Abstract
Expansion of certain trinucleotide repeats causes several types of human diseases, and such tracts are associated with the formation of deletions and other types of genetic rearrangements in Escherichia coli, yeast, and mammalian cells. Below, we show that long (230 repeats) tracts of the trinucleotide associated with Friedreich’s ataxia (GAA•TTC) stimulate both large (>50 bp) deletions and point mutations in a reporter gene located more than 1 kb from the repetitive tract. Sequence analysis of deletion breakpoints indicates that the deletions reflect non-homologous end joining of double-stranded DNA breaks (DSBs) initiated in the tract. The tract-induced point mutations appear to reflect a different mechanism involving single-strand annealing of DNA molecules generated by DSBs within the tract, followed by filling-in of single-stranded gaps by the error-prone DNA polymerase zeta.
Keywords: Friedreich’s ataxia, GAA•TTC triplet repeats, genome instability, mutations, Saccharomyces cerevisiae
1. Introduction
In humans, certain genetic diseases are caused by expansions of tri-, tetra-, penta- or dodecanucleotide repeats [1–5]. The types of repeats associated with expansions are usually, although not always [6], capable of forming “hairpins” or other types of non-canonical DNA structures. For example, the triplet repeat associated with Friedreich’s ataxia [2], (GAA•TTC), is capable for forming triplex DNA/H-DNA [4].
The effects of long (>100 repeats) trinucleotide tracts on genome stability have been examined in bacteria, yeast, and cultured mammalian cells. These tracts tend to have high levels of alterations, with deletions exceeding expansions in most yeast and bacteria studies [7]. These alterations are likely to reflect the processing of non-canonical DNA structures by various cellular repair proteins [8] or errors made during replicative DNA synthesis of these unconventional repetitive sequences [6].
In E. coli, plasmid-borne tracts of (GAA•TTC)n exhibit length- and orientation-dependent instability [9]. The longer (>100 repeat) tracts tend to contract, particularly when oriented such that the GAA sequences are on the lagging strand template during replication; henceforth, we will refer to this orientation as the GAA orientation and the reverse orientation as the TTC orientation. Similar orientation- and length-dependence effects are observed in yeast [10]. In yeast, replication forks stall at the (GAA•TTC)n tracts in the GAA, but not the TTC, orientation. This result argues that one cause of tract instability is likely to be related to stalled or broken DNA replication forks. In the experiments described below, all yeast strains had tracts in the GAA orientation based on our previous analysis of tract-associated replication fork blockage [11].
(GAA•TTC)n tracts also stimulate recombination and chromosome rearrangements. Plasmid-borne tracts in E. coli elevate the frequency of both intra- and interchromosomal recombination [12]. Kim et al. [13] showed that (GAA•TTC)n tracts increased the frequency of deletions, translocations, and ectopic recombination in the GAA orientation with a smaller effect in the TTC orientation. In strains with a (GAA•TTC)230 tract in the GAA orientation, a double-stranded DNA break (DSB) occurs within the tract; this break is not observed for tracts in the TTC orientation. The (GAA•TTC)230 tracts in both orientations act as strong mitotic recombination hotspots in diploid yeast cells [11]. In addition, in these strains, a tract-associated DSB was observed with (GAA•TTC)230 tracts in both orientations. Based on these results, it is likely that there are two types of tract-associated DSBs: those that occur in exponentially-growing cells (orientation-dependent) and those that occur in stationary phase cells (orientation-independent).
