Abstract
Modified thrombin-binding aptamers (TBAs) carrying uridine (U), 2′-deoxy-2′-fluorouridine (FU) and North-methanocarbathymidine (NT) residues in the loop regions were synthesized and analyzed by UV thermal denaturation experiments and CD spectroscopy. The replacement of thymidines in the TGT loop by U and FU results in an increased stability of the antiparallel quadruplex structure described for the TBA while the presence of NT residues in the same positions destabilizes the antiparallel structure. The substitution of the thymidines in the TT loops for U, FU and NT induce a destabilization of the antiparallel quadruplex, indicating the crucial role of these positions. NMR studies on TBAs modified with uridines at the TGT loop also confirm the presence of the antiparallel quadruplex structure. Nevertheless, replacement of two Ts in the TT loops by uridine gives a more complex scenario in which the antiparallel quadruplex structure is present along with other partially unfolded species or aggregates.
Keywords: Quadruplex, Thrombin-binding aptamer, NMR, Oligonucleotide synthesis, Uridine, 2′-Deoxy-2′-fluorouridine, North-methanocarbathymidine
1. Introduction
Nucleic acid sequences rich in guanines are predisposed to form higher order structures because of the capacity of guanine bases to self associate via Hoogsteen hydrogen bonds to form planar tetrads. Several tetrads placed next to another stack in a very effective G-quadruplex structure.1,2 G-rich DNA or RNA sequences have been identified in biologically relevant regions of the genome.3 These sequences are present in telomeres4 and in promoter regions of oncogenes such as c-myc5, c-Kit6, K-ras7 and others.
Quadruplex structures differ in the number of associated strands and their orientation, and in the topology and conformation of the loops that connect the G-tetrads. When the guanine strands are oriented in opposite directions with loops above and below the terminal G-tetrad, the quadruplex is “antiparallel”. In this type of quadruplex, half of the guanines are oriented in syn around the glycosyl bond and the other half in anti.8,9 In contrast, when all the guanines are oriented in the same direction with loops located on the side of the G-tetrads, the quadruplex is “parallel”. In this case, the glycosyl bonds of nucleosides adopt an anti-orientation.2,8 Mixed parallel/antiparallel quadruplexes have also been observed.10 Moreover intramolecular quadruplexes can exhibit several G-quadruplex conformations. For example, a number of studies have demonstrated that human telomere regions forms various G-quadruplex structures.11–14
While many of the studies have addressed modifications in the guanines of the G-tetrad and their implication in folding topology and molecularity,15,16 less attention has been paid to the conformation of the nucleosides at the loop regions.
Loops play a key role in determining the nature of the folding and stability of the G-quadruplex.17–19 Both loop length and sequence are crucial in the folding and can either stabilize or destabilize the quadruplex structure.17
Several authors have studied nucleotide substitution in quadruplex forming oligonucleotides containing a single base loop20 or different loop length compositions.18,21,22 In addition studies involving quadruplexes with non-nucleosidic linkers instead of loops revealed the formation of parallel quadruplexes.23 Another important point is the loop-loop interactions such as hydrogen bonding and stacking, which directly affect quadruplex topology.24,25 In addition to these factors, cations also make a key contribution to the folding and stability of the quadruplex.26,27
Although DNA G-quadruplex structure has been studied intensely, little is known about the stability and folding of RNA quadruplex forming oligonucleotides. These RNA quadruplexes are characterized by a parallel orientation of strands, which results from the preferential anti-orientation of their glycosyl bonds.28
Several authors studied quadruplex formation by DNA/RNA hybrid analogues of G4T4G4, in particular the role of the dT4 loop and the effect of its substitution for rU4.29 Moreover, the substitution of a T for U in the tetramolecular TG4T quadruplex, was studied demonstrating that it has a parallel orientation of the four strands.30 This substitution favored the formation of dimeric structures d[(UG4T)4]2 in the presence of K+ and NH4+. In addition, the stabilization of 2′-deoxy-2′-fluoro-D-arabinonucleic acids (2′-F-ANA) on various positions including the loops of G-quadruplex structures has been studied.31
Normally, the ring conformation or puckering can be conveniently described by two parameters, namely the pseudorotation angle and the amplitude of the puckering. In nucleosides and nucleotides structures, as well as in oligonucleotides, the sugar pseudorotation angle is populated essentially by two types of conformation, referred to as the North and South.32,33 In the B-form of DNA, 2′-deoxyribose sugars exhibit a preferential 2′-endo/3′-exo (South) conformation, while A-form helices and RNA tend to have 2′-exo/3′-endo (North) puckering.
Thrombin-binding aptamer (TBA) is a 15-mer DNA structure d(GGTTGGTGTGGTTGG) that specially binds to human thrombin.34,35 NMR spectroscopy reveals that this structure adopts a uniquely folded structure with two stacked G-tetrads connected through edge-loops (one TGT and two TT loops) involving antiparallel alignment of the adjacent strands.36–38 The TBA loops are crucial for the thrombin-binding and recognition. NMR shows that the two TT loops interact with the fibrinogen recognition site (exosite I) of one thrombin molecule and the TGT loop interacts with the heparin-binding site (exosite II) of a second thrombin molecule.35
TBA is characterized by nucleotides with a South (S) sugar pucker. The loops of the aptamer adopt the anti orientation while the guanines on the same G-tetrad plane display alternating syn/anti conformations with respect to the glycosyl torsion angle.36 The introduction of a nucleotide with opposite glycosyl orientation directly affects the stability of TBA.15,16 Several authors have also studied the influence of the loop length and composition of the loops on the folding of the aptamer.39 In addition, cation-binding to TBA is important for driving TBA stability40 and recently it has been described that a multiple pathway process is affected by the type of cation.27 According to NMR experiments, TBA denaturation begins with the opening of the G-G base pairs that are not protected by a loop, followed by the opening the TGT loop.41
Here we studied the conformational changes produced in TBA when the thymidines located in the loops are substituted by ribonucleosides (uridine and 2′-deoxy-2′-fluorouridine) or by the conformationally-restricted pseudonucleoside North-methanocarbathymidine.42
2′-Fluoro-2′-deoxy ribonucleosides show a strong preference >90% for the North type conformer in solution.43–45 North-methanocarbathymidine is a conformationally-restricted pseudonucleoside with a carbocyclic bicyclo[3.1.0]hexane ring. North or South-locked platforms can be prepared by shifting the position of the cyclopropane ring.42,46,47
We found that the North conformation of these derivatives directly interferes with TBA folding and stability, thereby confirming the relevance of loop flexibility in TBA folding.
