Abstract
In mammalian cells, POLQ (pol θ) is an unusual specialized DNA polymerase whose in vivo function is under active investigation. POLQ has been implicated by different experiments to play a role in resistance to ionizing radiation and defense against genomic instability, in base excision repair, and in immunological diversification. The protein is formed by an N-terminal helicase-like domain, a C-terminal DNA polymerase domain, and a large central domain that spans between the two. This arrangement is also found in the Drosophila Mus308 protein, which functions in resistance to DNA interstrand crosslinking agents. Homologs of POLQ and Mus308 are found in multicellular eukaryotes, including plants, but a comparison of phenotypes suggests that not all of these genes are functional orthologs. Flies defective in Mus308 are sensitive to DNA interstrand crosslinking agents, while mammalian cells defective in POLQ are primarily sensitive to DNA double-strand breaking agents. Cells from Polq−/− mice are hypersensitive to radiation and peripheral blood cells display increased spontaneous and ionizing radiation-induced levels of micronuclei (a hallmark of gross chromosomal aberrations), though mice apparently develop normally. Loss of POLQ in human and mouse cells causes sensitivity to ionizing radiation and other double strand breaking agents and increased DNA damage signaling. Retrospective studies of clinical samples show that higher levels of POLQ gene expression in breast and colorectal cancer are correlated with poorer outcomes for patients. A clear understanding of the mechanism of action and physiologic function of POLQ in the cell is likely to bear clinical relevance.
Keywords: DNA polymerase, POLQ, Translesion synthesis, Thymine glycol, Abasic site, Genomic Instability, Ionizing Radiation
Introduction
The functions of the 15 known mammalian DNA polymerases are currently being explored [1, 2]. DNA polymerases (pols) play pivotal roles not only in DNA replication (pols α, δ, ε), but also in base excision repair (pol β), mitochondrial replication and repair (pol γ), non-homologous end-joining and immunological diversity (pols λ, μ, and terminal-deoxynucleotidyl transferase), and DNA damage tolerance including translesion synthesis (η, κ, ζ, Rev1). Some DNA polymerases have roles in more than one pathway of DNA processing.
Identification of mammalian POLQ initially arose from interest in the Mus308 gene product of the fruit fly Drosophila melanogaster. Mus308 mutants are hypersensitive to agents that cause DNA interstrand cross-links (ICL), with only modest sensitivity to monofunctional alkylating agents [3]. This suggested that Mus308 might play a specific role in repair of ICLs in DNA. Characterization of the Mus308 gene showed that it encodes an unusual domain configuration, with the C-terminal part of the protein encoding a DNA polymerase, and the N-terminal part of the protein encoding a DNA helicase [4] (Figure 1).
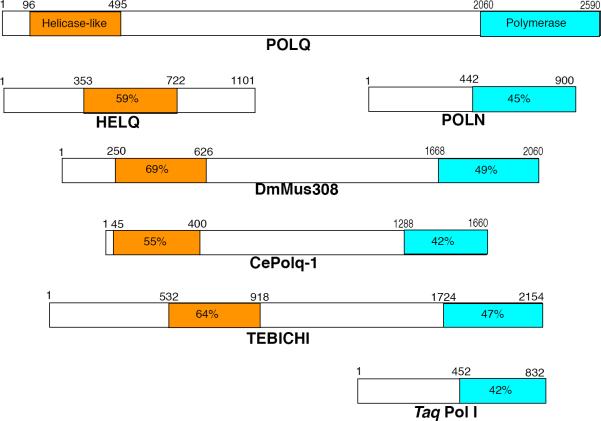
Figure 1. Structural comparison and related grouping of POLQ gene family members.
The orange regions and blue shaded regions are affiliated with the respective helicase and polymerase domains of the proteins. Numbers above the proteins indicate amino acid lengths. Percentages indicating the individual % similarity between the helicase-like and polymerase domains of POLQ were generated in MacVector.
A cDNA encoding the DNA polymerase domain of a homologous enzyme, designated DNA polymerase θ (POLQ), was identified by Sharief et al. [5]. The predicted protein of 1762 amino acids did not include a helicase-like domain, and is now recognized to correspond to the C-terminal portion of full-length POLQ. Independently, Abbas and Linn assembled a larger 9.1 kb cDNA encoding both polymerase and helicase domains and predicted it to encode a protein of 2724 amino acids [6]. Seki et al. independently isolated the first complete and functional POLQ cDNA, showing that it encodes a 2590 amino acid protein with both a DNA polymerase and helicase-like domain [7]. An orthologous mouse POLQ of 2544 amino acids was predicted from the genomic sequence by Shima et al. [8].
The polymerase domains of POLQ and Mus308 belong to the “A” family of DNA polymerases, showing sequence similarity to E. coli DNA polymerase I [4, 9]. As described in more detail below, however, the polymerase domain of POLQ contains three unique “insert” regions within the DNA polymerase sequence (Figure 2A and B) which determine some of its biochemical properties [10]. While much is known about the enzymatic properties of POLQ in vitro, further study is needed to better determine its functions in cells. Here we review data on the roles of POLQ in the defense against DNA damage.
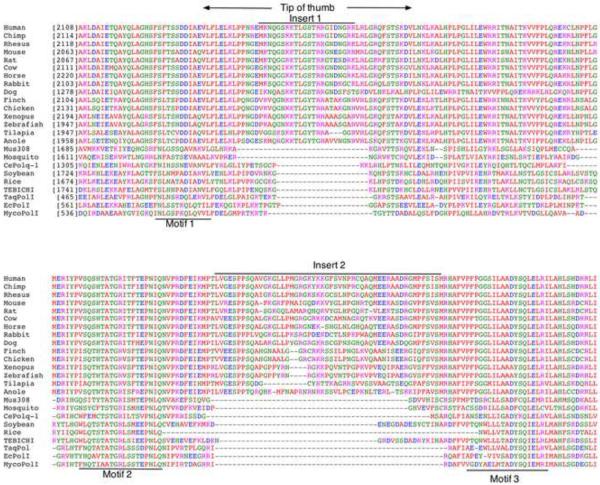
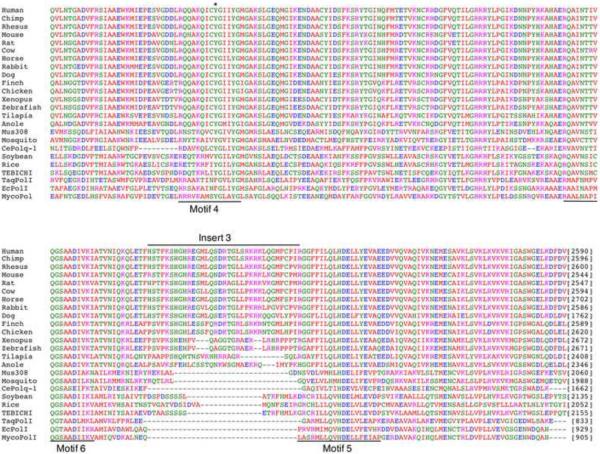
Figure 2. Map and alignment of POLQ polymerase domain.
