Abstract
Background
Samples collected from CF patient airways often contain large amounts of host-derived nucleic acids that interfere with recovery and purification of microbial and viral nucleic acids. This study describes metagenomic and metatranscriptomic methods that address these issues.
Methods
Microbial and viral metagenomes, and microbial metatranscriptomes, were successfully prepared from sputum samples from five adult CF patients.
Results
Contaminating host DNA was dramatically reduced in the metagenomes. Each CF patient presented a unique microbiome; in some Pseudomonas aeruginosa was replaced by other opportunistic bacteria. Even though the taxonomic composition of the microbiomes are very different, the metabolic potentials encoded by the community are very similar. The viral communities were dominated by phages that infect major CF pathogens. The metatranscriptomes reveal differential expression of encoded metabolic potential with changing health status.
Conclusions
Microbial and viral metagenomics combined with microbial transcriptomics characterize the dynamic polymicrobial communities found in CF airways, revealing both the taxa present and their current metabolic activities. These approaches can facilitate the development of individualized treatment plans and novel therapeutic approaches.
Keywords: cystic fibrosis, viruses, microbes, metagenomics, metatranscriptomics
Introduction
In the lungs of cystic fibrosis (CF) patients, the defective cystic fibrosis transmembrane regulator (CFTR) protein affects transepithelial ion transport, consequently hindering the normal airway clearance mechanisms(1,2). The resultant static mucoid environment is colonized by a dynamic and complex community of microbes, viruses, and fungi (reviewed in LiPuma et al. 2010 (3)).
While standard microbial culture techniques had identified the key pathogens, more recent culture-independent approaches based on 16S rRNA gene sequencing revealed a much wider range of microbial species associated with CF lungs (4–9). However, 16S rRNA-based methods are limited in taxonomic resolution and are subject to biases (summarized in Claesson et al. 2010(10)); their predictions of metabolic activities are confined to those general functions known for the taxa, thus overlooking potentially important strain-specific variants. Metagenomics can overcome those limitations.
The metagenomic approach has been used to study viruses in human-associated environments such as blood (11), feces (12–14), and the lungs (15). It has also been successfully used to characterize the viral communities in sputum samples from CF and non-CF individuals (15). The presence of phages in CF airways is of particular relevance for clinical treatment, as environmental stress from the CF mucus and frequent antibiotic treatment is known to enhance phage mobility and promote the phage-mediated spread of antibiotic resistance genes in CF lungs (16,17).
On the other hand, it is challenging to generate microbial metagenomes from CF samples. One reason for this is that microbial DNA isolated from CF sputum or lung tissue samples usually contains a large amount of human DNA, often greater than 99% of the total DNA recovered (18–20). Although some intact human cells may be present in the original sample, most of the contaminating DNA is extracellular and adsorbed to the surface of microbes, making isolation of pure microbial DNA particularly difficult.
Complementing metagenomics, metatranscriptomics characterizes the microbial genes expressed in an environment and can monitor shifts in their transcription or stability in response to perturbations, e.g., antibiotic treatments in CF patients. This approach has been used to investigate microbial community metabolism in marine (21–23) and soil (24,25) environments, but its application to host-associated microbes has been limited to a few instances (26,27) due to technical challenges (Supplementary Table 1). One such challenge is that messenger RNAs (mRNAs) account for only ~5% of total cellular RNA. Various rRNA depletion methods have been developed to enrich samples for mRNA (Supplementary Table 2). Concurrent application of multiple methods (e.g., mRNA-ONLY™, MICROBExpress™ and MessageAmp™) can remove more of the rRNA in some instances, but efficacy remains limited, especially when working with partially degraded rRNA(28).
Metatranscriptomics of host-associated communities is particularly difficult. Amplification of the microbial RNA by methods that utilize synthetic polyadenylation is not applicable for samples that contain large amounts of eukaryotic mRNA. The appended poly-A tails reduce the amount of useful sequence data, especially when pyrosequencing technologies such as Roche/454 (29) are used. Due to the short half-life and small quantity of mRNA, sample filtration and manipulation with buffer should be avoided prior to RNA extraction. This inevitably causes an increase in host RNA contamination when dealing with host-associated microbial samples. In the recent microbial metatranscriptomic study of mule deer lymph nodes by Wittekindt et al. (2010) (26), 99.3% of the taxonomically assigned reads were host-derived and <0.01 % were microbial.
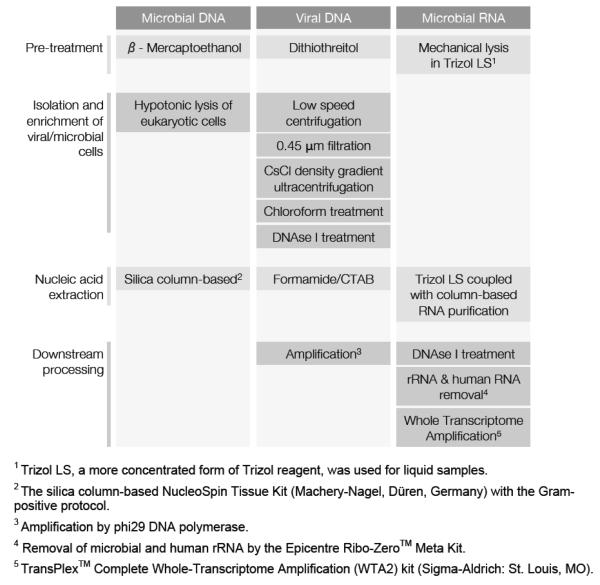
Here we describe protocols to generate viral and microbial metagenomes, as well as microbial metatranscriptomes, from fresh CF sputum using 454 GS FLX Titanium pyrosequencing (Fig. 1). These methods target and enrich for viral and microbial DNA, as well as microbial mRNA, while minimizing contamination with host nucleic acids. This is the first study to simultaneously survey the microbiome, virome, and community metatranscriptome in any ecosystem.
FIGURE 1.
Workflow for the preparation of CF sputum samples for microbiome, virome, and metatranscriptome sequencing.
Materials and Methods
Note
A detailed standard protocol describing each step can be downloaded from www.coralandphage.org.
