Abstract
Due to the critical role of Foxp3+ regulatory T cells (Tregs) in the regulation of immunity and the enrichment of Tregs within many human tumors, a number of emerging therapeutic strategies for the treatment of cancer involve the depletion or modulation of Tregs, with the aim of eliciting enhanced anti-tumor immune responses. Here, we review recent advances in understanding of the fundamental biology of Tregs, and discuss the implications of these findings for current models of tumor-associated Treg biology. In particular, we discuss the context-dependent functional diversity of Tregs, the developmental origins of these cells, and the nature of the antigens that they recognize within the tumor environment. In addition, we highlight critical areas of focus for future research.
Regulatory T cells and the immunotherapy of cancer
Recent trials demonstrating the efficacy of cancer immunotherapies, including antibody blockade of inhibitory molecules [1], adoptive T cell transfer [2], and autologous cell-based vaccines [3], have provided strong evidence that the immune system can be manipulated for clinical benefit in human cancer patients. In this regard, a number of emerging therapies for the treatment of cancer involve the ablation or modulation of CD4+Foxp3+ regulatory T cells (Tregs) concomitant with the administration of cancer vaccines or adoptive cell therapy, in an effort to induce robust anti-tumor immune responses capable of mediating cancer regression [4,5]. Tregs, characterized by expression of the transcription factor Foxp3, are critical for the prevention of autoimmunity, the maintenance of immune homeostasis, and the regulation of immune responses to foreign and self antigens in mice and man [6,7]. Mice with a loss-of-function mutation in Foxp3 lack functional Tregs, and die of organ-specific autoimmunity and lymphoproliferative disease at an early age [8–11]. Likewise, human patients harboring hypomorphic mutations in FOXP3 succumb to immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) [12,13]. Tregs are often found at elevated frequencies in the peripheral blood and tumors of human patients, and for many cancers, a high density of Tregs correlates with poor disease outcome [14]. Therefore, despite their essential role in maintaining the integrity of the host, Tregs may also play an insidious role in the promotion of cancer development and progression in some types of malignancies. An improved understanding of the fundamentals and complexities of regulatory T cell biology may enable the selective modulation of Tregs for the treatment of cancer.
In this review, we highlight recent progress in the field of regulatory T cell biology, and discuss the potential implications of these findings on our understanding of tumor-associated Tregs. Strategies for the therapeutic targeting of Tregs have been reviewed extensively elsewhere [15–17], and will not be discussed in depth here. In addition, while many T cell subsets exhibiting regulatory activity have been described [18], we will focus exclusively on CD4+Foxp3+ Tregs, due to their prominent role in immune regulation.
Tregs in human cancers: correlative or causal?
Many studies indicate that a high density of tumor-infiltrating Tregs correlates with disease outcome in a variety of human cancers (reviewed recently by [14]). For example, multiple reports have demonstrated that in human hepatocellular cancer, high Treg density is correlated with poor prognosis, while in human colorectal carcinoma, high Treg density is associated with improved outcome. Given these associations, it is important to determine whether Tregs play a direct, causal role in either promoting or suppressing disease progression in different types of cancer, as well as the biology underlying these distinct outcomes. For example, Tregs are commonly hypothesized to play a causal role in promoting cancer development via the suppression of anti-tumor immune responses. However, depending on the context, it is also possible that Tregs suppress tumor development by dampening inflammation, which is thought to promote tumor progression in many cancers [19]. Here, we consider three possible scenarios to explain an association between Treg density and disease outcome. First, Tregs may play a causal role in modulating tumorigenesis. Second, Treg density may correlate with that of another cell type or factor that plays a causal role in tumor development. Third, it is possible that high Treg density is associated with cancers of a particular stage and grade, but Tregs play no major functional role in modulating cancer development. In this case, Tregs would have prognostic significance but little functional significance. Addressing the key issue of correlation vs. causality will require loss-of-function in vivo evidence demonstrating that the selective depletion or modulation of Tregs causes significant changes in tumorigenesis or tumor progression in genetically engineered mice that are predisposed to the development of indigenous cancer. Such evidence has been elusive due to the fact that sustained Treg depletion in mice induces severe autoimmunity and other secondary adverse effects [20,21]. Thus, it is difficult to induce complete, sustained Treg ablation over the prolonged time periods required for tumor development in many mouse models of primary cancer, thereby limiting the use of this experimental approach. The most striking available data come from multiple studies demonstrating that Treg depletion can induce cancer regression in mouse sarcomas induced by treatment with the chemical carcinogen methylcholanthrene [22–24]. However, data demonstrating a role for Tregs in the modulation of cancer development in genetically-driven mouse models of primary carcinomas are necessary to demonstrate the generality of this concept in different types of cancer (Box 2). Furthermore, in order to elucidate the impact of Tregs on tumorigenesis, it is important to consider recent advances in our understanding of the complexities and specialized roles of Tregs within various inflammatory contexts.
Context-dependent Treg function
Tregs regulate a multitude of inflammatory processes [7], but the mechanisms by which these cells exhibit such diverse functions are not well understood. In recent years, a number of studies have helped to illuminate basic principles underlying the diversity of Treg function (also reviewed by [25]). In one such study, the authors demonstrated that Treg expression of T-bet, a transcription factor that specifies the differentiation of T helper type 1 (Th1) T cells, was required for the regulation of Th1 inflammation [26]. In other studies, it was shown that Treg expression of IRF4 and STAT3, transcription factors that are essential for the differentiation of Th2 and Th17 T cells, respectively, were critical for the suppression of the respective classes of inflammatory responses [27,28]. More recently, these concepts have been extended by the findings of a study of Tregs associated with the visceral adipose tissue (VAT) [29]. The authors demonstrated that PPAR-γ, a nuclear receptor essential for adipocyte differentiation, was highly expressed by VAT-associated Tregs and was required for optimal Treg maintenance and/or infiltration within adipose tissue.
Taken together, these and other studies demonstrate that in order to regulate a particular inflammatory state or anatomical site, Tregs make context-dependent adaptations, adopting transcriptional programs that are central to key cell types operating within the inflammatory milieu. Thus, Tregs can exhibit chameleon-like properties, changing “color” to persist and function within the “foliage” of a given environment.