In mammalian cells, DNA sequences capable of forming triplex DNA, including (GAA•TTC)n tracts, can induce mutations of flanking sequences. Wang and Vasquez [14] showed that a triplex-forming sequence derived from the c-myc promoter stimulated mutations in an immediately-adjacent reporter gene by about 20-fold; over 98% of these mutations were deletions or complex rearrangements. Wojciechowska et al. [15] found that (GAA•TTC)200 tracts stimulated deletions and rearrangements of an adjacent reporter gene 3- to 10-fold. No significant tract-associated increases in point mutations were observed in either of these mammalian cell studies. In contrast, Bidichandani et al. [16] reported a three-fold elevation of point mutations induced in the X25 gene by a (GAA•TTC) tract located within the intron in serially-passaged lymphoblasts. In addition, point mutations could be induced about 10-fold in mammalian cells by transformation with triplex-forming oligonucleotides [17]. In yeast, Shishkin et al. [18] showed that (GAA•TTC) tracts located within an intron of the URA3 stimulated point mutations and small deletions in the coding sequences flanking the intron; subsequent studies showed that these mutations were generated independently of the error-prone DNA polymerase zeta (K. Shah and S. Mirkin, personal communication). Large deletions that included the tracts as well as the flanking URA3 coding sequences were also observed. In addition, expansions of the (GAA•TTC) tracts were observed. These expansions were much more frequent with long (≥ 125 repeats) than short tracts (≤ 100 repeats) and were not affected significantly by the orientation of the tract.
In both E. coli [19] and S. cerevisiae [20–23], DSBs can elevate the frequency of mutations in sequences located near the break. These studies utilize site-specific endonucleases, HO or I-SceI, to efficiently generate DSBs. Strathern et al. [20] showed that an HO-induced DSB stimulated reversion of a point mutation in a tightly-linked reporter gene; this effect was largely dependent on Rev3p [24], encoding the catalytic subunit of DNA polymerase zeta. In a system that allowed detection of both point mutations and deletions, most of the mutations induced by HO were point mutations or frameshifts with large deletions representing a minor class [25]. Yang et al. [21] showed that long single-stranded regions adjacent to the site of an I-SceI-generated DSB were hypermutable by ultraviolet light and methyl methanesulfonate, and this hypermutability was dependent on DNA polymerase zeta. In contrast to the studies of Holbeck and Strathern [24] and Yang et al. [21], Hicks et al. [22] found that mutations introduced during MATα-HMR gene conversion events were independent of DNA polymerase zeta. Mutations introduced during break-induced replication in yeast were also only modestly (two-fold) reduced in cells lacking DNA polymerase zeta [23].
In the studies described in the paragraph above, site-specific endonucleases were used to generate the mutagenic DSB. In our study, we examine the mutagenic effects of sequences that are prone to DSB formation in the absence of a site-specific endonuclease. Below, we investigate mutations induced by a (GAA•TTC)230 tract located about 1 kb from the reporter gene. We show that long (230 repeats), but not short (20 repeats) significantly elevate the frequency of both large deletions and point mutations within the reporter gene. The large deletions usually delete part or all of the (GAA•TTC) tract in addition to part or all of the URA3 gene, and sequence analysis demonstrates that these deletions are a consequence of non-homologous end-joining events. The tract-induced point mutations require Rev3p and are usually associated with reductions in the length of the (GAA•TTC)230 tract. We suggest that deletions and point mutations reflect two different outcomes of the cellular repair of tract-induced DSBs. In a related study in yeast, elevated levels of mutations were observed in a reporter gene located about 8 kb from DSB-prone sites generated by either a quasi-palindrome or a (GAA•TTC)230 tract (N. Saini, Y. Zhang, Y. Nishida, and K. Lobachev, personal communication). These induced mutations were also dependent on DNA polymerase zeta.
2. Materials and methods
2.1 Yeast strains
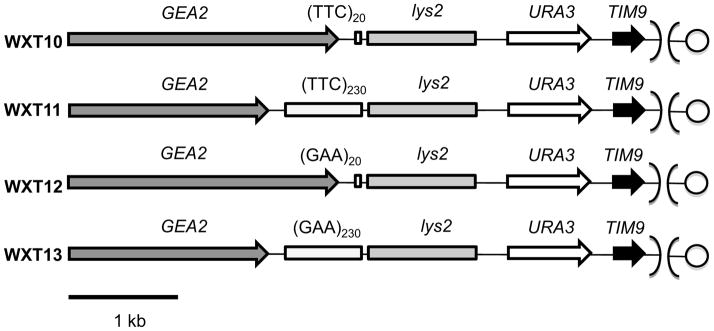
We used four haploid strains with two different tract sizes (20 and 230 repeats) of GAA•TTC tracts in two different orientations (termed “GAA” and “TTC”): WXT10[(TTC)20], WXT11 [(TTC)230], WXT12 [(GAA)20], and WXT13 [(GAA)230] [11]. WXT10-13 had an insertion of the wild-type URA3 gene located 1.2 kb centromere-proximal to the tract (Table S1; Fig. 1, Fig. S1). These strains are derived from the haploid strain PSL5 [26] that is derived from the YJM789 genetic background [27]. The DNA sequences flanking the tracts are shown in Fig. S2. The details of the constructions and genotypes of these strains are given in Supplementary Material (Text S1, Table S1, and Table S2). Standard yeast procedures were used for transformations [28]. Media were prepared as described previously [26,29].