2. Results and discussion
2.1. Synthesis of modified thrombin-binding aptamer oligonucleotides
Thymidines present in the loop regions of TBA were replaced by several uridine (U), 2′-deoxy-2′-fluorouridine (FU) or North-methanocarbathymidine (NT) residues (Figure 1A). To this end, several oligonucleotide sequences were prepared by solid-phase phosphoramidite synthesis. U, FU and NT phosphoramidites were used for the synthesis of modified TBA sequences. The 2′-hydroxyl group of uridine was protected with the t-butyldimethylsilyl (TBDMS) group.
Figure 1.
(A) North-Methanocarbathymidine, uridine and 2′-deoxy-2′-fluorouridine residues used for the synthesis of modified thrombin-binding aptamers (TBAs). (B) Scheme of the modified TBAs showing the changes in the TGT loop. (C) Scheme of the modified TBAs showing the changes in the TT loops.
First, the two thymidines located in the central loop of the aptamer formed by the T7G8T9 sequence were replaced by U (TBA-U7,U9), FU (TBA-FU7,FU9) or NT (TBA-NT7,NT9) (Figure 1B).
Two or four thymidines present in the two TT loops were then substituted by U, FU or NT. All four positions were modified by U, (TBA-U3,U4,U12,U13) or by FU (TBA-FU3,FU4,FU12,FU13). Two U residues were introduced at different positions TBA-U3,U4 and TBA-U4,U13. The NT monomer was introduced in positions 4 and 13 (TBA-NT4,NT13) (Figure 1C).
Oligonucleotide sequences were synthesized by standard protocols with an increasing coupling time for ribonucleotides and NT residues. After the assembly of the sequences, supports were treated with ammonia in order to remove the protecting groups and release the oligonucleotide from the support. In addition, the TBA sequences containing U were treated with fluoride solution to remove the TBDMS group. The DMT-on oligonucleotides were purified by HPLC. The modified aptamers were characterized by mass spectrometry (Table 1).
Table 1.
Mass spectra of modified thrombin-binding aptamers
| Oligonucleotide | Mass (found) | Mass (expected) |
|---|---|---|
| TBA-U7,U9 | 4727.7 | 4729.7 |
| TBA- FU7,FU9 | 4731.7 | 4733.7 |
| TBA-NT7,NT9 | 4743.9 | 4746.0 |
| TBA-U3,U4,U12,U13 | 4731.6 | 4733.7 |
| TBA-FU3,FU4,FU12,FU13 | 4739.8 | 4741.7 |
| TBA-U3,U4 | 4726.6 | 4729.7 |
| TBA-U4,U13 | 4726.9 | 4729.7 |
| TBA-NT4,NT13 | 4743.8 | 4746.0 |
2.2. Thermal UV experiments
Folding and unfolding of modified TBAs was estimated by heating/cooling experiments recording the UV absorbance as a function of temperature. This biophysical technique relies on the observation that the absorbance of guanines at 295 nm is higher in a G-quadruplex structure than in the denatured form. Here we determined the denaturation-renaturation profiles and the melting temperature of several modified aptamers at a range of concentrations 30, 15, 3 and 1.5 μM (see supplementary data, table 1S). In most of the cases, the aptamers did not present hysteresis, thus the transitions were reversible and the melting and annealing curves were almost equivalent. The modified TBAs were classified into three groups on the basis of their melting profile (I, II, III, Table 2). The first category (I) included those with a similar stability to TBA and no melting dependence on the concentration. We interpreted this profile as a reversible G-quartet formation with relatively rapid kinetics. The second category (II) was characterized by a low reversible melting temperature that was affected by the concentration of the aptamer, thereby indicating a potential association of the strands. The third category (III) indicated a destabilized G-quadruplex structure as a result of a low melting temperature independent of the concentration. The midpoints of the melting transitions (Tm) and the difference with unmodified TBA were calculated for all oligonucleotides (Table 2). In particular, the central T7G8T9 loop modifications showed a distinct behaviour. The introduction of U7 and U9 and FU7 and FU were well tolerated in the TBA structure. TBA-U7,U9 had a higher melting temperature (54.8 °C) while TBA-FU7,FU9 showed a small increase in melting temperature to TBA (51.9 °C). No dependence of the melting temperature on the concentration was observed. Both aptamers were placed in group I with similar structural properties as unmodified TBA.
Table 2.
UV denaturation data of modified thrombin-binding aptamersa
| Oligonucleotide | Profile | Tm (°C) | ΔTm (°C) |
|---|---|---|---|
| TBA | I | 49.9 | -- |
| TBA-U7,U9 | I | 54.8 | +4.9 |
| TBA- FU7,FU9 | I | 51.9 | +2 |
| TBA-NT7,NT9 | II | 32.6 | −17.3 |
| TBA-U3,U4,U12,U13 | III | 32.2 | −17.7 |
| TBA-FU3,FU4,FU12,FU13 | III | 42.8 | −7.1 |
| TBA-U3,U4 | III | 41.0 | −8.9 |
| TBA-U4,U13 | II | 32.2 | −17.7 |
| TBA-NT4,NT13 | III | 31.0 | −18.9 |
Uncertainty ± 0.5° C, ODN concentration: 3 μM. Buffer. 10 mM sodium cacodylate 100 mM KCl pH 7
At 3 μM TBA-NT7,NT9 had a low melting temperature 32.6 °C. The melting profile was unusually high and broad at concentration of 30 μM, whereas at low concentration it was decreased. This TBA was classified in group II. As described in the CD section, this modified TBA partially lost the antiparallel structure.
