Abstract
The bacterial species Serratia marcescens secretes both beneficial and cytotoxic proteins. Here we report that a crp mutant exhibited elevated secreted protease activity. A genetic screen revealed that the gene coding for the metalloprotease serralysin was necessary for the elevated proteolysis, and this was confirmed by western blot analysis. Proteomic analysis of secreted proteins corroborated increased secretion of serralysin protease by crp mutants compared to the wild type. The crp-mutant-secreted fractions also contained less chitinase and chitin binding protein. These data support the hypothesis that cAMP-CRP is an upstream indirect regulator of serralysin production and they provide novel insight into the S. marcescens secretome.
Keywords: Secretome, Metalloprotease, Proteomics, Cyclic AMP, Chitinase, Flagella
1. Introduction
Serratia marcescens has been used as a model organism for the study of bacterial protein secretion and causes opportunistic and ocular infections (Binet et al., 1997; Hejazi and Falkiner, 1997; Hume and Willcox, 2004; Letoffe et al., 1991; Marty et al., 2002; Murdoch et al., 2011; Rao et al., 2011). This member of the Enterobacteriaceae family secretes a number of exoenzymes including a nuclease, chitinase, lipase/esterases, phospholipases and proteases (Akatsuka et al., 1995; Benedik and Strych, 1998).
Many of the characterized proteins that are secreted by S. marcescens are exported through type one secretion systems (T1SSs) that are characterized by an inner-membrane spanning ATP binding cassette protein that complexes with a protein that spans the periplasm and an outer membrane protein of the TolC family (Binet et al., 1997). Some of the proteins that are secreted this way are the HasA hemophore that is secreted by a T1SS composed of the HasD, HasE and HasF proteins. HasA interacts with the SecB chaperone prior to being secreted by HasDEF, but does not use the Sec system to cross the inner membrane (Sapriel et al., 2003). The lipase LipA, surface-layer protein (SlaA) and metalloprotease PrtS (also referred to as PrtA and serralysin) are secreted through another T1SS apparatus composed of LipB, LipC and LipD (Akatsuka et al., 1997).
The large surface-associated S. marcescens pore-forming hemolysin, ShlA, is secreted by the two partner secretion system (type V secretion) and is cotranscribed with its secretory partner ShlB (Hertle, 2005). S. marcescens has a type VI secretion system that may be of key importance in microbe-microbe interactions (Murdoch et al., 2011; Rao et al., 2011).
Secretion mechanisms of other S. marcescens exoenzymes are incompletely characterized. Chitinase A (ChiA) has an N-terminal signal sequence, whereas chitinase B (ChiB) does not, suggesting that these proteins may be secreted through different pathways (Watanabe et al., 1997). There is evidence that ChiB is exported to the periplasm and is only detected in culture medium through cell lysis (Brurberg et al., 1995). Data support the theory that the nuclease protein, NucA, crosses the inner membrane by the Sec system, but it is unknown how it passes through the outer membrane (Suh et al., 1995). The phoshoplipase A (PhlA) protein was reported to be secreted only in cells with an intact flagellum assembly system, suggesting that PhlA may be secreted by flagellar components (Givskov and Molin, 1993). Furthermore, Sec and Twin-Argnine translocation system proteins have not been characterized from S. marcescens.
Whereas some of the secretion machinery for S. marcescens has been characterized, very little is known about the transcription factors that regulate secreted enzymes by S. marcescens (Ball et al., 1990). Our study of the role of cAMP-CRP in controlling S. marcescens surface structures revealed that mutation of crp conferred a hyper-protease phenotype that had not been previously reported. CRP is a global regulator and a member of the highly conserved CRP-FNR family of transcription factors. CRP-FNR family regulators act mainly as positive regulators of gene expression, but can also act as negative regulators (Green et al., 2001). The DNA binding affinity and specificity of CRP is regulated through binding of cAMP (Heyduk and Lee, 1989) that is made by adenylate cyclase (CyaA) in response to environmental input, especially the extracellular glucose concentration (Ishizuka et al., 1993; Kalivoda et al., 2008).
We have previously shown that S. marcescens with mutations in cyaA and crp have highly elevated levels of type one pili and severely reduced flagella production and that these effects are mediated at the transcriptional level (Kalivoda et al., 2008; Kalivoda et al., 2010; Stella et al., 2008). It has also been shown that transcription from the S. marcescens phlA promoter is regulated by cAMP-CRP in Escherichia coli (Givskov and Molin, 1992), and there are reports in the literature of other CRP-FNR family members regulating individual secreted proteins by other organisms (Huang et al., 2007; Martìnez-Cadena et al., 1981; Reverchon et al., 1989; West et al., 1994).
In this study we report that crp and cyaA mutants demonstrated elevated levels of secreted proteolysis. The goal of this study was to determine the identity of the protease upregulated in the crp and cyaA mutants.
2. Materials and methods
2.1. Bacterial strains and growth conditions
All bacteria were grown in lysogeny broth (LB) or brain heart infusion (BHI) as noted. Experiments were performed at 30 °C. Gentamicin was used at ten μg/ml, kanamycin was used at 100 μg/ml and tetracycline was used at ten μg/ml where appropriate. All S. marcescens strains were derived from PIC 361 (Presque Island Culture Collection) and are listed in Table 1. The cyaA and crp mutants described herein grow at nearly identical rates as the wild-type (WT) strain in LB broth (Kalivoda et al. 2008).