Figure 1.
Depictions of genes flanking the GAA•TTC tracts on chromosome V in strains used in the study (not drawn to scale). In all strains, the tracts (shown by white rectangles) are located about 1.2 kb from the URA3 gene that is used as a reporter of mutagenesis. The GEA2 and TIM9 genes flank the tract and the URA3 gene; the centromere is shown as a circle. A 1 kb spacer fragment containing a 3’ fragment of the LYS2 gene is inserted between the tracts and the URA3 gene. The nearest essential genes flanking the tract are TIM9 and SNU13 (located about 14 kb centromere-distal to the tract).
2.2 Measurements of mutation rates and analysis of ura3 mutations
We measured the rate of ura3 mutations in the WXT strains by measuring the frequency of 5-fluoroorotic acid (5-FOA) resistant derivatives in multiple independent cultures (>20). The methods used to determine the rates of ura3 mutations in various strains were similar to those described previously [30] except for the drug used for selection. A detailed description of rate measurements is given in Supplementary Material (Text S1).
Independent 5-FOAR resistant strains were analyzed by PCR to determine the ratio of ura3 point mutations to deletions. In two separate PCR reactions, we examined the sizes of the GAA•TTC tracts and the sizes of the URA3 gene. In about 150 strains with a point mutation of URA3, we sequenced the PCR fragment to locate the point mutation. Strains that lacked a GAA•TTC tract and/or a URA3 gene were further examined by other PCR reactions in order to determine the deletion breakpoint. Details of this analysis, including the sequence of the diagnostic primers, are described in Supplementary Material. Sequences of deletions are in Table S3, and sequences of point mutations are in Tables S4 and S5.
2.3 Statistical analysis
Analysis of the statistical significance of the numbers of events of different classes was done using the Fisher exact test on the VassarStats Website (http://faculty.vassar.edu/lowry/VassarStats.html). Calculation of mutation rates and 95% confidence limits on these rates were done as described previously [31].
3. Results
3.1 Experimental system
In our previous study in this genetic background [11], we showed that in one orientation (defined as the GAA orientation) the (GAA•TTC)230 tract resulted in a replication fork block; this block was not observed for the (GAA•TTC)230 tract in the opposite orientation or for the (GAA•TTC)20 tract in either orientation. We also showed that the (GAA•TTC)230 tracts stimulated reciprocal mitotic crossovers in an orientation-independent manner, and that tract-associated double-stranded DNA breaks (DSBs) occurred in about 1% of stationary phase cells with long tracts.
Our experiments involved four closely-related haploid strains (WXT10-13) that had either long (230 repeats) or short (20 repeats) (GAA•TTC) tracts in both orientations placed about 1 kb from a wild-type URA3 gene on chromosome V (Fig. 1). Strains with a wild-type URA3 gene are sensitive to 5-fluoro-orotate (5-FOA) [32]. Thus, we could determine whether the (GAA•TTC) tracts stimulated nearby mutations by measuring the frequencies of 5-FOA-resistant colonies in multiple independent cultures and converting those frequencies into rates by the method of the median [30,33]. Details of the constructions of these strains are in Supplementary Material (Text S1, Tables S1 and S2) and Lee et al. [26].