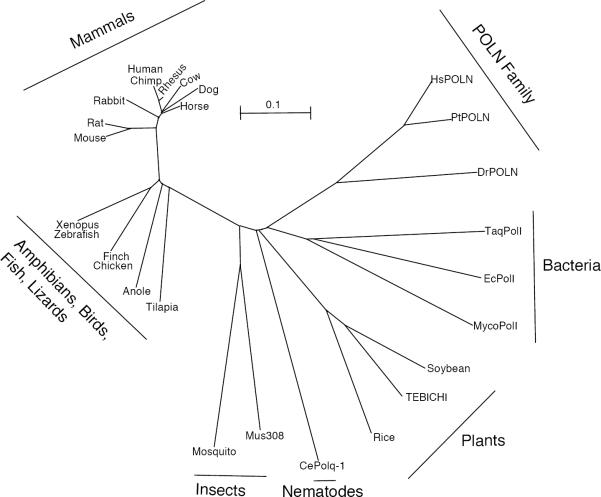
(A) The C-terminal portion of POLQ necessary for polymerase activity is displayed. The polymerase domain of POLQ is displayed in blue. Conserved A-family DNA polymerase motifs are noted in black. The unique sequence inserts found in POLQ are shown in yellow. An inactive DnaQ-like 3'→5' exonuclease domain and a fragment similar to the E. coli 5'→3' exonuclease domain and the 5'-dRP lyase activity of POLQ mapped to the polymerase domain are noted. An asterisk notes the tyrosine residue in motif 4 of the polymerase domain that is responsible for ddNTP incorporation. The location of the serine to proline mutation found in the Chaos1 mutant mouse is noted in the central domain. (B) Sequence alignment of POLQ family members constructed in MacVector. Starting and ending amino acid numbers for each species in the alignment are notated in brackets. (C). The dendogram was created SeaView. The scale bar indicates a distance of 0.1 amino acid substitution per site.
MycoPolI: Mycobacterium tuberculosis pol I; EcPolI: Escherichia coli pol I; TaqPolI: Thermus aquaticus pol I; TEBICHI: Arabidopsis thaliana TEBICHI; Rice: Oryza sativa pol I; Soybean: Glycine max POLQ-like; CePolq-1: Caenorhabditis elegans Polq-1; Mosquito: Aedes aegypti POLQ; Mus308: Drosophlia melanogaster Mus308; Anole: Anolis carolinensis POLQ-like; Tilapia: Oreochromis niloticus POLQ; Zebrafish: Danio rerio POLQ; Xenopus: Xenopus tropicalis POLQ; Chicken: Gallus gallus POLQ; Finch: Taeniopygia guttata POLQ; Dog: Canis lupus POLQ; Rabbit: Oryctolagus cuniculus; Horse: Equus caballus POLQ; Cow: Bos taurus POLQ; Rat: Rattus norvegicus POLQ; Mouse: Mus musculus POLQ; Rhesus: Macaca mulatta POLQ; Chimp: Pan troglodytes POLQ; Human: Homo sapiens POLQ; HsPOLN Homo sapiens POLN; PtPOLN: Pan troglodytes POLN; MmPOLN: Mus musculus POLN; DrPOLN: Danio rerio POLN.
POLQ gene family distribution in nature
Genes with similarity to POLQ and Mus308 are present in planate, protists, and multicellular eukaryotes (Figure 1), but not in yeast or other fungi. It remains to be determined whether the POLQ-like genes are true orthologs, as it appears that there are functional differences between species. Mutations in the polq-1 gene of C. elegans confer sensitivity to ICL-inducing agents [11]. TEBICHI, a POLQ-like protein found in Arabidopsis, is important for normal plant development and contributes to alleviating DNA replication stress and perhaps recombination. Tebichi mutant plants are sensitive to both the ICL-inducing agent mitomycin C (MMC) and to the monofunctional alkylating agent methyl methane sulfonate (MMS) [12, 13].
As described below, there are apparent functional differences in POLQ/Mus308 related genes from different species. The availability of numerous high quality sequences allows comparison by primary alignment, revealing several evolutionary patterns in the POLQ family. The length of the central domain of POLQ is about 1500–1600 amino acids long in vertebrates, but comprises only about 800 residues in plant homologs and 800–1000 in invertebrate animals (Table 1A). Further, the positions of the polymerase domain “inserts” are conserved within the entire POLQ family, but the inserts are much shorter in the non-vertebrate family members (Table 1B). Expansion of the size of the central domain and the inserts apparently arose contemporaneously in a common vertebrate ancestor. The variations in the central domain and polymerase domain insert lengths may account for some of the differences in damage sensitivities associated with defects in POLQ family genes in different organisms (Table 1A and 1B, Figure 2C).
Table 1.
Conservation of POLQ central domain length and polymerase domain inserts in evolution. Length of the (A) central domain and (B) polymerase domain inserts in POLQ gene family members. All lengths are in amino acids.
| A Length of POLQ central domain | ||
|---|---|---|
| Species | Length (aa) | |
| Human | 1565 | Vertebrates |
| Chimp | 1596 | |
| Mouse | 1552 | |
| Cow | 1593 | |
| Horse | 1599 | |
| Chicken | 1610 | |
| Finch | 1581 | |
| Xenopus | 1557 | |
| Zebrafish | 1457 | |
| Tilapia | 1271 | |
| Anole | 1364 | |
|
|
||
| Pea Aphid | 905 | Invertebrates |
| Western Mite | 1018 | |
| Fruit Fly | 1042 | |
| Mosquito | 801 | |
| C. elegans | 888 | |
|
|
||
| Arabidopsis | 806 | Plants |
| Soybean | 817 | |
| B Length of POLQ polymerase domain inserts (aa) | ||||
|---|---|---|---|---|
| Species | Insert 1 | Insert 2 | Inset 3 | |
| Human | 22 | 52 | 33 | Vertebrates |
| Chimp | 22 | 52 | 33 | |
| Mouse | 22 | 52 | 33 | |
| Chicken | 22 | 52 | 33 | |
| Dog | 22 | 52 | 33 | |
| Xenopus | 22 | 48 | 28 | |
| Zebrafish | 19 | 51 | 24 | |
| Tilapia | 19 | 42 | 25 | |
|
|
||||
| Fruit Fly | 4 | 9 | 9 | Invertebrates |
| Mosquito | 0 | 10 | 7 | |
| C. elegans | 3 | 9 | 0 | |
|
|
||||
| Soybean | 5 | 18 | 13 | Plants |
| Rice | 5 | 0 | 16 | |
| Arabidopsis | 5 | 17 | 13 | |
Two other genes in multicellular eukaryotes show significant and intriguing relationships with POLQ. HELQ (formerly HEL308) is a DNA repair helicase with amino acid sequence similarity to the helicase-like domain of POLQ. HELQ homologs are found in both animals [14] and archaea [15] but not in fungi or bacteria. POLN is a 900 amino acid protein in human cells, harboring an A-family DNA polymerase domain related to that of Mus308 and POLQ [9, 16, 17]. In contrast with POLQ, the phylogenetic distribution of POLN appears to be limited to the deuterostome lineage of eukaryotes, including vertebrates. Phylogenetic analysis suggests that POLN and POLQ were divergent from one another before the onset of the vertebrate lineage (Figure 2C).
