Abstract
Differences in replication fidelity, as well as mutator and antimutator strains, suggest that virus mutation rates are heritable and prone to natural selection. Human immunodeficiency virus type 1 (HIV-1) has many distinct advantages for the study of mutation rate optimization given the wealth of structural and biochemical data on HIV-1 reverse transcriptase (RT) and mutants. In this study, we conducted parallel analyses of mutation rate and viral fitness. In particular, a panel of 10 RT mutants – most having drug resistance phenotypes – were analyzed for their effects on viral fidelity and fitness. Fidelity differences were measured using single-cycle vector assays, while fitness differences were identified using ex vivo head-to-head competition assays. As anticipated, virus mutants possessing either higher or lower fidelity had a corresponding loss in fitness. While the virus panel was not chosen randomly, it is interesting that it included more viruses possessing a mutator phenotype rather than viruses possessing an antimutator phenotype. These observations provide the first description of an interrelationship between HIV-1 fitness and mutation rate and support the conclusion that mutator and antimutator phenotypes correlate with reduced viral fitness. In addition, the findings here help support a model in which fidelity comes at a cost of replication kinetics, and may help explain why HIV-1 and RNA viruses maintain replication fidelity near the extinction threshold.
Keywords: retrovirus, lentivirus, evolution, mutation, fitness
Introduction
Mutation rates among viruses and other microbes range between 1.1 × 10−3 – 5.0 × 10−10 mutations per site per generation. Much of this range can be explained by an inverse relationship between mutation rate and genome size. DNA-based microbes, for example, possess a near-constant mutation rate (0.003–0.004 mutations/genome/generation) over a genome size of 10,000 base pairs. However, RNA viruses mutate at a considerably higher rate (10–100 fold) than DNA viruses of comparable size.. The observed high mutation rates of RNA viruses makes them convenient model systems to investigate selection pressures on the viral mutation rate.
Human immunodeficiency virus type 1 (HIV-1) has a high mutation rate, determined to be 1.2 – 3.4 × 10−5 mutations per target base per round of replication. Many of these mutations are deleterious or lethal, and therefore have limited long-term contribution to genetic diversity. In general, the high mutation rate of RNA viruses has been hypothesized to push the virus to the brink of either error catastrophe or extinction 12. In fact, drugs and host proteins that increase the HIV-1 mutation rate possess antiviral activity.
The high mutation rate of HIV-1 is often thought to be important for adaptation, including the ability infect new host cells and the emergence antiviral drug resistance. Mutator phenotypes have been found in bacteria and lytic RNA viruses 31, as well as HIV-1. They commonly have an advantage in new or fluctuating environments. Mutations that increase mutation rate can become associated with adaptive changes and therefore ‘hitchhike’ to a greater frequency within a population. Experimental data on selection for a high mutation rate in viruses is limited. Studies in vesicular stomatitis virus (VSV) and HIV-1 found that anti-mutator strains did not impede the ability of these viruses to adapt to novel environments or to develop drug resistance. On the other hand, a poliovirus variant with a viral polymerase possessing 3 times higher fidelity than wt was far less virulent in mice, suggesting a reduced ability for virus adaptation. HIV-1 adaptation may be important for avoidance from the host immune response that allows viral replication in long-term non-progressors. To date, studies of the differences in the relationship of mutation rate and phenotypic variation among retroviruses and RNA viruses to that of DNA viruses are limited 50.
An alternative explanation is that the high mutation rate of HIV-1 is due to some level of genetic constraint. RNA-dependent RNA polymerases of RNA viruses, as well as HIV-1 reverse transcriptase (RT), lack intrinsic 3′-to-5′ proofreading activity. Nonetheless, there is a wide range of mutation rates across RNA and retroviruses 3. The mutation rates of these viruses can be elevated by drugs that either increase mutation rate or terminate polymerase extension with 2′,3′-dideoxynucleoside analogues. Drug resistant variants have been found to have lower mutation rates in RNA viruses 46 as well as in HIV-1.
If HIV-1 has the ability to evolve to a lower mutation rate, increasing replication fidelity is likely limited by polymerase kinetics or processivity. Evidence with VSV anti-mutator strains suggest that direct fitness costs are imposed as measured by viral replication 55. A meta-analysis approach combining eleven previous studies found that the HIV-1 mutation rate correlated with the biochemical parameter kcat−1, i.e., the rate at which a polymerase extends past a terminal base mispair 56. This and other studies suggest that polymerase fidelity is intrinsically linked to replication rate – that is, more energy and/or time for base recognition, nucleotide synthesis, and/or polymerase translocation is required for the polymerase to incorporate the correct base. According to this model, the higher mutation rate of RNA viruses and retroviruses may be due to a relatively greater cost to the virus in increasing replication fidelity. To date, it is unclear how generally applicable the link between polymerase fidelity and replication rate may be among RNA viruses and retroviruses.
HIV-1 provides a powerful system for the study of constraints on the viral mutation rate. In this study, a library of ten HIV-1 mutants with varying fidelity (mainly amino acid substitutions in HIV-1 RT that confer drug resistance to nucleoside RT inhibitors) was used to investigate the relationship of mutation rate to viral fitness. Most of the RT variants are not only clinically relevant, but define a range in viral fidelity that maintains viral infectivity. By measuring mutation rate and fitness of HIV-1 under near identical conditions, a cost of replication fidelity was identified. Viruses with both higher and lower mutation rates – relative to wt – led to reductions in viral fitness, suggesting that wt HIV-1 may be close to an optimum. These studies provide the first demonstration of the relationship between HIV-1 mutation rate and viral fitness and aid in gaining greater insight into their interaction.
Results
In the present study, a panel of ten viruses encoding RT variants was generated to test the hypothesis that both lower and higher mutation rates reduce HIV-1 fitness below that of wt (Fig. 1a–b). Of these ten mutants, eight confer resistance to one or more clinically approved nucleoside reverse transcriptase inhibitors (nRTIs), while the remaining two (i.e., Y115A and V148I) have the largest magnitude impact on HIV-1 fidelity (Fig. 5 and Table S1). Table S1 provides a comprehensive summary of previous fitness, processivity, and fidelity measures for each mutant. Although there is some general agreement regarding the findings with any particular RT variant, the phenotypic measurements of the RT mutants vary greatly between studies, which is likely due to discrepancies in reagents, assays, and laboratories. Additionally, the table includes viruses with RT variants having previously uncharacterized fitness and/or mutation rates. In order to investigate the potential interrelationship between viral fitness and mutation rate, direct parallel comparisons of fitness and mutation rate were conducted here in this study under standardized experimental conditions.
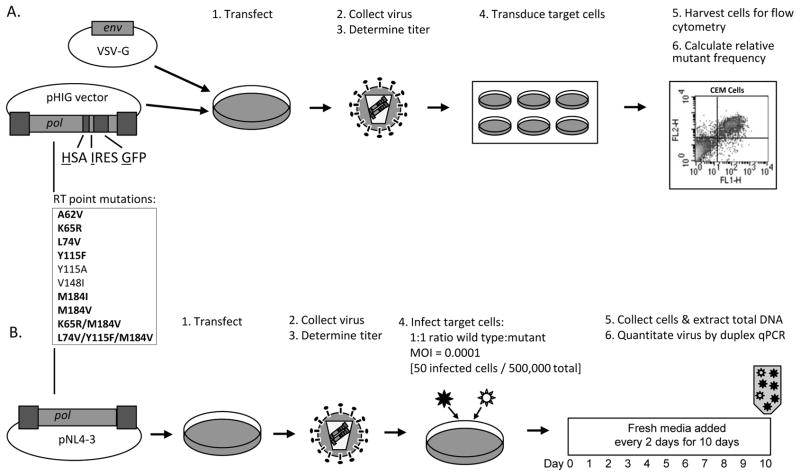
Figure 1. Experimental approach.
