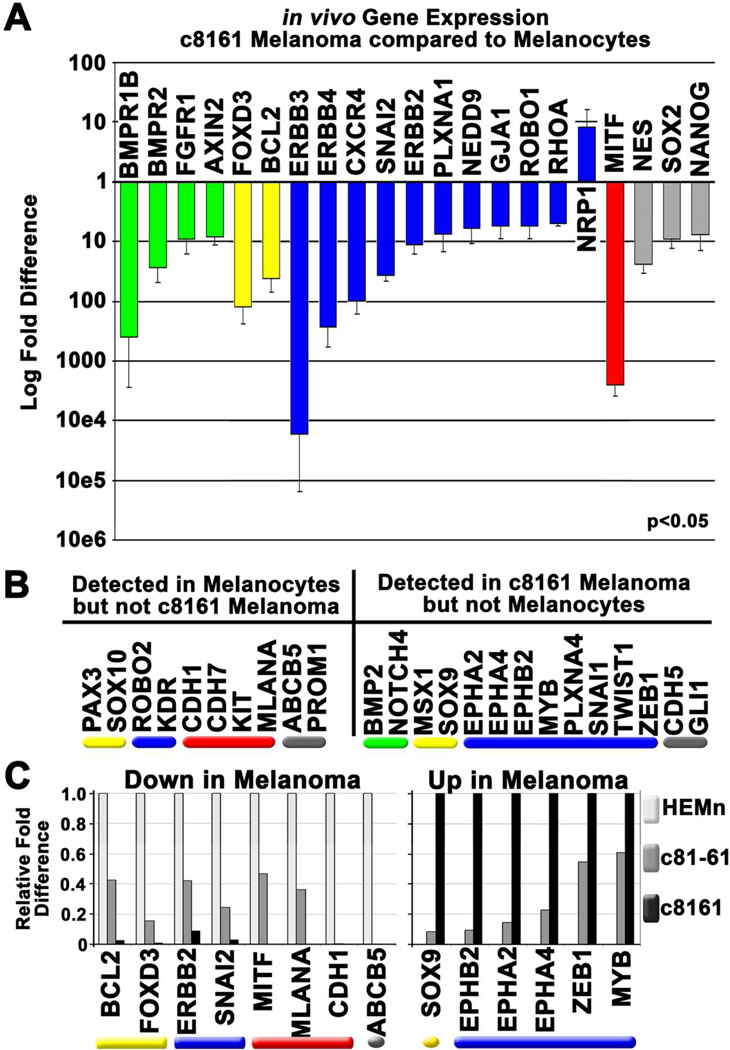
Are neural crest-related genes up-regulated in melanoma versus normal melanocytes? Of the neural crest-related genes expressed by both melanocytes and c8161 cells, only NRP1 showed significantly higher expression in c8161 cells (Figure 5A). By contrast, 20 genes showed a significant reduction in c8161 cells, including many involved in EMT, migration, and pluripotency (Figure 5A).
Figure 5. Expression of some neural crest-related genes correlates with disease progression.
A) A graph showing in vivo differential gene expression observed in transplanted c8161 melanoma cells compared to transplanted melanocytes. Error bars represent the standard deviation of three biological replicates. All changes shown in the graph are statistically significant (p<0.05). B) A list of genes expressed by melanocytes but undetected in c8161 and vice versa. C) A graph showing in vivo differential gene expression of genes that show intermediate expression in poorly invasive, more differentiated melanoma cells (c81-61) compared to both melanocytes and c8161 melanoma cells. Genes reduced in c81-61 cells compared to melanocytes are shown in the left panel while those that show an induction compared to melanocytes are shown in the right panel.
Importantly, c8161 cells did re-express 12 neural crest-related genes not detected in primary melanocytes (Figure 5B). 66% of these genes are associated with EMT and migration, including SNAI1, TWIST1, ZEB1, and three Eph family members, EPHA2, EPHA4, and EPHB2. This re-expression was accompanied by the aberrant silencing of genes associated with melanocyte differentiation (Figure 5B).
EPHA2, EPHA4, and EPHB2 were also re-expressed in the poorly aggressive c81-61 cells, albeit at significantly lower levels (Figure 5C). Likewise, several genes expressed by melanocytes but undetected in c8161 cells showed intermediate expression in c81-61 cells (Figure 5C). These included melanocyte differentiation markers MITF, MLANA, and CDH1. Thus, the differential expression of these genes could aid in the identification of an intermediate tumor stage and facilitate prognosis.