Derivatives with modifications in the T3,T4 and T12,T13 loops were the most sensitive to the substitution and most of them were classified in group III. The stability of aptamers modified in the two TT loops was lower than unmodified TBA and structures with modifications with uridine or 2′-deoxy-2′-fluorouridine in the central T7G8T9 loop. The oligonucleotides with the four T positions modified by U (TBA-U3,U4,U12,U13) or by FU (TBA-FU3,FU4,FU12,FU13) showed melting temperatures of 32.2 and 42.8 °C respectively. The melting temperatures of these two aptamers were independent of the aptamer concentration and these structures were classified in group III. TBA-U3,U4,U12,U13 was observed to be a highly destabilized TBA derivative. Remarkably, TBA-FU3,FU4,FU12,FU13 had the highest melting temperature of this group and the melting/association processes differed slightly. In addition, at high concentration the melting temperature profile broadened.
TBA-U4,U13 and TBA-NT7,NT9 displayed an important decrease in stabilitity with a melting temperature around 32 °C. These oligonucleotides did not show hysteresis in the heating/cooling process. These two aptamers showed a similar profile with high and broad melting temperatures at concentrations of 15–30 μM. In contrast, at low concentrations the melting temperatures decreased. TBA-U4,U13 and TBA-NT7,NT9 were categorized in group II.
Hydrogen bonding between T4 and T13 and base stacking interactions of these two residues on the adjacent G quartet are essential for the aptamer stability, thus any modification in these positions directly affects TBA folding.36,48
TBA-U3,U4 containing two continuous U residues showed a moderate decrease in the stability and the melting temperature was not affected by the concentration of the aptamer. This oligonucleotide was placed in the group III.
To our knowledge, the analog FU has not been introduced into TBA to date. In contrast, 2′-fluoroarabinothymidine (2′-F-araT) has been substituted in some TBA loop positions. This modified nucleoside has a DNA-like (South/East) conformation. The replacement of some loop thymidines caused a moderate increase in thermal stability with a concentration-independent melting profile and no hysteresis in the heating/cooling processes31.
The introduction of FU in the TGT loop produced a moderate increase in TBA stability. In contrast, TBA-FU3,FU4, FU12,FU13 have the highest melting temperature of the aptamers modified in the TT loops.
The NT substitution studied here is similar to LNA (Locked Nucleic Acid). LNA is also a bicyclo derivative with a 2′-O-4′-C-methylene-linked ribonucleoside. The sugar is also forced to be in the 3′-endo conformation and the conformational preference of the glycosyl bond is anti. LNA substitution in both loops decreased the stability of the TBA.49 In particular, the replacement of the thymidines by the corresponding LNA derivative in position 4 resulted in the destabilization of the quadruplex. In the present study, the aptamers modified with North-methanocarbathymidine had a low melting temperature; thus, this analogue produced a comparable destabilizing effect on aptamer stability. The restricted conformation of this analog may interfere with the quadruplex folding of the TBA aptamer. Although a recent study shows that North-methanocarbathymidine is flexible and it can also adopt a North/East (C4′-exo) conformation when present in a DNA loop.50
The introduction of nucleoside derivatives that are flexible and have no sugar restrictions has been reported in various loop positions of TBA.48,51 Generally, these derivatives are well tolerated. Nevertheless, Unlocked Nucleic Acids51 in positions 4, 9 or 13 of the TBA are less stable than the unmodified TBA. Surprisingly, the acyclic nucleoside N1-(3-hydroxy-2-hydroxymethyl-2-methylpropyl)-thymine is tolerated and the stability of the loop-modified TBA was similar to unmodified TBA.
Thermodynamic analysis of the UV melting curves52 for those sequences forming unimolecular antiparallel quadruplex (TBA, TBA-U7,U9, TBA-FU7,FU9) are presented in Table 3. Thermodynamic parameters for other modified TBAs were not calculated because the required two-state equilibrium is not clear for these compounds.
Table 3.
Thermodynamic parameters of the thermal denaturation of unimolecular quadruplex
| Oligonucleotide | ΔH° (Kcal/mol) | ΔS° (cal/Kmol) | ΔG° (25°C) (Kcal/mol) |
|---|---|---|---|
| TBA | −45.53 ± 0.41 | −140.94 ± 1.29 | −3.50 |
| TBA-U7,U9 | −47.51 ± 0.69 | −144.84 ± 2.12 | −4.32 |
| TBA-FU7,FU9 | −44.19 ± 0.88 | −135.94 ± 2.72 | −3.66 |
ODN concentration: 3 μM. Buffer. 10 mM sodium cacodylate 100 mM KCl pH 7
TBA-U7,U9 appears to have a greater enthalpic contribution to the stability of their folded structure. The enthalpic differences between this aptamer and TBA and TBA-FU7,FU9 may be due to sugar pucker or other specific effects. TBA-U7, U9 is clearly more stable than TBA. On the other hand, TBA-FU7,FU9 is only slightly more stable than TBA.
We then analyzed the thermal difference spectra (TDS) of the aptamers. The curves are the result of the arithmetic difference between high (80°C) and low (15°C) temperature absorbance spectra in the 230–340 nm wavelength range. The TDS of the modified aptamers matched the spectra of TBA, with two positive peaks at ~240 and ~275 nm and two negative peaks ~260 nm and ~295 nm. TBA-NT7,NT9 showed only one positive maxima ~275 nm and one negative peak ~295 nm (see supplementary data Figure 1S).