Measurement of A) viral fidelity and B) viral fitness, among panel of 10 HIV-1 RT mutants. A) An HIV-1 vector (pHIG) sensitive to changes in fidelity was manipulated to consist of NL4-3 wt or one of ten RT mutants. Each of these vectors was co-transfected with a VSV-G envelope expression plasmid into a producer cell line. Cell culture supernatants were collected, filtered to remove cellular debris, and titered for downstream application. Viral equivalents were used to transduce CEM target cells to calculate relative mutant frequencies by flow cytometry analysis. B) The same 10 RT mutants were introduced into the NL4-3 molecular clone. Together with wt, these clones were generated and titered, as described above. Each of the mutant viruses was competed against wt by infecting 5 × 105 CEM-GFP target cells at an equal MOI of 0.0001 (i.e., 50 infected cells). Virus was passaged every other day, for ten days, and then relative amounts of viral nucleic acid were quantified by duplex qPCR. The RT point mutations listed in bold text are nucleoside RT inhibitor mutations. Abbreviations: HSA, murine heat stable antigen; IRES, internal ribosome entry site; GFP, green fluorescence protein; VSV-G, vesicular stomatitis virus-glycoprotein; FL1-H, fluorescence channel 1, height of intensity; FL2-H, fluorescence channel 2, height of intensity; pol, HIV-1 gene consisting of protease, RT, and integrase; MOI = multiplicity of infection; qPCR, quantitative PCR.
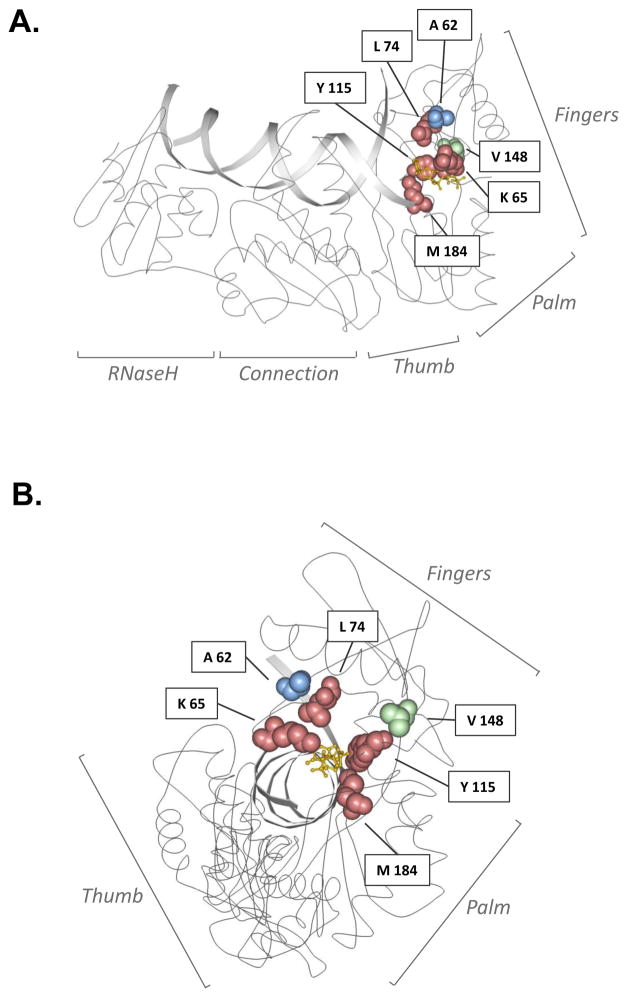
Figure 5. Locations of amino acid residues in HIV-1 reverse transcriptase (RT) associated with drug resistance and/or enzyme fidelity.
A ribbon diagram of HIV-1 RT is shown in side-view (A) or head-on (B) view of the catalytic domain, with interlaid primer:template DNAs, including a space-filling of amino acid residues investigated in this study. HIV-1 RT subdomains are indicated, and amino acid residues investigated in this study are represented as space-filled and are color-coded to indicate phenotype. Red denotes primary drug-resistant mutation site; blue denotes secondary drug-resistant mutation site; green denotes non-drug-resistant site. The yellow molecule depicts a dideoxynucleotide at the polymerase active site. Image adapted from of Huang, H. et al.96 and created with Protein Workshop software 97. Image obtained from the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB) 98; IPDB ID: 1RTD.
Drug-resistance conferring substitutions in HIV-1 RT influence the fidelity of replication
A single-cycle vector assay was used to measure differences in HIV-1 mutant frequency relative to the NL4-3 wt reference strain. This assay uses flow cytometry analysis to score target cells for expression of a pair of marker genes (Fig. 1a). Cells expressing only one of the two marker genes are interpreted as being infected yet harboring a mutated virus. This assay increases the ability of identifying mutations that arise during HIV-1 replication. Since the marker genes have not been exhaustively studied to determine precisely the target bases that result in a scorable phenotypic difference, the changes in mutation rate that can be determined are analyzed relative to that of the wt HIV-1 reference vector. This dual reporter system has been previously established to quantify increased mutational load in the presence of known viral mutagens (i.e. 5-azacytidine, decitabine, and APOBEC3G), and validated by sequencing analysis.
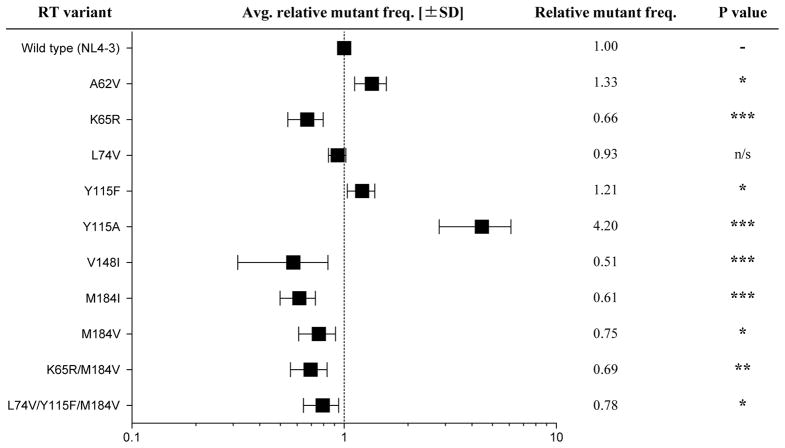
In parallel analyses, each HIV-1 vector harboring a RT variant was used to transduce the CEM T-cell line. The results from the single-cycle assay predictably led to the grouping of the RT variant viruses into 3 main categories: 1) mutant frequency higher than wt; 2) mutant frequency lower than wt; 3) mutant frequency comparable to wt virus. Three of the RT variant viruses (i.e., A62V, Y115F/A) had a higher mutant frequency than wt, six had a lower mutant frequency (i.e., K65R, V148I, M184I/V, K65R/M184V, and L74V/Y115F/M184V), and one RT variant (L74V) was found to have comparable mutant frequency to wt (Fig. 2 and Table S1). These results are generally consistent with previous work, though there are some inconsistencies noted (see supplementary material).
Figure 2. HIV-1 RT mutants influence virus mutant frequencies.
Each of the 10 RT mutants was measured for differences in fidelity relative to the wt reference strain. Mutants are displayed in ascending order of their RT position. Mutant frequencies were calculated relative to wt and are depicted on log-scale. The actual measurements are listed in the adjacent column. Values represent at least 4 biological replicates that were experimentally repeated 4 to 8 times. SD is standard deviation and represented by error bars. * P value < 0.05; ** P value < 0.01; *** P value < 0.001.
Drug-resistant conferring substitution mutations in HIV-1 RT reduce viral fitness
In order to assess the fitness of the panel of RT variant viruses under investigation, each virus was subjected to a head-to-head competition assay against wt (Fig. 1b). This assay was performed in a close derivative of CEM (i.e., CEM-GFP), so we could measure fitness and fidelity under similar cellular conditions. Viral stocks were titered on CEM-GFP cells to determine infectious unit equivalents of mutant and wt virus. Each assay was initiated with a 1:1 ratio of wt and mutant virus and passaged for 10 days before quantification of each clone by qPCR. The low MOI used in these assays (i.e., 0.0001) and the limited number of replication cycles during the assay is expected to minimize the likelihood of recombination.