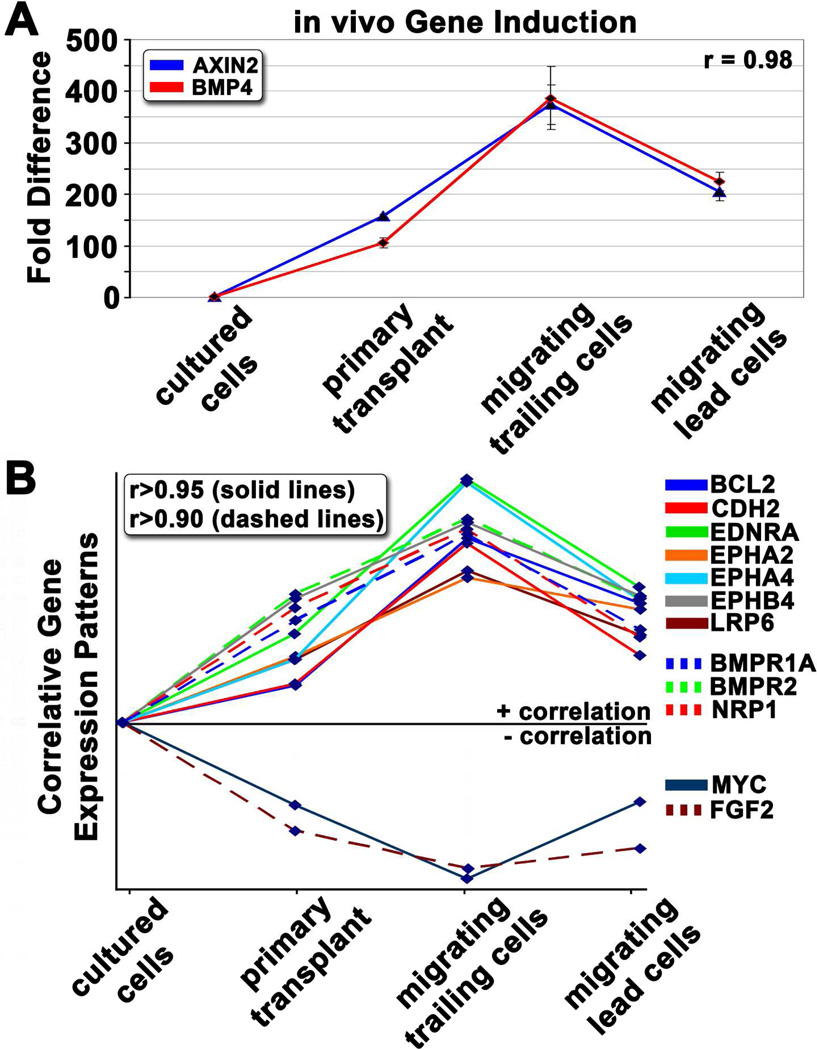
To examine whether differential gene expression could be used to identify covariant expression patterns (suggestive of a mutual regulatory element or genetic interaction), we clustered genes based on their overall expression patterns. This identified several highly correlative gene expression patterns. The most striking similarity was observed between BMP4 and AXIN2 (Figure 6A). Both genes were minimally expressed in cultured cells, but were significantly induced upon exposure to the embryonic neural crest microenvironment. Further induction was observed in cells that had recently exited the neural tube, whereas cells that had migrated away from the neural tube began to down-regulate these two genes. We further identified 12 other genes whose expression patterns correlated with BMP4 and AXIN2 (Figure 6B). Multiple signaling families were represented, including Wnt and Bmp receptors LRP6 and BMPR2, and genes associated with adhesion (CDH2) and survival (BCL2).
Figure 6. Correlative expression patterns highlight important signaling pathways and novel gene interactions.
A) A graph depicting relative gene expression patterns for BMP4 and AXIN2 in cells harvested from four distinct microenvironments: cultured cells, primary transplant site, trailing migratory cells, and lead migratory cells (see Figure 1 cartoon for location descriptions). Pearson correlation was used to generate r-values. B) Gene expression patterns showing high correlation (r>0.95 depicted by solid lines, r>0.90 depicted by dashed lines) to AXIN2 and BMP4.
Evaluation of Eph/Ephrin Gene Expression in Transplanted c8161 Melanoma Cells
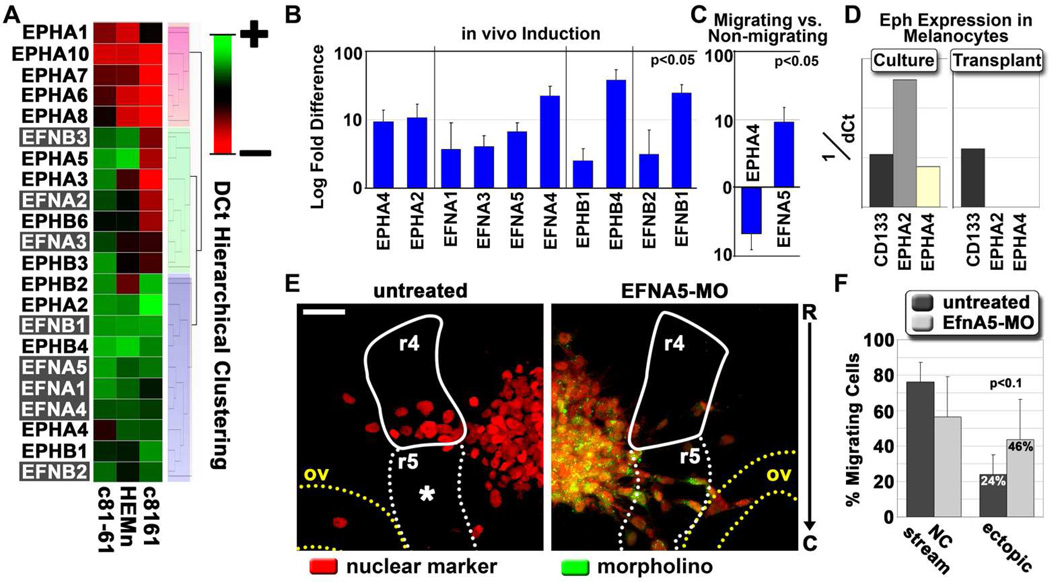
Three of the genes showing a high correlation to AXIN2 and BMP4 were Eph family members. As several Eph/ephrin family members demonstrated dynamic regulation by the chick embryo microenvironment, we evaluated the expression of all Eph receptors and ephrin ligands (Eph/ephrins) in our transplant assay. Gene expression for all known human Eph/ephrins was compared in c8161, c81-61, and melanocytes by hierarchical clustering. This analysis revealed three primary clusters (Figure 7A). Two of these clusters represented Eph/ephrins either commonly expressed or absent from all cell types. However, the third cluster defined 7 family members, including both class A and class B Ephs and ephrins, that were specifically highly down-regulated or undetected in c8161 cells compared to both c81-61 cells and melanocytes. In contrast, only EPHB2 appeared highly expressed by the melanoma cells but undetected in melanocytes (Figure 7A). Thus, silencing of Eph/ephrins is more common than Eph/ephrin overexpression in c8161.