2.3. Circular Dichroism experiments
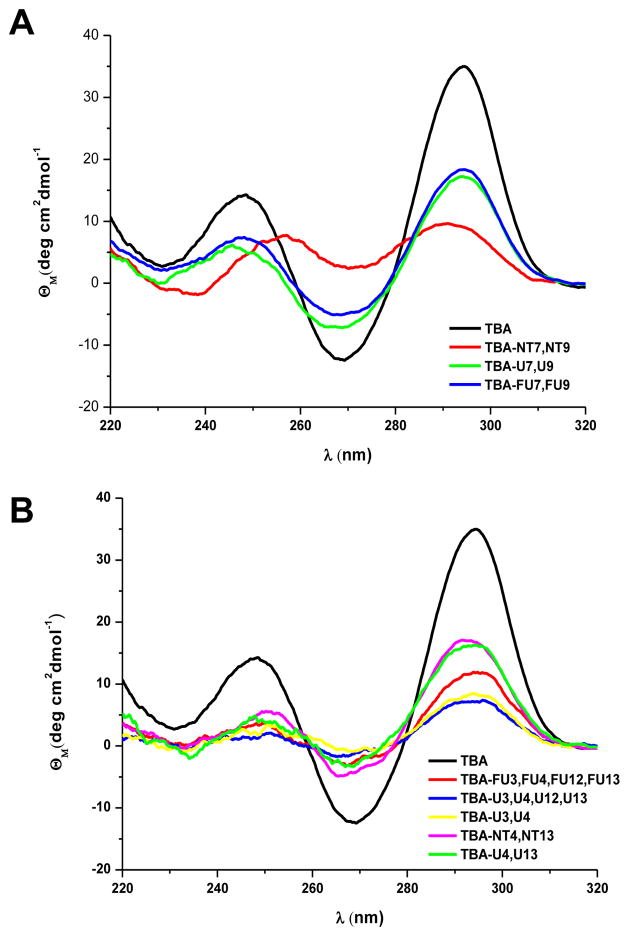
Circular Dichroism (CD) spectra of all modified thrombin-binding aptamers were recorded (Figure 2). The CD spectra of these derivatives exhibited the characteristics described for the antiparallel quadruplex structure with two positive bands at ~245 and ~295 nm and a negative band at ~270 nm. This profile was similar to unmodified TBA but the magnitude of the band at ~270 nm was less negative than the TBA at the same oligonucleotide concentration. Nevertheless, CD spectra of TBA-NT7,NT9 differed from the other modified thrombin-binding aptamers. The signature of this derivative contained two similar broad bands at ~255 nm and ~290 nm which were shifted compared to an antiparallel quadruplex structure. This observation indicates partial loss of the antiparallel structure.
Figure 2.
CD spectra of the modified thrombin-binding aptamers (TBAs). A) TBAs modified in the central TGT loop. B) TBAs modified in the TT loops.
2.4. NMR studies on modified thrombin-binding aptamer oligonucleotides
TBA-U4,U13 and TBA-U7,U9 were selected for NMR studies as they are examples of a destabilizing or stabilizing uridine TBA modification, respectively, with respect to the unmodified TBA. 2D NOESY experiments were performed on TBA-U4,U13 and TBA-U7,U9. Completed sequential assignments were obtained only for the latter following the known strategy for the quadruplex structure36,37. The proton chemical shifts for TBA-U7,U9 are reported in Table 4.
Table 4.
Proton chemical shift of TBA-U7,U9 in 10 mM potassium phosphate buffer and 5 mM KCl (pH 6.7), T=5°C
| H8/H6 | H1′ | H2′ | H2″ | H3′ | Me6/H5 | NH | |
|---|---|---|---|---|---|---|---|
| G1 | 7.43 | 6.11 | 2.87 | 2.87 | 4.91 | -- | 12.10 |
| G2 | 8.21 | 6.11 | 3.04 | 2.35 | 5.15 | -- | 12.12 |
| T3 | 7.90 | 6.03 | 2.22 | 2.60 | 4.92 | 2.00 | 11.31 |
| T4 | 7.16 | 6.05 | 2.05 | 2.74 | 4.89 | 1.05 | 11.43 |
| G5 | 7.50 | 6.10 | 3.56 | 2.86 | 4.91 | -- | 12.31 |
| G6 | 7.72 | 6.05 | 2.93 | 2.59 | 5.18 | -- | 12.30 |
| U7 | 8.03 | 6.24 | 4.18 | -- | 4.55 | 6.03 | 11.31 |
| G8 | 7.50 | 5.73 | 1.98 | 2.31 | 4.89 | -- | 10.71 |
| U9 | 7.41 | 5.98 | 4.43 | --- | 4.43 | 5.98 | 10.40 |
| G10 | 7.43 | 6.07 | 3.72 | 2.99 | 4.92 | -- | 12.11 |
| G11 | 8.23 | 6.03 | 3.05 | 2.32 | 5.16 | -- | 12.11 |
| T12 | 7.91 | 6.19 | 2.23 | 2.67 | 4.92 | 2.00 | 11.43 |
| T13 | 7.17 | 6.09 | 2.07 | 2.61 | 4.95 | 0.98 | 11.43 |
| G14 | 7.50 | 6.07 | 3.39 | 2.83 | 4.90 | -- | 12.30 |
| G15 | 8.10 | 6.15 | 2.70 | 2.44 | 4.81 | -- | 12.37 |
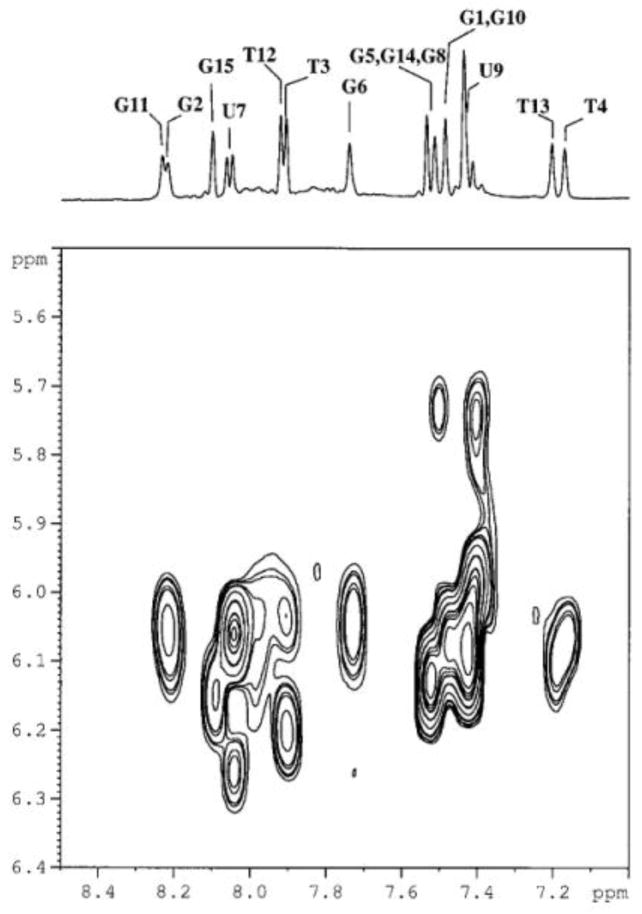
The hydrogen-bonded imino protons gave rise to resonances in the 10.5–12.0 ppm range of the 1D 1H NMR spectra of both aptamers. TBA-U7,U9 showed NMR features similar to unmodified TBA such as the number of signals, protons chemical shift and NOe patterns. Strong intraresidue NOe cross peaks between aromatic protons H6/H8 and anomeric protons H1′ were observed together with other non-sequential interactions. For example, H8, H1′, H2′ and H2″ of G2 showed interactions with methyl, H2′ and H2″ of T4. Moreover, aromatic and ribose protons of G11 showed an interaction with methyl and ribose protons of T13. Figure 3 displays the H6/8-H1′ region of the NOESY spectra. In addition, the presence of eight sharp hydrogen-bonded G imino protons signals and four imino proton signals belonging to the oligonucleotide units of the loops indicated that a unique species with antiparallel structure was present in solution (Figure 4) as proved also by CD measurements.