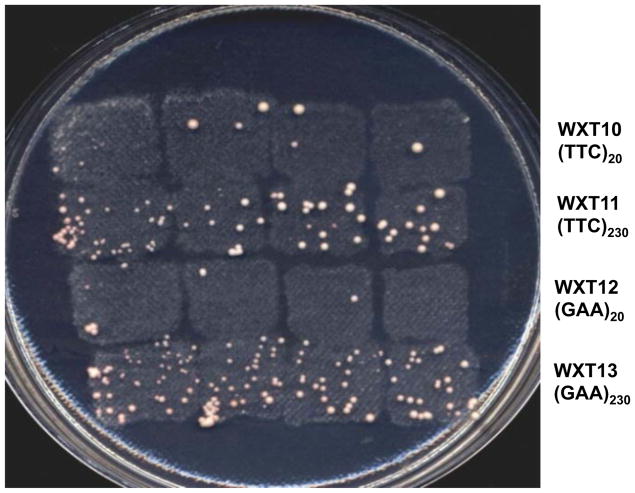
Before detailed measurements of mutation rates were done, we replica-plated “patches” of WXT10-13 grown in rich medium (allowing the growth of both Ura+ and Ura− cells) to 5-FOA-containing medium. It was apparent that the long (GAA•TTC) tracts in both orientations strongly elevated ura3 mutations (Fig. 2). The rates of ura3 mutations are shown in Table 1. Relative to an isogenic strain without any tract (MD415), the short tracts (strains WXT10 and WXT12) were not mutagenic. In contrast, 230-repeat (GAA•TTC) tracts stimulated mutation rates more than 30-fold in both orientations (strains WXT11 and WXT13). Although there is no strong orientation-dependence of the mutagenic effects of the 230-repeat tracts, the strain with the replication-fork-blocking GAA orientation (WXT13) had an approximately two-fold stronger mutagenic effect than the strain with the non-blocking TTC orientation (WXT11, Table 1).
Figure 2.
Induction of ura3 mutations, leading to 5-FOAR strains, by (GAA•TTC)230 tracts. Single colonies derived from the strains WXT10 (TTC)20, WXT11 (TTC)230,WXT12 (GAA)20, and WXT13 (GAA)230 were used to make patches of cells on rich growth medium. These patches were replica-plated to medium containing 5-fluoro-orotate. This medium allows the growth of ura3 mutant strains, but not wild-type strains. The elevated number of 5-FOAR colonies in strains with the long tracts (WXT11 and WXT13) relative to those with the short tracts (WXT10 and WXT12) is evident.
Table 1.
Rates of the GAA•TTC tract-associated ura3 mutations
| Relevant genotype (strain name)1 | Tract orientation, size, and URA3 location2 | Rate of 5-FOAR (95% confidence limits) [normalized rate]3 |
|---|---|---|
|
| ||
| Wild-type (MD416) | No tract | 3.5×10−8 (2.1–4.8) [1] |
| Wild-type (WXT10) | (TTC)20 | 2.5×10−8 (1.8–3.8) [0.7] |
| Wild-type (WXT11) | (TTC)230 | 1.4×10−6 (0.9–1.5) [40] |
| Wild-type (WXT12) | (GAA)20 | 3.0×10−8 (1.8–3.0) [0.9] |
| Wild-type (WXT13) | (GAA)230 | 3.2×10−6 (2.4–3.6) [91] |
| rev3 (WXT81) | (TTC)230 | 9.6×10−8 (4.7–14) [2.7] |
| rev3 (WXT83) | (GAA)230 | 4.4×10−8 (2.6–13) [1.3] |
All strains are isogenic with PSL5 (MATα ade2-1 ura3 can1Δ::SUP4-o gal2 ho::hisG; [29]) except for alterations introduced by transformation as described in Supplementary Material.
(GAA•TTC) tracts, 20 or 230 repeats in length, were inserted in all strains except for MD416. The GAA and TTC orientations are the same as those defined previously [11].
Rates and 95% confidence limits were calculated as described in Stone et al. [30]. Rates were normalized (shown in brackets) by dividing the observed rate for the strain with the GAA or TTC tracts by the rate observed in the strain without any tract.