Figure 7. Eph and ephrin expression and regulation in melanoma cells and primary melanocytes.
A) Hierarchical cluster analysis based on dCt (cycle threshold normalized for input amount) values. All known human Eph and ephrin family members were analyzed for expression in c8161, c81-61 and primary melanocytes (HEMn). Three primary clusters are highlighted by red, green and gray boxes. B) A graph depicting the induction of Eph/ephrins expressed by c8161 cells following transplantation into the chick embryo, compared to gene expression values from cultured cells. All reported gene expression changes were statistically significant with an adjusted p-value < 0.05. Error bars represent the standard deviation of three biological replicates. C) A graph depicting log-fold changes in Eph/ephrin gene expression observed in migrating c8161 melanoma cells compared to non-migrating cells. D) A graph depicting the expression of EPHA2 and EPHA4 in primary melanocytes harvested from culture or from transplants. Normalized values are provided as 1/dCt. CD133 was used as a positive expression control. E) Microscope images acquired from chick embryos transplanted with wild-type c8161 melanoma cells or c8161 cells pre-treated with a translation-blocking morpholino against ephrin-A5. Cells were labeled with an H2B:cerulean fusion construct (pseudo-colored red). Morpholinos were tagged with lissamine and pseudo-colored green. Embryonic tissue boundaries and fiducial points are provided, including rhombomere 4 in the hindbrain (r4, solid white line), rhombomere 5 (dashed white line), and the otic vesicle (ov, dashed yellow line). The image is viewed rostral (R) to caudal (C) along the y-axis. Fluorescent images represent projected z-stacks, based on a maximum intensity projection. Images were acquired using a Zeiss 20X NA 1.0 objective. Scale bar is 50 microns. F) A graph depicting the percent of c8161 migratory cells that emigrated along the canonical neural crest rhombomere 4 migratory pathway versus those that breached typical migratory pathway boundaries. Values are given as a percent of total migrating cells. Error bars represent the standard deviation of four biological replicates. The statistical analysis was performed using a Student’s t-test.
To determine which Eph/ephrins expressed by c8161 cells were responsive to exposure to the chick embryonic neural crest microenvironment, transplanted c8161 cells were harvested by LCM and analyzed for changes in Eph/ephrin expression. We found that ten out of thirteen (77%) Eph/ephrins expressed by c8161 cells were significantly induced following transplantation while none were down-regulated (Figure 7B). Additionally, gene expression changes were also observed in migrating versus non-migrating c8161 melanoma cells. EFNA5 exhibited a ten-fold induction in migratory cells, while EPHA4 was significantly reduced, suggesting possible reciprocal roles for these genes (Figure 7C).
Because of the significant induction observed for EFNA5 in migrating c8161 cells (Figure 7C), we knocked down ephrin-A5 protein expression in c8161 cells using translation-blocking morpholinos. c8161 cells were loaded with a fluorophore-conjugated morpholino and transplanted into the chick embryo. Untreated c8161 cells predictably emigrated adjacent to rhombomere 4, avoiding the non-permissive neural crest cell-free zone associated with the sub-region adjacent to rhombomere 5. In contrast, we observed that c8161 cells lacking ephrin-A5 were significantly less constrained to migrate lateral to rhombomere 4 and freely navigated into the neural crest cell-free zone adjacent to rhombomere 5 and toward the otic vesicle (Figure 7E,F). This suggested a role for ephrin-A5 in migratory pathfinding of metastatic melanoma cells.
Our hierarchical cluster analysis also revealed that primary melanocytes expressed moderately high levels of EPHA2 and EPHA4 under standard culture conditions (Figure 7A). This conflicted with our previous data that showed EPHA2 and EPHA4 were undetected in transplanted melanocytes (Figure 5B). Further evaluation revealed that melanocytes expressed EPHA2 and EPHA4 in culture, but silenced EPHA2 and EPHA4 following transplantation (Figure 7D). The expression of CD133, which was expressed at similar levels to EPHA2 and EPHA4 in culture, was used to verify that this loss of expression was not due to decreased sensitivity resulting from lower cDNA input amounts. This suggests that primary melanocytes respond to the chick embryonic neural crest microenvironment by repressing the expression of some Eph/ephrin signaling.