Table 1. S. marcescens.
strains, plasmids and primers used in this study.
| Name | Description | Source |
|---|---|---|
| CMS376 | Wild-type strain PIC 361 (Presque Island Culture Collection) | (Kalivoda et al., 2008) |
| CMS524 | CMS376 with cyaA-2 mutation, transposon insertion | (Kalivoda et al., 2008) |
| CMS613 | CMS376 with crp-1 mutation, insertion mutation | (Kalivoda et al., 2008) |
| CMS786 | CMS376 with crp-23 mutation, transposon insertion | (Kalivoda et al., 2008) |
| CMS1687 | CMS376 with crp-34 mutation, deletion mutation | (Kalivoda et al., 2010) |
| JF1C6 | CMS613 with prtS::tn, transposon insertion | This study |
| JF1C6r | JF1C6 with crp gene restored to wild-type | This study |
| C23M13 | CMS with lipD::tn, transposon insertion | This study |
| JF2E10r | CMS376 with hyper-PrtS phenotype | This study |
| Plasmids | ||
| pBT20 | Mariner transposon-delivery vector | (Kulasekara et al., 2005) |
| pMQ131 | oripBBR1, ahpA-3, oriT, URA3, CEN6/ARSH4 | (Shanks et al., 2009) |
| pMQ157 | pMQ131 + cyaA | (Kalivoda et al., 2008) |
| pMQ166 | pMQ131 + crp | (Kalivoda et al., 2010) |
| Primer number | Sequence | Target |
| 2638 | AACTGGAGGAAGGTGGGGAT | 16S-rDNA |
| 2639 | AGGAGGTGATCCAACCGCA | 16S-rDNA |
| 2900 | TACAACGTTGCGCAGAACTC | chiA |
| 2901 | GTAGTCCACGATGCCGTTCT | chiA |
| 2903 | AAAACTTCCCGTACCCTGCT | chp21 |
| 2904 | GTTCGGCTTGGTGATGAAAT | chp21 |
| 2970 | CTCTCCGTAGAAGGCGTGAC | lipB |
| 2971 | GTTAGGGAAACGCAGGATCA | lipB |
| 2973 | TGTTCGCCTTTTTCGATACC | hasD |
| 2974 | TGTTCGCCTTTTTCGATACC | hasD |
Transposon mutagenesis and mapping were performed as previously described (Kalivoda et al., 2008). Briefly, a crp mutant strain was mutagenized with a mariner-based transposon and the resulting mutants were selected on protease detection plates containing gentamicin (ten μg/ml) and tetracycline (ten μg/ml). One mutant strain, JF1C6, with severely reduced extracellular protease activity was identified.
2.2. Secreted protein fractions
To obtain secreted protein fractions, bacteria were grown in LB in 5 ml aliquots in test tubes on a TC-7 tissue culture roller (New Brunswick Inc.) set at a speed setting of 8. The culture turbidity (OD600 nm) was measured and cultures were normalized to an OD600 of 2.0 using LB. Cells were then removed by centrifugation and filtration through a 0.22 micron filter (Millex GV, Millipore). Bradford analysis was performed to measure protein concentration using bovine serum albumin (BSA, Fisher Scientific BP1600) as a standard. The protein concentration from medium alone was negligible (data not shown). Four independent biological replicates were used per genotype per experiment, and the experiment was performed twice on different days.
Two-dimensional difference gel electrophoresis (2D-DIGE) was performed on pooled secreted fractions as follows. Ten single colonies for each genotype were grown in ten different test tubes, each with 5 ml of LB. After 24 h, cultures were vortexed for 5–10 s at maximum speed, pooled, gently sonicated for 15 s to break up any cell clumps (Fisher Scientific Sonic Dismembrator Model 100, power level three), the culture turbidity was determined (OD600 nm), and the cultures were normalized by the addition of LB to OD600 nm = 2.0. Bacteria were pelleted by centrifugation and the supernatant was passed through a 0.22 micron filter. BSA (5 μg/ml) was added to WT and crp mutant filtered supernatants as an internal loading control. Protein fractions were packed on dry ice and sent to Applied Biomics (Hayward, CA) for processing. 2D-DIGE and protein identification were performed as previously described (Tshala-Katumbay et al., 2008). Protein spot analysis was performed with DeCyder 2D software and the ratio for each protein was determined. Proteins were identified by MALDI-TOF mass spectrometry. The experiment was performed on three different occasions.
2.3. Extracellular enzyme assays
Protease detection plates consisted of BHI agar supplemented with skim milk at 1% (w/v). To measure secreted protease activity from liquid cultures, sterile supernatants were prepared. Bacteria were grown for 18 h at 30 °C with aeration in LB (with or without cAMP that was dissolved directly in LB medium to the desired concentration and filter-sterilized). Stationary phase cultures (OD600 = 5–6) were normalized to OD600=2.0 using fresh LB, and were tested for protease activity using azocasein according to the method of Cruz-Romero et al. (Cruz-Romero et al., 2008). Protease zymograms with casein as a substrate were obtained from Bio-Rad and performed according to the specifications of the manufacturer using 15 μl of filter-sterilized stationary phase cultures as noted above.
DNase activity was tested using DNase detection plates containing methyl green (Difco). Plates were incubated at 30 °C for 24 h and clear zones were measured.
Secreted chitinase activity was assessed using chitin-azure that was acid-hydrolyzed (Shen et al., 2010) and suspended in LB agar to 0.08% (w/v). Bacteria grown on LB plates were patched onto the chitin-azure plates and incubated for seven days at 30 °C. Zones of clearing around colonies indicate secreted chitinase activity.
Secreted lipase activity was tested using LB agar supplemented with tributyrin and polysorbate 80 (Tween-80) at 1% (v/v), similar to Akatsuka, et. al. (Akatsuka et al., 2003).
2.4. RNA purification and semi-quantitative RT-PCR
Cultures were grown overnight in LB medium, subcultured to OD600 = 0.1, grown to OD600 = 0.8, subcultured to OD600 = 0.1 and allowed to grow to OD600 = ~4.0. RNA was isolated as described by Wargo et al, (Wargo et al., 2008). RNA (250 ng) was used in each reverse transcriptase (RT) reaction using Superscript III RT (Invitrogen) as prescribed by the manufacturers. A standard PCR reaction with 30 cycles of 20 s at 94 °C, 20 s at 55 °C and 30 s at 72 °C was performed to amplify products. No-reverse transcriptase and no-RNA controls were performed and showed no contaminating DNA (data not shown). The experiment was repeated 3 times with independent RNA samples with consistent results. Primer sequences are listed in Table 1, with 16S primer sequences taken from Lin and colleagues (Lin et al., 2010).
2.5. Immunoblots
Western blot analysis was performed as previously described (Shanks et al., 2012), using filtered secreted fractions from spent supernatant that had been normalized to OD600 = 2.0. A polyclonal anti-serralysin (PrtS) antibody was used to detect serralysin (Letoffe et al., 1991). Horseradish peroxidase-conjugated secondary antibodies were used for detection of primary antibodies and exposed to X-ray film for 60 s. Immunoblot experiments were performed three times on different days using independent samples. Exposed film was scanned, and the density of bands was quantified using Image-J software (NIH).