This concept of Treg “adaptation” could help to explain the potentially paradoxical role of Tregs in human colorectal carcinoma (CRC). Contrary to many other solid tumors, a high density of Tregs in CRCs is generally associated with either a neutral or favorable prognosis [14,30]. Additionally, it was recently reported that high frequencies of IL-17-producing cells were predictive of poor prognosis in CRC [31]. Moreover, complicating the matter, multiple groups have reported that a substantial fraction of Foxp3+ Tregs isolated from CRC samples produce IL-17 [32–34]. How can these findings be explained? A recent review suggested a model in which Tregs dampen tumor development by suppressing pro-inflammatory Th17 responses, which are thought to promote CRC [30]. However, if Tregs are dampening inflammation, why are they producing “pro-inflammatory” cytokines such as IL-17? In this light, it is possible that Tregs are adapting to contextual cues within a Th17-skewed inflammatory environment, and adopting a Th17-like phenotype in order to suppress inflammation.
The principles of Treg diversity and adaptation have significant implications for our understanding of tumor-associated Tregs. In future work, it will be important to determine the gene expression profiles of tumor-infiltrating Tregs relative to Tregs isolated from lymphoid organs or blood (Box 2), in an effort to identify specific transcriptional programs that may provide insight into the specialized in situ function(s) of Tregs infiltrating tumor lesions. In addition, the diversity of Treg function in varying inflammatory contexts suggests the possibility of therapeutic modulation of properties that are unique to tumor-associated Tregs. With this strategy in mind, it was demonstrated that administration of Pio, a synthetic agonist of PPAR-γ, induced the specific accumulation of Tregs in the adipose tissue of mice on a high-fat diet, leaving splenic Treg numbers unchanged [29]. Thus, if transcriptional programs that are specifically expressed by tumor-associated Tregs can be identified, it may be possible to selectively modulate these cells, leaving Tregs located elsewhere in the body unaffected. Finally, the versatility of Treg function raises the possibility that Tregs may perform multiple functions within the tumor environment, independently of a role in the suppression of inflammation or adaptive immune responses.
Newly defined roles for tumor-associated Tregs?
Given the principles of context-dependent diversity of Treg function discussed above, it is important to consider the following questions. What are the environmental cues sensed by tumor-associated Tregs, and what functional responses are elicited under these circumstances? In this regard, emerging evidence suggests that Tregs may exhibit specialized functions that impact both angiogenesis and metastasis within the tumor environment.
A recent study presented evidence suggesting a direct role for Tregs in the induction of angiogenesis under hypoxic conditions [35]. In experiments utilizing a transplantable ovarian cancer cell line, it was demonstrated that overexpression of the chemokine CCL28 resulted in increased angiogenesis and VEGFA production, which was diminished following Treg depletion. In other experiments, human Tregs cultured in hypoxic conditions produced VEGFA and promoted the in vitro differentiation of endothelial cells. Thus, Tregs may play a functional role in the promotion of angiogenesis through the production of VEGFA. In future work, it will be important to determine whether Treg-derived VEGFA is critical for angiogenesis in vivo in this tumor model and in other mouse models of genetically-driven cancer.
Another study reported evidence suggesting a role for RANK ligand (RANKL) expressed by regulatory T cells in the promotion of breast cancer metastasis [36]. In experiments in which a mammary carcinoma cell line was implanted in the mammary gland, and metastasis to the lung was monitored, metastasis did not occur in mice lacking T and B cells. Transfer of CD4+CD25+ Tregs restored metastasis, and this effect was diminished by blockade of RANKL. The authors concluded that Tregs promote cancer metastasis in a RANKL-dependent manner. In a different study, analysis of bone marrow cells from prostate cancer patients demonstrated that RANK-expressing dendritic cells induced Treg expansion in vitro, suggesting a role for RANK-RANKL signaling in the proliferation or maintenance of Treg populations [37]. Building from these observations, it will be interesting to determine whether conditional ablation of RANKL expression on Tregs modulates tumor progression or metastasis in murine cancer models, and to elucidate the cellular and molecular mechanisms driving metastasis downstream of RANKL.
Together, these studies present intriguing evidence suggesting that Tregs respond to diverse contextual cues within the tumor environment, and develop functional responses accordingly (Figure 1). These functions appear to be independent of the well-established role for Tregs in the suppression of adaptive immune responses and the regulation of inflammation, and therefore extend our understanding of Treg functional diversity. Thus, heterogeneity of Treg function in different contexts may help to explain the differing prognostic significance of Tregs infiltrating various types of cancer [14].
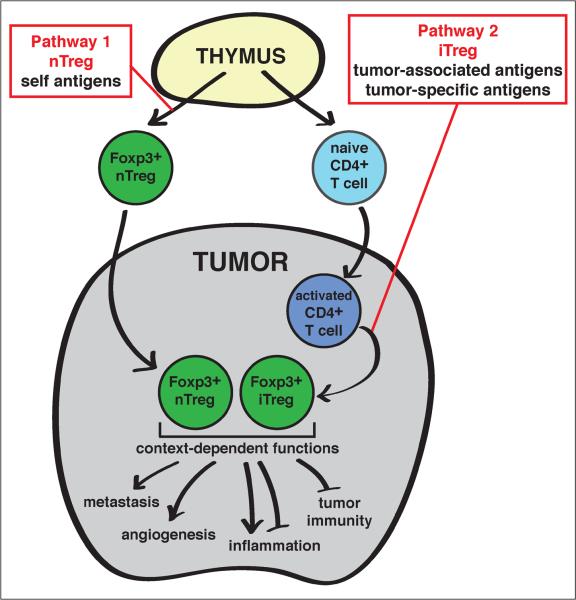
Figure 1. Conceptual model describing the biology of tumor-associated Tregs.
Tumor-associated Tregs are thought to follow one of two developmental pathways in order to enter the Foxp3+ Treg lineage. First, a developing thymocyte may recognize self antigen presented within the thymus during T cell maturation (Pathway 1, referred to as natural Tregs (“nTregs”)). Alternatively, a conventional CD4+ T cell may encounter a tumor-associated (self) or tumor-specific (“neo”) antigen in the tumor environment, become activated, and under the influence of an immunosuppressive tumor microenvironment, differentiate into a Foxp3+ Treg (Pathway 2, referred to as induced Tregs “iTregs”)). Next, within the tumor environment, Tregs may respond to context-dependent inflammatory signals (e.g. Th1, Th2, or Th17 inflammation), the tissue or organ type (e.g. colon, breast, or prostate) and even the immediate proximal microenvironment (e.g. stroma, tumor bed, or lymphoid cluster). From these environmental cues, Tregs are capable of mediating distinct functions, which may include promotion of angiogenesis or metastasis, regulation of inflammation, and suppression of anti-tumor adaptive immune responses.