DISCUSSION
We have applied our understanding of the embryonic neural crest and the power of our chick embryo transplant model to better understand the molecular mechanisms of melanoma metastasis, including cell dissemination and invasion. Previous work from our laboratory has shown that aggressive melanoma cells transplanted into the chick embryo respond to embryonic environmental cues and migrate along stereotypical neural crest emigration routes (Kulesa et al., 2006). Furthermore, a subset of migrating melanoma cells re-expressed the melanocyte differentiation marker Melan-A. Together, these findings suggested that aggressive melanoma cells display a highly plastic neural crest-like phenotype and may respond to potent embryonic signals that promote migration or differentiation. Here we report results that build on these initial findings and support the hypothesis that melanoma cells are predisposed to being metastatic based on their intrinsic relationship to the neural crest and suggest a model in which melanoma cells exploit portions of the neural crest developmental program to facilitate metastasis.
Our experimental design combined laser capture microdissection with highly sensitive RT-qPCR to directly test to what extent c8161 aggressive melanoma cells utilize neural crest-related signaling pathways for invasion (see Table S2 for a summary of the results). We found that c8161 cells have a much higher propensity to respond to the embryonic neural crest microenvironment, as manifest by the induction of genes associated with the neural crest developmental program. Poorly aggressive c81-61 cells were less responsive and primary melanocytes did not significantly induce any of the analyzed genes (Figure 2A). Responsive genes primarily included those involved in the early stages of the chick neural crest developmental program: induction, specification, EMT and survival. Importantly, this responsiveness did not appear to be driven by the up-regulation of common tumor-promoting genes. Rather, the majority of genes assayed were expressed at lower levels in c8161 cells compared to primary melanocytes. This included several genes commonly associated with cancer pathogenesis such as the EGF receptor family members ERBB2, ERBB3, and ERBB4, the purported melanoma oncogenes MITF and NEDD9, and several markers of pluripotency, including NES, SOX2, and NANOG (Figure 5). Yet many of these genes also exhibited dynamic regulatory properties in response to the chick embryonic neural crest microenvironment (Figures 2,3). The ability of c8161 cells to respond to chick embryonic neural crest microenvironmental signals suggests that receptors associated with the neural crest developmental program are functional and active. Interestingly, none of the receptors that were expressed by both c8161 cells and primary melanocytes showed increased expression in the melanoma cells. Rather, key receptors associated with the neural crest developmental program (BMPR1B, BMPR2, FGFR1) showed decreased expression in c8161 cells relative to primary melanocytes (Figure 5). It is possible that pathway components downstream of these receptors may enhance receptor activity and thereby promote melanoma cell responsiveness to the chick embryonic microenvironment. Recent evidence suggests a predominant role for BMP4 in melanoma metastasis, in part through regulation of SNAI2 (Gupta et al., 2005; Rothhammer et al., 2005). Although we did not observe significant changes in SNAI2 gene expression, our data support a role for BMP4 in the invasion process. BMP4, along with the Wnt target gene AXIN2, displayed the highest induction in c8161 cells following transplantation into the chick embryonic neural crest microenvironment (106- and 157-relative fold increases respectively, Figure 2). Wnt and Bmp are important signals in neural crest development, and are currently thought to be antagonistic, with dominant Wnt signaling driving melanocyte specification while BMP4 promotes neural and glial differentiation at the expense of pigment cells (Jin et al., 2001). The observed up-regulation of BMP4 suggests that Bmp signaling is dominant in c8161 melanoma cells. The activation of neuronal-fate-promoting Bmp signaling supports previous findings that melanomas frequently acquire neuronal-like characteristics and may explain the high proficiency with which melanomas metastasize to the brain (Iyengar and Singh, 2010; Madajewicz et al., 1984). Additionally, because Axin2 is a component of the beta-catenin destruction complex, the high AXIN2 levels may result in reduced Wnt/beta-catenin signaling. The significant negative correlation observed between AXIN2 and MYC, a known target of Wnt/beta-catenin signaling, supports this possibility. Thus, overactive Bmp signaling combined with inhibition of differentiation-promoting Wnt signals may correlate with the heightened plasticity observed in c8161 cells.
Three Eph receptors also showed high correlation with BMP4 and AXIN2 (Figure 6). Significantly, two of them (EPHA2 and EPHA4) were also genes expressed by c8161 cells but not detected in primary melanocytes. When combined with the notable up-regulation observed for BMP4, it is intriguing to surmise that BMP4 may directly facilitate the up-regulation of some Eph family members, although a direct link between BMP4 and Ephs has not been reported. Yet, our data clearly highlight the dynamic regulation of the Eph/ephrin families following exposure to the chick embryonic neural crest microenvironment. We demonstrated that 10 out of 13 Eph/ephrins expressed in c8161 cells diplayed inductive responses resulting from exposure to the chick neural crest embryonic microenvironment (Figure 7). Importantly, this contrasts with primary melanocytes, which appear to actively repress gene expression of EPHA2 and EPHA4 following transplantation.