3. Results
3.1. cAMP-CRP control of secreted protease activity and identification of the protease
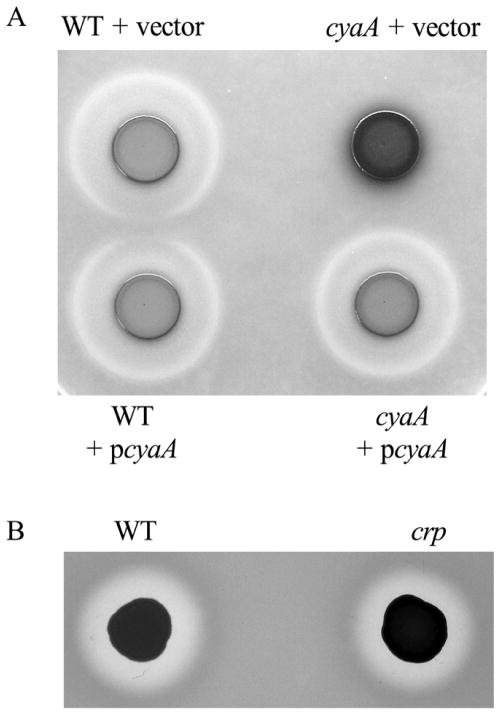
Compared to the WT strain, crp and cyaA mutant strains produced larger zones of activity on BHI-based protease detection plates (Fig. 1A). The cyaA mutant used in this study was previously shown to be deficient in cAMP production and the crp mutant was unable to respond to cAMP (Kalivoda et al., 2008; Kalivoda et al., 2010; Stella et al., 2008). As with the plate-based proteolysis assay, more proteolytic activity was evident using filtered supernatants from cultures grown in culture medium from the crp (Fig. 1B–lane 3) and cyaA (Fig. 1C) mutants than from the wild-type (Fig. 1B, lane 1 and Fig. 1C). Addition of wild-type crp and cyaA genes on a plasmid to the crp and cyaA mutant, respectively, or addition of exogenous cAMP to the cyaA growth medium restored wild type levels of proteolysis (data not shown and Fig. 1C). These data support the notion that S. marcescens secreted protease production is inhibited by cAMP-CRP.
Fig. 1. Control of secreted protease activity by cAMP-CRP.
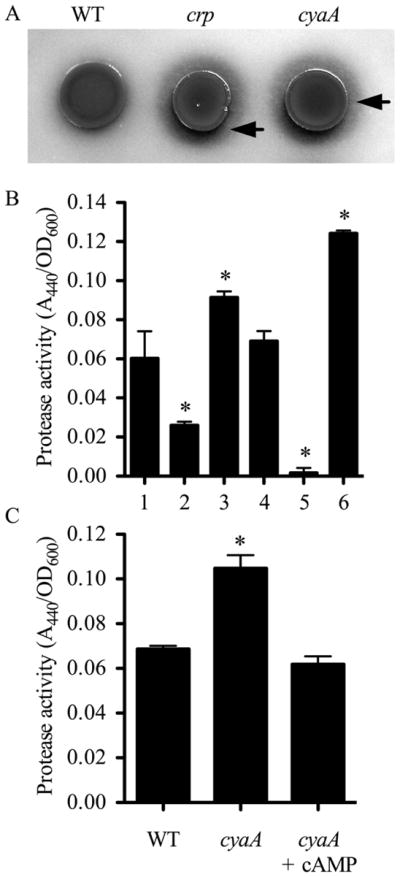
A. Protease detection plate shows larger zone of clearing (arrows) around the cyaA (CMS524) and crp mutants (CMS786) compared to the isogenic WT strain (CMS376). B. Digestion of azocasein by normalized filtered supernatants. 1. WT (CMS376); 2. prtS (JF1C6r); 3. crp (CMS613); 4. crp prtS (JF1C6); 5. crp lipD (C23M13); 6. prtS over-expression strain (JF2E10r). C. Digestion of azocasein by normalized filtered supernatants. The cyaA mutant (CMS524) was grown in growth medium without or with cAMP (5 mM). Exogenous cAMP restored WT levels of proteolysis. Asterisks indicate a significant difference from WT (CMS376), ANOVA with Tukey’s post-test (p<0.05). Error bars equal one standard deviation.
S. marcescens strains have been reported to secrete multiple proteases of different classes ( Matsumoto et al., 1984; Matsumoto, 2004). To identify the protease with increased activity in the crp mutant, a genetic screen was performed. One mutant, JF1C6, was identified with reduced extracellular protease activity (Fig. 1B-lane 4) compared to the crp mutant (Fig. 1B-lane 3). The transposon in JF1C6 was mapped to the gene for metalloprotease serralysin prtS at base pair 711 out of 1515. Serralysin is a cytotoxic protease that is sufficient to cause keratitis in an animal model (Marty et al., 2002; Matsumoto, 2004). Targeted mutation of the serralysin gene in the crp mutant background similarly reduces secreted protease activity (data not shown). Mutation of prtS in the wild-type background yielded a defect in secreted protease activity (Fig. 1B-lane 2, strain JF1C6r). Another protease-defective mutant, C23M13, had a transposon insertion in the lipD gene, which codes for part of the type 1 secretion system that secretes serralysin (Fig. 1B-lane 5). The crp lipD double mutant exhibited even lower extracellular protease activity than the prtS mutant, suggesting that the LipBCD T1SS secretes multiple proteases and this prediction was supported by zymogram analysis (data not shown).