Treg lineage stability
The stability of Foxp3+ Tregs has recently become a topic of substantial debate, and could represent yet another source of Treg functional heterogeneity. Several early studies demonstrated that under certain conditions, Foxp3+ Tregs can differentiate into Foxp3neg cells exhibiting effector function [38–41], suggesting that Tregs may undergo “reprogramming” in response to particular inflammatory stimuli. Potential instability of the Treg lineage raises the possibility of therapeutic intervention to “disarm” tumor-associated Tregs by driving their differentiation into Foxp3neg cells lacking suppressor function. On the other hand, several recent reports have challenged this idea by providing evidence that Foxp3+ Tregs constitute a stable lineage. First, using inducible labeling to track the stability of Foxp3 expression in adult mice, it was found that Foxp3+ Tregs exhibited a high degree of stability under steady-state and inflammatory conditions [42]. In addition, two studies demonstrated the presence of a minor population of uncommitted nonregulatory T cells that exhibit transient, promiscuous Foxp3 expression [40,43], suggesting that the instability of Foxp3+ Tregs observed by other groups may in fact reflect the presence of this nonregulatory Foxp3-expressing population. Given the conflicting findings in the field, the stability and plasticity of Treg populations remain active areas of investigation. Further work will be needed to define the factors regulating promiscuous expression of Foxp3 by nonregulatory T cells, and to determine whether the concepts of Treg instability and reprogramming are relevant in the context of cancer.
Treg trafficking to tumors
In recent years, there has been considerable interest in understanding the inflammatory signals that drive Treg recruitment into tumors, in an effort to develop strategies for the selective blockade of Treg trafficking. An early study implicated the chemokine CCL22 and its receptor CCR4 as important factors driving Treg trafficking to human ovarian cancers [44]. In this study, it was shown that tumor ascites contained high amounts of CCL22, and that CCL22 induced Treg migration in vitro. Furthermore, in a xenotransplant tumor model, administration of anti-CCL22 antibody reduced the trafficking of adoptively transferred human Tregs to the transplanted tumor. Following this initial report, a growing number of chemokine ligand-receptor pairs, often referred to as chemokine-receptor “axes”, have been implicated in the trafficking of Tregs to different types of cancer (reviewed recently by [45]).
The importance of chemokine-driven Treg trafficking to tumors lesions raises the possibility of therapeutic intervention based on the blockade of this process. In this regard, a number of considerations may influence the feasibility and efficacy of such approaches. First, while many studies have demonstrated that a particular chemokine is sufficient to induce Treg trafficking, due to the redundancy and promiscuity of many chemokine ligand-receptor pairs [46], it will be important to determine whether the chemokine-receptor axis in question is essential for Treg trafficking (Box 2). If Tregs are able to use alternate chemokine-receptor axes, blockade of a specific axis is unlikely to be effective. In this regard, a recent study demonstrated that treatment of mice bearing established pancreatic tumors with a CCR5 antagonist reduced Treg infiltration within the tumor and slowed tumor growth [47], demonstrating a non-redundant role for CCR5 signaling that could be exploited to slow tumor growth in this model. Second, as discussed above, the ability of Tregs to adopt transcriptional programs similar to the target cells under regulation suggests that a given chemokine receptor may not be expressed only by tumor-infiltrating Tregs, but may also be expressed by other immune cell types within the tumor. Thus, disruption of a given axis may inhibit the trafficking of both Tregs and immune effector cells, potentially reducing therapeutic efficacy. Third, in treating established tumors, it will be critical to determine whether blockade of a chemokine-receptor axis will impact tumor-infiltrating Treg populations that are already present within the tumor. Future work will be required to address these concepts for chemokine-receptor axes that have been implicated in Treg trafficking to different types of cancer.
Treg localization within the tumor
Our heightened appreciation of the diversity of Treg function suggests that following recruitment into the tumor environment, Tregs may play multiple roles within a single tumor. For example, it is possible that Tregs localized in different regions within a cancer tissue may have distinct functions. Consistent with this idea, a survey of human mammary carcinomas [48] revealed that a high density of Foxp3+ cells within lymphoid-enriched areas of the tumor was associated with poor survival, while a high density of Foxp3+ cells distributed elsewhere in the tumor exhibited no association with disease outcome. As an additional example, a survey of colorectal carcinoma revealed that elevated Treg density within normal mucosa was associated with poor prognosis, while a high Treg density within tumor tissue correlated with improved outcome [49]. These and other studies highlight the importance of quantifying Foxp3+ cells in different areas of a tissue sample (Box 2), including the tumor bed, the stroma, clusters or aggregates of lymphoid cells, preinvasive lesions, the zone of tumor invasion, and normal tissue adjacent to a malignant region. In-depth analyses of the relationship between disease outcome and the density of Tregs within these different regions may provide important clues as to the function and impact of tumor-infiltrating Tregs, and may serve as useful diagnostic markers.
Treg development and antigen specificity
If Treg heterogeneity exists within a given tumor, what are the drivers of functional diversity? The antigen specificity of a T cell, conferred by expression of the T cell antigen receptor (TCR), is an important determinant of many aspects of T cell biology. For tumor-infiltrating Tregs, antigen recognition is likely to drive development into the Foxp3+ lineage, activation and differentiation within the tumor environment, and co-localization with cellular interaction partners (antigen presenting cells) within the tumor. Thus, an understanding of the nature of the antigens recognized by tumor-associated Tregs would provide important insights into the biology of these cells.
First, to review the necessary conceptual framework, we will discuss recent advances in our understanding of the developmental origins of Tregs, and the nature of the antigens recognized by these cells, two concepts that are closely interrelated (Figure 1). Foxp3+ Tregs are thought to develop by two pathways. “Natural” Tregs (nTregs), also referred to as thymic Tregs (tTregs), originate in the thymus, the major site of T cell development and maturation [50]. nTregs are thought to recognize self antigens [51–53], although little is known regarding the identity or nature of these antigens. “Induced” Tregs (iTregs), also called adaptive Tregs (aTregs) or peripherally-induced Tregs (pTregs), develop from conventional T cell precursors at extrathymic sites throughout the body, in a process that is augmented by exposure to TGF-β and retinoic acid [54]. Whether iTregs recognize a similar array of self antigens as nTregs, or whether they recognize a distinct set of antigens has been unclear. In addition, it has been unclear whether nTregs and iTregs serve redundant functions, or whether there is a “division of labor” between these two subsets [54,55].