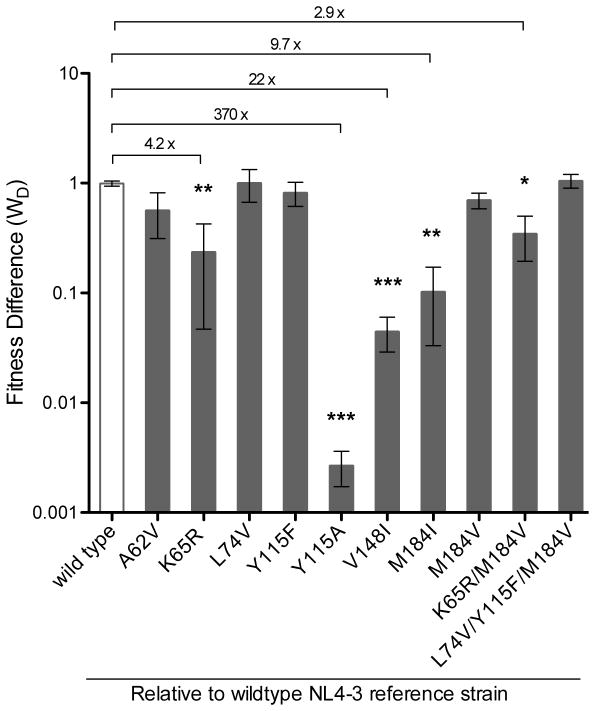
A significant decrease in viral fitness relative to wt virus was observed for 5 variants (i.e., K65R, Y115A, V148I, M184I, and K65R/M184V) (Fig. 3). Three of the other variants analyzed (i.e., A62V, Y115F, and M184V) appeared to have somewhat lower fitness (i.e., 18%–43% less fit than wt, Table S2), but these and the remaining 2 mutant viruses were not significantly different than wt. In control experiments, wt virus competed against a wt reference strain showed no discernable fitness differences (Fig. 3 and Table S2).
Figure 3. Impact of HIV-1 RT mutants on viral fitness.
Each of the 10 RT mutants, and a wt control, was passaged with a wt reference strain in a head-to-head competition assay. Mutants are assembled by RT position, and respective fitness differences (WD) are displayed on a log-scale. Asterisk symbols denote level of significance with numerical values along horizontal line showing fold difference from wt. Error bars represent SEM, standard error of the mean. * P value < 0.05; ** P value < 0.01; *** P value < 0.001.
Interconnection between HIV-1 fitness and mutation rate
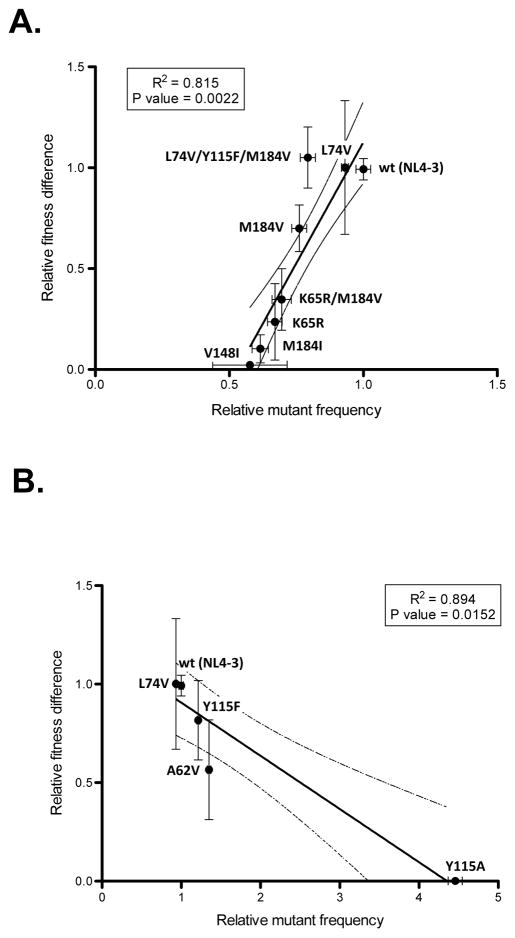
An optimality model for the HIV-1 mutation rate in a constant environment would suggest that RT variant viruses with higher or lower mutation rates would be associated with reduced fitness. To investigate this, mutant viruses were grouped by their fidelity relative to wt and the relationship between fitness and fidelity was analyzed by Pearson’s correlation and linear regression. Analysis of the 6 RT variant viruses with lower mutation rate than wt, along with L74V (which has no effect on mutation rate) and the wt reference strain, demonstrated a significant correlation between lower mutation rates and lower fitness (two-tailed Pearson correlation coefficient r = 0.903, P value = 0.0022, Fig. 4a). These same analyses were performed without wt to confirm that the correlation was not driven merely by a high fitness of wt RT and low fitness of mutants; the pattern was unchanged (two-tailed Pearson correlation coefficient r = .9244, P value = 0.0028). Exclusion of the K65R/M184V and L74V/Y115F/M184V mutants, to avoid retesting of the same mutations in a different genetic background, also did not affect results (two-tailed Pearson correlation coefficient r = 0.980, P value = 0.0032). These analyses support the conclusion that there is a fitness cost to high fidelity.
Figure 4. Correlative influence of HIV-1 RT variants on both virus mutant frequency and fitness.
Assembly of RT mutants that have A) higher, or B) lower, fidelity relative to wt and the corresponding fitness difference measurements. L74V and wt are included in both panel A) and B). Lines represent the linear regression calculated from best-fit values. Dashed lines are 95% confidence intervals of best-fit line. Error bars represent SEM, standard error of the mean.
The effect of viral variants with higher mutation rates (including L74V) were next tested for their effect on fitness. Inclusion of wt into analysis resulted in a significant correlation (two-tailed Pearson correlation coefficient r = −0.945, P value = 0.0152, Fig. 4b). This correlation was no longer significant after removal of wt from the analysis (two-tailed Pearson correlation coefficient r = −0.948, P value = 0.0518). Taken together, while the total number of mutants in this study with a high mutation rate is relatively small, the results from our analysis suggest that the wt virus has a mutation rate near the optimum, at least under these culture conditions, and in the absence of strong adaptive pressures.
Discussion
In this study, the relationship between HIV-1 fitness and mutation rate was investigated in parallel analyses. Advantages to performing this analysis with HIV-1 include its high mutation rate (which has clinical implications) and the wealth of data published on HIV-1 RT – including in vitro biochemical measurements (i.e., fidelity and processivity) as well as many measurements of viral fitness (Fig. 5 and Table S1). While these previous measurements have clearly enhanced the field, there are significant advantages to using a single data set from parallel analyses to explore the interaction between fitness and mutation rate. For instance, the fitness assay methodology used here in this study (i.e., direct ex vivo competition measurements) is more accurate than in vitro studies of catalytic rates; the rate of polymerization during viral replication may not always be a limiting factor to viral reproductive rate 56. Most importantly, the parallel analyses allow for more direct comparisons between fitness and mutation rate, which is important for deciphering the relationships between these two important biological properties.
There is evidence supporting at least three evolutionary factors that govern mutation rate. A low mutation rate may reduce the rate of adaptation (though this will be the limiting factor for adaptation only under certain conditions). A high mutation rate can cause an increase in mutational load in the population, or decrease individual fitness by causing lethal mutations in progeny. Finally, there may be physiological limits on polymerase extension as fidelity is improved. The first of these forces varies greatly as adaptive conditions change; but if the latter two dominate, a relatively stable and optimal fidelity may exist.
As demonstrated in this study, a decrease in mutation rate (i.e., increased replication fidelity) comes at a cost to viral fitness (Fig. 4a). This observation is consistent with a recent meta-analysis of HIV-1, in which an inverse correlation between the enzymatic incorporation rate Kcat−1 and viral fidelity was shown, suggesting that the fitness cost of increased fidelity is tied to energetic constraints of nucleotide synthesis 56. Such a fidelity-cost model has been postulated by enzymologists over the past few decades to explain the impaired enzyme kinetic observations of antimutator polymerases of the DNA bacteriophage T4. Similar biochemical studies with HIV-1 mutants support this model summarized by the correlation between increased fidelity and impaired enzyme processivity – a biochemical measure for how long a polymerase is able to engage and extend an oligonucleotide before the complex dissociates (Table S1). Additionally, steady-state and pre-steady state biochemical kinetics assays have been performed on some of the RT mutants discussed. These assays measure binding affinities (KM and Kd, respectively) and catalytic turnover rates (kcat and kpol, respectively) for both wt and mutant polymerases. These parameters can then be used to calculate relative polymerase efficiencies for misinsertion (catalytic efficiency of incorrect nucleotide incorporatoin) and mismatch-extension (catalytic efficiency of correct nucleotide synthesis downstream a mismatched primer-template). Several independent enzyme kinetics analyses found that K65R mutants had decreased misinsertion and mismatch-extension efficiencies along with a compromised ability to utilize natural nucleotide substrates. These studies have also been performed on M184I/V mutants and the findings follow a similar theme, although the effect size is greater. Specifically, measurements for M184I show an increased misinsertion fidelity relative to M184V, while both mutants suffer processivity defects and impaired nucleotide incorporation efficiencies (Table S1 and 68). These biochemical studies lend additional mechanistic support that strengthen the HIV-1 fidelity-fitness cost model invoked by our study and others (Fig. 4A and 56). Additional studies with RNA-dependent RNA polymerases also support this model. If the cost of increased fidelity is greater in RNA viruses, this might explain the lower fidelity of these viruses overall, even in the absence of a strict constraint.