3.2 Analysis of the mutational spectra in WXT10-WXT13
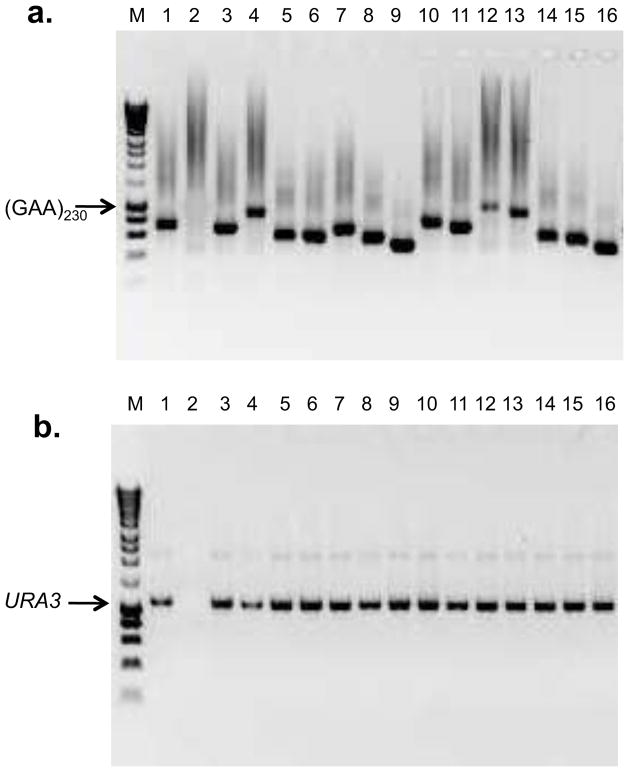
We characterized the types of ura3 mutations in strains with short and long tracts. The initial characterization involved two PCR reactions. One primer pair (Double-CHK-R and Big-CHK-F; sequences in Table S2) flanked the tract and was used to monitor deletions or expansions of repeats. The second pair (Del-R and Del-F; Table S2) closely flanked the URA3 insertion. Using these primer pairs, we examined 84, 154, 84, and 176 5-FOAR colonies from strains WXT10 [(TTC)20], WXT11 [(TTC)230], WXT12 [(GAA)20], and WXT13 [(GAA)230,] respectively. An example of the analysis for 5-FOAR colonies derived from WXT13 is shown in Fig. 3.
Figure 3.
Gel analysis of alterations in tract length and alterations in the size of the URA3 gene in ura3 mutants with a (GAA)230 tract. Independent 5-FOAR mutants were isolated from the haploid strain WXT13. Using primers that closely flank the tract (Doub-CHK-R and Big-CHK-F) or closely flank the URA3 gene (URA-F and URA3-R), we performed PCR and then looked for size changes of the tract (Fig. 3a) or the URA3 gene (Fig. 3b) by gel electrophoresis in 16 independent 5-FOAR derivatives.
a. Tract size. Most of the tracts in the 5-FOAR derivatives are shorter than the tract in the parental WXT13 strain (position indicated by the arrow). Derivative 2 lacks a PCR fragment, indicating deletion of one or both primer sites.
b. URA3 size. Most strains have a band that is approximately the size of the original wild-type URA3 gene, with the exception of derivative 2. The lack of a PCR fragment in derivative 2 by this analysis and that shown in Fig. 3a suggests that this strain has a deletion that includes both tract and URA3 sequences, a conclusion supported by mapping and sequencing the deletion breakpoints.