Low fidelity and translesion synthesis activities of POLQ
Human POLQ has been purified as a recombinant protein from a baculovirus-infected insect cell expression system [7, 10]. The protein is active on substrates including oligonucleotide primer-templates, hairpin primer-templates, activated calf thymus DNA and poly (dA)-oligo (dT) [10, 18]. The activity of recombinant POLQ is relatively resistant to aphidicolin, a potent inhibitor of eukaryotic replicative DNA polymerases (pols α,δ,ε) [7, 19–21]. POLQ is sensitive to dideoxynucleoside triphosphate (ddNTP), consistent with the presence of a tyrosine residue in motif 4 of family-A DNA polymerases which facilitates ddNTP incorporation (Figure 2B) [18, 22]. Sequence alignment shows that POLQ contains a fragment of a 5'→3' exonuclease domain found in E. coli pol I and an inactive DnaQ-like 3'→5' exonuclease that is lacking conservation in the key HxAxxD ExoIIIε metal binding motif. Purified POLQ lacks a 3'→5' exonuclease activity, which is a common feature of low fidelity DNA polymerases (Figure 2A). Using a primer extension assay to measure DNA polymerase fidelity, POLQ was found to frequently misincorporate a G or T across from a T in the template [7, 10]. In a gap-filling assay to measure mutation frequency, human POLQ generated single base errors at a 10 to 100-fold higher rate than the other A family DNA polymerase members (γ, ν) [23]. POLQ adds single nucleotides to homopolymeric runs at particularly high rates during gap filling, exceeding 1% in certain sequence contexts. The enzyme generates single base substitutions at an average rate of 2.4 × 10−3, comparable in rate to the inaccurate family Y human pol κ (5.8 × 10−3). This is unusual, as the most studied low fidelity polymerases belong to the Y family (η, ι, κ, Rev1) rather than the A family of polymerases [24].
Maga and co-workers purified an activity from HeLa cell nuclear extracts that could efficiently utilize abasic site-containing DNA as a template. The purified preparation contained several polypeptides of about 100 kDa, one of which was recognized by an antibody raised against a fragment of recombinant POLQ [25]. The active fraction exhibited relatively high fidelity of DNA polymerization, and a 3' → 5' exonuclease activity but no ATPase or helicase activities [25]. It seems likely that this preparation contained an active fragment of POLQ, co-purifying with a 3' → 5' nuclease activity. The identity of this exonuclease is currently unknown but it would be of interest to isolate it, as it may be physiologically relevant to the activity of POLQ.
POLQ has the ability to catalyze DNA synthesis across specific forms of endogenous and exogenous DNA damage in a process called translesion DNA synthesis (TLS). A notable POLQ enzymatic trait is the ability to incorporate an A residue opposite an abasic (AP) sites and extend from the incorporated nucleotide. Although many DNA polymerases tend to insert an A opposite an AP site (known as the “A” rule [26–28]), the highly efficient extension of the A opposite an AP site is unique to POLQ [10]. For example, POLH (η) can insert a base opposite an AP site but cannot efficiently extend from the lesion, a property common to human Y family polymerases [10, 29–33]. About 18,000 AP sites arise in each diploid mammalian cell per day by hydrolytic depurination, but insertion of an A opposite such a site would be mutagenic[34]. Abasic sites are also generated frequently as intermediates in the base excision repair of uracil following accidental incorporation of dUTP during genomic DNA replication [35–37]. If a DNA replication fork encounters such an AP site before it is repaired, introduction of an A would reconstitute the original undamaged template and be physiologically useful. It remains to be determined whether this AP site bypass activity is a function of POLQ in vivo. Thymine glycol (Tg) is a common product of reactive oxygen species-mediated damage to DNA. POLQ and POLN can efficiently bypass both enantiomers (5R,5S) of Tg, whereas pol η extends efficiently only from the 5R-Tg [10, 38, 39]. POLQ cannot insert a base opposite the common UV-radiation induced cyclobutane pyrimidine dimers or (6–4) photoproducts. However, if pol ι [40] is used to incorporate bases opposite a 6–4PP, POLQ can extend the poorly matched primer-terminus [10, 41]. POLQ was also able to efficiently extend mismatched A:G, A:T, and A:C termini [41].
The ability of POLQ to extend from mismatched, poorly matched or unmatched termini is unusual and will be understood better after high-resolution structural features of the active site are determined. The three “inserts” in the catalytic domain are unique to POLQ (Figure 2A and B), and some specific deletions of these inserts have been analyzed with respect to activity. Insert 1 comprises 22 amino acids between the first and second conserved motifs in the “thumb” subdomain and its sequence is highly conserved throughout vertebrates [10]. In other A-family DNA polymerases, the thumb region influences DNA binding and frameshift fidelity. Deletions in the thumb region of E. coli Pol I can cause errors in processivity and an increased use in misaligned primer/template complex [22, 42, 43]. Insert 2 is 52 amino acids long and falls between the second and third motif and has the least sequence conservation among the inserts while insert 3 is 33 amino acids lying between the fifth and sixth motif (Fig. 2A and B). A fragment of POLQ (residues 1792–2590) containing the pol domain and a portion of the central domain (Figure 2A) is active as a DNA polymerase when produced as a recombinant protein in E. coli. Shorter fragments are much less active, or inactive [44, 45] indicating that this region of the central domain is required for efficient polymerase activity. The active fragment is able to bypass AP sites and Tg adducts, showing that these activities are independent of the N-terminal helicase-like domain. Elimination of Insert 1 of this POLQ fragment reduced processivity of the enzyme but had little, if any, bearing on the translesion synthesis properties of the enzyme. However, removal of either inserts 2 or 3 reduced activity on undamaged DNA and eliminated the ability of POLQ to bypass AP sites or Tg adducts [46].
The helicase-like domain of POLQ has 7 conserved motifs of the superfamily II DNA and RNA helicase family [7]. The POLQ helicase domain exhibited some single-stranded DNA-dependent ATPase activity, though lower in comparison to that of HELQ. No overt helicase activity of POLQ has been reported [7], and it is possible that the enzyme only displays helicase activity with an as yet untested substrate or requires accessory factors. Several cellular functions have been suggested for POLQ, and these are summarized in the next sections.
Hematopoiesis & somatic hypermutation
Some investigations have explored the possibility of a specialized immunological function for POLQ, either in somatic hypermutation (SHM) or class switch recombination (CSR) of immunoglobulin genes. POLQ is well-expressed in hematopoieiic cells, including germinal center B cells [47]. SHM acts to introduce point mutations in the variable (V) region of antibodies to alter their affinities while CSR changes the immunoglobulin constant region. Both processes are set in motion by the AID enzyme which can deaminate cytosine to uracil in DNA [48]. Mutations may then arise from low-fidelity repair synthesis or misincorporation opposite an AP site [49, 50].
In one study, Polq−/− mice were reported to have decreased mutations of C:G sequences at SHM hotspots while mutations at A:T sequences were unaffected [51]. POLH is important for A:T mutagenesis in SHM and Polh−/− Polq−/− mice do not exhibit a decrease in A:T mutations when compared to Polh−/− mice [52, 53]. POLQ extends beyond mispairs with A or T and can extend weakly from mispaired termini generated by POLH, suggesting that POLQ and POLH might cooperate during A:T mutagenesis [53]. In an independent set of experiments mice carrying a gene disruption that completely eliminates the POLQ enzyme had a frequency of SHM that was reduced by 60–80% [54]. There was no overall change in the proportion of events at A:T and C:G positions. Finally, Martomo et al. found similar mutation frequencies of V genes in Peyer's patches of Polh−/− Polq+/+ and Polh−/− Polq−/− mutant mice, suggesting that POLQ has a minor role, if any, in SHM [55].