Figure 3.
Selected region of monodimensional 1H-NMR (top) and 2D-NOESY spectrum (below) of TBA-U7,U9 acquired at 5°C
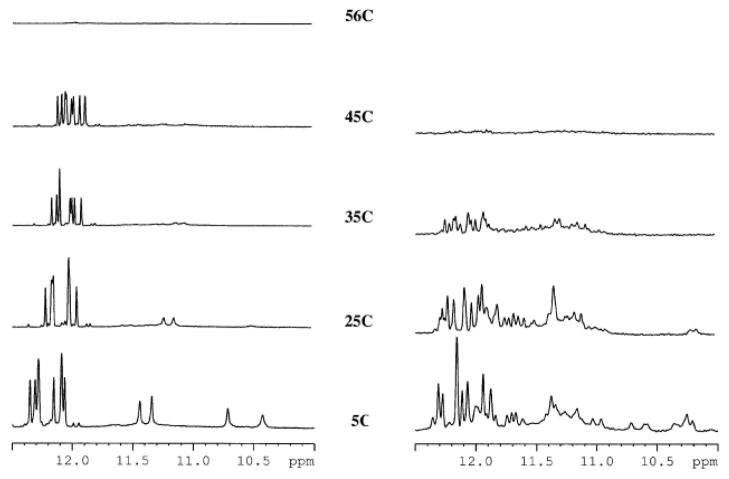
Figure 4.
Imino regions of the 1H-NMR spectra of TBA-U7,U9 (left) and of TBA-U4,U13 (right) at a range of temperatures.
For TBA-U7,U9 the increase in temperature from 5°C to 60°C resulted in a melting temperature of 56°C, as previously found with UV experiments, with the proton signals from 10.5 to 11.5 ppm disappearing first. The substitution of T4 and T13 by U4 and U13 affected the NMR spectra: the imino protons resonances between 10.5 and 12.5 ppm were broad and in superior number as will be expected for a unique species, thereby indicating that the chemical equilibria present in solution are complicated and that multiple species could be present in solution under the same experimental conditions at the ionic strength and temperature used for TBA-U7,U9. These resonances were temperature-sensitive and were fully exchanged at 35°C (Figure 4). In order to better study the molecular species of the TBA analogues, we performed NMR DOSY experiments at room temperature. The diffusion coefficients (D) obtained were 10−9,7979 m2s-1 for both aptamers. This value corresponds to a molecular weight of a monomeric species. The NMR results for these two TBA derivatives were consistent with those from polyacrylamide gel electrophoresis (PAGE). TBA-U7,U9 and TBA have a simple NMR spectra and appeared in the gel as a single band whereas TBA-U4,U13 showed a complex NMR spectrum and have two or more bands in the gel (see supplementary material).
3. Conclusions
In summary, here we have studied the substitution of several thymidines located at the loop positions for derivatives with a North conformation (uridine, 2′-deoxy-2′-fluorouridine and North-methanocarbathymidine). Our findings demonstrate that changes in the conformation of the sugar puckering of the nucleotides present in loop regions are important for determining the folding and stability of TBA.
The central TGT loop of TBA is more tolerant to modifications (except for NT) than the two lateral TT loops. The introduction of U and FU modifications into the TGT loop has a stabilizing effect on the antiparallel quadruplex structure of TBA.
The thymidines located in the two TT loops are very sensitive to modification. This study and others demonstrate that the presence of thymidines in TT loops is essential to induce the folding of TBA into the antiparallel quadruplex. These thymidines cannot be replaced by ribonucleosides without a severe destabilization and this may explain why RNA quadruplex have a preference for parallel quadruplexes. Substitutions of positions 4 and 13 are crucial as there is a hydrogen bond between these two thymines that is critical for TBA folding.37,38 The introduction of modifications in the two TT loops generates the formation of several species. Although it is difficult to assess the composition and structure of these species, data are provided to assign them to partially destabilized antiparallel structures or, in some cases, to dimers and other high-molecular aggregates, especially at high aptamer concentration.