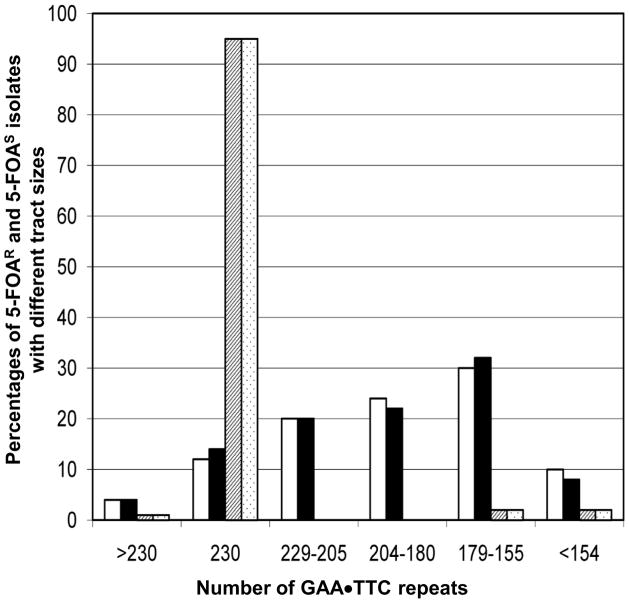
We observed that most of the 5-FOAR derivatives in the strains with 230 repeats (138 of 154 for WXT11 and 158 of 176 for WXT13) retained the repetitive tract, but the sizes of the repetitive tracts were usually smaller than the original tract size. As shown in Fig. 4, less than 20% of the tracts in the 5-FOAR strains with 230 repeats in either orientation retained a tract of the original size. In contrast, when we examined Ura+ derivatives of WXT11 or WXT13, more than 90% tracts were of the original size, and none of the tracts lacked the PCR fragment generated by primers flanking the tract. One interpretation of this result is that the mechanism that produces a mutation in ura3 in the strain with the long tracts is a consequence of a tract-associated DNA lesion. In our study, as in previous studies of the behavior of long trinucleotide tracts in yeast [7], tract expansion events are rare relative to deletions (Fig. 4).
Figure 4.
Tract alterations observed in independent isolates of the URA3 parental strains WXT11 ([GAA•TTC]230 tract in TTC orientation) and WXT13 ([GAA•TTC]230 tract in GAA orientation) compared to tract alterations in 5-FOAR mutants derived from WXT11 and WXT13. The white and black rectangles show tract sizes in the 5-FOAR isolates of WXT11 and WXT13, respectively. The striped and stippled rectangles indicate tract sizes in individual URA3 (5-FOAS) isolates of WXT11 and WXT13, respectively. The numbers of isolates examined for each strain were: WXT11 ura3 strains (50), WXT13 ura3 (50), WXT11 URA3 (100), and WXT13 URA3 (100).
About 10% of the 5-FOAR derivatives lacked a PCR fragment indicative of the trinucleotide tract, indicating deletion one or both of the primer sites flanking the tract (derivative 2, Fig. 3A); these same strains also usually lacked the PCR fragment characteristic of the URA3 gene (derivative 2, Fig. 3B). We interpret this result as indicating that these strains had a deletion that extended from the tract to the URA3 gene, a conclusion supported by subsequent DNA sequence analysis described below. No deletions of tracts or of the URA3 genes were observed in strains with the 20-repeat tracts.
A summary of the frequency of point mutations and large deletions (classification based on PCR analysis) is shown in Table 2. In this analysis, a large deletion is defined as a deletion that removes primer-binding sites for both the tract and the URA3 gene. As discussed below, by subsequent DNA sequence analysis, we found a small number of small (< 20 bp) deletions within URA3; these are included as point mutations in Table 2. About 10% of the ura3 mutations obtained in the (TTC)230 strain WXT11 were large deletions versus 0% in the (TTC)20; by Fisher exact test, the numbers of events in these two categories were significantly different (p<0.002). Similarly, the number of large deletions in the (GAA)230 5-FOAR strain WXT13 was significantly in excess of that observed in the (GAA)20 strain WXT12 (p<0.003).
Table 2.
Numbers of ura3 deletions and ura3 point mutations recovered in strains with insertions of (GAA • TTC) tracts.
| Relevant genotype(strain name)1 | Tract orientation and size2 | # 5-FOAR strains examined3 | # strains with ura3 deletions (%)4 | # strains with ura3 point mutations (%) |
|---|---|---|---|---|
|
| ||||
| Wild-type (WXT10) | (TTC)20 | 84 | 0 | 84 |
| Wild-type (WXT11) | (TTC)230 | 154 | 16 (10%) | 138 (90%) |
| Wild-type (WXT12) | (GAA)20 | 84 | 0 | 84 |
| Wild-type (WXT13) | (GAA)230 | 176 | 18 (10%) | 158 (90%) |
| rev3 (WXT81) | (TTC)230 | 103 | 32 (31%) | 71 (69%) |
| rev3 (WXT83) | (GAA)230 | 98 | 25 (26%) | 73 (74%) |
All strains are isogenic with PSL5 (MATα ade2-1 ura3 can1Δ::SUP4-o gal2 ho::hisG; [29]) except for alterations introduced by transformation as described in Supplementary Material
(GAA•TTC) tracts, 20 or 230 repeats in length, were inserted in all strains. The GAA and TTC orientations are the same as those defined previously [11].