The Polq gene was disrupted in the DT40 chicken B cell line by Yoshimura et al. using gene targeting [56, 57]. Strikingly, a PolnPolq double mutant showed a five-fold decrease in immunoglobulin (Ig) gene conversion, the primary method of V region diversity in avian species, and a triple PolhPolnPolq mutant failed to exhibit any Ig gene conversion [57]. In mammals, Ig class switching occurs by a different mechanism. The absence of POLQ does not seem to impair CSR in mammals as the mouse CH12F3 B-cell line defective for Polq can efficiently switch from IgM to IgA after appropriate stimulation [58].
Base excision repair
POLQ might serve as a backup polymerase for base excision repair (BER). In mammalian cells, single-nucleotide BER is performed by POLB (pol β)[45, 59]. A fragment of human POLQ displayed 5'-deoxyribose-phosphate lyase (dRp-lyase) activity, an activity necessary for single-nucleotide BER [60]. This dRp-lyase activity is present within a 24 kDa region of the POLQ polymerase domain (Figure 2). This is a contrast to the much stronger dRp-lyase activity of pol β, which is present in a distinct domain separate from the polymerase domain. 5'-dRp lyase activity was still present in a catalytically inactive 98 kDa polymerase fragment of POLQ [60].
In chicken DT40 cells, Polq and Polq Polb mutants were examined with regard to BER [56]. The Polq single mutant was sensitive to hydrogen peroxide (which can cause oxidative damage to bases) but not to MMS, although both agents cause damage that is repaired by BER (Table 2A). The Polq Polb mutant was more sensitive to MMS than either of the single mutants. Extracts from the Polq mutant DT40 cell line appeared to have a reduced capacity for both single-nucleotide and long patch BER [56].
Table 2.
Sensitivity of POLQ-defective cells to DNA damaging agents. Sensitivity of POLQ (A) vertebrate knockouts and mutants, (B) siRNA-depleted cell lines, and (C) lower eukaryote and plants knockouts to various DNA damaging agents. Sensitivity of cells to damaging agents were described as (+) hypersensitive, (−) no sensitivity, or (ND) not described.
| A Vertebrate POLQ knockouts and mutants | |||||
|---|---|---|---|---|---|
| Agent | Chaos1 | BMSC | CH12 Pol | CH12 TKO | DT40 |
| IR | + | ++ | + | + | − |
| Bleomycin | ND | ++ | ND | ND | ND |
| VP-16 | ND | ND | + | + | ND |
| Peroxide | ND | − | ND | ND | + |
| Paraquat | ND | − | ND | ND | ND |
| Paraquat | ND | − | ND | ND | ND |
| MMS | ND | ND | − | − | − |
| MMC | − | ND | ND | ND | − |
| Cisplatin | ND | ND | +/− | +/− | − |
| UVC | ND | + | + | ND | − |
| B POLQ-depleted human cells | ||||
|---|---|---|---|---|
| Agent | siPOC | siMRC5 | SiQ20B | siHeLa |
| IR | − | − | + | ++ |
| Temozolomide | ND | ND | − | ND |
| C POLQ knockouts in Drosophila, C. elegans, and Arabidopsis | |||
|---|---|---|---|
| Agent | Mus308 | polq-1 | TEBICHI |
| IR | − | +/− | ND |
| MMS | − | + | + |
| MMC | + | ND | + |
| Cisplatin | + | + | ND |
| HN2 | + | + | ND |
| UVC | ND | − | ND |
Mouse bone marrow stromal cells lacking Polq did not show increased sensitivity to hydrogen peroxide or paraquat in comparison to controls [61]. Human SQ20B head and neck cancer cells depleted for POLQ with siRNA were not sensitive to treatment with temozolomide, an alkylating agent that is related to MMS (Table 2B) [62]. CH12 mouse B lymphoma cells containing a deletion in an exon required for POLQ catalytic activity were somewhat more sensitive to MMS in the presence or absence of methoxyamine, which binds to AP sites and prevents single-nucleotide BER by pol β [52]. Taken together, it appears that POLQ is not a major contributor to BER. POLQ may be a backup enzyme for BER in the nematode C. elegans, which lacks pol β, although at least one additional DNA polymerase also appears to function in BER in this organism [63].
Interstrand Crosslink Repair
A recent report implicating Mus308, the Drosophila ortholog of POLQ, in an alternative form of end joining that may contribute to ICL repair [64]. Drosophilamus308, C. elegans polq-1, and Arabidopsis TEBICHI are all sensitive to the ICL-inducing agent MMC (Table 2C) [3, 11, 12]. In vertebrate cells, there are conflicting reports concerning the sensitivity of POLQ to ICL-inducing agents. DT40 cells lacking POLQ were not hypersensitive to cisplatin, a clinically used ICL agent [56, 57]. However deletion of Polq slightly sensitized murine CH12B-lymphoma cells to cisplatin and MMC [58]. The chaos1 mutant mice and cells derived from them exhibited no hypersensitivity to MMC, suggesting that POLQ does not participate in the repair of ICLs in vivo [65].
Tolerance of Double Strand Breaks and Alternative End-Joining of Breaks
While multiple functions have been offered, the strongest suggested role for POLQ is in the defense of radiation-mediated genomic instability. The chaos1 (chromosome aberration occurring spontaneously 1) mouse was produced in an phenotype-driven screen [8]. Male mice were periodically dosed with N-ethyl-N-nitrosourea (ENU) to induce germline mutations before mating. The generation 1 male offspring were backcrossed with generation 2 females to create generation 3 offspring that were potentially homozygous. The frequency of micronuclei (MN) was measured in male generation 3 mice. MN arise from acentric chromosome fragments (due to chromosome breakage) or whole chromosomes that were not packaged into the nucleus at the time of cell division [66]. DNA damaging agents induce MN, and those that form shortly before the nucleus is ejected can be assayed in peripheral blood reticulocytes by flow cytometry [67]. The chaos1 mice show increased spontaneous MN in blood samples, and the level is further elevated by ionizing radiation (IR) and MMC [8]. The chaos1 recessive mutation was mapped to a region of chromosome 16 that included the Polq gene, and determined to be a T to C point mutation in exon 19 of mouse Polq. This missense mutation converts Ser to Pro at amino acid 1932, located near the C-terminal end of the central domain, adjacent to the polymerase domain of POLQ (Figure 2A). Expression of wild type Polq from a bacterial artificial chromosome was able to suppress the chaos1 phenotype, suggesting that chaos1 mice possess a mutant allele of Polq [65].
A Polq−/− knockout mouse was then generated by Shima et al., introducing a point mutation in exon 1 to create a premature stop codon and replacing exons 2–5 with a neomycin resistance cassette [65]. A lack of complementation was observed between the chaos1 and Polq null alleles demonstrating that chaos1 is a mutant allele of Polq. The authors also noted a marked developmental disadvantage for Atm−/− Polq−/− double mutant mice. The few mice that did survive displayed signs of severe DNA damage stress, including increased chromosomal instability, decreased body weight and interestingly a delayed onset of thymic lymphomas in comparison to Atm−/− mice [65]. Goff et al. found that Polq−/− mice have increased levels of IR-induced MN and confirmed that the levels of spontaneous MN formation were nearly identical to those reported for chaos1 mice [61, 65]. Goff et al. further found that bone marrow stromal cells from the Polq−/− mice were hypersensitive to gamma irradiation and to bleomycin, a drug that causes DNA double-strand breaks (Table 2A) [61]. Inhibition of ATM activation with the drug KU55933 increased radiosensitivity in wild type but not in Polq−/− cells. A Polq-null mutant was also constructed by Li et al. in the mouse CH12F3 B cell line, and was more sensitive than normal cells to gamma-irradiation and etoposide [58]. POLQ was also found as a hit in an siRNA screen of genes affecting IR sensitivity of human cells. Depletion of POLQ in SQ20B and HeLa cells caused an increase in IR-induced γH2AX foci and sensitized the cells to gamma-irradiation (Table 2B) [62]. These observations indicate that POLQ has a role in the tolerance or repair of DNA double strand breaks.