A role for Eph/ephrin signaling has been reported for many types of cancers, including melanoma (Pasquale, 2010; Vogt et al., 1998). Likewise, Eph/ephrin interactions have proven important for migration and sorting of embryonic neural crest cells (Kasemeier-Kulesa et al., 2006; Krull et al., 1997; Smith et al., 1997), including melanoblasts, the melanocytic precursor (Harris et al., 2008; Santiago and Erickson, 2002). Many of the Eph/ephrins known to be involved in neural crest cell migration and pathfinding are expressed by c8161 cells. However, hierarchical clustering of the Eph/ephrins revealed seven different Eph/ephrins undetected in c8161 cells compared to c81-61 cells and primary melanocytes (Figure 7). Although the effects of the loss of these Eph/ephrins in c8161 cells remains under investigation, it is tempting to speculate that aberrant Eph/ephrin expression could result in deregulated signaling and facilitate metastasis by allowing tumor cells to overcome repulsive cues found in the microenvironment. As an example, EFNA5 has been shown to regulate trunk neural crest cell migration (McLennan and Krull, 2002). Our work demonstrated that knockdown of ephrin-A5 in c8161 cells allowed for their ectopic invasion into the region adjacent to rhombomere 5, an area of the head that typically excludes neural crest cell migration (Sechrist et al., 1993).Also, EFNA5 exhibited a nearly ten-fold increase in the migrating c8161 population (Figure 7C), suggesting that ephrin-A5 may be a regulator of melanoma metastasis.
Lastly, our data revealed the re-expression of twelve neural crest-related genes in c8161 cells that were undetected in melanocytes (Figure 5). The majority of these genes, including three members of the Eph receptor tyrosine kinase family, are involved in microenvironmental sensing and migratory pathfinding (EPHA2, EPHA4, EPHB2, PLXNA4), and EMT (SNAI1, TWIST1, ZEB1, MYB). Thus, the aberrant reacquisition of neural crest-related genes typically silenced during melanocyte differentiation may contribute to the enhanced environmental sensing and plasticity displayed by c8161 cells. Interestingly, we observed that several of these genes exhibited intermediate levels of expression in the poorly aggressive c81-61 melanoma cells (Figure 5C). Intermediate expression was also observed in c81-61 cells for several genes expressed by primary melanocytes but undetected in c8161 cells. Together, this suggested that moderate expression of these genes may highlight a transitory phase during transformation from melanocyte to malignant melanoma. We conclude that melanoma cells reacquire some neural crest-related genes associated with EMT and migration while silencing differentiation genes.
In summary, this study has revealed important insights into the molecular pathogenesis of metastatic melanoma. We postulate that malignant melanoma cells reacquire aspects of the embryonic neural crest program during neoplastic transformation, which promotes the malignant phenotype. Our data revealed the re-expression of multiple neural crest-related genes in c8161 cells that were undetected in melanocytes. The majority of these genes are involved in microenvironmental sensing, migration, and EMT. Further work will be required to decipher which of the observed transcriptional changes will be relevant to human disease. Because of the similarities between the embryonic neural crest developmental program and tumor metastasis, we postulate that many of the transcriptional changes identified by our model system may reflect novel aspects of the complex metastatic process. Our data suggest that c8161 melanoma cells have a much higher propensity to respond to the embryonic neural crest microenvironment than poorly aggressive c81-61 melanoma cells or primary melanocytes. Although we cannot rule out the possibility that pre-existing heterogeneities within the c8161 tumor cell population conferred the differing migratory abilities observed, the molecular and behavioral reproducibility of c8161 migration in our chick embryo transplant model support the premise that powerful embryonic signaling cues promote the dynamic gene expression patterns observed in the highly plastic c8161 cells. Furthermore, the fact that so few changes were observed in the poorly aggressive melanoma cells or the primary melanocytes suggests that the ability to regulate transcription may be a readout of cellular plasticity and thus may reflect enhanced metastatic potential. Together, these data suggest that a profile of metastasis, based on the expression patterns of neural crest-related genes, may be established for melanoma cells and that the relative expression of some neural crest-related genes may assist in monitoring melanotic lesions and facilitating accurate diagnosis and prognosis, while also providing novel therapeutic targets.
METHODS
Cell Culture
c8161 human malignant melanoma cells and c81-61 human poorly aggressive melanoma cells (c8161 and c81-61 are isogenic) were provided by Dr. Mary Hendrix, Children’s Memorial Research Center, Chicago, IL. Cells were maintained in RPMI supplemented with 10% fetal bovine serum. Primary melanocytes (HEMn-LP) were obtained from Cascade Biologics and were maintained in Medium 254 supplemented with human melanocyte growth supplement as per manufacturer’s instructions. For drop culture conditions, cells were resuspended in the appropriate growth medium at a concentration of 4e6 cells/ml. A 20ul drop was hung from the lid of a 35mm culture dish and incubated for 40 hours under standard culture conditions.
Morpholino Knockdown
Morpholino design and production were performed by Gene Tools, LLC. The sequence ACATCTCCACGTGCAACATCACGCC was used to target ephrin-A5. c8161 cells were loaded with lissamine-conjugated morpholino by scrape delivery as per manufacturer’s instructions. Following morpholino delivery, cells were sorted using a MoFlo high speed cell sorter. Sorted cells were set into our drop culture system in preparation for transplantation.