Recent work has provided considerable insight into these issues [56,57]. These studies demonstrated that CNS1, a conserved non-coding regulatory element in the Foxp3 locus, is dispensable for nTreg development in the thymus, but is crucial for iTreg generation in the periphery [56,57]. Importantly, mice deficient in iTregs due to a targeted mutation in CNS1 exhibited spontaneous pathology exclusively at mucosal sites such as the lungs and gastrointestinal tract [56]. Extending these findings, a recent study utilizing CNS1-mutant mice demonstrated a role for iTregs in the maintenance of fetal-maternal tolerance during pregnancy, in a process involving the recognition of paternal antigens [58]. Together, these findings provide strong support for a model in which iTregs serve a crucial non-redundant function in the maintenance of immune tolerance and homeostasis, by functioning primarily at surfaces that interface with the external environment, with iTreg differentiation driven by recognition of foreign antigens that are not presented in the thymus.
This model of iTreg distribution and function is consistent with recent findings showing that a substantial fraction of colonic Tregs recognize antigens that derive from commensal microorganisms [59]. Expression of colonic Treg-derived TCRs did not facilitate Treg development in the thymus, but instead drove the accumulation of Tregs in the colon, consistent with an extrathymic origin of these Tregs.
Integrating these new findings with existing evidence, we can envision a simplified model of Treg development and specificity. This model predicts that nTregs recognize self antigens and function to maintain organ-specific immune tolerance to self tissues, while iTregs recognize foreign antigens (such as those derived from commensal bacteria, food, or allogeneic tissue), and function to maintain immune and tissue homeostasis at surfaces that interface with the external environment. Thus, iTregs extend the Treg repertoire to include Tregs specific for antigens that are not presented for recognition in the thymus during T cell development [60]. In addition, iTreg development enables new Treg specificities to be generated throughout the lifetime of an organism in response to changes in the environment.
What are the implications of these concepts on our understanding of the developmental pathways and antigen specificities of tumor-associated Tregs? We consider two potential scenarios (Figure 1), which are not mutually exclusive. First, a developing tumor may recruit pre-existing thymic-derived nTregs reactive to self antigens (Figure 1, Pathway 1). Second, tumors may induce the de novo generation of Tregs by driving the differentiation of CD4+ effector T cells reactive to tumor-associated antigens into the Foxp3+ iTreg lineage (Figure 1, Pathway 2). This second process is a potential mechanism by which tumors evade immune destruction, due to the simultaneous diversion of effector T cells into a different fate and the generation of new Tregs with suppressive activity. In order to determine the relative contributions of these two pathways in the context of cancer, it is important to elucidate not only the developmental origins of Treg populations, but also the nature of the antigens recognized by these cells.
It has been difficult to assess the developmental origins of endogenous tumor-associated Treg populations for two reasons. First, it is not possible to recapitulate thymic and peripheral development of human Tregs using in vitro approaches. Second, the lack of definitive markers for the delineation of nTregs and iTregs (Box 2) precludes direct determination of the developmental origin of these cells. In the absence of such information regarding Treg development, there is considerable interest in determining the nature of the antigens recognized by Tregs. The rationale is that the antigens recognized by nTregs must be presented in the thymus for recognition during T cell development. If Tregs are found to recognize tumor-associated or tumor-specific antigens that are aberrantly or uniquely expressed by tumor cells following cancer development, these antigens are unlikely to be presented in the thymus to drive nTreg development. Therefore, Tregs recognizing tumor-associated or tumor-specific antigens are likely to be iTregs (Figure 1).
Two recent reports present indirect evidence that provide insight into Treg antigen specificity. In these studies, sequence analyses of TCRs expressed by purified T cell subsets isolated from transplantable [61] or chemically-induced murine tumors [62] revealed that the TCR repertoire of CD4+Foxp3+ Tregs was distinct from the repertoire expressed by CD4+Foxp3neg T effector cells. These data suggest that the antigens recognized by Tregs within the tumor are different than those recognized by T effector cells. Furthermore, since TCR sequence overlap was not observed between Foxp3+ and Foxp3neg subsets, these data suggest that the in situ differentiation of T effector cells into iTregs is negligible within the tumor. If this is a common process, one would anticipate at least a small, but detectable, overlap between the TCR repertoires of each subset, representing a “snapshot” of effector T cell populations undergoing conversion to the iTreg lineage. An important caveat of these studies is that due to the low number of TCR sequences analyzed in these studies, it is unclear whether the sampling of sequences represents the true breadth of the TCR repertoire. In future work, it will be important to extend these findings to other murine cancer models, including models of oncogene-induced malignancies in other tissues, as well as CD4+FOXP3+ and CD4+FOXP3neg T cell subsets isolated from primary human tumors (Box 2).
A number of reports provide data suggesting the existence of Tregs reactive to tumor-associated antigens (TAAs) in human cancer patients. In this regard, two studies have utilized in vitro culture-based assays to detect TAA-specific Tregs in the peripheral blood of melanoma patients [63,64]. In these experiments, antigen-specific T cells are first expanded by extended in vitro culture with peptides derived from TAAs, and the presence of antigen-specific Tregs is then determined by assessing proliferation [63] or CD3 downregulation [64]. However, in these assays, it is unclear whether the antigen-specific T cells detected are derived from bona fide Tregs, or originate from naïve T cell precursors that expand and develop regulatory characteristics following in vitro culture. In another approach, a short-term in vitro culture assay was developed to examine the antigen specificities of Tregs isolated from the blood of human colorectal carcinoma (CRC) patients [65]. Using this assay, the authors demonstrated that culture of CD4+CD25+ T cells with dendritic cells pulsed with defined TAA peptides conferred the capacity to suppress the proliferation of polyclonally-activated effector T cells. This activity was observed for a percentage of samples from CRC patients, but was not observed in samples from healthy donors. Collectively, data from these and other studies suggest the possibility that Tregs reactive to peptides derived from TAAs are present at detectable levels in the blood of cancer patients.