The mechanism by which POLQ aids in the defense against IR-mediated genomic instability is under investigation. Recent studies of Drosophila Mus308 function point towards a role of this gene in an alternative form of end joining which relies upon annealing of microhomologies in order for DNA end joining to occur. Yu et al. investigated the inaccurate repair of DSBs mediated by the I-SceI nuclease in Mus308 mutants and found a decrease in a process termed synthesis-dependent microhomology-mediated end joining (SD-MMEJ) [68]. This repair pathway is independent of Ku70, Lig4, and Rad51 and distinct from “canonical” non-homologous end-joining (NHEJ) of DNA breaks. SD-MMEJ is related to or identical to pathways defined in other biological systems as alternative end joining (alt-EJ) or micro-homology-mediated end-joining (mm-EJ). Analysis of mus308 mutants showed a decrease in the use of long micro-homologies at the junction site and abrogation of complex T-nucleotide insertion events, and suggested that this alt-EJ pathway is occurring in parallel to LIG4-mediated canonical end-joining [64]. In Drosophila, Mus308 does not appear to participate in homology-directed repair. Mus308 mutant larvae are not more sensitive to IR than wild type, but they are synergistically sensitized to radiation by addition of a spn-A (Rad51) mutation, indicating that Mus308-dependent repair is independent of homologous recombination in Drosophila [69]. This is in contrast to the Polq mutant in chicken DT40 cells, which showed decreased levels of homologous recombination repair of I-Sce-I-mediated DNA DSBs [57, 64]. Human and mouse POLQ mutants are radiosensitive, suggesting some divergence of POLQ and Mus308 functions. In a biochemical study, an active DNA polymerase fragment of POLQ performed template-independent extension of single-stranded DNA or duplex DNA with protruding or mismatched 3'-hydroxyl termini [70]. Such activities would be useful in alt-EJ. It will be interesting to see whether genetic data supports a role for mammalian POLQ in the repair of DNA double strand breaks by alt-EJ. The ability of POLQ to bypass Tg, AP sites, or other lesions might also contribute to tolerance of IR-induced DNA damage.
Functions at the DNA replication fork
There are some suggestions that POLQ might work at the DNA replication fork or in close connection with DNA replication proteins. As described below, POLQ gene expression correlates with expression of genes involved in cell cycle control and DNA replication [71]. MRC5VA cells were constructed that stably express recombinant POLQ, and this resulted in an increase in γH2AX foci, an increase in chromosomal abnormalities and defective DNA replication fork progression [72]. These data show that it is imperative for cells to keep levels of POLQ tightly controlled to avoid detrimental effects on the cell. In the same screen that identified the chaos1 allele of POLQ, a second mutant termed chaos3 showed a similar phenotype of increased micronuclei. Chaos3 has been identified as a hypomorphic allele of Mcm4, a component of the helicase operating during semiconservative replication [73]. In Arabidopsis, TEBICHI gene mutants show striking defects in regulated cell division and differentiation of the meristem [12].
POLQ expression in mammalian & cancer tissue
The POLQ gene is ubiquitously expressed in human and mouse tissues and cell lines, as measured by northern blot analysis, reverse transcriptase polymerase chain reaction (RT-PCR), and microarrays. Levels of expression appear to be highest in the testis, human placental tissue and hematopoietic cells [7, 65, 74]. POLQ is a member of a cluster of genes in which higher levels of expression correlate with lower patient survival for several cancers. This cluster includes genes involved in DNA replication, repair, recombination and cell cycle control. Higher expression of POLQ mRNA was observed in some tumor samples from stomach, lung, and particularly colon cancers compared to matched control nontumor tissue [74]. Stratification of colon cancers based on POLQ expression alone indicated that the “POLQ high” group had poorer survival than the “POLQ low” group by an average of ~24 months. Another study of colorectal cancer patients indicated that tumors with higher expression of a group of 47 DNA replication-related genes (including POLQ) were significantly correlated with poorer patient survival [71]. POLQ expression levels alone were correlated with poor patient survival [71]. In an analysis focusing on the expression of DNA polymerases in human breast cancers Lemée et al. found that of all 14 nuclear DNA polymerases, only POLQ expression was significantly higher in the cancer in comparison to normal tissues. Stratification according to expression showed that higher levels of POLQ expression were correlated with poorer survival outcomes [72]. Another study of POLQ expression in early breast cancer indicated POLQ had the highest expression in ER negative and high-grade tumors, and that higher expression was correlated with poor relapse-free survival [75]. Considering that radiation therapy is a frontline treatment for breast cancer and that human cancer cells depleted for POLQ are sensitive to ionizing radiation, POLQ is a molecule of interest in the study of breast cancers and a potentially druggable target.
In some non-tumor cells, suppression of POLQ by siRNA had little effect on radiation sensitivity [62]. It would be clinically beneficial if tumor cell but not normal cells could be sensitized by POLQ suppression, but it remains to be seen whether this is generally the case. As described above, genetic ablation of POLQ from normal mouse cell lines leads to radiation hypersensitivity. Despite this radiation sensitivity, the hematopoietic system of Polq−/− mice seems resilient. Mice irradiated with a sub-lethal dose of 7 Gy of γ-irradiation showed a subsequent depression of counts for white blood cells, red blood cells, lymphocytes and platelets. Recovery to normal levels took place in Polq−/− mice as well as wild-type mice, showing that the hematopoietic system remains resilient even in Polq-defective animals [61]. Due to its unique properties and clinical interest, further work to understand this enzyme and its true role in the cell is pivotal.
Conclusion
POLQ has unique translesion synthesis characteristics influenced by inserts within the polymerase domain. The detailed structure of the protein should be informative in determining the exact origin of the novel biochemical properties of POLQ. Further study needs to be given to the ability of POLQ to catalyse template-independent DNA synthesis.
There is an accumulation of evidence that POLQ is preventing radiation-induced genomic instability in cells as both murine non-tumorigenic and human tumorigenic cell lines defective for POLQ are sensitive to radiation. The mechanism as to how POLQ is contributing to genomic stability still needs to be sorted out. POLQ may be preventing this instability through the bypass of IR-induced lesions or through an alternative end joining mechanism. Additional study is needed in order to see if mammalian POLQ participates in this repair pathway and may answer the question of whether Mus308 is a paralog or true ortholog of POLQ. Also of interest is whether POLQ acts at the replication fork and what are the cellular consequences of its presence. The elevated levels of POLQ expression in breast cancers coupled with the radiosensitivity of POLQ-depleted cancer cells makes the enzyme an intriguing target for radiosensitization in breast cancer.
Acknowledgements
We thank Kei-ichi Takata, Francesca Cole, and Sylvie Doublié for helpful comments on the manuscript. This research was supported by NIH Training Grant T32 CA09480, Grant P01 CA09717 from the National Cancer Institute and by NIH Cancer Center Support Grant P30-CA016672 (University of Texas M. D. Anderson Cancer Center).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement The authors declare that they have no financial, personal or professional competing interests that could be construed to have influenced this paper.