TBA loops are directly implicated in the interaction with and inhibition of thrombin. Normally, aptamers are DNA or RNA structures. Nevertheless, the synthesis of hybrid DNA/RNA TBAs could provide a new strategy to prepare aptamers with improved inhibitory properties. Research in these directions is currently underway and results will be presented in due course.
4. Experimental Section
4.1. Materials
All the standard phosphoroamidites and reagents for DNA synthesis were purchased from Applied Biosystems and from Link Technologies. The remaining chemicals and solvents were from commercial sources (Sigma, Aldrich, Fluka) and were used without further purification.
4.2. Oligonucleotide synthesis and purification
The assembly of the modified TBAs was performed on a DNA/RNA synthesizer by solid-phase 2-cyanoethylphosphoroamidite chemistry. Commercially available controlled pore glass functionalized with 5′-O-DMT-2′-deoxyguanosine was used as the solid support and tetrazole as the activator. Phosphoramidites of 2′-deoxynucleosides, 2′-O-TBDMS-protected uridine, and 2′-F-uridine were obtained from commercial sources and the North-methanocarbathymidine monomer was obtained as described.42 Coupling time was increased to 15 minutes for ribonucleotides and for North-methanocarbathymidine phosphoramidites. Oxidation using tert-butyl hydroperoxide was used after the introduction of the North-methanocarbathymidine derivative.42 The following oligonucleotides were synthesized in 0.2 μmol scale: TBA-FU7,FU9, TBA-NT7,NT9, TBA-FU3,FU4, FU12,FU13, TBA-U3,U4, TBA-U3,U4,U12,U13, TBA-4U, and TBA-NT4,NT13. Oligonucleotides TBA-U7,U9 and TBA-U4,U13 were synthesized in 1 μmol scale.
After synthesizing the aptamers using the DMT-on strategy, the solid supports were treated with an ammonia solution in order to cleavage the oligonucleotides from the solid support and remove the phosphate-protecting and base-protecting groups. In the next step, the 2′-O-TBDMS-protecting group of oligonucleotides containing uridine derivatives was removed by treatment with a mixture of triethylamine hydrofluoride/triethylamine/1-methy-2-pyrrolidone (4:3:6). Fully deprotected oligonucleotides were purified by reverse phase semi-preparative HPLC in the DMT-on mode. Solvent A: 5% MeCN in 100 mM triethylammonium acetate (pH 6.5) and solvent B: 70% MeCN in 100 mM triethylammonium acetate (pH 6.5). Column: Nucleosil 120C18 (10μm, 200 × 10mm). Flow rate: 3 ml/min. Conditions: 20 min linear gradient from 15–80% B. Finally, the DMT group was removed by a 30-minute treatment with 80% acetic acid. The aqueous phase was extracted with diethyl ether and the products were desalted on NAP-10 (Sephadex G-25) columns and characterized by mass spectrometry. Oligonucleotides used for NMR studies (TBA-U7,U9 and TBA-U4,U13) were passed through a DOWEX (Na+) resin to exchange triethylammonium to sodium cations. The desired oligonucleotides were obtained in satisfactory overall yields (60–70 OD260 for 1 μmol and 15–20 OD260 for 0.2 μmol). Oligonucleotides were characterized by mass spectrometry (MALDI-TOF, Table 1). Conditions: matrix 2,4,6-trihydroxyacetophenone (THAP) 10 mg/ml in acetonitrile/water (1:1) using ammonium citrate (50 mg/ml) as an additive.
4.3. UV Absorbance and Circular Dichroism measurements
The thermal melting curves were obtained following the absorption change at 295 nm for TBA aptamers from 15°C to 80°C, with a linear temperature ramp of 0.5°/min in Teflon-stoppered 1-cm path-length quartz cells on a JASCO V-650 spectrophotometer equipped with a Peltier temperature control. For TBA derivatives all the measurements were repeated twice, conducted in 10 mM sodium cacodylate buffer, and 100 mM KCl (pH 7.0). Thermal melting curves of the aptamers were analyzed in a range of concentrations (1.5, 3, 15 and 30 μM) using different cuvettes with 1, 5 and 10 mm path length. The denaturation-renaturation profiles (hysteresis) were studied at 3 μM. Samples were denatured at 85°C and left to slowly cool overnight prior to analysis. Melting curves of TBA, TBA, TBA-U7,U9, TBA-FU7,FU9 were analyzed as described in the literature52 to extract thermodynamic parameters using Matlab program (R2009b version, Math-Works, Natick, MA, USA).
The CD spectra were recorded on a JASCO spectropolarimeter J-810. CD spectra were registered between 220–320 nm. The samples (3 μM) were dissolved in 10 mM sodium cacodylate buffer, 100 mM KCl (pH 7.0) and annealed by heating to 85 °C and slowly cooling to room temperature before recording spectra.
4.4. NMR Spectroscopy
The oligonucleotides were dissolved in 0.55 ml of H2O/D2O (90:10 v/v), corresponding to 0.8 mM of quadruplex concentration, in the presence of 10 mM potassium phosphate buffer and 5 mM KCl (pH 6.7).
The NMR spectra were recorded on a Bruker AV600 spectrometer operating at a frequency of 600.10 MHz, for 1H nucleus. 1H spectra were recorded at variable temperature ranging from 5 °C to 65°C. Chemical shifts (δ) were measured in ppm. 1H NMR spectra were referenced to external DSS (2,2-dimethyl-2-silapentane-5-sulfonate sodium salt) set at 0.00 ppm. Estimated accuracy for protons is within 0.02 ppm.