In order to solidify the concept that human cancer patients harbor Tregs specific for TAAs, two types of studies will be critical in future work. First, the ex vivo detection of TAA-specific Tregs using peptide/MHC multimer reagents [66–68] would provide direct evidence that Tregs reactive to tumor antigens are present at detectable frequencies in human cancer patients (Box 2). In one such study [69], ex vivo staining of samples from melanoma patients revealed that HLA-DQ multimers bearing a peptide from the Melan-A antigen stained a CD4+ T cell population comprising <0.1% of CD4+ T cells in the peripheral blood, a fraction of which expressed FOXP3. Thus, while FOXP3+CD4+ T cells of this specificity are detectable in some patients, they do not appear to contribute substantially to the polyclonal Treg repertoire in these patients. Second, it will be important to characterize individual TCRs expressed by human Tregs reactive to tumor-associated antigens, in order to evaluate the fine specificity of these receptors (Box 2). This could be achieved by the generation of T cell clones expressing Treg-derived TCRs [70,71], or by retroviral or lentiviral transduction of Treg-derived TCRs into immortalized cell lines [72]. These approaches would allow determination of whether TCRs expressed by tumor-infiltrating Tregs are “specific” for tumor-associated antigens, or whether these TCRs are cross-reactive, conferring promiscuous recognition of multiple peptide antigens.
In all, despite the available evidence highlighted above, much remains to be learned about the developmental origins and antigen specificities of tumor-associated Tregs. Substantial advances in these areas will require the identification of definitive markers of Treg origin, and means to directly characterize and enumerate antigen-specific Tregs isolated from primary tumors (Box 2).
Concluding remarks
The studies discussed in this review exemplify our emerging appreciation for the multi-faceted role that Tregs may play in the context of cancer. Recent progress in the field reveals that Tregs, once thought to be of “single-minded” purpose of dampening innate and adaptive immunity, instead exhibit substantial versatility, responding to varying contextual cues and adopting specialized functions accordingly. This functional diversity may be a critical factor underlying the observed differences in the prognostic significance of Treg density in different types of cancer, and illustrates the importance of studying Tregs contextually. We anticipate that the complexity of Treg biology will provide a fertile area for future research, and may reveal novel means for the selective therapeutic targeting of tumor-associated Tregs.
Box 1. Markers for the analysis of Tregs.
As described below, progress in the field has been slowed by the lack of definitive markers for the identification of Tregs and the delineation of Tregs of thymic (nTreg) or extrathymic (iTreg) origin.
Identification of Tregs
Foxp3
Foxp3, a transcription factor that is essential for Treg development and lineage specification [8–11], is considered to be the most definitive marker of CD4+ regulatory T cells. However, in humans, FOXP3 can be expressed by activated T cells lacking regulatory activity [13,74], complicating its use as a Treg marker in human studies. Approaches incorporating the use of additional cell-surface markers have been used to more precisely define the phenotypic subsets of human FOXP3+ cells that exhibit suppressive properties in vitro. For example, in experiments utilizing the cell surface marker CD127 (the IL-7 receptor α-chain), it was found that suppressive activity was restricted to the CD4+Foxp3+CD127low subset [75]. In another study using the markers CD45RA (an isoform of the CD45 phosphatase) and CD25 (the IL-2 receptor α-chain), it was found that CD45RA+ FOXP3low and CD45RA− FOXP3high subsets exhibited suppressive activity, while CD45RA− FOXP3low cells produced cytokine and were not suppressive [74].
CD25
Seminal work by Sakaguchi and colleagues [76] demonstrated that regulatory T cell activity in mice was highly enriched in the CD4+CD25+ T cell subset. Based on these and other studies, CD25 was the most commonly used Treg marker until the development of anti-Foxp3 antibodies [77] and Foxp3 reporter mice [78] in 2005. However, subsequent work has revealed the presence of suppressive FOXP3+ T cells lacking CD25 expression in both mice [78] and humans [75].
Delineation of nTregs and iTregs
Helios
While initial reports suggested that the transcription factor Helios is a marker of thymic-derived nTregs [79], subsequent studies have revealed that Helios can also be expressed by iTregs generated experimentally [80–82]. Therefore, while Helios may serve as a marker of activated Tregs of increased suppressive capacity [82,83], it is not considered a definitive marker of Treg origin.
Methylation status of Foxp3 gene
Recently, several assays have been developed for the epigenetic analysis of the methylation status of CpG motifs within conserved elements of the Foxp3 locus [84–89]. Complete demethylation of these regions is associated with stable Foxp3 expression by T cells, and is a hallmark of thymic-derived nTregs [85–88]. However, while Foxp3-expressing cells generated by experimental manipulation often exhibit partial demethylation and unstable Foxp3 expression [85–88], it is unclear whether naturally occurring iTregs (existing in the absence of experimental manipulation) exhibit complete or partial demethylation of this region. In addition, available assays rely on the analysis of bulk populations, and do not provide information at the single-cell level.
Box 2. Important areas of future research
Loss-of-function evidence demonstrating that Tregs modulate tumor progression in vivo in genetically-driven murine cancer models
Gene expression profiling of tumor-associated Tregs to identify unique transcriptional programs
Quantification of Tregs localized in different regions of a cancer, and multivariate analysis of association with disease outcome
Identification of definitive markers to delineate and isolate Tregs in human samples
Identification of definitive markers to delineate nTregs and iTregs
Direct identification and enumeration of antigen-specific Tregs using peptide/MHC multimers
Characterization of the fine specificity / cross-reactivity of TCRs expressed by Tregs
Analysis of TCR repertoire overlap for CD4+FOXP3+ and CD4+FOXP3neg T cell subsets isolated from human tumors
Characterization of chemokine receptors and homing receptors critical for Treg function and trafficking to the tumor
Acknowledgements
We thank Fabiola Rivas for critical reading of the manuscript. P.A.S. is partially supported by the following sources: R01 (#1R01CA160371-01), a Cancer Research Institute Investigator Award, a Cancer Research Foundation Young Investigator Award, an American Cancer Society Institutional Research Grant (#IRG-58-004), and the University of Chicago Comprehensive Cancer Center (including Support Grant #P30 CA14599).