Sample collection
Eight sputum samples were collected from five CF volunteers (CF1 through CF5) at the Adult CF Clinic (San Diego, CA, United States) by expectoration into a sterile cup, with the exception of sample CF4-A that was a tracheal aspirate. All collection was in accordance with the University of California Institutional Review Board (HRPP 081500) and San Diego State University Institutional Review Board (SDSU IRB#2121). Clinical status at the time of collection was designated as on treatment (during systemic antibiotic treatment), exacerbation (prior to systemic antibiotic treatment), post treatment (upon completion of antibiotic treatment) or stable (when clinically stable and at their clinical and physiological baseline). Each sample was syringe-homogenized and divided into aliquots for metagenomic and metatranscriptomic analyses, culturing, and storage.
Virome protocol
(Supplementary Note 1) Dithiothreitol was added to the diluted sputum to aid mucus dissolution. Viral particles were purified by cesium chloride (CsCl) density gradient ultracentrifugation as described in Thurber et al. (2009)(30). For one sample (CF4-A), the density gradient purification step was omitted for comparison. Viral DNA was extracted using CTAB/phenol:chloroform, and amplified with Phi29 DNA polymerase.
Microbiome protocol
(Supplementary Note 1) Sputum samples were treated with β-mercaptoethanol to disrupt mucus. Pelleted bacterial cells were repeatedly washed and centrifuged, then treated with DNase to remove human DNA.
Microbial metatranscriptome protocol
(Supplementary Note 1) Microbial cells in the sputum aliquots were mechanically lysed by vortexing with zirconia beads in TRIzol® LS (Life Technologies, NY). RNA was extracted using the Zymo Clean & Concentrator™ 25 kit (Zymo Research, Irvine, CA) with the small RNA removal protocol variation and treated with RNase-free DNase I (Ambion, Life Technologies: Grant Island, NY).
cDNA was generated using the WTA-2 kit (Sigma-Aldrich). The effect of nebulization on transcript length was assessed as described in Supplementary Note 2. Similarly, two rRNA depletion methods were tested: (i) the ‘Ambion’ method, i.e., MICROBEnrich™ and MICROBExpress™, that removes bacterial rRNA as well as human rRNA and mRNA; and (ii) the Ribo-Zero™ method, i.e., Ribo-Zero™ rRNA Removal kit (Epidemiology version) (Epicentre, an Illumina company, Madison, WI) that removes bacterial and human rRNA.
Sequencing and data preprocessing/analysis
All samples were sequenced using the GS-FLX Titanium chemistry system. Primer tags in WTA amplified samples were removed using TagCleaner (31). All datasets were preprocessed to remove duplicates and reads of low quality using PRINSEQ (32) (Supplementary Note 1). Metagenomic datasets were further screened and human-derived reads were removed using DeconSeq (33).
The preprocessed metagenomes were annotated using BLASTn against the NCBI nucleotide database. Sequences assigned to the phylum Chordata and to vector or synthetic sequences were identified and removed. Virome sequences were then compared against an in-house boutique viral database containing 4,019 unique complete viral genomes using tBLASTx and normalized viral abundances were calculated. In the preprocessed metatranscriptomes, rRNA-like and non-rRNA reads were identified using BLASTn against the SILVA database (34). Non-rRNA reads were annotated using BLASTx against the NCBI non-redundant protein database. For details of database generation and content, normalization, as well as BLAST parameters, see Supplementary Note 1.
Taxonomic assignments
The best hit was assigned to the alignment with the highest coverage, identity, and score values. Query sequences with no BLAST hits above the defined threshold were designated as unassigned. The diversity of microbiomes was calculated based on the number of bacterial species identified in the datasets (Supplementary Note 1).
Metabolic pathways
Sequences from the metagenomes and metatranscriptomes (excluding all Chordata, vector, and synthetic sequences) were compared against the KEGG protein database using BLASTx. (The CF1-A metatranscriptome was omitted due to insufficient data.) For each pathway, the best hits and their abundances were identified and normalized using HUMAnN (35). Normalized pathway abundance values were used to calculate similarities between samples based on random forests (36) and to partition the samples by Partitioning Around Medoids (PAM) clustering (37).
Data accessibility
Sequence data was deposited in the NCBI Short Read Archive (SRA) with accession numbers SRP007749, SRP009392, and SRP009438.
Results and Discussion
This is the first study to describe a comprehensive workflow (Fig. 1) for the generation of viromes, microbiomes, and microbial metatranscriptomes from any environment. The coupling of metagenomic and metatranscriptomic approaches provides an overview of both who is there and what they are doing, i.e., community taxonomy combined with the community’s encoded and expressed functional diversity. For this work, viral metagenomes (viromes), microbial metagenomes (microbiomes), and microbial metatranscriptomes were generated from twelve fresh sputum samples that had been collected from five adult CF patients (Table 1).
TABLE 1.
Results summary for all viromes, microbiomes, and metatranscriptomes.
| Metagenomes | Metatranscriptomes | |||||
|---|---|---|---|---|---|---|
|
|
||||||
| Patient ID |
Time Point |
Date of Collection |
Health Status |
Microbial | Viral | Microbial |
| % Microbial sequences (Number of microbial sequences) |
% Viral sequencesa VLPs Observedb |
% non-rRNA sequences (Number of non-rRNA sequences) |
||||
| CF1 | A | 09/02/2010 | Stable | N/Af | N/Af | 10.7% d (738) |
| B | 10/18/2010 | Stable | N/Af | N/Af | 30.3%d (41,789) |
|
| C | 11/12/2010 | On Treatment |
N/Af | N/Af | 30.6%d (38,532) |
|
| D | 02/11/2011 | Exacerbation | 9% (14,691) |
31.99% Yes |
N/Af | |
| F | 03/14/2011 | Post Treatment |
79% (40,825) |
6.07% No |
31.6%e (7,971) |
|
| CF2 | A | 11/10/2010 | On Treatment |
N/Af | N/Af | 87.6%d (68,976) |
| CF3 | B | 11/10/2010 | On Treatment |
N/Af | N/Af | 89.6%d (93,375) |
| CF4 | A | 01/22/2011 | Exacerbation | 2% (3,834) |
0.90%c No |
86.8% d (59,394) |
| B | 02/01/2011 | Post Treatment |
0.2% (405) |
6.77% No |
99.1 %e (32,446) |
|
| C | 03/20/2011 | Stable | 23% (41,636) |
1.93% No |
95.1%e (34,411) |
|
| CF5 | A | 10/07/2011 | Exacerbation | 2% (1,034) |
3.00% Yes |
N/Af |
| B | 10/21/2011 | Post Treatment |
1% (247) |
3.78% Yes |
N/Af | |
Based on tBLASTx against viral genome database (threshold of 40% identity over at least 60% of the query sequence).