The proposed fidelity-fitness cost model implies a lower limit to RNA virus mutation rates; therefore, by extension, RNA virus populations likely balance the fitness gains associated with increased replication speed against the fitness costs associated with the inherent loss in fidelity. This interplay may have driven such viral populations to exist near the ‘extinction threshold’ – a theoretical transitory phase that, once surpassed by excessive mutational load, initiates a catastrophic meltdown of genomic stability. Existence near this threshold may be tolerated by RNA virus populations because they are structured to limit increased mutation rates through (i) large population sizes that tolerate forward and reversion mutations at the vast majority of nucleotide positions and (ii) diminished fitness effects as mutations accumulate, i.e., positive epistasis. A detailed analysis of RT mutant behavior during single-cycle reverse transcription events would likely be informative; our results cannot distinguish between, for example, longer generation time and a decreased rate of successful infection.
Our findings also suggest that higher mutation rates were associated with lower fitness (Fig. 4b). Some studies have found mutator strains to have higher fitness under conditions where adaptation was important. However, when adaptation is less important, mutators are generally at a disadvantage. In addition, numerous studies have shown that viral mutagens (i.e., 5-azacytosine and ribavirin) and the APOBEC3 host proteins can reduce infectivity of HIV-1 and other RNA viruses. This current study with HIV-1 supports the general notion that even relatively small increases to the mutation rates of RNA viruses and retroviruses decrease fitness, reinforcing the promise of lethal mutagenesis as an antiviral intervention strategy. Additionally, of the RT mutants found to increase HIV-1 mutation rates, Y115A/F have been biochemically interrogated for their effects on misinsertion and mismatch-extension efficiencies, as described above. Mutations at this position decrease fidelity measures, while Y115A seems to also suffer greater processivity defects (Table S1).
Much of our mutation rate data is in good agreement with previously published studies using various methodologies (Table S1 and Fig. 2). For example, the measured mutation rates for the K65R, L74V, Y115A, V148I, M184I, and M184V RT variants generally coincided with previous findings. Detailed analysis of the HIV-1 mutation rate using the lacZα gene as a mutation target led to similar but not identical determinations. In the current study, a more qualitative assay was utilized with a mutation target that has not been as extensively characterized. One limitation of this is not being able to calculate mutation rates with high precision. Nonetheless, this assay provides an appropriate surrogate measure of relative differences in mutation rate, as has been previously validated.
Of the four remaining RT variants analyzed in this study, A62V and K65R/M184V have, to our knowledge, never been analyzed for their influence on the HIV-1 mutation rate, while minor discrepancies were noted with Y115F and L74V/Y115F/M184V (Fig. 2 versus Table S1). The Y115F RT mutant had previously been shown to slightly decrease viral fidelity, but these trends were not found to be significant; in this study this RT variant was shown to have a 20% (1.2-fold) higher mutant frequency (Fig. 2 and Table S1). Previous analysis of the triple mutant L74V/Y115F/M184V showed an almost 2-fold increase in frameshift mutations 35, while current analysis revealed a 30% (1.3-fold) decrease in mutant frequency (Fig. 2 and Table S1). This discrepancy is likely due to the inherent differences between mutation assays. For instance, the previously published in vivo single-cycle frameshift assay measures the effect of an RT variant to cause frameshifts at a poly-thymidine tract 35. The fidelity assay used in this study, however, provides a more global analysis of both substitution mutations as well as frameshift mutations.
While many of our analyses of HIV-1 fitness among the RT variant viruses are consistent with previous observations, there were discrepancies (Fig. 3 and Table S1). These differences can be attributed to either: 1) the type of fitness assay (i.e., parallel growth versus single-cycle versus dual competition versus in vivo reversion); 2) the end-point assay measure and its representation (i.e., p24 capsid levels versus viral copy number and percent difference versus fold difference); and 3) the type of reagents used (i.e., molecular clones versus cloned patient samples and cell lines versus primary cells). Although there is no consensus fitness assay in the field, several labs have popularized the co-culturing dual competition assay as a sensitive and internally controlled assay.
Of the ten HIV-1 RT variant viruses that were analyzed in the dual competition assay, seven had been previously assayed for their effects on viral fitness (Fig. 5, Table S1). The fitness difference measures of these 7 mutants (i.e., A62V, K65R, L74V, Y115A, M184I, M184V, and K65R/M184V) were generally similar to previously reported ranges (Fig. 3 and Table S1). Two minor discrepancies were L74V and M184V. The fitness analysis in this study indicated that L74V had no difference from wt (Fig. 3), while 3 of 5 previous studies found a subtle, 11% to 2-fold, defect to viral fitness (Table S1). The current analysis revealed that the M184V mutant had an insignificant 1.4-fold decrease in viral fitness, while 7 of 8 previous studies found this variant’s fitness loss to range from 4% to 14-fold relative to wt (Table S1). These inconsistencies may be explained by any number of the aforementioned differences among fitness assays. It is also possible that certain cell types, especially with low dNTP pool concentrations, may exacerbate subtle replication defects. It may be that the CEM cell line, chosen to limit variability when comparing RT variant measures between the two assays, minimizes fitness differences present in other cell types.
In summary, the data presented in this study support a relatively simple optimality model for the HIV-1 mutation rate, in which extreme high and low mutation rates are selected against. While this mutational optimum may depend on the niche (e.g., transmission event, virus spread in the lymph node), and may be easily disrupted in the presence of novel adaptive conditions, it is interesting that in this study the mutational optimum was near that of the reference wt strain. In summary, this is the first study to directly compare fidelity and fitness measures in HIV-1, and the first description of an interrelationship between HIV-1 fitness and mutation rate.
Materials and Methods
Plasmid constructs and cell lines
The HIV-1 vector pHIG, used in the mutant frequency assays, has been previously described 13. Briefly, a ~2.0-kbp dual reporter cassette composed of the murine heat stable antigen CD24 (HSA), an internal ribosome entry site (IRES), and enhanced green fluorescent protein (eGFP) was placed in-frame and 3′ to the NL4-3 nef start codon. The original vector pNL4-3.HSA.R+.E- was obtained from the AIDS Research and Reference Reagent Program (Division of AIDS, NIAID, NIH, Germantown, MD) and contributed by N. Landau 59. The HIV-1 NL4-3 molecular clones used in the fitness assays were a kind gift from E. Arts and described here 60. To generate the 10 drug-resistance mutants in HIV-1 pol, the 2100–5983 region was subcloned into pCR®2.1-TOPO® (Invitrogen). Site-directed mutagenesis was performed to introduce point mutations using site-directed mutagenesis (QuickChange II Site-Directed Mutagenesis, Stratagene Santa Clara, CA). Correct clones were confirmed by sequencing analysis and cloned back into the pHIG vector, using SbfI (2844) and AgeI (3486) restriction enzymes (New England Biolabs, NEB, Ipswich, MA); or back into the NL4-3 molecular clone using MscI sites (2683 and 4545). Inserts were, again, sequenced confirmed for orientation and quality.
The G glycoprotein of vesicular stomatitis virus (VSV-G) envelope expression plasmid HCMV-G was used to pseudotype virions and was a kind gift from J. Burns (University of California, San Diego). The phycoerythrin (PE)-conjugated antibody to HSA (anti-CD24) was purchased form BD Pharmingen (San Diego, CA). The human embryonic kidney (HEK 293T) cell line was purchased from ATCC (Manassas, VA) and maintained in Cellgro DMEM (Manassas, VA) plus 10% HyCLone FetalClone III (FC3; Thermo-Scientific). The CEM-GFP cell line was obtained from the AIDS Research and Reference Reagent Program (Division of AIDS, NIAID, NIH, Germantown, MD), contributed by J. Corbeil 61. The CEM cell line was a kind gift from M. Malim. Both CEM and CEM-GFP lines were maintained in RPMI (Gibco, Life Technologies Invitrogen, Grand Island, NY) plus 10% FC3.