Chick Embryo Transplants
Fertile White Leghorn chick eggs were acquired from Centurion Poultry. Eggs were incubated at 38°C for approximately 39 hours until the 9-somite stage of development. Eggs were windowed and embryos were visualized following the injection of 10% India ink in Howard Ringer's solution below the blastodisc. Embryos were staged according to the criteria of Hamburger and Hamilton (Hamburger and Hamilton, 1951). Transplantation of the embryos was performed as previously described (Kulesa et al., 2006).
Static Imaging and Image Processing
Individual embryos were removed from eggs with paper rings, rinsed with Ringer's solution and placed dorsal side up within a thin ring of high-vacuum grease on 22×75mm microslides. Embryos were imaged using a Zeiss 710 multi-photon upright microscope. 2-photon z-stacks were acquired at a wavelength of 850nm using a W Plan-Apochromat 20×/1.0 DIC objective (Zeiss). All image processing was performed using Zeiss AIM software.
Tissue Processing and Laser Capture Microdissection
Tissue of interest was removed from the embryo, submerged in chilled Tissue-Tek OCT Compound (Sakura Finetek) and immediately flash frozen in 2-methylbutane at −60°C. Frozen tissue was cryosectioned at 20um, under RNase-free conditions, onto Zeiss NF 1.0 PEN Membrane Slides (Carl Zeiss MicroImaging). Following cryosectioning, slides were dipped in cold 0.1% DEPC water to remove excess OCT and fixed by cold, gradient ethanol dehydration. Slides were stored at −80°C. Upon removal from the freezer, slides were immediately placed in desiccation chambers containing anhydrous calcium sulfate (W.A. Hammond Drierite) and allowed to dry completely before LCM. LCM was conducted on a Zeiss PALM Microbeam system using a focused 355nm UV laser. Samples were collected in 200ul adhesive caps (Carl Zeiss MicroImaging). RNA integrity numbers from representative slides were determined to be greater than 7.5.
RT-qPCR
For consistency, all samples collected by LCM and from cell culture contained between 10 and 100 cells. Each sample was lysed at room temperature for 20 minutes in 10ul of Ambion’s Cells-to-Ct lysis solution as per manufacturer’s instructions. Lysates were stored at −80°C. cDNA was synthesized directly from cell lysates in 20ul reactions using ABI’s High Capacity cDNA Reverse Transcription kit (Applied Biosystems Inc.) per the manufacturer’s instructions, except the reactions were incubated for 60 minutes at 37°C. Linear, gene-specific amplification of cDNA was performed with Ambion’s Cells-to-Ct kit in 25ul reactions using validated Taqman Gene Expression Assays (Applied Biosystems Inc.). Assay IDs of all Taqman Gene Expression Assays are listed as supplemental data (Table S1) per MIQE guidelines (Bustin et al., 2009). cDNA was diluted 1:20 with 1X TE per manufacturer’s recommendations for downstream qPCR analysis. All qPCR was performed in technical and biological triplicate and assembled on either a CAS4200 or QIAgility (Qiagen/Corbett Life Science) PCR setup robot. qPCR was performed on an ABI7900 HT (Applied Biosystems Inc.) in MicroAmp Optical 384-well reactions plates using the following cycling conditions: 50°C for 2 minutes, 95°C for 10 minutes & 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. Each 10ul qPCR reaction included 4ul pre-amplified and diluted cDNA, 5ul 2X Gene Expression Master Mix (Applied Biosystems Inc.), 0.5ul 20X validated Taqman Gene Expression Assay and 0.5ul 0.1% DEPC water. All amplification plots generated were analyzed collectively and a Ct threshold was manually set at 0.2. “No Template” negative qPCR controls were run for each assay analyzed and proved to be clean.
Statistics, Data Analysis and Processing
Integromics’ RealTime StatMiner bioinformatics software was used for Ct data analysis and fold-change calculations via the ddCt method. All samples were normalized using a three-gene normalization strategy. The variance modeling strategy Limma was employed for all gene expression analysis (Jeanmougin et al., 2010). Where noted in the figure legends, the Benjamini-Hochberg adjustment was applied to further reduce the false discovery rate. Data compilation and graphing were performed in Microsoft Excel. Figure processing was performed with Adobe Photoshop CS3.
Supplementary Material
SIGNIFICANCE.
The rate of misdiagnosis of malignant melanoma is abnormally high due to complex tumor heterogeneity, a poor understanding of disease pathogenesis, and a lack of molecular markers associated with metastatic disease. We have developed an in vivo model system to study the molecular attributes of melanoma. Our system exploits the ancestral link between melanoma and an invasive, multi-potent embryonic cell population called the neural crest. We established a molecular profile of invasion for melanoma cells based on the expression pattern of neural crest-related markers. Deciphering the molecular signature of melanoma will facilitate accurate diagnosis and evaluation of metastatic potential.
ACKNOWLEDGEMENTS
CMB thanks the National Institutes of Health Ruth L. Kirschtein Postdoctoral Fellowship Program for funding and partial support from the Stowers Institute for Medical Research. PMK thanks the kind generosity of the Stowers Institute for Medical Research for support. We gratefully acknowledge the laboratory of Dr. Mary Hendrix for providing the c8161 and c81-61 melanoma cell lines used in these experiments and for thoughtful and constructive discussion.