Observation by epifluorescence microscopy of the viral fraction collected following cesium chloride density gradient centrifugation.
Sample collected by filtration through 0.45 μm filter without cesium chloride density gradient ultracentrifugation.
Following rRNA depletion by the Ambion kits.
Following rRNA depletion by the Ribo-Zero™ (Epidemiology) kit.
Sample not available.
Viruses in CF sputum
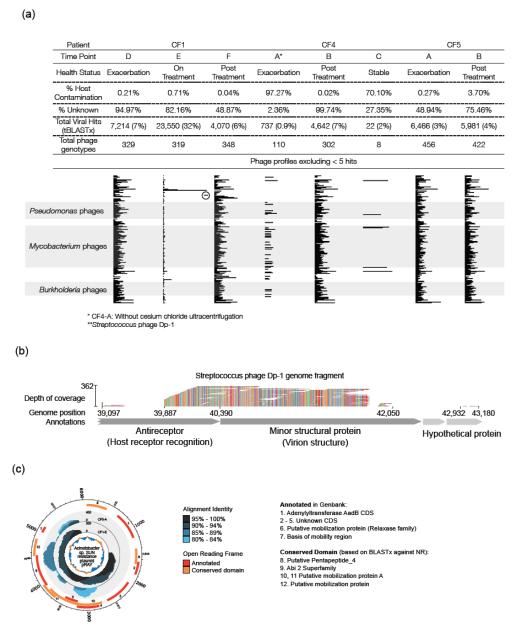
The metagenomic approach was successfully used previously to characterize viruses in CF lungs (15). In this study, viromes were generated from eight sputum samples obtained from three CF patients (Table 1, Supplementary Table 3). Two methods for purifying virus-like particles (VLPs) from sputum were compared. Seven samples were purified by filtration and cesium chloride density gradient ultracentrifugation, followed by chloroform and DNase I treatment. This procedure yielded viromes that contained little (0.02% – 3.7%) host-derived sequence (with one exception due possibly to its exceptionally high amount of mucins and free DNA, thus more viscous sputum; Supplementary Table 3). Omission of the density gradient ultracentrifugation step for the eighth sample (see Methods) resulted in a virome with 97% host-derived sequence. Cesium chloride density gradient centrifugation, previously shown to recover the majority of known phages (25) remains the method of choice for reducing host contamination when isolating viruses from complex samples such as CF sputum.
Analysis of the seven cesium chloride density-purified viromes using tBLASTx against the viral genome database identified more than 450 viral genotypes with each virome containing 319 – 456 genotypes (except CF4-C that contained only eight; Fig. 2a). Unknowns accounted for 49% to >99% of the total reads in most of the viromes (Supplementary Table 3), which is typical for viral metagenomes (15). The exceptions were those samples highly contaminated by host sequences (CF4-A and CF4-C; Fig. 2a). The high number of “unknown” sequences implies the presence of novel viruses that cannot be identified by database similarity, as had been found in previous studies (11,15).
FIGURE 2.
Taxonomic analysis of CF viromes. (a) Putative host range profiles for phage communities. Each bar represents the sum of the normalized abundance values for all phage genotypes with the same putative bacterial host. Only the top 21 hosts are shown. (b) Nucleotide-level alignment of CF1-E virome sequences against a region of the Streptococcus phage Dp-1 genome. Depth of coverage was based on 90% nucleotide identity. The colors represent different nucleotides and demonstrate alignment quality. (c) Coverage plot of Acinetobacter sp. SUN resistance plasmid pRAY recovered from CF1-E and CF5-A.
The majority of the viruses identified were phage, predominantly those that infect known CF pathogens. Their predicted bacterial hosts were tallied and the top 21 were used to construct predicted host range profiles (Fig. 2a). The profiles were highly similar between patients, and even more so for multiple samples from the same patient. They were dominated by phages that infect major CF pathogens such as Streptococcus, Burkholderia, Mycobacterium, Enterobacteria, and Pseudomonas genera. Streptococcus phage (particularly Dp-1) were found in high abundance in the samples with the greatest abundance (>30%) of Streptococcus spp. (i.e., CF1-D and CF1-E; Fig. 4a).
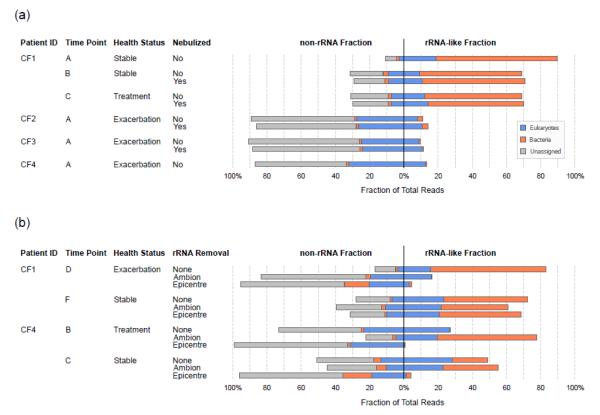
FIGURE 4.
Evaluation of the effects of nebulization and rRNA depletion on the relative amounts of rRNA and non-rRNA in metatranscriptomes. (a) Effects of nebulization. All samples were treated by the Ambion rRNA depletion kits. (b) Comparison of rRNA depletion methods. “Ambion” method uses a combination of MICROBEnrich™ + MICROBExpress™; “Epicentre” method uses Ribo-Zero™ rRNA Removal kit (Epidemiology version).
Streptococcus phage Dp-1 had been first isolated in 1975 from patients presenting with upper respiratory symptoms and was described as a virulent phage (38). Here tBLASTx analysis detected phage Dp-1 genes for DNA replication and packaging, host-receptor recognition, tail and capsid structural proteins, and host lysis (endolysin). The endolysin suggests possible top-down control of the Streptococcus spp. by lytic phage predation in these patients (39).