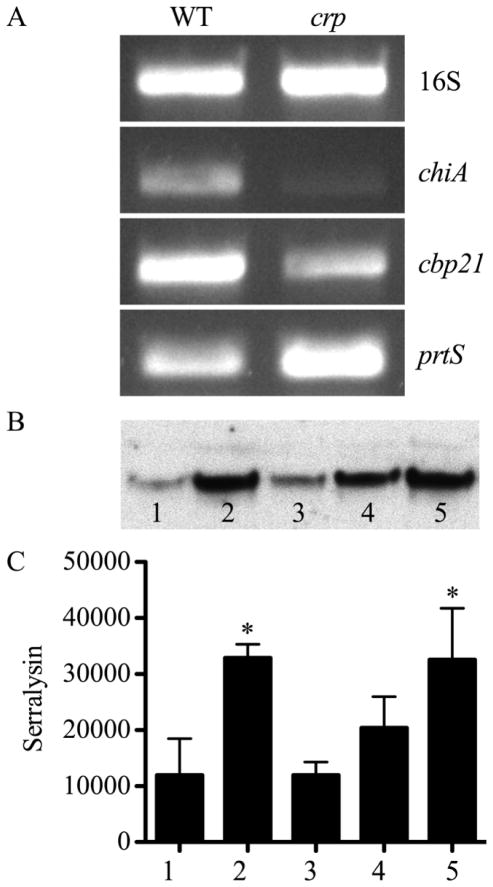
Analysis of the prtS transcript supports the fact that it is more abundant in the crp mutant (Fig. 2A). To verify that the altered levels of prtS transcript resulted in increased serralysin levels, immunoblot analysis was performed using antibodies against serralysin (Fig. 2B–C). A significant increase in serralysin production was observed relative to WT (Fig. 2B and C, lane 1) in both cyaA (Fig. 2B and C, lane 2) and crp (Fig. 2B and C, lane 5) mutants. Complementation of the cyaA mutant with the wild-type cyaA gene expressed from a plasmid (Fig. 2B and C, lane 3) reduced the hyper-serralysin levels of cyaA mutants to levels indistinguishable from the WT. Similarly, exogenous cAMP (5 mM) decreased the serralysin levels in the cyaA mutant (Fig. 2B and C, lane 4), but not to wild-type levels. Together, these data support the model that cAMP-CRP inhibits production of serralysin.
Fig. 2. Control of serralysin production by cAMP-CRP.
A. Representative semi-quantitative RT-PCR of WT (CMS376) and crp (CMS1687) cultures including the 16S rDNA amplicon to indicate equal starting RNA concentrations. B. Immunoblot of normalized secreted fractions with anti-serralysin antibody. C. Quantification of anti-serralysin immunoblots with bands measured by densitometry. B–C. 1. WT with empty vector (pMQ131); 2. cyaA mutant with empty vector (pMQ131); 3. cyaA mutant with cyaA on a plasmid (pMQ157); 4. cyaA mutant grown with exogenous cAMP (5 mM); 5. crp mutant. Asterisks indicate a significant difference from WT with empty vector (pMQ131), ANOVA with Tukey’s post-test (p<0.05). The average and standard deviation of three independent experiments are shown.
3.2. Impact of the crp mutation on protein secretion
To differentiate whether mutation of crp confers a serralysin-specific effect or a pleiotropic effect on secreted protein production, we measured total protein levels in WT and a Δcrp strain grown in LB broth for 24 h. Importantly, there was very little difference in growth between these wild-type and the crp mutant strains in LB medium (Kalivoda et al., 2008). The secreted protein concentration was found to be significantly higher in the WT than in the isogenic Δcrp mutant (Fig. 3A). It is possible that the reduction in secreted protein concentration of the crp mutant relative to the WT is due to increased production of serralysin that hydrolyzes other extracellular proteins. To test this prediction, protein concentrations were measured from a prtS mutant and an isogenic crp prtS double mutant, such that secreted PrtS could not influence the outcome. We observed a similar pattern to that of the PrtS-positive strains where the prtS supernatants had significantly more protein than the crp prtS strain (Fig. 3B).
Fig. 3. Effect of crp mutation on secreted proteins.
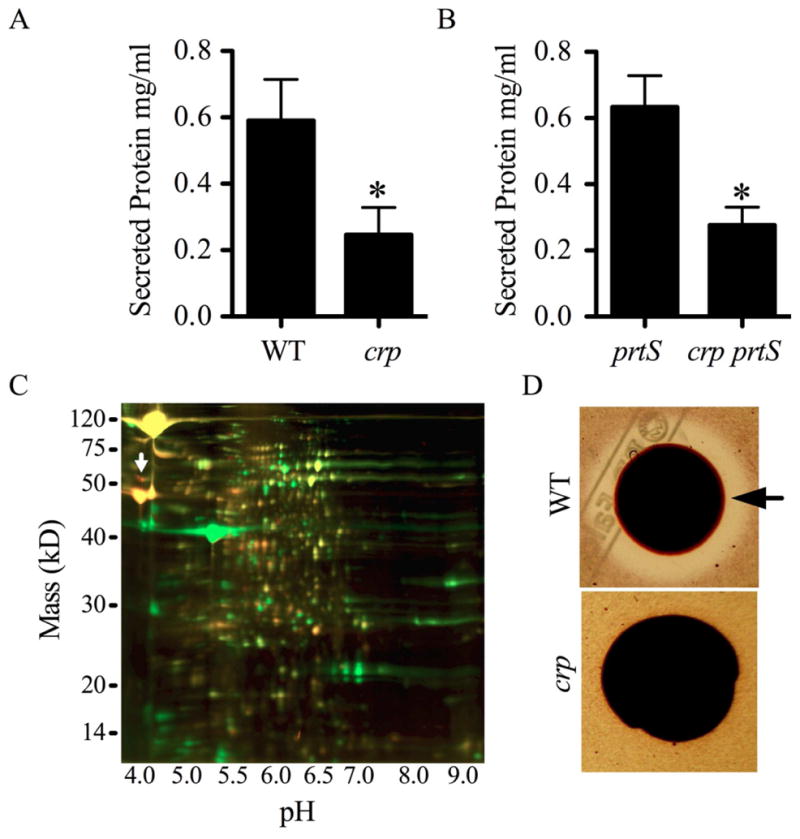
A. Concentration of secreted proteins in spent supernatants normalized by culture density. B. Protein concentrations measured from secreted fractions of isogenic prtS (JF1C6r) and crp prtS (JF1C6) mutant strains. C. Representative 2-D DIGE of secreted proteins; WT = green, crp = red. The white arrow indicates serralysin protease (PrtS). D. Representative chitinase detection plate shows a large zone of clearing around the WT (CMS376) colony (arrow), and an absence of detectable chitinase activity by the crp mutant (CMS1687). Asterisks indicate a significant difference from the WT or prtS by two-tailed Student’s T-test (p<0.05). Error bars equal one standard deviation.