All mutations were independently isolated.
This column includes only deletions that are detectable by the PCR analysis described
in the text. Deletions <50 bp would not be detectable.
The extent of the deletions was determined by PCR analysis using additional primers in the region containing the tract and the URA3 gene (details in Supplementary Material). The expected extent of the deletions is limited by the essential centromere-proximal TIM9 gene located <300 bp downstream of URA3 and the centromere-distal SNU13 gene located about 14 kb upstream of URA3. We sequenced 34 deletions, one example of which is shown in Fig. 5. In this example and in all of the other sequenced deletions (data in Table S3), there are no short repetitive DNA sequences at the deletion breakpoints. Thus, the deletions reflect non-homologous end-joining (NHEJ) events. The summary of the mapping of deletions in WXT11 and WXT13 is shown in Fig. 6. All deletions removed part or all of the tract, and all or part of URA3.
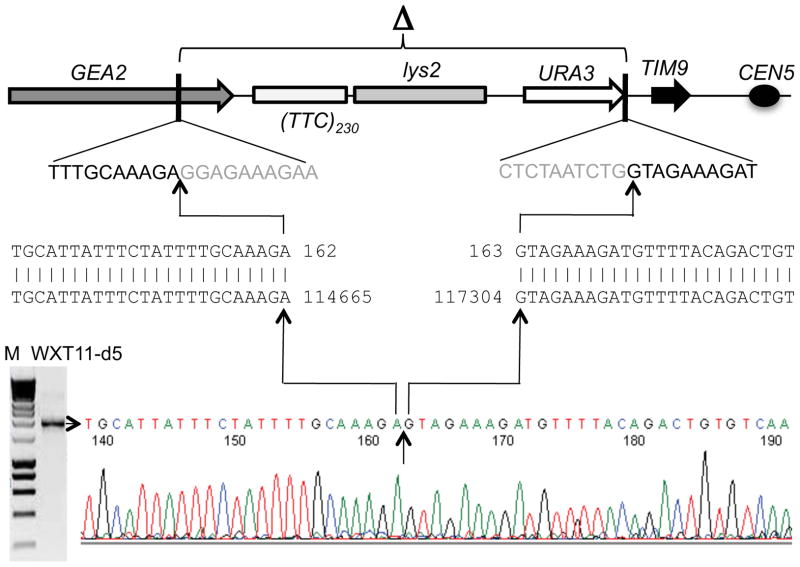
Figure 5.
Sequence analysis of an ura3 deletion derived from WXT11 (WXT11-d5). PCR analysis indicated that this 5-FOAR isolate lacked at least one primer site flanking the tract and one primer site flanking the URA3 gene. PCR analysis using one primer located about 1.5 kb upstream of the tract (Del-R) and a second located about 1 kb downstream of URA3 indicated that this strain had a 2.7 kb deletion. Sequence analysis of the PCR product (shown at the bottom part of the figure) demonstrated that the deletion was between SGD coordinates 114665 and 117304. The sequences displayed with clustered vertical lines are the SGD sequence of chromosome V (on the bottom) and the sequence of the PCR fragment shown on the top. The sequences shown in gray below the line drawing of chromosome V are the deleted sequences at the breakpoints. The comparison of these sequences indicates that the deletion reflects a non-homologous end-joining event.