When reads from the CF1-E virome were mapped against the Dp-1 reference genome (GI:327198314), high depth of coverage was observed for a Dp-1 genome fragment in a 3 kbp region that codes for an antireceptor and a minor structural protein (Fig. 2b). Similarly, high coverage of regions of the Acinetobacter sp. SUN resistance plasmid pRAY was also observed in three samples (CF1-E, CF5-A, and CF5-B), including regions encoding a domain of the Abi-2 superfamily (proteins that confer resistance to phage infection) and mobilization proteins (Fig. 2c). Finding these short sequences from these two genomes highly enriched in the viromes implies that they must be present in many other genomes, as well, likely the result of active horizontal gene transfer (HGT) in CF lungs. HGT is an important mechanism by which microbes evolve and adapt to the CF lung environment (40), and phage can potentially facilitate this process.
The archaeal virus BJ1 (GI: 119756985) was identified in every virome—the first finding of an archaeal virus in the lungs. The hypersaline surface liquid in CF airways may be ecologically similar to the hypersaline lake in Inner Mongolia where the virus was isolated (41). Archaea were identified in low abundance in one microbiome (CF4-C; <0.1%) and all metatranscriptomes (<1%; data not shown), suggesting that they could play a role in the CF lung ecosystem. However, since more than 71% of the predicted ORFs for this archaeal virus show no similarity to any known genes, its genome sequence provides no clues as to what that role might be.
Eukaryotic DNA viruses in CF individuals have been shown to be dominated by a few viral genomes (15) that could potentially cause persistent infections, exacerbations, tumorigenesis, and poor clinical outcomes (42–44). The eukaryotic viruses identified in a previous study of the lungs of CF patients included torque teno virus (TTV), retroviruses, and human herpesviruses (15). In the current study, eukaryotic viruses, including human herpesviruses and retroviruses, were found in all samples (Supplementary Table 4), with torque teno viruses in high abundance in one sample (CF1-D).
Because RNA viruses are involved in the majority of respiratory infections, a filtration-based method was used to isolate RNA viruses from CF sputum (data not shown). However, this method was unable to recover identifiable RNA viruses, likely due their low abundances and technical challenges in their isolation.
Microbes in CF sputum
When characterizing a microbial community, metagenomics surpasses a 16S rRNA-based approach in that it (1) frequently permits high-confidence species-level taxonomic assignment; (2) allows prediction of specialized functional capabilities of the adapted community, rather than inferring function from taxonomy; and (3) avoids the bias inherent in the selection of any universal target for PCR amplification. However, preparations of ‘microbial’ DNA derived from CF sputum or lung tissue are typically dominated by human DNA that was extracellular or adsorbed to the microbes. Several standard methods, including separation of human and microbial cells by percoll gradients (45), treatment with DNase I, selective degradation of human DNA by ethidium bromide monoazide (46), and use of the MolYsis kit (Molzym Life Science), have failed to reduce human DNA in CF samples (personal communications). In this study, the most effective procedure was found to be a modification of the method described by Breitenstein et al. (1995)(18) that employs a combination of β-mercaptoethanol to reduce biofilm disulfide bonds, hypotonic lysis of eukaryotic cells, and DNase I treatment of soluble DNA (Supplementary Fig. 1; Supplementary Table 5). Sufficient microbial DNA was extracted by this procedure to make amplification prior to sequencing unnecessary, thus avoiding potential amplification bias.
The amount of human contamination (13% – 97% of total preprocessed reads) was highly dependent on the sample properties. Samples collected from patients during exacerbations might be expected to contain higher levels of host DNA due to greater inflammation and neutrophil activity than those collected during and immediately following treatment. However, our metagenomic data showed no significant correlation between the fraction of host DNA and a patient’s health status even though the amount of host DNA varies markedly among the metagenomes.
With high-throughput pyrosequencing, even a relatively small proportion of non-host sequence data can be sufficient to provide significant information. After the removal of eukaryotic reads, the microbiomes contained >75% bacterial reads (Table 1) and 2% – 12% unknowns, with the remainder being artificial and cloning vectors or synthetic constructs (Supplementary Table 5). The number of bacterial species identified, including aerobes and anaerobes, ranged from 24 to 256 (Supplementary Table 6).
Each patient presented a unique microbial profile (Fig. 3a). The predominant groups persisted across the time points assayed but the relative abundance varied with exacerbations and antibiotic treatments. This suggests complex community dynamics in which the predominant groups adapt and persist, while others come and go in response to antibiotic treatment or other perturbations.
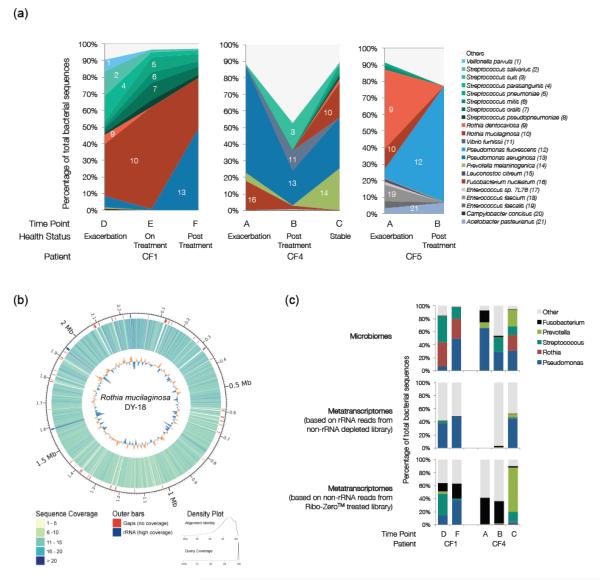
FIGURE 3.
Taxonomic analysis of the microbial communities in three CF patients across multiple time points. (a) Species-level comparisons between microbiomes. Identification was based on unique best hits using BLASTn against the NCBI nucleotide database. All species shown from the same genus are assigned similar colors. (b) Sequence coverage of the Rothia mucilaginosa DY-18 genome by reads from the CF1-E microbiome. (c) Genus-level comparisons between microbiomes and metatranscriptomes. (The CF4-A taxonomy is not shown here because an rRNA removal kit was used during metatranscriptome preparation.)