Phase sensitive NOESY spectra were acquired in TPPI mode, with 4 K × 1024 complex FIDs, spectral width of 15.000 Hz, recycling delay of 1.5 s, and 96 scans. Mixing times ranged from 50 ms to 300 ms. TOCSY spectra were acquired with the use of an MLEV-17 spin-lock pulse (field strength 10.000 Hz, 60 ms total duration). All spectra were transformed and weighted with a 90° shifted sine-bell squared function to 4 K _ 4 K real data points. The sequential assignments of the modified TBAs were performed by applying well established procedures for the analysis of quadruplex DNA. The Sparky program was used to assign the NOESY cross-peaks
Pseudo two-dimensional DOSY experiments53 were performed using the pulse-program‘stebpgp1s’, diffusion delay: 0.12–0.40 s; gradient pulse: 1.5 ms; number of increments: 64. Raw data were processed using the standard DOSY software present in the Bruker library (TOPSPIN v. 1.3). A calibration curve was obtained using the following standards: glucose (MW 180; 3 mM in D2O 99.9%; NaCl 0.1 M, phosphate buffer 10 mM, pH 6.7), d(CGTACG)2 (MW 3754; 3 mM in D2O 99.9%; NaCl 0.1 M, phosphate buffer 10 mM, pH6.7), aprotinin (MW 6500; 1 mM in D2O 99.9%; pH 6.8), lysozyme (MW 14400; 1 mM in D2O 99.9%; pH 6.9), trypsin (MW 23500; 1 mM in D2O 99.9%; pH 3.0) and LCTI (Lens culinaris trypsin inhibitor, MW 7447; 1 mM in D2O 99.9%; pH 3.0).
Supplementary Material
Acknowledgments
This work was partially supported by grants from the Spanish Ministerio de Ciencia e Innovación MICINN (CTQ2010-20541), the Generalitat de Catalunya, (2009/SGR/208), the University of Milano (PUR 2009 Funds) and PRIN09 (2009Prot.2009J54YAP_005). RF is a recipient of a FPI predoctoral contract (MICINN). Collaborative research was funded by a Cost action (G4net, MP0802) and an Italian-Spanish collaborative action (IT2009-0067). CIBER-BBN is an initiative funded by the VI National R&D&i Plan 2008-2011, Iniciativa Ingenio 2010, Consolider Program, CIBER Actions and financed by the Instituto de Salud Carlos III with assistance from the European Regional Development Fund. Partial support from the Intramural Research Program of the NIH, National Cancer Institute for Cancer Research is also acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Anna Aviñó, Email: aaagma@cid.csic.es.
Stefania Mazzini, Email: stefania.mazzini@unimi.it.
Ruben Ferreira, Email: ruben.ferreira@irbbarcelona.org.
Raimundo Gargallo, Email: raimon_gargallo@ub.edu.
Victor E. Marquez, Email: marquezv@mail.nih.gov.
Ramon Eritja, Email: recgma@cid.csic.es.
References and notes
- 1.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Nucleic Acids Res. 2006;34:5402. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neidle S, Balasubramanian S. Quadruplex Nucleic Acids. Royal Society of Chemistry; London: 2006. [Google Scholar]
- 3.Shafer RH, Smirnov I. Biopolymers. 2001;56:209. doi: 10.1002/1097-0282(2000/2001)56:3<209::AID-BIP10018>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 4.Oganesian L, Bryan TM. BioEssays. 2007;29:155. doi: 10.1002/bies.20523. [DOI] [PubMed] [Google Scholar]
- 5.Simonsson T, Kubista M. Nucleic Acids Res. 1998;26:1167. doi: 10.1093/nar/26.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernando H, Reszka AP, Huppert J, Ladame S, Rankin S, Venkitaraman AR, Neidle S, Balasubramanian S. Biochemistry. 2006;45:854. doi: 10.1021/bi0601510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cogoi S, Luigi E, Xodo LE. Nucleic Acids Res. 2006;34:2536. doi: 10.1093/nar/gkl286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simonsson T. Biol Chem. 2001;382:621. doi: 10.1515/BC.2001.073. [DOI] [PubMed] [Google Scholar]
- 9.Patel DJ, Bouaziz S, Kettani A, Wang Y. In: Oxford Handbook of Nucleic Acid Structures. Neidle S, editor. Oxford University Press; Oxford: 1988. p. 389. [Google Scholar]
- 10.Dai J, Dexheimer TS, Chen D, Carver M, Ambrus A, Jones RA, Yang D. J Am Chem Soc. 2006;128:1096–1098. doi: 10.1021/ja055636a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xue Y, Kan ZY, Wang Q, Yao Y, Liu J, Hao YH, Tan Z. J Am Chem Soc. 2007;129:11185. doi: 10.1021/ja0730462. [DOI] [PubMed] [Google Scholar]
- 12.Phan AT, Luu KN, Patel DJ. Nucleic Acids Res. 2006;34:5715. doi: 10.1093/nar/gkl726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rujan IN, Meleney JC, Bolton PH. Nucleic Acids Res. 2005;33:2022. doi: 10.1093/nar/gki345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ying L, Green JJ, Li H, Klenerman D, Balasubramanian S. Proc Natl Acad Sci USA. 2003;100:14629. doi: 10.1073/pnas.2433350100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saneyoshi H, Mazzini S, Aviñó A, Portella G, González C, Orozco M, Marquez VE, Eritja R. Nucleic Acids Res. 2009;37:5589. doi: 10.1093/nar/gkp598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang C-F, Shafer RH. J Am Chem Soc. 2006;128:5966. doi: 10.1021/ja0603958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bugaut A, Balasubramanian S. Biochemistry. 2008;47:689. doi: 10.1021/bi701873c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smargiasso N, Rosu F, Hsia W, Colson P, Baker ES, Bowers MT, De Pauw E, Gabelica V. J Am Chem Soc. 2008;130:10208. doi: 10.1021/ja801535e. [DOI] [PubMed] [Google Scholar]