References
- [1].Hübscher U, Maga G, Spadari S. Eukaryotic DNA polymerases. Annu RevBiochem. 2002;71:133–163. doi: 10.1146/annurev.biochem.71.090501.150041. [DOI] [PubMed] [Google Scholar]
- [2].Lange SS, Takata K, Wood RD. DNA polymerases and cancer. Nature Reviews Cancer. 2011;11:96–110. doi: 10.1038/nrc2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Aguirrezabalaga I, Nivard MJ, Comendador MA, Vogel EW. The cross-linking agent hexamethylphosphoramide predominantly induces intra-locus and multi-locus deletions in postmeiotic germ cells of Drosophila. Genetics. 1995;139:649–658. doi: 10.1093/genetics/139.2.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Harris PV, Mazina OM, Leonhardt EA, Case RB, Boyd JB, Burtis KC. Molecular cloning of Drosophila mus308, a gene involved in DNA cross-link repair with homology to prokaryotic DNA polymerase I genes. Mol Cell Biol. 1996;16:5764–5771. doi: 10.1128/mcb.16.10.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Sharief FS, Vojta PJ, Ropp PA, Copeland WC. Cloning and chromosomal mapping of the human DNA polymerase theta (POLQ), the eighth human DNA polymerase. Genomics. 1999;59:90–96. doi: 10.1006/geno.1999.5843. [DOI] [PubMed] [Google Scholar]
- [6].Abbas AR. Ph.D. thesis. 2000. I. Cloning and characterization of human DNA polymerase theta. II. Protein interactions of human damaged DNA binding protein; p. 3002618. UMI: [Google Scholar]
- [7].Seki M, Marini F, Wood RD. POLQ (Pol theta), a DNA polymerase and DNA-dependent ATPase in human cells. Nucleic Acids Res. 2003;31:6117–6126. doi: 10.1093/nar/gkg814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Shima N, Hartford SA, Duffy T, Wilson LA, Schimenti KJ, Schimenti JC. Phenotype-based identification of mouse chromosome instability mutants. Genetics. 163(2003):1031–1040. doi: 10.1093/genetics/163.3.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Marini F, Kim N, Schuffert A, Wood RD. POLN, a nuclear PolA family DNA polymerase homologous to the DNA cross-link sensitivity protein Mus308. J Biol Chem. 2003;278:32014–32019. doi: 10.1074/jbc.M305646200. [DOI] [PubMed] [Google Scholar]
- [10].Seki M, Masutani C, Yang LW, Schuffert A, Iwai S, Bahar I, Wood RD. High-efficiency bypass of DNA damage by human DNA polymerase Q. EMBO J. 23(2004):4484–4494. doi: 10.1038/sj.emboj.7600424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Muzzini DM, Plevani P, Boulton SJ, Cassata G, Marini F. Caenorhabditis elegans POLQ-1 and HEL-308 function in two distinct DNA interstrand cross-link repair pathways. DNA Repair (Amst) 2008;7:941–950. doi: 10.1016/j.dnarep.2008.03.021. [DOI] [PubMed] [Google Scholar]
- [12].Inagaki S, Suzuki T, Ohto MA, Urawa H, Horiuchi T, Nakamura K, Morikami A. Arabidopsis TEBICHI, with helicase and DNA polymerase domains, is required for regulated cell division and differentiation in meristems. Plant Cell. 2006;18:879–892. doi: 10.1105/tpc.105.036798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Inagaki S, Nakamura K, Morikami A. A link among DNA replication, recombination, and gene expression revealed by genetic and genomic analysis of TEBICHI gene of Arabidopsis thaliana. PLoS Genet. 2009;5:e1000613. doi: 10.1371/journal.pgen.1000613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Marini F, Wood RD. A human DNA helicase homologous to the DNA cross-link sensitivity protein Mus308. J Biol Chem. 2002;277:8716–8723. doi: 10.1074/jbc.M110271200. [DOI] [PubMed] [Google Scholar]
- [15].Woodman IL, Bolt EL. Molecular biology of Hel308 helicase in archaea. Biochemical Society transactions. 37(2009):74–78. doi: 10.1042/BST0370074. [DOI] [PubMed] [Google Scholar]
- [16].Takata K, Arana ME, Seki M, Kunkel TA, Wood RD. Evolutionary conservation of residues in vertebrate DNA polymerase N conferring low fidelity and bypass activity. Nucleic Acids Res. 2010;38:3233–3244. doi: 10.1093/nar/gkq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Takata KI, Shimizu T, Iwai S, Wood RD. Human DNA polymerase N (POLN) is a low-fidelity enzyme capable of error-free bypass of 5S-thymine glycol. J Biol Chem. 2006;281:23445–23455. doi: 10.1074/jbc.M604317200. [DOI] [PubMed] [Google Scholar]
- [18].Seki M, Marini F, Wood RD. POLQ (Pol θ), a DNA polymerase and DNA-dependent ATPase in human cells. Nucleic Acids Res. 2003;31:6117–6126. doi: 10.1093/nar/gkg814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Sheaff R, Ilsley D, Kuchta R. Mechanism of DNA polymerase alpha inhibition by aphidicolin. Biochemistry. 1991;30:8590–8597. doi: 10.1021/bi00099a014. [DOI] [PubMed] [Google Scholar]
- [20].Dresler SL, Gowans BJ, Robinson-Hill RM, Hunting DJ. Involvement of DNA polymerase delta in DNA repair synthesis in human fibroblasts at late times after ultraviolet irradiation. Biochemistry. 1988;27:6379–6383. doi: 10.1021/bi00417a028. [DOI] [PubMed] [Google Scholar]
- [21].Cheng CH, Kuchta RD. DNA polymerase epsilon: aphidicolin inhibition and the relationship between polymerase and exonuclease activity. Biochemistry. 1993;32:8568–8574. doi: 10.1021/bi00084a025. [DOI] [PubMed] [Google Scholar]
- [22].Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- [23].Arana ME, Seki M, Wood RD, Rogozin IB, Kunkel TA. Low-fidelity DNA synthesis by human DNA polymerase theta. Nucleic Acids Res. 2008;36:3847–3856. doi: 10.1093/nar/gkn310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Goodman MF. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu Rev Biochem. 2002;71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- [25].Maga G, Shevelev I, Ramadan K, Spadari S, Hubscher U. DNA polymerase theta purified from human cells is a high-fidelity enzyme. J Mol Biol. 2002;319:359–369. doi: 10.1016/S0022-2836(02)00325-X. [DOI] [PubMed] [Google Scholar]
- [26].Strauss BS. The “A” rule revisited: polymerases as determinants of mutational specificity. DNA Repair (Amst) 2002;1:125–135. doi: 10.1016/s1568-7864(01)00014-3. [DOI] [PubMed] [Google Scholar]
- [27].Avkin S, Adar S, Blander G, Livneh Z. Quantitative measurement of translesion replication in human cells: evidence for bypass of abasic sites by a replicative DNA polymerase. Proc Natl Acad Sci U S A. 2002;99:3764–3769. doi: 10.1073/pnas.062038699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Strauss B, Rabkin S, Sagher D, Moore P. The role of DNA polymerase in base substitution mutagenesis on non-instructional templates. Biochimie. 1982;64:829–838. doi: 10.1016/s0300-9084(82)80138-7. [DOI] [PubMed] [Google Scholar]