Virus production and virus titer assays
Both the pHIG vector virus and the NL4-3 molecular clones were produced by transfection of HEK 293T cells. The calcium phosphate method was used to transfect 2 × 106 cells with 20 ug plasmid viral DNA and 2.5 ug plasmid HCMV-G envelop. Viral supernatants were collected 48 h post-transfection and passed through 0.2-um filter. To determine infectious units (IU) of virus per mL supernatant the tissue culture infectious dose (50% TCID50) end-point dilution assays was used. For each wildtype (wt) and mutant virus stock, 100 uL of 10-fold serial diluted supernatant was added to 50,000 CEM-GFP indicator cells in 250 uL total volume of a 96-well plate. Each dilution series consisted of n=6. Cells were maintained every other day for 10 days. At day 10, the number of GFP-positive wells was determined by visual inspection. To calculate the TCID50, the number of GFP-positive wells was multiplied by 1/6 and summed with 0.5; 10 to the negative power of this sum is the TCID50, and the TCID50 divided by 0.1 mL (100 uL added at each dilution) is the number of IU in each mL of supernatant. An MOI of 0.0001 can then be computed by calculating the amount of supernatant with 50 IU (MOI = IU/# of cells; 0.0001 × 500,000 cells = 50 IU).
Mutant frequency analysis by flow cytometry
Vector virus, both wt and the panel of 10 RT variants, was used to transduce 50,000 CEM cells by 2 h spinoculation at 1200 × g. Each experimental replicate consisted of 4 to 6 biological replicates. Viral stocks were titered on the CEM line to maintain 15–30% transduction efficiency. This range of infection ensures an MOI of less than 1 to reduce the probability of co-infection but also was not low enough to be subject to background anti-HSA staining (see below). Cells were prepared and stained as previously described 13. Briefly, 48 h post-transduction, cells were resuspended in 75 uL PBS/2% FC3 with 1:250 anti-HSA-PE and incubated for 20-min at 4 °C. Cells were then washed with 1 mL PBS/2% FC3 and finally resuspended in 200 uL PBS/2% FC3 for flow cytometry analysis. Cells were analyzed with a FACScan (BD Biosciences). Gates were chosen based on live cell morphology of forward scatter channel (FSC) and side scatter channel (SSC) of 10,000 cells. Excitation with the 488 nm Argon laser provided detection of GFP emission at 507 nm in fluorescence channel 1 (FL-1), and detection of PE-anti-HSA at 578 nm in fluorescence channel 2 (FL-2). Compensation was set based on single-color controls to eliminate spillover and re-verified based on the geometric mean of the single-color positive to negative-detected populations. All flow cytometry data was analyzed by FlowJo software v 9.2 (Ashland, OR). Mutant frequency calculations were determined from the percentage of target cells expressing a single reporter gene relative to the percentage of total infected cells (i.e., %[HSA+/GFP−] plus %[HSA−/GFP+] divided by the % of total infected cells). Mutant frequencies were set relative to wt for each experimental replicate.
Dual competition assay
Each of the 10 RT variants was replicated in the presence of the isogenic wt NL4-3 clone. In each head-to-head competition assay, the wt and RT mutant virus clone can be independently quantified by qPCR based on probes designed to a specific region in vif; either the wt subtype B consensus denoted as vifA or the same region consisting of 11 synonymous point mutations denoted as vifB. Briefly, 5 × 105 CEM-GFP cells were infected with a 1:1 ratio of wt to RT mutant virus at an MOI of 0.0001. Virus growth was maintained for 10 days, while cells were split and fresh culture media replenished every 2 days. Each pair-wise competition was repeated 3 independent times consisting of 4 biological replicates.
TaqMan Duplex qPCR assay
Competition experiments were analyzed using a duplex qPCR assay with modifications from here 60. At day 10 in the competition assay, cells were collected and resuspended in 200 uL PBS. Total genomic DNA was extracted using the Roche High Pure PCR Template Preparation Kit (Roche Applied Science) and eluted into 50 uL total volume. First, 5 uL of extracted DNA were subjected to a brief, external PCR amplification reaction in 50 uL, using VifOut+ (5′-GCA AAG CTC CTC TGG AAA GGT GAA GGG-3′) and VifOout- (5′-CTT CCA CTC CTG CCC AAG TAT CCC-3′) primers. Reactions were performed with Platinum PCR Supermix (Invitrogen) under the following cycling conditions: 1 cycle at 94 °C for 2 min; 10 cycles at 94 °C for 30 s, 55 °C for 30 s, and 68 °C for 45 s; and 1 cycle at 68° C for 5 min. The reaction was then purified with GenElute PCR Clean-Up Kit (Sigma) and eluted into 35 uL.
The TaqMan assay utilized probes to differentiate between the two forms of the NL4-3 backbone (vifA and vifB). The vifA probe sequence was 5′-AGG ATC TCT ACA GTA CTT GGC ACT AGC A-3′ and vifB probe sequence was 5′-AGG AAG CTT GCA ATA TCT AGC GTT AGC A-3′. The probes were labeled with Cy5 and Black Hole Quencher 2 (vifA) and FAM and Black Hole Quencher 1 (vifB) at 5′- and 3′-ends, respectively. The reactions were run on a BioRad CFX96 Touch Real-Time PCR Detection System (BioRad). For the duplex qPCR reaction, the forward primer was VifIn+ (5′-GAA AGA GAC TGG CAT TTG GGT CAG GG-3′) and reverse primer was VifIn- (5′-GTC TTC TGG GGC TTG TTC CAT CTG TCC-3′). Primers were optimized and added at a final concentration of 375 nM each, while vifA probe added at 250nM and vifB probe added at 125nM. To this, 2 uL of PCR product from above, was added to the primer/probe in 10X TaqMan® Gene Expression Master Mix (Applied Biosystems) to a 20 uL total reaction. Reaction conditions were: 95 °C for 10 min, the 95 °C for 15 s and 53°C for 1 min for 40 cycles. All reactions were performed in duplicate, including a 9-log range in plasmid DNA template (5 × 109 to 5 × 101 copies), as well as a negative control (no-template). The PCR efficiency, E, of each standard curve (vifA or vifB) was calculated based on the curves slope, E = (10−1/s-1) × 100%. To maximize sample data confidence, E of VifA and VifB standard curves were within 5% and R2 of each standard curve was ≥99%. Because the competing strains were added at an equal ratio, differences in relative amounts of each clone after appropriate viral growth were used to calculate fitness differences. Co-cultured viral clones were quantified using duplex qPCR, which relies on mutant- and wt-specific probes to specifically differentiate the two strains. This assay was both specific and sensitive over a 6-log range in viral copy numbers, well within the range of viral detection (Fig. S1).
Calculation of viral fitness
Fitness differences (WD) were calculated for each wt versus RT mutant co-culturing competition assay. For each of the 4 biological replicates, the frequency of the mutant strain was calculated as follows [mutant copy #/(mutant copy # + wt copy #)]. This frequency is then arcsine transformed [arcsine (√mutant frequency)] to convert the proportional data to a normal distribution. Next, the mean of the arcsine transformed data is calculated and untransformed by squaring the sine of each mean. Data from the 3 experimental replicates were compiled to show the fitness differences.
Statistical analyses
All statistical analysis and graphical representation was done using GraphPad Prism version 5.0 (La Jolla, CA). A one-way ANOVA was used to determine differences between wt HIV-1 and the 10 RT mutants for both mutant frequency and fitness. Relative mutant frequency differences were analyzed by first log transforming the data. A Dunnett’s multiple comparison post-hoc test was used to compare differences between wt and each mutant RT measure. A two-tailed Pearson’s coefficient of correlation was used on the mutant frequency and fitness measures to determine the chance that there was zero correlation between the parameters. An analysis by the nonparametric two-tailed Spearmen’s rank correlation, robust to the effect of outliers, had similar results and is generally not reported. Linear regression was used to identify R-squared values.
Supplementary Material
Highlights.