CF4 presented a classic CF lung microbiome where P. aeruginosa was one of the main players at all time points. In contrast, CF1 was colonized mainly by Rothia mucilaginosa and Streptococcus spp. during exacerbation. Effective treatments decreased R. mucilaginosa, thereby increasing the proportion of P. aeruginosa. The Rothia dentocariosa that colonized CF5 during exacerbations was eliminated by treatments, and the patient was subsequently colonized by Pseudomonas fluorescence instead of the common CF pathogen, P. aeruginosa. The microbiome profiles also showed that P. aeruginosa can be replaced as the main player by other opportunistic bacteria from the environment, as evidenced here by the colonization of (i) CF5, a landscape architect, by soil-dwelling P. fluorescens instead of the more common CF pathogen, P. aeruginosa, and (ii) CF1 by the oral flora R. mucilaginosa (Fig. 3a). By going beyond the traditional tracking of particular recognized CF pathogens, metagenomics offers the possibility of personalized clinical treatment plans.
In some situations, metagenomics can yield in-depth genomic analysis (47,48). In this study, the CF1-E microbiome provided 7.8X average coverage over 93.56% of the genome of the most abundant species, R. mucilaginosa. Mapping of short reads against the reference genome DY-18 (GI: 283133067) (Fig. 3b) revealed only 41 gaps that were >1,000 bp. Of those gaps, almost 20% were located in non-coding regions, 20% in regions annotated as hypothetical proteins, and the rest in genes of known function (Supplementary Table 7). In-depth analysis and interpretation of the genomic changes will be presented elsewhere (manuscript in preparation).
Evaluation of microbial metatranscriptome preparation
A high quality metatranscriptome contains relatively few rRNA reads and an unbiased sampling of RNAs of various lengths. Nebulization, the first step during preparation of a sequencing library, is a potential source of transcript size-induced bias since the size of our cDNA ranged from 50 to 4,000 bp (Supplementary Fig. 2). While nebulization of high molecular weight DNA creates random fragments, application to lower molecular weight cDNA may result in non-uniform coverage or the loss of short transcripts (49).
Here, the effect of nebulization on transcript length was tested on four samples (Supplementary Note 1). There was no difference in the relative translated protein length profiles with and without nebulization (Supplementary Fig. 3). The median translated polypeptide length (345 – 412 amino acids; Supplementary Table 8) is in the high range of previously described microbial protein lengths (50), possibly due to the use of the small RNA removal protocol in the RNA Clean & Concentrator™ kit (Zymo).
Nebulization also did not affect the relative proportion of rRNA-like and non-rRNA reads, although the proportion of rRNA varied from patient to patient (9.4% to 70.8%; Fig. 4a). A reduced rRNA fraction was often associated with an increased proportion of eukaryote and unidentified reads.
Effective mRNA enrichment by rRNA removal using subtractive hybridization (e.g., the MICROBEnrich™ from Ambion) had been previously demonstrated on synthetic microbial communities (28). However, efficacy depended on the integrity of the RNA and community composition. Here, four samples (CF1-D, CF1-F, CF4-B, and CF4-C) were used to compare two hybridization-based commercial rRNA removal kits. Each sample was divided into three aliquots for alternative treatments: (A) MICROBEnrich™ + MICROBExpress™ (Ambion); (E) Early access version of Ribo-Zero™ rRNA removal kit - Epidemiology (Epicentre); and (N) No treatment. In total, these twelve metatranscriptomes yielded 425,523 reads (105 Mbp), 48 – 77% of which were retained after data preprocessing (average read length = 252 bp).
With either treatment, the proportion of rRNA was reduced significantly for CF1-D but minimally for CF1-F (Fig. 4b, Supplementary Table 9). Of the two treatment methods, the Ribo-Zero™ is more effective in eukaryotic rRNA removal as evidenced by CF4-B and CF4-C. In these samples that contained a larger proportion of eukaryotic reads, the Ribo-Zero™ treatment removed 96% and 90% of the rRNA, while the Ambion treatments increased the relative rRNA content.
Notably these rRNA removal methods were markedly less effective for sample CF1-F. Even with Ribo-Zero™, the most effective for all other samples, the treatment yielded only a 5% reduction in total rRNA. This inter-sample variation in rRNA removal could reflect differences in the microbial community present, the quality of extracted RNA, the accessibility of rRNAs for probe hybridization, and/or the degree of homology between the designed rRNA probes and the unknown community members.
Use of either rRNA depletion treatment precludes subsequent rRNA-based analysis of the sample because both methods distort the apparent relative abundances of microbial taxa (Supplementary Fig. 4). Bias was apparent for all samples except CF1-F; neither rRNA depletion method had significant effect on that sample.
Microbial taxonomy three ways
Three methods were used to determine the relative abundances of the microbial genera present within a patient sample: (1) annotation of microbiomes; (2) 16S rRNA-based annotation of metatranscriptomes; and (3) annotation of metatranscriptomes based on encoded protein sequences. The marked differences observed among the three (Fig. 3c) indicate that some community members are more transcriptionally active, thus contribute more to community metabolism than their relative numbers would predict. This is further evidenced by functional characterization (see below).