- 19.Phan AT, Kuryavyi V, Patel DJ. Curr Opin Struct Biol. 2006;16:288. doi: 10.1016/j.sbi.2006.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rachwal PA, Brown T, Fox KR. FEBS Lett. 2007;581:1657. doi: 10.1016/j.febslet.2007.03.040. [DOI] [PubMed] [Google Scholar]
- 21.Hazel P, Huppert J, Balasubramanian S, Neidle S. J Am Chem Soc. 2004;126:16405. doi: 10.1021/ja045154j. [DOI] [PubMed] [Google Scholar]
- 22.Guédin A, De Cian A, Gros J, Lacroix L, Mergny JL. Biochimie. 2008;90:686. doi: 10.1016/j.biochi.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 23.Risitano A, Fox KR. Nucleic Acids Res. 2004;32:2598–2606. doi: 10.1093/nar/gkh598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keniry MA, Owen EA, Shafer RH. Nucleic Acids Res. 1997;25:4389. doi: 10.1093/nar/25.21.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dai J, Punchihewa C, Ambrus A, Chen D, Jones RA, Yang D. Nucleic Acids Res. 2007;35:2440. doi: 10.1093/nar/gkm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Engelhart AE, Plavec J, Persil O, Hud NV. In: Nucleic Acid-Metal Ion Interactions. Hud NV, editor. Chapter 4. Royal Society of Chemistry; London: 2008. pp. 118–153. [Google Scholar]
- 27.Reshetnikov RV, Sponer J, Rassokhina OI, Kopylov AM, Tsvetkov PO, Makarov AA, Golovin AV. Nucleic Acids Res. 2011;39:9789. doi: 10.1093/nar/gkr639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joachimi A, Benza A, Hartig JS. Bioorg Med Chem. 2009;17:6811. doi: 10.1016/j.bmc.2009.08.043. [DOI] [PubMed] [Google Scholar]
- 29.Vondrušková J, Kypr J, Kejnovská I, Fialová M, Vorlíčková M. Biopolymers. 2008;89:797. doi: 10.1002/bip.21015. [DOI] [PubMed] [Google Scholar]
- 30.Sket P, Plavec J. J Am Chem Soc. 2010;132:12724. doi: 10.1021/ja104889t. [DOI] [PubMed] [Google Scholar]
- 31.Peng CG, Damha MJ. Nucleic Acids Res. 2007;35:4977. doi: 10.1093/nar/gkm520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dickerson RE, Drew HR, Conner BN, Wing RM, Fratini AV, Kopka ML. Science. 1982;216:475. doi: 10.1126/science.7071593. [DOI] [PubMed] [Google Scholar]
- 33.Altona C, Sundaralingam M. J Am Chem Soc. 1972;94:8205. doi: 10.1021/ja00778a043. [DOI] [PubMed] [Google Scholar]
- 34.Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Nature. 1992;355:564. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 35.Padmanabhan K, Padmanabhan KP, Ferrara JD, Sadler JE, Tulinsky A. J Biol Chem. 1993;268:17651. doi: 10.2210/pdb1hut/pdb. [DOI] [PubMed] [Google Scholar]
- 36.Schultze P, Macaya RF, Feigon J. J Mol Biol. 1994;235:1532. doi: 10.1006/jmbi.1994.1105. [DOI] [PubMed] [Google Scholar]
- 37.Wang KY, McCurdy S, Shea RG, Swaminathan S, Bolton PH. Biochemistry. 1993;32:1899. doi: 10.1021/bi00059a003. [DOI] [PubMed] [Google Scholar]
- 38.Wang KY, Krawczyk SH, Bischofberger N, Swaminathan S, Bolton PH. Biochemistry. 1993;32:11285. doi: 10.1021/bi00093a004. [DOI] [PubMed] [Google Scholar]
- 39.Smirnov I, Shafer RH. Biochemistry. 2000;39:1462. doi: 10.1021/bi9919044. [DOI] [PubMed] [Google Scholar]
- 40.Trajkovski M, PrimoŠket P, Plavec J. Org Biomol Chem. 2009;7:4677. doi: 10.1039/b914783g. [DOI] [PubMed] [Google Scholar]
- 41.Mao X, Gimeiner WH. J Mol Biol. 2005;256:417. [Google Scholar]
- 42.Maier MA, Choi Y, Gaus H, Barchi JJ, Marquez VE, Manoharan M. Nucleic Acids Res. 2004;32:3642. doi: 10.1093/nar/gkh667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Catlin JC, Guschlbauer W. Biopolymers. 1975;14:51. doi: 10.1002/bip.1975.360140105. [DOI] [PubMed] [Google Scholar]
- 44.Guschlbauer W, Jankowski K. Nucleic Acids Res. 1980;8:1421. doi: 10.1093/nar/8.6.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cushley RJ, Codington JF, Fox JJ. Can J Chem. 1968;46:1131. doi: 10.1021/jo01265a115. [DOI] [PubMed] [Google Scholar]
- 46.Marquez VE, Siddiqui MA, Ezzitouni A, Russ P, Wang J, Wagner RW, Matteucci MD. J Med Chem. 1996;39:3739. doi: 10.1021/jm960306+. [DOI] [PubMed] [Google Scholar]
- 47.Maderia M, Shenoy S, Van QN, Marquez VE, Barchi JJ. Nucleic Acids Res. 2007;35:1978. doi: 10.1093/nar/gkm025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coppola T, Varra M, Oliviero G, Galeone A, D’Isa G, Mayol L, Morelli B, Buccic RM, Vellecco V, Cirino G, Borbone N. Bioorg Med Chem. 2008;17:8244. doi: 10.1016/j.bmc.2008.07.040. [DOI] [PubMed] [Google Scholar]
- 49.Bonifacio L, Church FC, Jarstfer MB. Int J Mol Sci. 2008;9:422. doi: 10.3390/ijms9030422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pallan PS, Marquez VE, Egli M. Biochemistry. 2012;51:2639. doi: 10.1021/bi300215k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pasternak A, Hernandez FJ, Rasmussen LM, Vester B, Wengel J. Nucleic Acids Res. 2011;39:1155. doi: 10.1093/nar/gkq823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Puglisi JD, Tinoco I. Methods Enzymol. 1989;180:304. doi: 10.1016/0076-6879(89)80108-9. [DOI] [PubMed] [Google Scholar]
- 53.Morris KF, Johnson CS., Jr J Am Chem Soc. 1992;114:3139. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.