Parallel analyses of HIV-1 mutation rate and viral fitness were conducted
A panel of 10 RT mutants were analyzed for their effects on viral fidelity and fitness
Virus mutants possessing either higher or lower fidelity had a corresponding loss in fitness
More viruses possessed a mutator phenotype rather than an antimutator phenotype
This is the first description of an interrelationship between HIV-1 fitness and mutation rate
Mutator and antimutator phenotypes correlated with reduced viral fitness
Acknowledgments
We thank Eric Arts and Michael Lobritz for reagents and technical advice pertaining to the fitness assay. We also acknowledge Michael Travasano and Scott Lunos and Cavan Reilly for help with statistical analysis of the fitness differences and mutant frequency measures. This research was supported by NIH grant R01 GM56615. M.J.D. was supported by NIH grant T32DA007097 and R.H.H. by T32DE007288.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Drake JW, Charlesworth B, Charlesworth D, Crow JF. Rates of spontaneous mutation. Genetics. 1998;148:1667–86. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drake JW, Holland JJ. Mutation rates among RNA viruses. Proc Natl Acad Sci. 1999;96:13910–13913. doi: 10.1073/pnas.96.24.13910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanjuan R, Nebot MR, Chirico N, Mansky LM, Belshaw R. Viral mutation rates. Journal of virology. 2010;84:9733–48. doi: 10.1128/JVI.00694-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lynch M. The origins of eukaryotic gene structure. Molecular biology and evolution. 2006;23:450–68. doi: 10.1093/molbev/msj050. [DOI] [PubMed] [Google Scholar]
- 5.Drake JW. Comparative rates of spontaneous mutation. Nature. 1969;221:1132. doi: 10.1038/2211132a0. [DOI] [PubMed] [Google Scholar]
- 6.Drake JW. A constant rate of spontaneous mutation in DNA-based microbes. Proc Natl Acad Sci USA. 1991;88:7160–7164. doi: 10.1073/pnas.88.16.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mansky LM, Temin HM. Lower in vivo mutation rate of human immunodeficiency virus type 1 than predicted from the fidelity of purified reverse transcriptase. J Virol. 1995;69:5087–5094. doi: 10.1128/jvi.69.8.5087-5094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abram ME, Ferris AL, Shao W, Alvord WG, Hughes SH. Nature, position, and frequency of mutations made in a single cycle of HIV-1 replication. Journal of Virology. 2010;84:9864–78. doi: 10.1128/JVI.00915-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerrish PJ, Colato A, Perelson AS, Sniegowski PD. Complete genetic linkage can subvert natural selection. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:6266–71. doi: 10.1073/pnas.0607280104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rouzine IM, Brunet E, Wilke CO. The traveling-wave approach to asexual evolution: Muller’s ratchet and speed of adaptation. Theoretical population biology. 2008;73:24–46. doi: 10.1016/j.tpb.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsimring LS, Levine H, Kessler DA. RNA virus evolution via a fitness-space model. Physical review letters. 1996;76:4440–4443. doi: 10.1103/PhysRevLett.76.4440. [DOI] [PubMed] [Google Scholar]
- 12.Bull JJ, Sanjuán R, Wilke CO. Theory of lethal mutagenesis for viruses. J Virol. 2007;81:2930–2939. doi: 10.1128/JVI.01624-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dapp MJ, Clouser CL, Patterson S, Mansky LM. 5-Azacytidine can induce lethal mutagenesis in human immunodeficiency virus type 1. Journal of virology. 2009;83:11950–8. doi: 10.1128/JVI.01406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dapp MJ, Holtz CM, Mansky LM. Concomitant Lethal Mutagenesis of Human Immunodeficiency Virus Type 1. Journal of molecular biology. 2012 doi: 10.1016/j.jmb.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson JP, DR, Loeb LA. Viral error catastrophe by mutagenic nucleosides. Annu Rev Microbiol. 2004;58:183–205. doi: 10.1146/annurev.micro.58.030603.123649. [DOI] [PubMed] [Google Scholar]
- 16.Loeb LA, Essigmann JM, Kazazi F, Zhang J, Rose KD, Mullins JI. Lethal mutagenesis of HIV with mutagenic nucleoside analogs. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:1492–7. doi: 10.1073/pnas.96.4.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith RA, LL, Preston BD. Lethal mutagenesis of HIV. Virus Res. 2005;107:215–28. doi: 10.1016/j.virusres.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 18.Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–9. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 19.Harris KS, Brabant W, Styrchak S, Gall A, Daifuku R. KP-1212/1461, a nucleoside designed for the treatment of HIV by viral mutagenesis. Antiviral Res. 2005;67:1–9. doi: 10.1016/j.antiviral.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 20.Yu Q, Konig R, Pillai S, Chiles K, Kearney M, Palmer S, Richman D, Coffin JM, Landau NR. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nature structural & molecular biology. 2004;11:435–42. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- 21.Nowak MA, Anderson RM, McLean AR, Wolfs TF, Goudsmit J, May RM. Antigenic diversity thresholds and the development of AIDS. Science. 1991;254:963–9. doi: 10.1126/science.1683006. [DOI] [PubMed] [Google Scholar]
- 22.Lee HY, Perelson AS, Park SC, Leitner T. Dynamic correlation between intrahost HIV-1 quasispecies evolution and disease progression. PLoS computational biology. 2008;4:e1000240. doi: 10.1371/journal.pcbi.1000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mansky LM. Retrovirus mutation rates and their role in genetic variation. J Gen Virol. 1998;79:1337–1345. doi: 10.1099/0022-1317-79-6-1337. [DOI] [PubMed] [Google Scholar]
- 24.Coffin JM. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–9. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 25.Perelson AS, Neumann AU, Markowitz M, Leonard JM, Ho DD. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–6. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 26.Mansky LM. HIV mutagenesis and the evolution of antiretroviral drug resistance. Drug Resist Updat. 2002;5:219–223. doi: 10.1016/s1368-7646(02)00118-8. [DOI] [PubMed] [Google Scholar]
- 27.Horst JP, Wu TH, Marinus MG. Escherichia coli mutator genes. Trends in microbiology. 1999;7:29–36. doi: 10.1016/s0966-842x(98)01424-3. [DOI] [PubMed] [Google Scholar]
- 28.Gross MD, Siegel EC. Incidence of mutator strains in Escherichia coli and coliforms in nature. Mutation research. 1981;91:107–10. doi: 10.1016/0165-7992(81)90081-6. [DOI] [PubMed] [Google Scholar]
- 29.LeClerc JE, Li B, Payne WL, Cebula TA. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science. 1996;274:1208–11. doi: 10.1126/science.274.5290.1208. [DOI] [PubMed] [Google Scholar]
- 30.Matic I, Radman M, Taddei F, Picard B, Doit C, Bingen E, Denamur E, Elion J. Highly variable mutation rates in commensal and pathogenic Escherichia coli. Science. 1997;277:1833–4. doi: 10.1126/science.277.5333.1833. [DOI] [PubMed] [Google Scholar]
- 31.Suarez P, Valcarcel J, Ortin J. Heterogeneity of the mutation rates of influenza A viruses: isolation of mutator mutants. Journal of Virology. 1992;66:2491–4. doi: 10.1128/jvi.66.4.2491-2494.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gutierrez-Rivas M, Menendez-Arias L. A mutation in the primer grip region of HIV-1 reverse transcriptase that confers reduced fidelity of DNA synthesis. Nucleic Acids Research. 2001;29:4963–72. doi: 10.1093/nar/29.24.4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mansky LM, Bernard LC. 3′-azido-3′-deoxythymidine (AZT) and AZT-resistant reverse transcriptase can increase the in vivo mutation rate of human immunodeficiency type 1. J Virol. 2000;74:9532–9539. doi: 10.1128/jvi.74.20.9532-9539.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mansky LM, Pearl DK, Gajary LC. Combination of drugs and drug-resistant reverse transcriptase results in a multiplicative increase of human immunodeficiency virus type 1 mutant frequencies. J Virol. 2002;76:9253–9259. doi: 10.1128/JVI.76.18.9253-9259.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen R, Yokoyama M, Sato H, Reilly C, Mansky LM. Human immunodeficiency virus mutagenesis during antiviral therapy: impact of drug-resistant reverse transcriptase and nucleoside and nonnucleoside reverse transcriptase inhibitors on human immunodeficiency virus type 1 mutation frequencies. J Virol. 2005;79:12045–57. doi: 10.1128/JVI.79.18.12045-12057.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sniegowski PD, Gerrish PJ, Lenski RE. Evolution of high mutation rates in experimental populations of E. coli. Nature. 1997;387:703–5. doi: 10.1038/42701. [DOI] [PubMed] [Google Scholar]