Community Metabolic Profiles
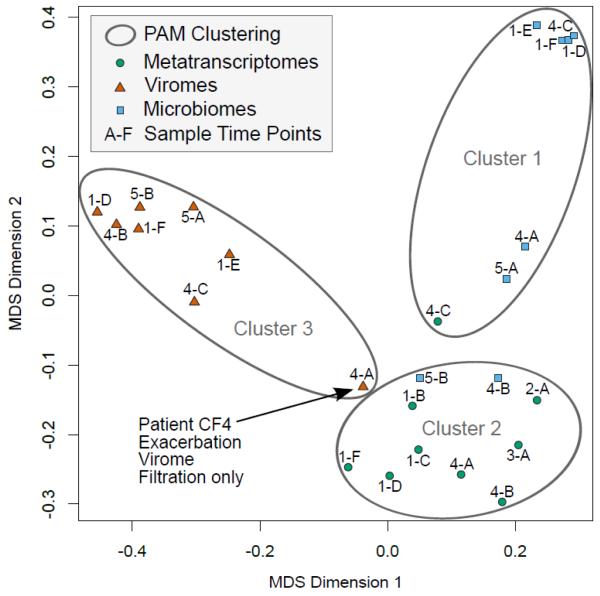
Whereas metagenomics surveys the functional capabilities encoded by members of the microbial commmunity, adding metatranscriptomic data provides insights into the current metabolic activities, insights that can assist in tailoring an effective treatment. To compare these approaches, the viromes, microbiomes, and metatranscriptomes were functionally annotated using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. A total of 216 metabolic pathways and 212 modules (collection of functional units) were identified. Multidimensional scaling (MDS) and clustering of all datasets based on the normalized abundance value (see Methods) yielded three distinct groups, thus showing that a different view of community metabolism can be obtained from each method (Fig. 5). The results demonstrate the internal consistency of each method. The few exceptions were (i) the clustering of the CF4-C metatranscriptome with the microbiomes; (ii) the clustering of the CF4-B and CF5-B microbiomes with the metatranscriptomes attributable to the low number of reads; (iii) the CF4-A virome, purified without the gradient utracentrifugation, appearing as an outlier in the virome cluster.
FIGURE 5.
Comparison of KEGG metabolic pathways identified in viromes, microbiomes, and metatranscriptomes as shown by multidimensional scaling (MDS). Grouping by Partitioning Around Medoids (PAM) clustering placed all samples in the appropriate cluster with the exception of the CF4-C metatranscriptome. CF1-A was omitted from both analyses due to insufficient data.
Overall, the metabolic profiles derived from the microbiomes were the most similar between patients as well as between time points for each patient (Supplementary Fig. 5), indicating a shared pool of metabolic genes required for survival in the CF environment. The greatest variation, likely reflecting specialized adaptations within the viral and microbial communities, is seen in the principal component analysis (PCA) plot (Supplementary Fig. 6).
Supplementary Material
Clinical implications.
The picture of dynamic and diverse polymicrobial communities presented here deviates from classic CF clinical profiles derived from culturing, thereby challenging one-size-fits-all treatment regimes. For example, current treatments targeting the classic CF pathogen, P. aeruginosa, would not be effective against P. fluorescence, R. mucilaginosa, or R. dentocariosa—all of which were abundant in these microbiomes. The ability to identify the resident viruses and microbes that could potentially trigger exacerbation events makes effective individual treatment plans a possibility, including intervention based on predicted disease progression. Ongoing surveillance can monitor inter-patient transmission and inform infection control measures. In addition, shifting the focus from pathogen taxonomy to the community metabolisms associated with periods of stability and exacerbation opens the door to novel therapeutic approaches that change the airway environment to favor less pathogenic communities.
Summary.
The combination of metagenomic and metatranscriptomic approaches demonstrated here can provide insight into the complex and dynamic interations between the host and both the microbial and viral communities present in CF lungs.
The methods described successfully recover viral DNA, microbial DNA, and microbial mRNA from CF sputum, while minimizing contamination with host nucleic acids.
Of the viruses identified in the virome reads, most are phage that infect major CF pathogens. These likely include vectors for clinically-significant microbe-microbe horizontal gene transfer. However, the majority of virome reads are “unknown,” thus potentially novel viruses.
To identify the microbes present, the microbiomes were annotated using BLASTn against the NCBI nucleotide database. Each CF patient possessed a unique microbial profile that shifted over time and sometimes reflected the acquisition of persistent opportunistic bacteria from the environment. High genome coverage for the most abundant species allowed in-depth genomic analysis.
The third concurrent approach, microbial metatranscriptomics, monitors the active community metabolism, as opposed to the metabolic potential encoded in the genomes. Of the three measures, the metatranscriptomes showed the greatest variation between patients and over time, thus is best able to capture the dynamic nature of these complex communities.
Acknowledgements
This work was supported by the National Institute of Health and Cystic Foundation Research Inc. through grants (1 R01 GM095384-01 and CFRI #09-002) awarded to Forest Rohwer. We thank Epicentre, an Illumina company for providing early access to Ribo-Zero™ Epidemiology kits. We thank Peter Salamon, Ben Felts, Katie Barott, Jeremy Barr, and Katrine Whiteson for critical readings and discussions of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Boucher RC. An overview of the pathogenesis of Cystic Fibrosis lung disease. Advanced Drug Delivery Reviews. 2002 Dec 5;54(11):1359–71. doi: 10.1016/s0169-409x(02)00144-8. [DOI] [PubMed] [Google Scholar]
- 2.Riordan JR. CFTR function and prospects for therapy. Annu. Rev. Biochem. 2008;77:701–26. doi: 10.1146/annurev.biochem.75.103004.142532. [DOI] [PubMed] [Google Scholar]