- [29].Masutani C, Kusumoto R, Iwai S, Hanaoka F. Mechanisms of accurate translesion synthesis by human DNA polymerase eta. EMBO J. 2000;19:3100–3109. doi: 10.1093/emboj/19.12.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Eukaryotic polymerases iota and zeta act sequentially to bypass DNA lesions. Nature. 2000;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- [31].Lawrence CW. Cellular roles of DNA polymerase zeta and Rev1 protein. DNA Repair (Amst) 2002;1:425–435. doi: 10.1016/s1568-7864(02)00038-1. [DOI] [PubMed] [Google Scholar]
- [32].Vaisman A, Frank EG, McDonald JP, Tissier A, Woodgate R. poliota-dependent lesion bypass in vitro. Mutat Res. 2002;510:9–22. doi: 10.1016/s0027-5107(02)00248-8. [DOI] [PubMed] [Google Scholar]
- [33].Zhang Y, Yuan F, Wu X, Taylor JS, Wang Z. Response of human DNA polymerase iota to DNA lesions. Nucleic Acids Res. 2001;29:928–935. doi: 10.1093/nar/29.4.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA repair and mutagenesis. 2nd edition ASM Press; Washington, DC: 2006. [Google Scholar]
- [35].Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- [36].Nakamura J, Walker VE, Upton PB, Chiang SY, Kow YW, Swenberg JA. Highly sensitive apurinic/apyrimidinic site assay can detect spontaneous and chemically induced depurination under physiological conditions. Cancer Res. 1998;58:222–225. [PubMed] [Google Scholar]
- [37].Guillet M, Boiteux S. Origin of endogenous DNA abasic sites in Saccharomyces cerevisiae. Mol Cell Biol. 2003;23:8386–8394. doi: 10.1128/MCB.23.22.8386-8394.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kusumoto R, Masutani C, Iwai S, Hanaoka F. Translesion synthesis by human DNA polymerase eta across thymine glycol lesions. Biochemistry. 2002;41:6090–6099. doi: 10.1021/bi025549k. [DOI] [PubMed] [Google Scholar]
- [39].Takata K, Shimizu T, Iwai S, Wood RD. Human DNA polymerase N (POLN) is a low fidelity enzyme capable of error-free bypass of 5S-thymine glycol. J Biol Chem. 2006;281:23445–23455. doi: 10.1074/jbc.M604317200. [DOI] [PubMed] [Google Scholar]
- [40].Vaisman A, Lehmann AR, Woodgate R. DNA polymerases eta and iota. Adv Protein Chem. 2004;69:205–228. doi: 10.1016/S0065-3233(04)69007-3. [DOI] [PubMed] [Google Scholar]
- [41].Seki M, Wood RD. DNA polymerase theta (POLQ) can extend from mismatches and from bases opposite a (6–4) photoproduct. DNA Repair (Amst) 2008;7:119–127. doi: 10.1016/j.dnarep.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Minnick DT, Astatke M, Joyce CM, Kunkel TA. A thumb subdomain mutant of the large fragment of Escherichia coli DNA polymerase I with reduced DNA binding affinity, processivity, and frameshift fidelity. J Biol Chem. 1996;271:24954–24961. doi: 10.1074/jbc.271.40.24954. [DOI] [PubMed] [Google Scholar]
- [43].Cannistraro VJ, Taylor JS. DNA-thumb interactions and processivity of T7 DNA polymerase in comparison to yeast polymerase eta. J Biol Chem. 2004;279:18288–18295. doi: 10.1074/jbc.M400282200. [DOI] [PubMed] [Google Scholar]
- [44].Hogg M, Seki M, Wood RD, Doublié S, Wallace SS. Lesion bypass activity of DNA polymerase theta (POLQ) is an intrinsic property of the pol domain and depends on unique sequence inserts. J Mol Biol. 2011;405:642–652. doi: 10.1016/j.jmb.2010.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Prasad R, Longley MJ, Sharief FS, Hou EW, Copeland WC, Wilson SH. Human DNA polymerase {theta} possesses 5'-dRP lyase activity and functions in single-nucleotide base excision repair in vitro. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Hogg M, Seki M, Wood RD, Doublie S, Wallace SS. Lesion bypass activity of DNA polymerase theta (POLQ) is an intrinsic property of the pol domain and depends on unique sequence inserts. J Mol Biol. 2011;405:642–652. doi: 10.1016/j.jmb.2010.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Kawamura K, Bahar R, Seimiya M, Chiyo M, Wada A, Okada S, Hatano M, Tokuhisa T, Kimura H, Watanabe S, Honda I, Sakiyama S, Tagawa M, O-Wang J. DNA polymerase θ is preferentially expressed in lymphoid tissues and upregulated in human cancers. Int J Cancer. 2004;109:9–16. doi: 10.1002/ijc.11666. [DOI] [PubMed] [Google Scholar]
- [48].Honjo T, Muramatsu M, Fagarasan S. AID: how does it aid antibody diversity? Immunity. 2004;20:659–668. doi: 10.1016/j.immuni.2004.05.011. [DOI] [PubMed] [Google Scholar]
- [49].Neuberger MS, Di Noia JM, Beale RC, Williams GT, Yang Z, Rada C. Somatic hypermutation at A.T pairs: polymerase error versus dUTP incorporation. Nat Rev Immunol. 2005;5:171–178. doi: 10.1038/nri1553. [DOI] [PubMed] [Google Scholar]
- [50].Seki M, Gearhart PJ, Wood RD. DNA polymerases and somatic hypermutation of immunoglobulin genes. EMBO Rep. 2005;6:1143–1148. doi: 10.1038/sj.embor.7400582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Masuda K, Ouchida R, Takeuchi A, Saito T, Koseki H, Kawamura K, Tagawa M, Tokuhisa T, Azuma T, J OW. DNA polymerase theta contributes to the generation of C/G mutations during somatic hypermutation of Ig genes. Proc Natl Acad Sci U S A. 2005;102:13986–13991. doi: 10.1073/pnas.0505636102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Ukai A, Maruyama T, Mochizuki S, Ouchida R, Masuda K, Kawamura K, Tagawa M, Kinoshita K, Sakamoto A, Tokuhisa T, J OW. Role of DNA polymerase theta in tolerance of endogenous and exogenous DNA damage in mouse B cells. Genes Cells. 2006;11:111–121. doi: 10.1111/j.1365-2443.2006.00922.x. [DOI] [PubMed] [Google Scholar]
- [53].Masuda K, Ouchida R, Hikida M, Kurosaki T, Yokoi M, Masutani C, Seki M, Wood RD, Hanaoka F, J OW. DNA polymerases eta and theta function in the same genetic pathway to generate mutations at A/T during somatic hypermutation of Ig genes. J Biol Chem. 2007;282:17387–17394. doi: 10.1074/jbc.M611849200. [DOI] [PubMed] [Google Scholar]