Glossary Box
- 1. “naturally occurring Treg”
any Foxp3+ Treg that exists naturally, in the absence of experimental manipulation (likely a mixture of nTregs and iTregs)
- 2. “natural Treg (nTreg)”
a Foxp3+ Treg that develops in the thymus
- 3. “induced Treg (iTreg)”
a Foxp3+ Treg that differentiates into the Foxp3+ lineage extrathymically
- 4. “tumor-specific (neo-) antigen”
a unique antigen that is exclusively expressed by tumor cells, usually generated by mutation or changes in post-translational modification [73]
- 5. “tumor-associated antigen”
a non-mutated self antigen that is aberrantly expressed or over-expressed by tumor cells, but is also expressed by some normal cells [73]
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hodi FS, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363(8):711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galluzzi L, et al. Trial Watch: Adoptive cell transfer immunotherapy. Oncoimmunology. 2012;1(3):306–315. doi: 10.4161/onci.19549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kantoff PW, et al. Sipuleucel-T immunotherapy for castration-resistant prostate cancer. N Engl J Med. 2010;363(5):411–422. doi: 10.1056/NEJMoa1001294. [DOI] [PubMed] [Google Scholar]
- 4.Colombo MP, Piconese S. Regulatory-T-cell inhibition versus depletion: the right choice in cancer immunotherapy. Nat Rev Cancer. 2007;7(11):880–887. doi: 10.1038/nrc2250. [DOI] [PubMed] [Google Scholar]
- 5.Curiel TJ. Regulatory T cells and treatment of cancer. Curr Opin Immunol. 2008;20(2):241–246. doi: 10.1016/j.coi.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sakaguchi S. Regulatory T cells: history and perspective. Methods Mol Biol. 2011;707:3–17. doi: 10.1007/978-1-61737-979-6_1. [DOI] [PubMed] [Google Scholar]
- 7.Josefowicz SZ, et al. Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol. 2012;30:531–564. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brunkow ME, et al. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet. 2001;27(1):68–73. doi: 10.1038/83784. [DOI] [PubMed] [Google Scholar]
- 9.Lyon MF, et al. The scurfy mouse mutant has previously unrecognized hematological abnormalities and resembles Wiskott-Aldrich syndrome. Proc Natl Acad Sci U S A. 1990;87(7):2433–2437. doi: 10.1073/pnas.87.7.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fontenot JD, et al. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4(4):330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 11.Khattri R, et al. An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat Immunol. 2003;4(4):337–342. [PubMed] [Google Scholar]
- 12.Bennett CL, et al. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet. 2001;27(1):20–21. doi: 10.1038/83713. [DOI] [PubMed] [Google Scholar]
- 13.Gavin MA, et al. Single-cell analysis of normal and FOXP3-mutant human T cells: FOXP3 expression without regulatory T cell development. Proc Natl Acad Sci U S A. 2006;103(17):6659–6664. doi: 10.1073/pnas.0509484103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deleeuw RJ, et al. The Prognostic Value of FoxP3+ Tumor-Infiltrating Lymphocytes in Cancer: A Critical Review of the Literature. Clin Cancer Res. 2012;18(11):3022–3029. doi: 10.1158/1078-0432.CCR-11-3216. [DOI] [PubMed] [Google Scholar]
- 15.Byrne WL, et al. Targeting regulatory T cells in cancer. Cancer Res. 2011;71(22):6915–6920. doi: 10.1158/0008-5472.CAN-11-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pere H, et al. Comprehensive analysis of current approaches to inhibit regulatory T cells in cancer. Oncoimmunology. 2012;1(3):326–333. doi: 10.4161/onci.18852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Menetrier-Caux C, et al. Targeting regulatory T cells. Targeted oncology. 2012;7(1):15–28. doi: 10.1007/s11523-012-0208-y. [DOI] [PubMed] [Google Scholar]
- 18.Shevach EM. From vanilla to 28 flavors: multiple varieties of T regulatory cells. Immunity. 2006;25(2):195–201. doi: 10.1016/j.immuni.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 19.Grivennikov SI, et al. Immunity, inflammation, and cancer. Cell. 2010;140(6):883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim J, et al. Cutting edge: depletion of Foxp3+ cells leads to induction of autoimmunity by specific ablation of regulatory T cells in genetically targeted mice. J Immunol. 2009;183(12):7631–7634. doi: 10.4049/jimmunol.0804308. [DOI] [PubMed] [Google Scholar]
- 21.Kim JM, et al. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat Immunol. 2007;8(2):191–197. doi: 10.1038/ni1428. [DOI] [PubMed] [Google Scholar]
- 22.Chamoto K, et al. 3-Methylcholanthrene-induced transforming growth factor-beta-producing carcinomas, but not sarcomas, are refractory to regulatory T cell-depletion therapy. Cancer Sci. 2010;101(4):855–861. doi: 10.1111/j.1349-7006.2009.01469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Betts G, et al. The impact of regulatory T cells on carcinogen-induced sarcogenesis. Br J Cancer. 2007;96(12):1849–1854. doi: 10.1038/sj.bjc.6603824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teng MW, et al. Conditional regulatory T-cell depletion releases adaptive immunity preventing carcinogenesis and suppressing established tumor growth. Cancer Res. 2010;70(20):7800–7809. doi: 10.1158/0008-5472.CAN-10-1681. [DOI] [PubMed] [Google Scholar]
- 25.Campbell DJ, Koch MA. Phenotypical and functional specialization of FOXP3+ regulatory T cells. Nat Rev Immunol. 2011;11(2):119–130. doi: 10.1038/nri2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koch MA, et al. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat Immunol. 2009;10(6):595–602. doi: 10.1038/ni.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng Y, et al. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature. 2009;458(7236):351–356. doi: 10.1038/nature07674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaudhry A, et al. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science. 2009;326(5955):986–991. doi: 10.1126/science.1172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cipolletta D, et al. PPAR-gamma is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature. 2012;486(7404):549–553. doi: 10.1038/nature11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ladoire S, et al. Prognostic role of FOXP3+ regulatory T cells infiltrating human carcinomas: the paradox of colorectal cancer. Cancer Immunol Immunother. 2011;60(7):909–918. doi: 10.1007/s00262-011-1046-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tosolini M, et al. Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, th2, treg, th17) in patients with colorectal cancer. Cancer Res. 2011;71(4):1263–1271. doi: 10.1158/0008-5472.CAN-10-2907. [DOI] [PubMed] [Google Scholar]