- 3.LiPuma JJ. The changing microbial epidemiology in Cystic Fibrosis. Clinical Microbiology Reviews. 2010 Apr;23(2):299–323. doi: 10.1128/CMR.00068-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rogers GB, Carroll MP, Serisier DJ, Hockey PM, Jones G, Bruce KD. Characterization of bacterial community diversity in Cystic Fibrosis lung infections by use of 16S ribosomal DNA terminal restriction fragment length polymorphism profiling. J. Clin. Microbiol. 2004 Nov 1;42(11):5176–83. doi: 10.1128/JCM.42.11.5176-5183.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harris JK, De Groote MA, Sagel SD, Zemanick ET, Kapsner R, Penvari C, et al. Molecular identification of bacteria in bronchoalveolar lavage fluid from children with Cystic Fibrosis. PNAS. 2007 Dec 18;104(51):20529–33. doi: 10.1073/pnas.0709804104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bittar F, Richet H, Dubus J-C, Reynaud-Gaubert M, Stremler N, Sarles J, et al. Molecular detection of multiple emerging pathogens in sputa from Cystic Fibrosis patients. PLoS ONE. 2008;3(8):e2908. doi: 10.1371/journal.pone.0002908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox MJ, Allgaier M, Taylor B, Baek MS, Huang YJ, Daly RA, et al. Airway microbiota and pathogen abundance in age-stratified Cystic Fibrosis patients. PLoS ONE. 2010 Jun 23;5(6):e11044. doi: 10.1371/journal.pone.0011044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guss AM, Roeselers G, Newton ILG, Young CR, Klepac-Ceraj V, Lory S, et al. Phylogenetic and metabolic diversity of bacteria associated with Cystic Fibrosis. ISME J. 2011 Jan;5(1):20–9. doi: 10.1038/ismej.2010.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao J, Schloss PD, Kalikin LM, Carmody LA, Foster BK, Petrosino JF, et al. Decade-Long Bacterial Community Dynamics in Cystic Fibrosis Airways. PNAS. 2012 Apr 10;109(15):5809–14. doi: 10.1073/pnas.1120577109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Claesson MJ, Wang Q, O’Sullivan O, Greene-Diniz R, Cole JR, Ross RP, et al. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. NAR. 2010 Dec 1;38(22):e200. doi: 10.1093/nar/gkq873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Breitbart M, Rohwer F. Method for discovering novel DNA viruses in blood using viral particle selection and shotgun sequencing. BioTechniques. 2005 Nov;39(5):729–36. doi: 10.2144/000112019. [DOI] [PubMed] [Google Scholar]
- 12.Breitbart M, Hewson I, Felts B, Mahaffy JM, Nulton J, Salamon P, et al. Metagenomic analyses of an uncultured viral community from human feces. J. Bacteriol. 2003 Oct;185(20):6220–3. doi: 10.1128/JB.185.20.6220-6223.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang T, Breitbart M, Lee WH, Run J-Q, Wei CL, Soh SWL, et al. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 2006 Jan;4(1):e3. doi: 10.1371/journal.pbio.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakamura S, Yang C-S, Sakon N, Ueda M, Tougan T, Yamashita A, et al. Direct metagenomic detection of viral pathogens in nasal and fecal specimens using an unbiased high-throughput sequencing approach. PLoS ONE. 2009;4(1):e4219. doi: 10.1371/journal.pone.0004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Willner D, Furlan M, Haynes M, Schmieder R, Angly FE, Silva J, et al. Metagenomic analysis of respiratory tract DNA viral communities in Cystic Fibrosis and non-Cystic Fibrosis individuals. PLoS ONE. 2009;4(10):e7370. doi: 10.1371/journal.pone.0007370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fothergill JL, Mowat E, Walshaw MJ, Ledson MJ, James CE, Winstanley C. Effect of antibiotic treatment on bacteriophage production by a Cystic Fibrosis epidemic strain of Pseudomonas aeruginosa. Antimicrobial Agents and Chemotherapy. 2011 Jan;55(1):426–428. doi: 10.1128/AAC.01257-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rolain J-M, Francois P, Hernandez D, Bittar F, Richet H, Fournous G, et al. Genomic analysis of an emerging multiresistant Staphylococcus aureus strain rapidly spreading in Cystic Fibrosis patients revealed the presence of an antibiotic inducible bacteriophage. Biology Direct. 2009;4:1. doi: 10.1186/1745-6150-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Breitenstein S, Tümmler B, Römling U. Pulsed field gel electrophoresis of bacterial DNA isolated directly from patients’ sputa. NAR. 1995 Feb 25;23(4):722–3. doi: 10.1093/nar/23.4.722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shak S, Capon DJ, Hellmiss R, Marsters SA, Baker CL. Recombinant human DNase I reduces the viscosity of Cystic Fibrosis sputum. PNAS. 1990 Dec 1;87(23):9188–9192. doi: 10.1073/pnas.87.23.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lethem M, James S, Marriott C, Burke J. The origin of DNA associated with mucus glycoproteins in Cystic Fibrosis sputum. European Respiratory Journal. 1990 Jan 1;3(1):19–23. [PubMed] [Google Scholar]
- 21.Hewson I, Poretsky RS, Tripp HJ, Montoya JP, Zehr JP. Spatial patterns and light-driven variation of microbial population gene expression in surface waters of the oligotrophic open ocean. Environmental Microbiology. 2010;12(7):1940–56. doi: 10.1111/j.1462-2920.2010.02198.x. [DOI] [PubMed] [Google Scholar]
- 22.McCarren J, Becker JW, Repeta DJ, Shi Y, Young CR, Malmstrom RR, et al. Microbial community transcriptomes reveal microbes and metabolic pathways associated with dissolved organic matter turnover in the sea. PNAS. 2010;107(38):16420–16427. doi: 10.1073/pnas.1010732107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poretsky RS, Hewson I, Sun S, Allen AE, Zehr JP, Moran MA. Comparative day/night metatranscriptomic analysis of microbial communities in the North Pacific subtropical gyre. Environmental Microbiology. 2009;11(6):1358–75. doi: 10.1111/j.1462-2920.2008.01863.x. [DOI] [PubMed] [Google Scholar]
- 24.Urich T, Lanzén A, Qi J, Huson DH, Schleper C, Schuster SC. Simultaneous assessment of soil microbial community structure and function through analysis of the meta-transcriptome. PLoS ONE. 2008 Jun 25;3(6):e2527. doi: 10.1371/journal.pone.0002527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW, et al. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature. 2006;442(7104):806–9. doi: 10.1038/nature04983. [DOI] [PubMed] [Google Scholar]