- [54].Zan H, Shima N, Xu Z, Al-Qahtani A, Evinger AJ, Iii, Zhong Y, Schimenti JC, Casali P. The translesion DNA polymerase θ plays a dominant role in immunoglobulin gene somatic hypermutation. EMBO J. 2005;24:3757–3769. doi: 10.1038/sj.emboj.7600833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Martomo SA, Saribasak H, Yokoi M, Hanaoka F, Gearhart PJ. Reevaluation of the role of DNA polymerase theta in somatic hypermutation of immunoglobulin genes. DNA Repair (Amst) 2008;7:1603–1608. doi: 10.1016/j.dnarep.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Yoshimura M, Kohzaki M, Nakamura J, Asagoshi K, Sonoda E, Hou E, Prasad R, Wilson SH, Tano K, Yasui A, Lan L, Seki M, Wood RD, Arakawa H, Buerstedde JM, Hochegger H, Okada T, Hiraoka M, Takeda S. Vertebrate POLQ and POLbeta cooperate in base excision repair of oxidative DNA damage. Mol Cell. 2006;24:115–125. doi: 10.1016/j.molcel.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Kohzaki M, Nishihara K, Hirota K, Sonoda E, Yoshimura M, Ekino S, Butler JE, Watanabe M, Halazonetis TD, Takeda S. DNA polymerases nu and theta are required for efficient immunoglobulin V gene diversification in chicken. J Cell Biol. 2010;189:1117–1127. doi: 10.1083/jcb.200912012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Li Y, Gao X, Wang JY. Comparison of two POLQ mutants reveals that a polymerase-inactive POLQ retains significant function in tolerance to etoposide and gamma-irradiation in mouse B cells. Genes Cells. 2011;16:973–983. doi: 10.1111/j.1365-2443.2011.01550.x. [DOI] [PubMed] [Google Scholar]
- [59].Yoshimura M, Kohzaki M, Nakamura J, Asagoshi K, Sonoda E, Hou E, Prasad R, Wilson SH, Tano K, Yasui A, Lan L, Seki M, Wood RD, Arakawa H, Buerstedde JM, Hochegger H, Okada T, Hiraoka M, Takeda S. Vertebrate POLQ and POL beta cooperate in base excision repair of oxidative DNA damage. Mol Cell. 2006;24:115–125. doi: 10.1016/j.molcel.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Prasad R, Longley MJ, Sharief FS, Hou EW, Copeland WC, Wilson SH. Human DNA polymerase theta possesses 5'-dRP lyase activity and functions in single-nucleotide base excision repair in vitro. Nucleic Acids Res. 2009;37:1868–1877. doi: 10.1093/nar/gkp035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Goff JP, Shields DS, Seki M, Choi S, Epperly MW, Dixon T, Wang H, Bakkenist CJ, Dertinger SD, Torous DK, Wittschieben J, Wood RD, Greenberger JS. Lack of DNA polymerase theta (POLQ) radiosensitizes bone marrow stromal cells in vitro and increases reticulocyte micronuclei after total-body irradiation. Radiat Res. 2009;172:165–174. doi: 10.1667/RR1598.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Higgins GS, Prevo R, Lee YF, Helleday T, Muschel RJ, Taylor S, Yoshimura M, Hickson ID, Bernhard EJ, McKenna WG. A small interfering RNA screen of genes involved in DNA repair identifies tumor-specific radiosensitization by POLQ knockdown. Cancer Res. 2010;70:2984–2993. doi: 10.1158/0008-5472.CAN-09-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Asagoshi K, Lehmann W, Braithwaite EK, Santana-Santos L, Prasad R, Freedman JH, Van Houten B, Wilson SH. Single-nucleotide base excision repair DNA polymerase activity in C. elegans in the absence of DNA polymerase beta. Nucleic Acids Res. 2012;40:670–681. doi: 10.1093/nar/gkr727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Chan SH, Yu AM, McVey M. Dual roles for DNA polymerase theta in alternative end-joining repair of double-strand breaks in Drosophila. PLoS Genet. 2010;6:e1001005. doi: 10.1371/journal.pgen.1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Shima N, Munroe RJ, Schimenti JC. The mouse genomic instability mutation chaos1 is an allele of Polq that exhibits genetic interaction with Atm. Mol Cell Biol. 2004;24:10381–10389. doi: 10.1128/MCB.24.23.10381-10389.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Nusse M, Miller BM, Viaggi S, Grawe J. Analysis of the DNA content distribution of micronuclei using flow sorting and fluorescent in situ hybridization with a centromeric DNA probe. Mutagenesis. 1996;11:405–413. doi: 10.1093/mutage/11.4.405. [DOI] [PubMed] [Google Scholar]
- [67].Dertinger SD, Torous DK, Tometsko KR. Simple and reliable enumeration of micronucleated reticulocytes with a single-laser flow cytometer. Mutat Res. 1996;371:283–292. doi: 10.1016/s0165-1218(96)90117-2. [DOI] [PubMed] [Google Scholar]
- [68].Yu AM, McVey M. Synthesis-dependent microhomology-mediated end joining accounts for multiple types of repair junctions. Nucleic Acids Res. 2010;38:5706–5717. doi: 10.1093/nar/gkq379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Chan SH, Yu AM, McVey M. Dual Roles for DNA Polymerase Theta in Alternative End-Joining Repair of Double-Strand Breaks in Drosophila. PLoS Genet. 2010;6:e1001005. doi: 10.1371/journal.pgen.1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Hogg M, Sauer-Eriksson AE, Johansson E. Promiscuous DNA synthesis by human DNA polymerase theta. Nucleic Acids Res. 2012;40:2611–2622. doi: 10.1093/nar/gkr1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Pillaire MJ, Selves J, Gordien K, Gourraud PA, Gentil C, Danjoux M, Do C, Negre V, Bieth A, Guimbaud R, Trouche D, Pasero P, Mechali M, Hoffmann JS, Cazaux C. A 'DNA replication' signature of progression and negative outcome in colorectal cancer. Oncogene. 2010;29:876–887. doi: 10.1038/onc.2009.378. [DOI] [PubMed] [Google Scholar]
- [72].Lemee F, Bergoglio V, Fernandez-Vidal A, Machado-Silva A, Pillaire MJ, Bieth A, Gentil C, Baker L, Martin AL, Leduc C, Lam E, Magdeleine E, Filleron T, Oumouhou N, Kaina B, Seki M, Grimal F, Lacroix-Triki M, Thompson A, Roche H, Bourdon JC, Wood RD, Hoffmann JS, Cazaux C. DNA polymerase theta up-regulation is associated with poor survival in breast cancer, perturbs DNA replication, and promotes genetic instability. Proc Natl Acad Sci U S A. 2010;107:13390–13395. doi: 10.1073/pnas.0910759107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Shima N, Alcaraz A, Liachko I, Buske TR, Andrews CA, Munroe RJ, Hartford SA, Tye BK, Schimenti JC. A viable allele of Mcm4 causes chromosome instability and mammary adenocarcinomas in mice. Nat Genet. 2007;39:93–98. doi: 10.1038/ng1936. [DOI] [PubMed] [Google Scholar]
- [74].Kawamura K, Bahar R, Seimiya M, Chiyo M, Wada A, Okada S, Hatano M, Tokuhisa T, Kimura H, Watanabe S, Honda I, Sakiyama S, Tagawa M, J OW. DNA polymerase theta is preferentially expressed in lymphoid tissues and upregulated in human cancers. Int J Cancer. 2004;109:9–16. doi: 10.1002/ijc.11666. [DOI] [PubMed] [Google Scholar]
- [75].Higgins GS, Harris AL, Prevo R, Helleday T, McKenna WG, Buffa FM. Overexpression Of POLQ Confers a Poor Prognosis In Early Breast Cancer Patients. Oncotarget. 2010;1:175–184. doi: 10.18632/oncotarget.124. [DOI] [PMC free article] [PubMed] [Google Scholar]