- 37.Cooper TF, Lenski RE. Experimental evolution with E. coli in diverse resource environments. I. Fluctuating environments promote divergence of replicate populations. BMC evolutionary biology. 2010;10:11. doi: 10.1186/1471-2148-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giraud A, Matic I, Tenaillon O, Clara A, Radman M, Fons M, Taddei F. Costs and benefits of high mutation rates: adaptive evolution of bacteria in the mouse gut. Science. 2001;291:2606–8. doi: 10.1126/science.1056421. [DOI] [PubMed] [Google Scholar]
- 39.Pal C, Macia MD, Oliver A, Schachar I, Buckling A. Coevolution with viruses drives the evolution of bacterial mutation rates. Nature. 2007;450:1079–81. doi: 10.1038/nature06350. [DOI] [PubMed] [Google Scholar]
- 40.Tenaillon O, Toupance B, Le Nagard H, Taddei F, Godelle B. Mutators, population size, adaptive landscape and the adaptation of asexual populations of bacteria. Genetics. 1999;152:485–93. doi: 10.1093/genetics/152.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson T. Beneficial mutations, hitchhiking and the evolution of mutation rates in sexual populations. Genetics. 1999;151:1621–31. doi: 10.1093/genetics/151.4.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dawson KJ. Evolutionarily stable mutation rates. Journal of theoretical biology. 1998;194:143–57. doi: 10.1006/jtbi.1998.0752. [DOI] [PubMed] [Google Scholar]
- 43.Lee CH, Gilbertson DL, Novella IS, Huerta R, Domingo E, Holland JJ. Negative effects of chemical mutagenesis on the adaptive behavior of vesicular stomatitis virus. J Virol. 1997;71:3636–40. doi: 10.1128/jvi.71.5.3636-3640.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keulen W, van Wijk A, Schuurman R, Berkhout B, Boucher CA. Increased polymerase fidelity of lamivudine-resistant HIV-1 variants does not limit their evolutionary potential. AIDS. 1999;13:1343–1349. doi: 10.1097/00002030-199907300-00011. [DOI] [PubMed] [Google Scholar]
- 45.Leigh EG. Natural selection and mutability. Amer Naturalist. 1970;104:301–305. [Google Scholar]
- 46.Pfeiffer JK, Kirkegaard K. Increased fidelity reduces poliovirus fitness and virulence under selective pressure in mice. PLoS pathogens. 2005;1:e11. doi: 10.1371/journal.ppat.0010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vignuzzi M, Wendt E, Andino R. Engineering attenuated virus vaccines by controlling replication fidelity. Nature Medicine. 2008;14:154–61. doi: 10.1038/nm1726. [DOI] [PubMed] [Google Scholar]
- 48.Williamson S. Adaptation in the env gene of HIV-1 and evolutionary theories of disease progression. Molecular biology and evolution. 2003;20:1318–25. doi: 10.1093/molbev/msg144. [DOI] [PubMed] [Google Scholar]
- 49.Sheppard HW, Lang W, Ascher MS, Vittinghoff E, Winkelstein W. The characterization of non-progressors: long-term HIV-1 infection with stable CD4+ T-cell levels. AIDS. 1993;7:1159–66. [PubMed] [Google Scholar]
- 50.Elena SF, Sanjuan R. Adaptive value of high mutation rates of RNA viruses: separating causes from consequences. J Virol. 2005;79:11555–8. doi: 10.1128/JVI.79.18.11555-11558.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mansky LM, Le Rouzic E, Benichou S, Gajary LC. Influence of reverse transcriptase variants, drugs, and Vpr on human immunodeficiency virus type 1 mutant frequencies. J Virol. 2003;77:2071–2080. doi: 10.1128/JVI.77.3.2071-2080.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bessman MJ, Muzyczka N, Goodman MF, Schnaar RL. Studies on the biochemical basis of spontaneous mutation. II. The incorporation of a base and its analogue into DNA by wild-type, mutator and antimutator DNA polymerases. Journal of molecular biology. 1974;88:409–21. doi: 10.1016/0022-2836(74)90491-4. [DOI] [PubMed] [Google Scholar]
- 53.Goodman MF, Gore WC, Muzyczka N, Bessman MJ. Studies on the biochemical basis of spontaneous mutation. III. Rate model for DNA polymerase-effected nucleotide misincorporation. Journal of molecular biology. 1974;88:243–35. doi: 10.1016/0022-2836(74)90492-6. [DOI] [PubMed] [Google Scholar]
- 54.Hopfield JJ. Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proceedings of the National Academy of Sciences of the United States of America. 1974;71:4135–9. doi: 10.1073/pnas.71.10.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Furio V, Moya A, Sanjuan R. The cost of replication fidelity in an RNA virus. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:10233–7. doi: 10.1073/pnas.0501062102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Furio V, Moya A, Sanjuan R. The cost of replication fidelity in human immunodeficiency virus type 1. Proceedings Biological sciences/The Royal Society. 2007;274:225–30. doi: 10.1098/rspb.2006.3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Back NK, Nijhuis M, Keulen W, Boucher CA, Oude Essink BO, van Kuilenburg AB, van Gennip AH, Berkhout B. Reduced replication of 3TC-resistant HIV-1 variants in primary cells due to a processivity defect of the reverse transcriptase enzyme. EMBO J. 1996;15:4040–4049. [PMC free article] [PubMed] [Google Scholar]
- 58.Pandey VN, Kaushik N, Rege N, Sarafianos SG, Yadav PN, Modak MJ. Role of methionine 184 of human immunodeficiency virus type-1 reverse transcriptase in the polymerase function and fidelity of DNA synthesis. Biochemistry. 1996;35:2168–79. doi: 10.1021/bi9516642. [DOI] [PubMed] [Google Scholar]
- 59.He J, Choe S, Walker R, Di Marzio P, Morgan DO, Landau NR. Human immunodefiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J Virol. 1995;69:6705–6711. doi: 10.1128/jvi.69.11.6705-6711.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anastassopoulou CG, Marozsan AJ, Matet A, Snyder AD, Arts EJ, Kuhmann SE, Moore JP. Escape of HIV-1 from a small molecule CCR5 inhibitor is not associated with a fitness loss. PLoS pathogens. 2007;3:e79. doi: 10.1371/journal.ppat.0030079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gervaix A, West D, Leoni LM, Richmond DD, Wong-Staal F, Corbeil J. A new reporter cell line to monitor HIV infection and drug susceptibility in vitro. Proc Natl Acad Sci U S A. 1997;94:4653–8. doi: 10.1073/pnas.94.9.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Clouser CL, Patterson SE, Mansky LM. Exploiting drug repositioning for discovery of a novel HIV combination therapy. Journal of virology. 2010;84:9301–9. doi: 10.1128/JVI.01006-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sniegowski PD, Gerrish PJ, Johnson T, Shaver A. The evolution of mutation rates: separating causes from consequences. BioEssays: news and reviews in molecular, cellular and developmental biology. 2000;22:1057–66. doi: 10.1002/1521-1878(200012)22:12<1057::AID-BIES3>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 64.Fisher RA. The genetical theory of natural selection. Oxford, England: Clarendon Press; 1930. [Google Scholar]
- 65.Kimura M. On the evolutionary adjustments of spontaneous mutation rates. Genetical Research. 1967;9:11. [Google Scholar]
- 66.Orr HA. The rate of adaptation in asexuals. Genetics. 2000;155:961–8. doi: 10.1093/genetics/155.2.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Deval J, Navarro JM, Selmi B, Courcambeck J, Boretto J, Halfon P, Garrido-Urbani S, Sire J, Canard B. A loss of viral replicative capacity correlates with altered DNA polymerization kinetics by the human immunodeficiency virus reverse transcriptase bearing the K65R and L74V dideoxynucleoside resistance substitutions. The Journal of biological chemistry. 2004;279:25489–96. doi: 10.1074/jbc.M313534200. [DOI] [PubMed] [Google Scholar]
- 68.Deval J, White KL, Miller MD, Parkin NT, Courcambeck J, Halfon P, Selmi B, Boretto J, Canard B. Mechanistic basis for reduced viral and enzymatic fitness of HIV-1 reverse transcriptase containing both K65R and M184V mutations. The Journal of biological chemistry. 2004;279:509–16. doi: 10.1074/jbc.M308806200. [DOI] [PubMed] [Google Scholar]