As another control for the effect of protease production on secreted protein levels, we utilized a mutant strain with highly elevated serralysin levels, JF2e10r (Fig. 1B –lane 6). This mutant strain, in the WT background, has a transposon just upstream of the prtS gene with an outward-facing Ptac promoter yielding elevated transcription of prtS. We tested the effect of this mutation in prtS transcription using a lacZ reporter and found that it was expressed ~30-fold higher in JF2e10r compared to the WT using a chromosomal lacZ fusion (data not shown). Secreted protein levels from JF2e10r were similar to the WT (0.63 mg/ml for the WT and 0.74 mg/ml for JF2e10r, p = 0.48, n = 6). Together, these data suggest that the crp-associated secreted protein defect is not due to elevated protease activity degrading extracellular proteins.
2D-DIGE was carried out on the stationary phase secreted protein fraction (secretome) of WT and Δcrp strains to analyze the difference in secretion with greater resolution. The experiment was independently performed three times with consistent results. Forty-three protein spots were measured with ≥2-fold elevated expression in the WT strain (green) relative to the Δcrp mutant (red) (Fig. 3C).
A subset of spots with the most highly altered expression patterns were excised and identified by matrix-assisted laser desorption/ionization-mass spectometry (MALDI-TOF-MS) (Table 2). The protein with the greatest difference between the WT and the Δcrp mutant was the flagellar subunit flagellin. While this structure is surface-attached, flagella are easily sheared off the surface. As noted above, it has previously been shown that S. marcescens crp mutants do not make flagella, as cAMP-CRP directly and positively regulates transcription of the flagella master regulator operon flhDC (Stella et al., 2008). Other proteins more highly expressed by the WT included three different chitinases, chitin binding protein 21 and the ShlA hemolysin, a known virulence factor (Lin et al., 2010). Several other spots were identified and are predicted to be transmembrane, periplasmic or cytoplasmic proteins, suggesting that some proteins from lysed bacteria were found in the spent supernatant that may be a result of sample preparation. These include the following predicted proteins with the relative expression of the protein found in the crp to WT strains shown in parentheses: urocanate hydratase (−4.0 fold), glutamate transporter subunit (−4.2 fold), histidine-binding periplasmic protein (−5.8 fold), arginine-binding periplasmic protein (−7.6 fold), uridine phosphorylase (−7.6 fold), phosphoenolypyruvate carboxykinase (−8.0 fold), putative trehalose-6-phosphate hydrolase (−8.5 fold), 2-nitropropane dioxygenase-like protein (−15.9 fold) and a putative aminotransferase protein (−17.8 fold).
Table 2.
Secreted and surface proteins with altered production in a crp mutant. Protein Fold change (crp/WT)a
| Protein | Fold change (crp/WT)a |
|---|---|
| Serralysin | +2.22 |
| Chitinase C1 | −4.66 |
| ShlA hemolysin | −5.76 |
| Chitinase A | −10.26 |
| Chitinase B | −9.84 |
| Chitinase binding protein CBP21 | −11.51 |
| Flagellin | −83.68 |
| Flagellar hook protein | −14.40 |
Relative abundance of proteins with differential expression, calculated from the average of three 2D-DIGE gels from independent samples with consistent results, except for ShlA hemolysin, where the protein spot was present in only one gel, and the flagellar hook protein, which is an average from two gels. The relative abundance of each protein was first normalized to an internal loading control.
Chitinase indicator plates were used to verify the result that more chitinases were found in the WT supernatant. Secreted chitinase activity was clearly observed around WT colonies and absent around the Δcrp mutant (Fig. 3D). To further verify these results, we tested whether crp mutants had reduced transcription of genes for two sample chitin-related proteins. We observed that crp mutants had reduced levels of RNA transcript for chitinase A (chiA) and chitinase binding protein 21 (cbp21) relative to the WT (Fig. 2A).
The secreted protein with the highest expression in Δcrp mutant filtrates compared to the WT was identified by mass spectrometry as the serralysin metalloprotease (Table 2 and Fig. 3C, white arrow). This result is consistent with the genetic screen, western blot analysis and semi-quantitative RT-PCR described above.
Other secreted enzymes were tested to determine whether mutation of crp modified their production. S. marcescens extracellular lipase activity is a product of the LipA protein that uses the same type I secretion apparatus (LipBCD) that mediates serralysin protease secretion (Akatsuka et al., 1997). We noted that, whereas secreted protease activity increases for the crp and cyaA mutant strains, extracellular lipase activity is absent (Fig. 4A and data not shown). RT-PCR analysis of lipB and hasD expression from cells at a culture density of OD600=1.5 and 3.5 suggested that there is no significant difference in the expression of type 1 secretion systems between WT and Δcrp (data not shown). Unlike protease and lipase activity, we observed identical zones of secreted nuclease activity around crp and WT colonies using DNase detection agar (Fig. 4B).
Fig. 4. Effect of crp mutation on secreted lipase and nuclease activity.
A. Secreted lipase detection agar. The cyaA mutant (CM524) is defective in lipase activity and complemented by the wild-type cyaA gene in trans (pMQ157). B. Secreted nuclease activity (clear zone around colonies) is indistinguishable between the WT (CMS376) and crp mutant (CMS786).
4. Discussion
The major finding of this study was that mutation of crp led to elevated production of secreted protease activity that was correlated with increased transcript and protein levels of the metalloprotease serralysin/PrtS. Genetic analysis indicated that the gene for the serralysin protease, prtS, is required for the hyper-protease phenotype of a crp mutant. Immunoblots confirm that secreted serralysin levels were indeed responsive to cAMP levels and the CRP status of the strain, and serralysin was found to be upregulated in 2-D gel analysis of secreted proteins in crp mutant secreted fractions. This finding is of interest because serralysin and serralysin-like metalloproteases have been identified and shown to mediate host-pathogen interactions in a number of medically and agriculturally relevant bacteria (Basu and Apte, 2008; Felfoldi et al., 2009; Kumeta et al., 1999; Louis et al., 1998; Maeda and Morihara, 1995; Marokhazi et al., 2007; Massaoud et al., 2011; Matsumoto et al., 1998). The S. marcescens serralysin was shown to be cytotoxic to mammalian cells, to facilitate the invasion of bacteria into mammalian cells and to be sufficient to induce keratitis in a rabbit ocular pathogenesis model (Kamata et al., 1985; Matsumoto, 2004). However, strains of S. marcescens that did not exhibit extracellular protease activity in vitro were still capable of causing keratitis (Hume et al., 1999), suggesting that either extracellular protease activity is not a strict requirement for corneal infection or that these strains produced protease activity under in vivo conditions. Furthermore, no transcriptional regulators of serralysin production have previously been reported for S. marcescens. Subsequent experiments will focus on characterizing regulation of serralysin production by cAMP-CRP.