- 26.Wittekindt NE, Padhi A, Schuster SC, Qi J, Zhao F, Tomsho LP, et al. Nodeomics: pathogen detection in vertebrate lymph nodes using meta-transcriptomics. PLoS One. 5(10) doi: 10.1371/journal.pone.0013432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gosalbes MJ, Durbán A, Pignatelli M, Abellan JJ, Jiménez-Hernández N, Pérez-Cobas AE, et al. Metatranscriptomic approach to analyze the functional human gut microbiota. PLoS ONE. 2011 Mar 8;6(3):e17447. doi: 10.1371/journal.pone.0017447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He S, Wurtzel O, Singh K, Froula JL, Yilmaz S, Tringe SG, et al. Validation of two ribosomal RNA removal methods for microbial metatranscriptomics. Nat Meth. 2010 Oct;7(10):807–12. doi: 10.1038/nmeth.1507. [DOI] [PubMed] [Google Scholar]
- 29.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437(7057):376–80. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thurber RV, Haynes M, Breitbart M, Wegley L, Rohwer F. Laboratory procedures to generate viral metagenomes. Nat. Protocols. 2009 Mar;4(4):470–83. doi: 10.1038/nprot.2009.10. [DOI] [PubMed] [Google Scholar]
- 31.Schmieder R, Lim Y, Rohwer F, Edwards R. TagCleaner: Identification and removal of tag sequences from genomic and metagenomic datasets. BMC Bioinformatics. 2010;11(1):341. doi: 10.1186/1471-2105-11-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmieder R, Edwards R. [cited 2011 Feb 12];Quality control and preprocessing of metagenomic datasets. Bioinformatics [Internet] doi: 10.1093/bioinformatics/btr026. Available from: http://bioinformatics.oxfordjournals.org/content/early/2011/01/27/bioinformatics.btr026.abstract. [DOI] [PMC free article] [PubMed]
- 33.Schmieder R, Edwards R. Fast identification and removal of sequence contamination from genomic and metagenomic datasets. PLoS ONE. 2011 Mar 9;6(3):e17288. doi: 10.1371/journal.pone.0017288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. NAR. 2007 Dec 1;35(21):7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abubucker S, Segata N, Goll J, Schubert A, Rodriguez-Mueller B, Zucker J, et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLos Computational Biology [Internet] 2012;8(6):e1002358. doi: 10.1371/journal.pcbi.1002358. Available from: http://www.citeulike.org/group/15097/article/9207270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Breiman L. Random forests. machine learning. Springer Netherlands. 2001 Oct 1;45(1573-0565):5–32. [Google Scholar]
- 37.Kaufman L, Rousseeuw P. Finding groups in data: An introduction to cluster analysis [Internet] John Wiley & Sons, Inc; Hoboken, NJ, USA: [cited 2011 Dec 23]. 2008. Available from: doi: 10.1002/9780470316801.ch1. [Google Scholar]
- 38.McDonnell M, Ronda-Lain C, Tomasz A. “Diplophage”: A bacteriophage of Diplococcus pneumoniae. Virology. 1975 Feb;63(2):577–82. doi: 10.1016/0042-6822(75)90329-3. [DOI] [PubMed] [Google Scholar]
- 39.Rodríguez-Cerrato V, García P, del Prado G, García E, Gracia M, Huelves L, et al. In vitro interactions of LytA, the major pneumococcal autolysin, with two bacteriophage lytic enzymes (Cpl-1 and Pal), cefotaxime and moxifloxacin against antibiotic-susceptible and -resistant Streptococcus pneumoniae strains. Journal of Antimicrobial Chemotherapy. 2007 Nov 1;60(5):1159–62. doi: 10.1093/jac/dkm342. [DOI] [PubMed] [Google Scholar]
- 40.Qiu X, Kulasekara BR, Lory S. Role of Horizontal Gene Transfer in the Evolution of Pseudomonas aeruginosa Virulence. Genome Dyn. 2009;6:126–39. doi: 10.1159/000235767. [DOI] [PubMed] [Google Scholar]
- 41.Pagaling E, Haigh RD, Grant WD, Cowan DA, Jones BE, Ma Y, et al. Sequence analysis of an Archaeal virus isolated from a hypersaline lake in Inner Mongolia, China. BMC Genomics. 2007;8:410. doi: 10.1186/1471-2164-8-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Winnie GB, Cowan RG. Association of Epstein-Barr virus infection and pulmonary exacerbations in patients with cystic fibrosis. Pediatr. Infect. Dis. J. 1992 Sep;11(9):722–6. doi: 10.1097/00006454-199209000-00010. [DOI] [PubMed] [Google Scholar]
- 43.van Ewijk BE, van der Zalm MM, Wolfs TFW, Fleer A, Kimpen JLL, Wilbrink B, et al. Prevalence and impact of respiratory viral infections in young children with cystic fibrosis: prospective cohort study. Pediatrics. 2008 Dec;122(6):1171–6. doi: 10.1542/peds.2007-3139. [DOI] [PubMed] [Google Scholar]
- 44.Klein F, Amin Kotb WFM, Petersen I. Incidence of human papilloma virus in lung cancer. Lung Cancer. 2009 Jul;65(1):13–8. doi: 10.1016/j.lungcan.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 45.Childs WC, Gibbons RJ. Use of percoll density gradients for studying the attachment of bacteria to oral epithelial cells. Journal of Dental Research. 1988 May 1;67(5):826–830. doi: 10.1177/00220345880670050601. [DOI] [PubMed] [Google Scholar]
- 46.Lee J-L, Levin RE. Use of ethidium bromide monoazide for quantification of viable and dead mixed bacterial flora from fish fillets by polymerase chain reaction. Journal of Microbiological Methods. 2006 Dec;67(3):456–62. doi: 10.1016/j.mimet.2006.04.019. [DOI] [PubMed] [Google Scholar]
- 47.Iverson V, Morris RM, Frazar CD, Berthiaume CT, Morales RL, Armbrust EV. Untangling genomes from metagenomes: revealing an uncultured class of marine Euryarchaeota. Science. 2012 Feb 3;335(6068):587–90. doi: 10.1126/science.1212665. [DOI] [PubMed] [Google Scholar]
- 48.Narasingarao P, Podell S, Ugalde JA, Brochier-Armanet C, Emerson JB, Brocks JJ, et al. De novo metagenomic assembly reveals abundant novel major lineage of Archaea in hypersaline microbial communities. ISME J. 2012 Jan;6(1):81–93. doi: 10.1038/ismej.2011.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Torres TT, Metta M, Ottenwälder B, Schlötterer C. Gene expression profiling by massively parallel sequencing. Genome Research. 2008 Jan 1;18(1):172–177. doi: 10.1101/gr.6984908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brocchieri L, Karlin S. Protein length in eukaryotic and prokaryotic proteomes. NAR. 2005;33(10):3390–400. doi: 10.1093/nar/gki615. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data was deposited in the NCBI Short Read Archive (SRA) with accession numbers SRP007749, SRP009392, and SRP009438.