- 69.Garforth SJ, Domaoal RA, Lwatula C, Landau MJ, Meyer AJ, Anderson KS, Prasad VR. K65R and K65A substitutions in HIV-1 reverse transcriptase enhance polymerase fidelity by decreasing both dNTP misinsertion and mispaired primer extension efficiencies. Journal of molecular biology. 2010;401:33–44. doi: 10.1016/j.jmb.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Back NK, Berkhout B. Limiting deoxynucleoside triphosphate concentrations emphasize the processivity defect of lamivudine-resistant variants of human immunodeficiency virus type 1 reverse transcriptase. Antimicrobial agents and chemotherapy. 1997;41:2484–91. doi: 10.1128/aac.41.11.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wainberg MA. Increased fidelity of drug-selected M184V mutated HIV-1 reverse transcriptase as the basis for the effectiveness of 3TC in HIV clinical trials. Leukemia. 1997;11(Suppl 3):85–8. [PubMed] [Google Scholar]
- 72.Berkhout B. HIV-1 evolution under pressure of protease inhibitors: climbing the stairs of viral fitness. J Biomed Sci. 1999;6:298–305. doi: 10.1007/BF02253518. [DOI] [PubMed] [Google Scholar]
- 73.Wainberg MA, Drosopoulos WC, Salomon H, Hsu M, Borkow G, Parniak MA, Gu Z, Song Q, Manne J, Islam S, Castriota G, Prasad VR. Enhanced fidelity of 3TC-selected mutant HIV-1 reverse transcriptase. Science. 1996;271:1282–1285. doi: 10.1126/science.271.5253.1282. [DOI] [PubMed] [Google Scholar]
- 74.Rezende LF, Drosopoulos WC, Prasad VR. The influence of 3TC resistance mutation M184I on the fidelity and error specificity of human immunodeficiency virus type 1 reverse transcriptase. Nucleic Acids Research. 1998;26:3066–72. doi: 10.1093/nar/26.12.3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Feng JY, Anderson KS. Mechanistic studies examining the efficiency and fidelity of DNA synthesis by the 3TC-resistant mutant (184V) of HIV-1 reverse transcriptase. Biochemistry. 1999;38:9440–8. doi: 10.1021/bi990709m. [DOI] [PubMed] [Google Scholar]
- 76.Korneeva VS, Cameron CE. Structure-function relationships of the viral RNA-dependent RNA polymerase: fidelity, replication speed, and initiation mechanism determined by a residue in the ribose-binding pocket. The Journal of biological chemistry. 2007;282:16135–45. doi: 10.1074/jbc.M610090200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chao L, Cox EC. Competition Between High and Low Mutating Strains of Escherichia coli. Evolution. 1983;37:125–134. doi: 10.1111/j.1558-5646.1983.tb05521.x. [DOI] [PubMed] [Google Scholar]
- 78.Loh E, Salk JJ, Loeb LA. Optimization of DNA polymerase mutation rates during bacterial evolution. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:1154–9. doi: 10.1073/pnas.0912451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gibson TC, Scheppe ML, Cox EC. Fitness of an Escherichia coli mutator gene. Science. 1970;169:686–8. doi: 10.1126/science.169.3946.686. [DOI] [PubMed] [Google Scholar]
- 80.Funchain P, Yeung A, Stewart JL, Lin R, Slupska MM, Miller JH. The consequences of growth of a mutator strain of Escherichia coli as measured by loss of function among multiple gene targets and loss of fitness. Genetics. 2000;154:959–70. doi: 10.1093/genetics/154.3.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Notley-McRobb L, Seeto S, Ferenci T. Enrichment and elimination of mutY mutators in Escherichia coli populations. Genetics. 2002;162:1055–62. doi: 10.1093/genetics/162.3.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Denamur E, Matic I. Evolution of mutation rates in bacteria. Mol Microbiol. 2006;60:820–7. doi: 10.1111/j.1365-2958.2006.05150.x. [DOI] [PubMed] [Google Scholar]
- 83.Crotty S, Cameron CE, Andino R. RNA virus error catastrophe: direct molecular test by using ribavirin. Proc Natl Acad Sci U S A. 2001;98:6895–900. doi: 10.1073/pnas.111085598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Graci JD, Cameron CE. Therapeutically targeting RNA viruses via lethal mutagenesis. Future virology. 2008;3:553–566. doi: 10.2217/17460794.3.6.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Martin-Hernandez AM, Gutierrez-Rivas M, Domingo E, Menendez-Arias L. Mispair extension fidelity of human immunodeficiency virus type 1 reverse transcriptases with amino acid substitutions affecting Tyr115. Nucleic Acids Research. 1997;25:1383–9. doi: 10.1093/nar/25.7.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Martin-Hernandez AM, Domingo E, Menendez-Arias L. Human immunodeficiency virus type 1 reverse transcriptase: role of Tyr115 in deoxynucleotide binding and misinsertion fidelity of DNA synthesis. The EMBO journal. 1996;15:4434–42. [PMC free article] [PubMed] [Google Scholar]
- 87.Cases-Gonzalez CE, Gutierrez-Rivas M, Menendez-Arias L. Coupling ribose selection to fidelity of DNA synthesis. The role of Tyr-115 of human immunodeficiency virus type 1 reverse transcriptase. J Biol Chem. 2000;275:19759–19767. doi: 10.1074/jbc.M910361199. [DOI] [PubMed] [Google Scholar]
- 88.Abraha A, Troyer RM, Quinones-Mateu ME, Arts EJ. Methods to determine HIV-1 ex vivo fitness. Methods in molecular biology. 2005;304:355–68. doi: 10.1385/1-59259-907-9:355. [DOI] [PubMed] [Google Scholar]
- 89.Ball SC, Abraha A, Collins KR, Marozsan AJ, Baird H, Quinones-Mateu ME, Penn-Nicholson A, Murray M, Richard N, Lobritz M, Zimmerman PA, Kawamura T, Blauvelt A, Arts EJ. Comparing the ex vivo fitness of CCR5-tropic human immunodeficiency virus type 1 isolates of subtypes B and C. J Virol. 2003;77:1021–38. doi: 10.1128/JVI.77.2.1021-1038.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Troyer RM, Collins KR, Abraha A, Fraundorf E, Moore DM, Krizan RW, Toossi Z, Colebunders RL, Jensen MA, Mullins JI, Vanham G, Arts EJ. Changes in human immunodeficiency virus type 1 fitness and genetic diversity during disease progression. Journal of virology. 2005;79:9006–18. doi: 10.1128/JVI.79.14.9006-9018.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu Y, McNevin J, Zhao H, Tebit DM, Troyer RM, McSweyn M, Ghosh AK, Shriner D, Arts EJ, McElrath MJ, Mullins JI. Evolution of human immunodeficiency virus type 1 cytotoxic T-lymphocyte epitopes: fitness-balanced escape. Journal of Virology. 2007;81:12179–88. doi: 10.1128/JVI.01277-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gao L, Hanson MN, Balakrishnan M, Boyer PL, Roques BP, Hughes SH, Kim B, Bambara RA. Apparent defects in processive DNA synthesis, strand transfer, and primer elongation of Met-184 mutants of HIV-1 reverse transcriptase derive solely from a dNTP utilization defect. The Journal of biological chemistry. 2008;283:9196–205. doi: 10.1074/jbc.M710148200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jamburuthugoda VK, Santos-Velazquez JM, Skasko M, Operario DJ, Purohit V, Chugh P, Szymanski EA, Wedekind JE, Bambara RA, Kim B. Reduced dNTP binding affinity of 3TC-resistant M184I HIV-1 reverse transcriptase variants responsible for viral infection failure in macrophage. The Journal of biological chemistry. 2008;283:9206–16. doi: 10.1074/jbc.M710149200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Diamond TL, Roshal M, Jamburuthugoda VK, Reynolds HM, Merriam AR, Lee KY, Balakrishnan M, Bambara RA, Planelles V, Dewhurst S, Kim B. Macrophage tropism of HIV-1 depends on efficient cellular dNTP utilization by reverse transcriptase. J Biol Chem. 2004;279:51545–53. doi: 10.1074/jbc.M408573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Weiss KK, Chen R, Skasko M, Reynolds HM, Lee K, Bambara RA, Mansky LM, Kim B. A role for dNTP binding of human immunodeficiency virus type 1 reverse transcriptase in viral mutagenesis. Biochemistry. 2004;43:4490–500. doi: 10.1021/bi035258r. [DOI] [PubMed] [Google Scholar]
- 96.Huang H, Chopra R, Verdine GL, Harrison SC. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–75. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 97.Moreland JL, Gramada A, Buzko OV, Zhang Q, Bourne PE. The Molecular Biology Toolkit (MBT): a modular platform for developing molecular visualization applications. BMC bioinformatics. 2005;6:21. doi: 10.1186/1471-2105-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Research. 2000;28:235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.