Another finding of this study is that CRP appears to be a major factor in regulation of secreted proteins, with a ~50% decrease in the amount of overall protein in the spent supernatants of crp mutants compared to the WT. Lower concentrations of secreted proteins were found in supernatants of crp mutants and two-dimensional elecrophoretic separation indicated that many proteins have differential production in the crp mutant. Whereas most proteins were measured at reduced concentrations in the crp mutant supernatant, the serralysin metalloprotease levels were increased, further supporting our genetic and immunoblot studies indicating that serralysin levels are increased in the crp mutant. Given the importance of the highly conserved CRP transcription factor, it was surprising that the effect of crp mutation on the general secretome has not been thoroughly explored. However, Fox and colleagues isolated secreted protease-defective mutants of Pseudomonas aeruginosa with a point mutation in the vfr gene, a homolog of crp (Fox et al., 2008). Interestingly, the vfr mutant exhibited reduced expression of chitin binding protein D and three protease/peptidases including elastase B and protease IV. These observations from both P. aeruginosa and S. marcescens suggest a conserved role for CRP family proteins in regulation of secreted proteins among bacterial species from different orders.
The observation that two different secreted enzymes (LipA and PrtS) that use the same type I secretion system exhibit either eliminated or elevated secretion, respectively, from the crp mutant relative to the WT suggests that the phenotypes are not due to an alteration of LipBCD-T1SS function. Consistently, RT-PCR analysis showed no difference in lipB expression between the WT and Δcrp mutant strain. It must be noted that the lipase zones present around the WT strain and absent around the cyaA mutant in Fig. 4 were not proven to be from LipA and may be due to the activity of another lipase.
Furthermore, the observation that the extracellular nuclease is secreted at similar levels in the WT and crp strains suggests that there is not a general defect in the Sec-based secretion across the inner membrane. Furthermore, the transcript levels of genes for several secreted proteins correlated with altered protein levels, e.g. there was a decrease in chitin binding protein 21 protein levels and cbp21 transcript levels. Together, our data support a model where cAMP-CRP is directly or indirectly responsible for the transcription of several secreted proteins. Non-exclusive alternative models include: 1) CRP could positively regulate a secretion system factor, the absence of which elicits lower overall secreted protein levels; and 2) crp mutants may be less likely to lyse, leading to reduced overall protein levels in culture supernatants.
Proteomic analysis confirmed previous studies indicating that flagellin requires CRP for expression (Stella et al. 2008), and it was expected that the ShlA hemolysin should be detected at reduced levels in a crp mutant, as it has been shown that CRP directly and positively regulates the transcription factor complex FlhDC (Stella et al. 2008; Kalivoda et al. 2010) and that FlhDC positively regulates expression of shlA (Lin et al., 2010).
A noteworthy outcome of this study was that several proteins involved in chitin degradation were isolated as differentially expressed in a crp mutant. Supporting the reduction in chitinase-metabolizing enzymes, we observed that crp mutants were defective in hydrolysis of chitin azure. The implication of this observation is that the production of chitin-degrading proteins, important in some biofuel production strategies, may be induced by manipulating the cAMP-CRP activity of chitinase-producing microorganisms.
Acknowledgments
The authors thank Kim Brothers, Daniel Kadouri and Roni Lahr for critical reading of the manuscript, Jean-Marc Ghigo and the Wandersman Laboratory for the kind gift of the protease antibody, and John Liao at Applied Biomics for 2D-DIGE analysis. This study was funded by NIH grant AI085570 and a Research to Prevent Blindness Career Development Award to RS. Other funding sources were NIH grant EY08098, the Eye and Ear Foundation of Pittsburgh and unrestricted funds from Research to Prevent Blindness.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akatsuka H, Binet R, Kawai E, Wandersman C, Omori K. Lipase secretion by bacterial hybrid ATP-binding cassette exporters: molecular recognition of the LipBCD, PrtDEF, and HasDEF exporters. J Bacteriol. 1997;179:4754–60. doi: 10.1128/jb.179.15.4754-4760.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akatsuka H, Kawai E, Omori K, Shibatani T. The three genes lipB, lipC, and lipD involved in the extracellular secretion of the Serratia marcescens lipase which lacks an N-terminal signal peptide. J Bacteriol. 1995;177:6381–9. doi: 10.1128/jb.177.22.6381-6389.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akatsuka H, Kawai E, Sakurai N, Omori K. The Serratia marcescens bioH gene encodes an esterase. Gene. 2003;302:185–92. doi: 10.1016/s0378111902011502. [DOI] [PubMed] [Google Scholar]
- Ball TK, Wasmuth CR, Braunagel SC, Benedik MJ. Expression of Serratia marcescens extracellular proteins requires recA. J Bacteriol. 1990;172:342–9. doi: 10.1128/jb.172.1.342-349.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu B, Apte SK. A novel serralysin metalloprotease from Deinococcus radiodurans. Biochim Biophys Acta. 2008;1784:1256–64. doi: 10.1016/j.bbapap.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Benedik MJ, Strych U. Serratia marcescens and its extracellular nuclease. FEMS Microbiol Lett. 1998;165:1–13. doi: 10.1111/j.1574-6968.1998.tb13120.x. [DOI] [PubMed] [Google Scholar]