- 32.Blatner NR, et al. In colorectal cancer mast cells contribute to systemic regulatory T-cell dysfunction. Proc Natl Acad Sci U S A. 2010;107(14):6430–6435. doi: 10.1073/pnas.0913683107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang S, et al. Foxp3+IL-17+ T cells promote development of cancer-initiating cells in colorectal cancer. J Leukoc Biol. 2010;89(1):85–91. doi: 10.1189/jlb.0910506. [DOI] [PubMed] [Google Scholar]
- 34.Ma C, Dong X. Colorectal cancer-derived Foxp3(+) IL-17(+) T cells suppress tumour-specific CD8+ T cells. Scand J Immunol. 2011;74(1):47–51. doi: 10.1111/j.1365-3083.2011.02539.x. [DOI] [PubMed] [Google Scholar]
- 35.Facciabene A, et al. Tumour hypoxia promotes tolerance and angiogenesis via CCL28 and T(reg) cells. Nature. 2011;475(7355):226–230. doi: 10.1038/nature10169. [DOI] [PubMed] [Google Scholar]
- 36.Tan W, et al. Tumour-infiltrating regulatory T cells stimulate mammary cancer metastasis through RANKL-RANK signalling. Nature. 2011;470(7335):548–553. doi: 10.1038/nature09707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao E, et al. Regulatory T cells in the bone marrow microenvironment in patients with prostate cancer. Oncoimmunology. 2012;1(2):152–161. doi: 10.4161/onci.1.2.18480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsuji M, et al. Preferential generation of follicular B helper T cells from Foxp3+ T cells in gut Peyer's patches. Science. 2009;323(5920):1488–1492. doi: 10.1126/science.1169152. [DOI] [PubMed] [Google Scholar]
- 39.Zhou X, et al. Instability of the transcription factor Foxp3 leads to the generation of pathogenic memory T cells in vivo. Nat Immunol. 2009;10(9):1000–1007. doi: 10.1038/ni.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Komatsu N, et al. Heterogeneity of natural Foxp3+ T cells: a committed regulatory T-cell lineage and an uncommitted minor population retaining plasticity. Proc Natl Acad Sci U S A. 2009;106(6):1903–1908. doi: 10.1073/pnas.0811556106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cong Y, et al. A dominant, coordinated T regulatory cell-IgA response to the intestinal microbiota. Proc Natl Acad Sci U S A. 2009;106(46):19256–19261. doi: 10.1073/pnas.0812681106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rubtsov YP, et al. Stability of the regulatory T cell lineage in vivo. Science. 2010;329(5999):1667–1671. doi: 10.1126/science.1191996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miyao T, et al. Plasticity of Foxp3(+) T cells reflects promiscuous Foxp3 expression in conventional T cells but not reprogramming of regulatory T cells. Immunity. 2012;36(2):262–275. doi: 10.1016/j.immuni.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 44.Curiel TJ, et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med. 2004;10(9):942–949. doi: 10.1038/nm1093. [DOI] [PubMed] [Google Scholar]
- 45.Mailloux AW, Young MR. Regulatory T-cell trafficking: from thymic development to tumor-induced immune suppression. Crit Rev Immunol. 2010;30(5):435–447. doi: 10.1615/critrevimmunol.v30.i5.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zlotnik A, Yoshie O. The chemokine superfamily revisited. Immunity. 2012;36(5):705–716. doi: 10.1016/j.immuni.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan MC, et al. Disruption of CCR5-dependent homing of regulatory T cells inhibits tumor growth in a murine model of pancreatic cancer. J Immunol. 2009;182(3):1746–1755. doi: 10.4049/jimmunol.182.3.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gobert M, et al. Regulatory T cells recruited through CCL22/CCR4 are selectively activated in lymphoid infiltrates surrounding primary breast tumors and lead to an adverse clinical outcome. Cancer Res. 2009;69(5):2000–2009. doi: 10.1158/0008-5472.CAN-08-2360. [DOI] [PubMed] [Google Scholar]
- 49.Salama P, et al. Tumor-infiltrating FOXP3+ T regulatory cells show strong prognostic significance in colorectal cancer. J Clin Oncol. 2009;27(2):186–192. doi: 10.1200/JCO.2008.18.7229. [DOI] [PubMed] [Google Scholar]
- 50.Hsieh CS, et al. Selection of regulatory T cells in the thymus. Nat Rev Immunol. 2012;12(3):157–167. doi: 10.1038/nri3155. [DOI] [PubMed] [Google Scholar]
- 51.Hsieh CS, et al. Recognition of the peripheral self by naturally arising CD25+ CD4+ T cell receptors. Immunity. 2004;21(2):267–277. doi: 10.1016/j.immuni.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 52.Jordan MS, et al. Thymic selection of CD4+CD25+ regulatory T cells induced by an agonist self-peptide. Nat Immunol. 2001;2(4):301–306. doi: 10.1038/86302. [DOI] [PubMed] [Google Scholar]
- 53.Apostolou I, et al. Origin of regulatory T cells with known specificity for antigen. Nat Immunol. 2002;3(8):756–763. doi: 10.1038/ni816. [DOI] [PubMed] [Google Scholar]
- 54.Bilate AM, Lafaille JJ. Induced CD4+Foxp3+ regulatory T cells in immune tolerance. Annu Rev Immunol. 2012;30:733–758. doi: 10.1146/annurev-immunol-020711-075043. [DOI] [PubMed] [Google Scholar]
- 55.Curotto de Lafaille MA, Lafaille JJ. Natural and adaptive foxp3+ regulatory T cells: more of the same or a division of labor? Immunity. 2009;30(5):626–635. doi: 10.1016/j.immuni.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 56.Josefowicz SZ, et al. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature. 2012;482(7385):395–399. doi: 10.1038/nature10772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zheng Y, et al. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 2010;463(7282):808–812. doi: 10.1038/nature08750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Samstein RM, et al. Extrathymic generation of regulatory T cells in placental mammals mitigates maternal-fetal conflict. Cell. 2012;150(1):29–38. doi: 10.1016/j.cell.2012.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lathrop SK, et al. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011 doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Haribhai D, et al. A requisite role for induced regulatory T cells in tolerance based on expanding antigen receptor diversity. Immunity. 2011;35(1):109–122. doi: 10.1016/j.immuni.2011.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sainz-Perez A, et al. The T-cell receptor repertoire of tumor-infiltrating regulatory T lymphocytes is skewed towards public sequences. Cancer Res. 2012 doi: 10.1158/0008-5472.CAN-12-0277. [DOI] [PubMed] [Google Scholar]