REFERENCES
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clinical chemistry. 2009;55:611–522. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Duband JL, Monier F, Delannet M, Newgreen D. Epithelium-mesenchyme transition during neural crest development. Acta anatomica. 1995;154:63–78. doi: 10.1159/000147752. [DOI] [PubMed] [Google Scholar]
- Gammill LS, Bronner-Fraser M. Genomic analysis of neural crest induction. Development (Cambridge, England) 2002;129:5731–5741. doi: 10.1242/dev.00175. [DOI] [PubMed] [Google Scholar]
- Gupta PB, Kuperwasser C, Brunet JP, Ramaswamy S, Kuo WL, Gray JW, Naber SP, Weinberg RA. The melanocyte differentiation program predisposes to metastasis after neoplastic transformation. Nature genetics. 2005;37:1047–1054. doi: 10.1038/ng1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL. A Series of Normal Stages in the Development of the Chick Embryo. Journal of Morphology. 1951;88:49–92. doi: 10.1002/aja.1001950404. [DOI] [PubMed] [Google Scholar]
- Harris ML, Hall R, Erickson CA. Directing pathfinding along the dorsolateral path - the role of EDNRB2 and EphB2 in overcoming inhibition. Development (Cambridge, England) 2008;135:4113–4122. doi: 10.1242/dev.023119. [DOI] [PubMed] [Google Scholar]
- Hendrix MJ, Seftor EA, Seftor RE, Kasemeier-Kulesa J, Kulesa PM, Postovit LM. Reprogramming metastatic tumour cells with embryonic microenvironments. Nature reviews. 2007;7:246–255. doi: 10.1038/nrc2108. [DOI] [PubMed] [Google Scholar]
- Iyengar B, Singh AV. Patterns of neural differentiation in melanomas. Journal of biomedical science. 2010;17:87. doi: 10.1186/1423-0127-17-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanmougin M, De Reynies A, Marisa L, Paccard C, Nuel G, Guedj M. Should we abandon the t-test in the analysis of gene expression microarray data: a comparison of variance modeling strategies. PloS one. 2010;5:e12336. doi: 10.1371/journal.pone.0012336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin EJ, Erickson CA, Takada S, Burrus LW. Wnt and BMP signaling govern lineage segregation of melanocytes in the avian embryo. Developmental biology. 2001;233:22–37. doi: 10.1006/dbio.2001.0222. [DOI] [PubMed] [Google Scholar]
- Kasemeier-Kulesa JC, Bradley R, Pasquale EB, Lefcort F, Kulesa PM. Eph/ephrins and N-cadherin coordinate to control the pattern of sympathetic ganglia. Development (Cambridge, England) 2006;133:4839–4847. doi: 10.1242/dev.02662. [DOI] [PubMed] [Google Scholar]
- Krull CE, Lansford R, Gale NW, Collazo A, Marcelle C, Yancopoulos GD, Fraser SE, Bronner-Fraser M. Interactions of Eph-related receptors and ligands confer rostrocaudal pattern to trunk neural crest migration. Current biology : CB. 1997;7:571–580. doi: 10.1016/s0960-9822(06)00256-9. [DOI] [PubMed] [Google Scholar]
- Kulesa P, Ellies DL, Trainor PA. Comparative analysis of neural crest cell death, migration, and function during vertebrate embryogenesis. Dev Dyn. 2004;229:14–29. doi: 10.1002/dvdy.10485. [DOI] [PubMed] [Google Scholar]
- Kulesa PM, Kasemeier-Kulesa JC, Teddy JM, Margaryan NV, Seftor EA, Seftor RE, Hendrix MJ. Reprogramming metastatic melanoma cells to assume a neural crest cell-like phenotype in an embryonic microenvironment. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:3752–3757. doi: 10.1073/pnas.0506977103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin NM, Creuzet S, Couly G, Dupin E. Neural crest cell plasticity and its limits. Development (Cambridge, England) 2004;131:4637–4650. doi: 10.1242/dev.01350. [DOI] [PubMed] [Google Scholar]
- Le Lievre CS, Le Douarin NM. Mesenchymal derivatives of the neural crest: analysis of chimaeric quail and chick embryos. Journal of embryology and experimental morphology. 1975;34:125–154. [PubMed] [Google Scholar]
- Madajewicz S, Karakousis C, West CR, Caracandas J, Avellanosa AM. Malignant melanoma brain metastases. Review of Roswell Park Memorial Institute experience. Cancer. 1984;53:2550–2552. doi: 10.1002/1097-0142(19840601)53:11<2550::aid-cncr2820531129>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Marghoob AA, Changchien L, Defazio J, Dessio WC, Malvehy J, Zalaudek I, Halpern AC, Scope A. The most common challenges in melanoma diagnosis and how to avoid them. The Australasian journal of dermatology. 2009;50:1–13. doi: 10.1111/j.1440-0960.2008.00496_1.x. quiz 14–5. [DOI] [PubMed] [Google Scholar]
- Mayor R, Guerrero N, Martinez C. Role of FGF and noggin in neural crest induction. Developmental biology. 1997;189:1–12. doi: 10.1006/dbio.1997.8634. [DOI] [PubMed] [Google Scholar]