- Binet R, Letoffe S, Ghigo JM, Delepelaire P, Wandersman C. Protein secretion by Gram-negative bacterial ABC exporters--a review. Gene. 1997;192:7–11. doi: 10.1016/s0378-1119(96)00829-3. [DOI] [PubMed] [Google Scholar]
- Brurberg MB, Eijsink VG, Haandrikman AJ, Venema G, Nes IF. Chitinase B from Serratia marcescens BJL200 is exported to the periplasm without processing. Microbiology. 1995;141:123–31. doi: 10.1099/00221287-141-1-123. [DOI] [PubMed] [Google Scholar]
- Cruz-Romero M, Kelly AL, Kerry JP. Influence of packaging strategy on microbiological and biochemical changes in high-pressure-treated oysters. J Sci Food Agric. 2008;88:2713–23. [Google Scholar]
- Felfoldi G, Marokhazi J, Kepiro M, Venekei I. Identification of natural target proteins indicates functions of a serralysin-type metalloprotease, PrtA, in anti-immune mechanisms. Appl Environ Microbiol. 2009;75:3120–6. doi: 10.1128/AEM.02271-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox A, Haas D, Reimmann C, Heeb S, Filloux A, Voulhoux R. Emergence of secretion-defective sublines of Pseudomonas aeruginosa PAO1 resulting from spontaneous mutations in the vfr global regulatory gene. Appl Environ Microbiol. 2008;74:1902–8. doi: 10.1128/AEM.02539-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givskov M, Molin S. Expression of extracellular phospholipase from Serratia liquefaciens is growth-phase-dependent, catabolite-repressed and regulated by anaerobiosis. Mol Microbiol. 1992;6:1363–74. doi: 10.1111/j.1365-2958.1992.tb00857.x. [DOI] [PubMed] [Google Scholar]
- Givskov M, Molin S. Secretion of Serratia liquefaciens phospholipase from Escherichia coli. Mol Microbiol. 1993;8:229–42. doi: 10.1111/j.1365-2958.1993.tb01567.x. [DOI] [PubMed] [Google Scholar]
- Green J, Scott C, Guest JR. Functional versatility in the CRP-FNR superfamily of transcription factors: FNR and FLP. Adv Microb Physiol. 2001;44:1–34. doi: 10.1016/s0065-2911(01)44010-0. [DOI] [PubMed] [Google Scholar]
- Hejazi A, Falkiner FR. Serratia marcescens. J Med Microbiol. 1997;46:903–12. doi: 10.1099/00222615-46-11-903. [DOI] [PubMed] [Google Scholar]
- Hertle R. The family of Serratia type pore forming toxins. Curr Protein Pept Sci. 2005;6:313–25. doi: 10.2174/1389203054546370. [DOI] [PubMed] [Google Scholar]
- Heyduk T, Lee JC. Escherichia coli cAMP receptor protein: evidence for three protein conformational states with different promoter binding affinities. Biochemistry. 1989;28:6914–24. doi: 10.1021/bi00443a021. [DOI] [PubMed] [Google Scholar]
- Huang XD, Yao K, Zhang H, Huang XJ, Xu ZK. Surface modification of silicone intraocular lens by 2-methacryloyloxyethyl phosphoryl-choline binding to reduce Staphylococcus epidermidis adherence. Clin Experiment Ophthalmol. 2007;35:462–7. doi: 10.1111/j.1442-9071.2007.01516.x. [DOI] [PubMed] [Google Scholar]
- Hume EB, Conerly LL, Moreau JM, Cannon BM, Engel LS, Stroman DW, Hill JM, O’callaghan RJ. Serratia marcescens keratitis: strain-specific corneal pathogenesis in rabbits. Curr Eye Res. 1999;19:525–32. doi: 10.1076/ceyr.19.6.525.5283. [DOI] [PubMed] [Google Scholar]
- Hume EB, Willcox MD. Emergence of Serratia marcescens as an ocular surface pathogen. Arch Soc Esp Oftalmol. 2004;79:475–7. [PubMed] [Google Scholar]
- Ishizuka H, Hanamura A, Kunimura T, Aiba H. A lowered concentration of cAMP receptor protein caused by glucose is an important determinant for catabolite repression in Escherichia coli. Mol Microbiol. 1993;10:341–350. doi: 10.1111/j.1365-2958.1993.tb01960.x. [DOI] [PubMed] [Google Scholar]
- Kalivoda EJ, Stella NA, Aston MA, Fender JE, Thompson PP, Kowalski RP, Shanks RM. Cyclic AMP negatively regulates prodigiosin production by Serratia marcescens. Res Microbiol. 2010;161:158–67. doi: 10.1016/j.resmic.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalivoda EJ, Stella NA, O’dee DM, Nau GJ, Shanks RM. The cyclic AMP-dependent catabolite repression system of Serratia marcescens mediates biofilm formation through regulation of type 1 fimbriae. Appl Environ Microbiol. 2008;74:3461–70. doi: 10.1128/AEM.02733-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamata R, Matsumoto K, Okamura R, Yamamoto T, Maeda H. The serratial 56K protease as a major pathogenic factor in serratial keratitis. Clinical and experimental study. Ophthalmology. 1985;92:1452–9. doi: 10.1016/s0161-6420(85)33855-1. [DOI] [PubMed] [Google Scholar]
- Kulasekara HD, Ventre I, Kulasekara BR, Lazdunski A, Filloux A, Lory S. A novel two-component system contols the expression of Pseudomonas aeruginosa fimbrial cup genes. Mol Microbiol. 2005;55:368–380. doi: 10.1111/j.1365-2958.2004.04402.x. [DOI] [PubMed] [Google Scholar]
- Kumeta H, Hoshino T, Goda T, Okayama T, Shimada T, Ohgiya S, Matsuyama H, Ishizaki K. Identification of a member of the serralysin family isolated from a psychrotrophic bacterium, Pseudomonas fluorescens 114. Biosci Biotechnol Biochem. 1999;63:1165–70. doi: 10.1271/bbb.63.1165. [DOI] [PubMed] [Google Scholar]
- Letoffe S, Delepelaire P, Wandersman C. Cloning and expression in Escherichia coli of the Serratia marcescens metalloprotease gene: secretion of the protease from E. coli in the presence of the Erwinia chrysanthemi protease secretion functions. J Bacteriol. 1991;173:2160–6. doi: 10.1128/jb.173.7.2160-2166.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CS, Horng JT, Yang CH, Tsai YH, Su LH, Wei CF, Chen CC, Hsieh SC, Lu CC, Lai HC. RssAB-FlhDC-ShlBA as a major pathogenesis pathway in Serratia marcescens. Infect Immun. 2010;78:4870–81. doi: 10.1128/IAI.00661-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis D, Bernillon J, Wallach JM. Specificity of Pseudomonas aeruginosa serralysin revisited, using biologically active peptides as substrates. Biochim Biophys Acta. 1998;1387:378–86. doi: 10.1016/s0167-4838(98)00144-7. [DOI] [PubMed] [Google Scholar]