- 62.Hindley JP, et al. Analysis of the T-cell receptor repertoires of tumor-infiltrating conventional and regulatory T cells reveals no evidence for conversion in carcinogen-induced tumors. Cancer Res. 2011;71(3):736–746. doi: 10.1158/0008-5472.CAN-10-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vence L, et al. Circulating tumor antigen-specific regulatory T cells in patients with metastatic melanoma. Proc Natl Acad Sci U S A. 2007;104(52):20884–20889. doi: 10.1073/pnas.0710557105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ebert LM, et al. A novel method for detecting antigen-specific human regulatory T cells. J Immunol Methods. 2012;377(1–2):56–61. doi: 10.1016/j.jim.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 65.Bonertz A, et al. Antigen-specific Tregs control T cell responses against a limited repertoire of tumor antigens in patients with colorectal carcinoma. J Clin Invest. 2009;119(11):3311–3321. doi: 10.1172/JCI39608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vollers SS, Stern LJ. Class II major histocompatibility complex tetramer staining: progress, problems, and prospects. Immunology. 2008;123(3):305–313. doi: 10.1111/j.1365-2567.2007.02801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ayyoub M, et al. Monitoring of NY-ESO-1 specific CD4+ T cells using molecularly defined MHC class II/His-tag-peptide tetramers. Proc Natl Acad Sci U S A. 2010;107(16):7437–7442. doi: 10.1073/pnas.1001322107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cecconi V, et al. Use of MHC class II tetramers to investigate CD4+ T cell responses: problems and solutions. Cytometry A. 2008;73(11):1010–1018. doi: 10.1002/cyto.a.20603. [DOI] [PubMed] [Google Scholar]
- 69.Jandus C, et al. Tumor antigen-specific FOXP3+ CD4 T cells identified in human metastatic melanoma: peptide vaccination results in selective expansion of Th1-like counterparts. Cancer Res. 2009;69(20):8085–8093. doi: 10.1158/0008-5472.CAN-09-2226. [DOI] [PubMed] [Google Scholar]
- 70.Wang HY, et al. Recognition of a new ARTC1 peptide ligand uniquely expressed in tumor cells by antigen-specific CD4+ regulatory T cells. J Immunol. 2005;174(5):2661–2670. doi: 10.4049/jimmunol.174.5.2661. [DOI] [PubMed] [Google Scholar]
- 71.Wang HY, et al. Tumor-specific human CD4+ regulatory T cells and their ligands: implications for immunotherapy. Immunity. 2004;20(1):107–118. doi: 10.1016/s1074-7613(03)00359-5. [DOI] [PubMed] [Google Scholar]
- 72.Frecha C, et al. Stable transduction of quiescent T cells without induction of cycle progression by a novel lentiviral vector pseudotyped with measles virus glycoproteins. Blood. 2008;112(13):4843–4852. doi: 10.1182/blood-2008-05-155945. [DOI] [PubMed] [Google Scholar]
- 73.Seremet T, et al. Tumor-specific antigens and immunologic adjuvants in cancer immunotherapy. Cancer J. 2011;17(5):325–330. doi: 10.1097/PPO.0b013e3182326004. [DOI] [PubMed] [Google Scholar]
- 74.Miyara M, et al. Functional delineation and differentiation dynamics of human CD4+ T cells expressing the FoxP3 transcription factor. Immunity. 2009;30(6):899–911. doi: 10.1016/j.immuni.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 75.Liu W, et al. CD127 expression inversely correlates with FoxP3 and suppressive function of human CD4+ T reg cells. J Exp Med. 2006;203(7):1701–1711. doi: 10.1084/jem.20060772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sakaguchi S, et al. Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J Immunol. 1995;155(3):1151–1164. [PubMed] [Google Scholar]
- 77.Roncador G, et al. Analysis of FOXP3 protein expression in human CD4+CD25+ regulatory T cells at the single-cell level. Eur J Immunol. 2005;35(6):1681–1691. doi: 10.1002/eji.200526189. [DOI] [PubMed] [Google Scholar]
- 78.Fontenot JD, et al. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 2005;22(3):329–341. doi: 10.1016/j.immuni.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 79.Thornton AM, et al. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J Immunol. 2010;184(7):3433–3441. doi: 10.4049/jimmunol.0904028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Verhagen J, Wraith DC. Comment on “Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells”. J Immunol. 2010;185(12):7129. doi: 10.4049/jimmunol.1090105. author reply 7130. [DOI] [PubMed] [Google Scholar]
- 81.Gottschalk RA, et al. Expression of Helios in peripherally induced Foxp3+ regulatory T cells. J Immunol. 2011;188(3):976–980. doi: 10.4049/jimmunol.1102964. [DOI] [PubMed] [Google Scholar]
- 82.Zabransky DJ, et al. Phenotypic and functional properties of Helios+ regulatory T cells. PLoS One. 2012;7(3):e34547. doi: 10.1371/journal.pone.0034547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Akimova T, et al. Helios expression is a marker of T cell activation and proliferation. PLoS One. 2011;6(8):e24226. doi: 10.1371/journal.pone.0024226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huehn J, et al. Epigenetic control of FOXP3 expression: the key to a stable regulatory T-cell lineage? Nat Rev Immunol. 2009;9(2):83–89. doi: 10.1038/nri2474. [DOI] [PubMed] [Google Scholar]
- 85.Floess S, et al. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol. 2007;5(2):e38. doi: 10.1371/journal.pbio.0050038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Janson PC, et al. FOXP3 promoter demethylation reveals the committed Treg population in humans. PLoS One. 2008;3(2):e1612. doi: 10.1371/journal.pone.0001612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim HP, Leonard WJ. CREB/ATF-dependent T cell receptor-induced FoxP3 gene expression: a role for DNA methylation. J Exp Med. 2007;204(7):1543–1551. doi: 10.1084/jem.20070109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Baron U, et al. DNA demethylation in the human FOXP3 locus discriminates regulatory T cells from activated FOXP3(+) conventional T cells. Eur J Immunol. 2007;37(9):2378–2389. doi: 10.1002/eji.200737594. [DOI] [PubMed] [Google Scholar]
- 89.Wieczorek G, et al. Quantitative DNA methylation analysis of FOXP3 as a new method for counting regulatory T cells in peripheral blood and solid tissue. Cancer Res. 2009;69(2):599–608. doi: 10.1158/0008-5472.CAN-08-2361. [DOI] [PubMed] [Google Scholar]