- Mclennan R, Krull CE. Ephrin-as cooperate with EphA4 to promote trunk neural crest migration. Gene expression. 2002;10:295–305. doi: 10.3727/000000002783992389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mclennan R, Teddy JM, Kasemeier-Kulesa JC, Romine MH, Kulesa PM. Vascular endothelial growth factor (VEGF) regulates cranial neural crest migration in vivo. Developmental biology. 2010;339:114–125. doi: 10.1016/j.ydbio.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulemans D, Bronner-Fraser M. Gene-regulatory interactions in neural crest evolution and development. Developmental cell. 2004;7:291–299. doi: 10.1016/j.devcel.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Morton CA, Mackie RM. Clinical accuracy of the diagnosis of cutaneous malignant melanoma. The British journal of dermatology. 1998;138:283–287. doi: 10.1046/j.1365-2133.1998.02075.x. [DOI] [PubMed] [Google Scholar]
- Motohashi T, Yamanaka K, Chiba K, Miyajima K, Aoki H, Hirobe T, Kunisada T. Neural crest cells retain their capability for multipotential differentiation even after lineage-restricted stages. Dev Dyn. 2011;240:1681–1693. doi: 10.1002/dvdy.22658. [DOI] [PubMed] [Google Scholar]
- Nesbit M, Setaluri V, Herlyn M. Biology of Melanocytes and Melanoma. In: Balch CM, editor. Cutaneous melanoma. St. Louis, Mo.: Quality Medical Publishing; 1998. pp. xxii–596. [Google Scholar]
- Pasquale EB. Eph receptors and ephrins in cancer: bidirectional signalling and beyond. Nature reviews. 2010;10:165–180. doi: 10.1038/nrc2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–598. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigel DS, Friedman RJ, Kopf AW. The incidence of malignant melanoma in the United States: issues as we approach the 21st century. J Am Acad Dermatol. 1996;34:839–847. doi: 10.1016/s0190-9622(96)90041-9. [DOI] [PubMed] [Google Scholar]
- Roesch A, Fukunaga-Kalabis M, Schmidt EC, Zabierowski SE, Brafford PA, Vultur A, Basu D, Gimotty P, Vogt T, Herlyn M. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell. 2010;141:583–594. doi: 10.1016/j.cell.2010.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothhammer T, Poser I, Soncin F, Bataille F, Moser M, Bosserhoff AK. Bone morphogenic proteins are overexpressed in malignant melanoma and promote cell invasion and migration. Cancer research. 2005;65:448–456. [PubMed] [Google Scholar]
- Santiago A, Erickson CA. Ephrin-B ligands play a dual role in the control of neural crest cell migration. Development (Cambridge, England) 2002;129:3621–3632. doi: 10.1242/dev.129.15.3621. [DOI] [PubMed] [Google Scholar]
- Sechrist J, Serbedzija GN, Scherson T, Fraser SE, Bronner-Fraser M. Segmental migration of the hindbrain neural crest does not arise from its segmental generation. Development (Cambridge, England) 1993;118:691–703. doi: 10.1242/dev.118.3.691. [DOI] [PubMed] [Google Scholar]
- Smith A, Robinson V, Patel K, Wilkinson DG. The EphA4 and EphB1 receptor tyrosine kinases and ephrin-B2 ligand regulate targeted migration of branchial neural crest cells. Current biology : CB. 1997;7:561–570. doi: 10.1016/s0960-9822(06)00255-7. [DOI] [PubMed] [Google Scholar]
- Sommer L. Generation of melanocytes from neural crest cells. Pigment Cell Melanoma Res. 2011;24:411–421. doi: 10.1111/j.1755-148X.2011.00834.x. [DOI] [PubMed] [Google Scholar]
- Steventon B, Carmona-Fontaine C, Mayor R. Genetic network during neural crest induction: from cell specification to cell survival. Seminars in cell & developmental biology. 2005;16:647–654. doi: 10.1016/j.semcdb.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Strizzi L, Hardy KM, Seftor EA, Margaryan NV, Kirschmann DA, Kirsammer GT, Bailey CM, Kasemeier-Kulesa JC, Kulesa PM, Seftor REB, et al. In: Lessons from Embryogenesis Melanoma Development. Bosserhoff A, editor. Vienna: Springer; 2011. pp. 281–296. [Google Scholar]
- Taneyhill LA, Bronner-Fraser M. Dynamic alterations in gene expression after Wnt-mediated induction of avian neural crest. Mol Biol Cell. 2005;16:5283–5293. doi: 10.1091/mbc.E05-03-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneyhill LA, Coles EG, Bronner-Fraser M. Snail2 directly represses cadherin6B during epithelial-to-mesenchymal transitions of the neural crest. Development (Cambridge, England) 2007;134:1481–1490. doi: 10.1242/dev.02834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt T, Stolz W, Welsh J, Jung B, Kerbel RS, Kobayashi H, Landthaler M, Mcclelland M. Overexpression of Lerk-5/Eplg5 messenger RNA: a novel marker for increased tumorigenicity and metastatic potential in human malignant melanomas. Clinical cancer research : an official journal of the American Association for Cancer Research. 1998;4:791–797. [PubMed] [Google Scholar]
- Welch DR, Bisi JE, Miller BE, Conaway D, Seftor EA, Yohem KH, Gilmore LB, Seftor RE, Nakajima M, Hendrix MJ. Characterization of a highly invasive and spontaneously metastatic human malignant melanoma cell line. International journal of cancer. 1991;47:227–237. doi: 10.1002/ijc.2910470211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.