- Maeda H, Morihara K. Serralysin and related bacterial proteinases. Methods Enzymol. 1995;248:395–413. doi: 10.1016/0076-6879(95)48026-9. [DOI] [PubMed] [Google Scholar]
- Marokhazi J, Mihala N, Hudecz F, Fodor A, Graf L, Venekei I. Cleavage site analysis of a serralysin-like protease, PrtA, from an insect pathogen Photorhabdus luminescens and development of a highly sensitive and specific substrate. FEBS J. 2007;274:1946–56. doi: 10.1111/j.1742-4658.2007.05739.x. [DOI] [PubMed] [Google Scholar]
- Martìnez-Cadena MG, Guzman-Verduzco LM, Steieglitz H, Kupersztoch-Portnoy YM. Catabolite repression of Escherichia coli heat-stable enterotoxin activity. J Bacteriol. 1981;145:722–728. doi: 10.1128/jb.145.2.722-728.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty KB, Williams CL, Guynn LJ, Benedik MJ, Blanke SR. Characterization of a cytotoxic factor in culture filtrates of Serratia marcescens. Infect Immun. 2002;70:1121–8. doi: 10.1128/IAI.70.3.1121-1128.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massaoud MK, Marokhazi J, Venekei I. Enzymatic characterization of a serralysin-like metalloprotease from the entomopathogen bacterium, Xenorhabdus. Biochim Biophys Acta. 2011;1814:1333–9. doi: 10.1016/j.bbapap.2011.05.008. [DOI] [PubMed] [Google Scholar]
- Matsumoto H, Noguchi H, Hayakawa Y. Primary cause of mortality in the armyworm larvae simultaneously parasitized by parasitic wasp and infected with bacteria. Eur J Biochem. 1998;252:299–304. doi: 10.1046/j.1432-1327.1998.2520299.x. [DOI] [PubMed] [Google Scholar]
- Matsumoto K. Role of bacterial proteases in pseudomonal and serratial keratitis. Biol Chem. 2004;385:1007–16. doi: 10.1515/BC.2004.131. [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Maeda H, Takata K, Kamata R, Okamura R. Purification and characterization of four proteases from a clinical isolate of Serratia marcescens kums 3958. J Bacteriol. 1984;157:225–32. doi: 10.1128/jb.157.1.225-232.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdoch SL, Trunk K, English G, Fritsch MJ, Pourkarimi E, Coulthurst SJ. The opportunistic pathogen Serratia marcescens utilizes type VI secretion to target bacterial competitors. J Bacteriol. 2011;193:6057–69. doi: 10.1128/JB.05671-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao VA, Shepherd SM, English G, Coulthurst SJ, Hunter WN. The structure of Serratia marcescens Lip, a membrane-bound component of the type VI secretion system. Acta Crystallogr D Biol Crystallogr. 2011;67:1065–72. doi: 10.1107/S0907444911046300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverchon S, Huang Y, Bourson C, Robert-Baudouy J. Nucleotide sequences of the Erwinia chrysanthemi ogl and pelE genes negatively regulated by the kdgR gene product. Gene. 1989;85:125–34. doi: 10.1016/0378-1119(89)90472-1. [DOI] [PubMed] [Google Scholar]
- Sapriel G, Wandersman C, Delepelaire P. The SecB chaperone is bifunctional in Serratia marcescens: SecB is involved in the Sec pathway and required for HasA secretion by the ABC transporter. J Bacteriol. 2003;185:80–8. doi: 10.1128/JB.185.1.80-88.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks RM, Dashiff A, Alster JS, Kadouri DE. Isolation and identification of a bacteriocin with antibacterial and antibiofilm activity from Citrobacter freundii. Arch Microbiol. 2012;194:575–587. doi: 10.1007/s00203-012-0793-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks RM, Kadouri DE, Maceachran DP, O’toole GA. New yeast recombineering tools for bacteria. Plasmid. 2009;62:88–97. doi: 10.1016/j.plasmid.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C, Chen Y, Yang C, Chen J, Liu C. Colloid chitin azure is a dispersible, low-cost substrate for chitinase measurements in a sensitive, fast, reproducible assay. J Biomol Screen. 2010;15:213–217. doi: 10.1177/1087057109355057. [DOI] [PubMed] [Google Scholar]
- Stella NA, Kalivoda EJ, O’dee DM, Nau GJ, Shanks RM. Catabolite repression control of flagellum production by Serratia marcescens. Res Microbiol. 2008;159:562–568. doi: 10.1016/j.resmic.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh Y, Alpaugh M, Krause KL, Benedik MJ. Differential secretion of isoforms of Serratia marcescens extracellular nuclease. Appl Environ Microbiol. 1995;61:4083–8. doi: 10.1128/aem.61.11.4083-4088.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tshala-Katumbay D, Monterroso V, Kayton R, Lasarev M, Sabri M, Spencer P. Probing mechanisms of axonopathy. Part I: Protein targets of 1,2-diacetylbenzene, the neurotoxic metabolite of aromatic solvent 1,2-diethylbenzene. Toxicol Sci. 2008;105:134–41. doi: 10.1093/toxsci/kfn103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wargo MJ, Szwergold BS, Hogan DA. Identification of two gene clusters and a transcriptional regulator required for Pseudomonas aeruginosa glycine betaine catabolism. J Bacteriol. 2008;190:2690–9. doi: 10.1128/JB.01393-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Kimura K, Sumiya T, Nikaidou N, Suzuki K, Suzuki M, Taiyoji M, Ferrer S, Regue M. Genetic analysis of the chitinase system of Serratia marcescens 2170. J Bacteriol. 1997;179:7111–7. doi: 10.1128/jb.179.22.7111-7117.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West SE, Sample AK, Runyen-Janecky LJ. The vfr gene product, required for Pseudomonas aeruginosa exotoxin A and protease production, belongs to the cyclic AMP receptor protein family. J Bacteriol. 1994;176:7532–42. doi: 10.1128/jb.176.24.7532-7542.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]