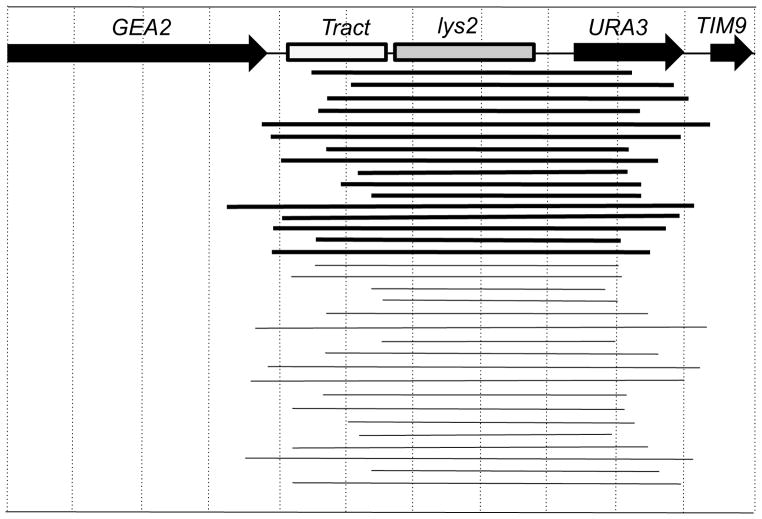
Figure 6.
Mapping of deletions induced in strains with (GAA•TTC)230 tracts. By sequence analysis, we mapped deletions in strains WXT11 and WXT13. In this figure, the distance between the vertical dotted lines represents 500 bp. Each horizontal line represents an independent deletion. Thick lines show deletions derived from WXT11 (TTC orientation) and thin lines shown deletions derived from WXT13 (GAA orientation).
As shown in Table 1, the frequencies of ura3 point mutations were substantially elevated by long, but not short, (GAA•TTC) tracts. We sequenced the mutant genes in 91 Ura− derivatives of strains with long tracts (WXT11 and WXT13) and 57 Ura− derivatives of strains with short tracts (WXT10 and WXT12). These data are shown in Tables S4 and S5. For both types of strains, almost all of the mutations (>97%) were single-base substitutions or single base in/dels. It was previously shown that most (>90%) of the 5-FOAR mutants derived from a wild-type strain have single base substitutions or single base in/dels [34]. One difference between the mutations observed in the long- and short-tract strains is that the long-tract strains had more double mutations. In the long-tract strains, 21 5-FOAR mutants had double mutations and 70 had one, whereas in the short-tract strains, only two strains had double mutations and 55 had single mutations; this difference is statistically significant by the two-tailed Fisher exact test (p=0.002).
3.3 Long (GAA)230 and (TTC)230 tracts do not elevate mutation rates throughout the genome
It is conceivable that the long (GAA)230 and (TTC)230 tracts could generate a DNA lesion that would activate error-prone DNA polymerases and stimulate mutations throughout the genome. To examine this possibility, we constructed isogenic CAN1 derivatives of WXT10, WXT11, WXT12, and WXT13, respectively MD573, MD574, MD577, and MD575. The mutation rates from canavanine-sensitivity to canavanine-resistance were calculated for each strain by fluctuation analysis. The rates of mutation for all strains were not significantly different (95% confidence intervals shown in parentheses): MD573 (TTC)20, 4.7 x 10−7 (3.7–6.0 x 10−7); MD574 (TTC)230, 3.8 x 10−7 (3.3–4.4 x 10−7); MD577 (GAA)20, 2.4 x 10−8 (2.0–3.0 x 10−7); MD575 (GAA)230, 3.9 x 10−7 (3.2–4.3 x 10−7). These results demonstrate that the long trinucleotide tracts do not elevate mutation rates throughout the genome and it is likely that they only act to stimulate mutation rates of genes located nearby.
3.4 Point mutations induced by the (GAA)230 and (TTC)230 tracts are dependent on Rev3p
REV3 encodes the catalytic subunit of the error-prone DNA polymerase zeta and, as discussed in the Introduction, the Rev3p is involved in translesion bypass and DSB repair. Since the mutations in a reporter gene induced by cleavage of a closely-linked HO recognition sequence are Rev3p-dependent [24,25], we examined the effect of the rev3 mutation on tract-stimulated mutations in URA3. The rev3 strains WXT81 and WXT83 are derived from WXT11 and WXT13, respectively. As shown in Table 1, most of the tract-induced ura3 mutations are Rev3p-dependent. In addition, for both WXT81 and WXT83, about 28% of the ura3 mutations were deletions compared to the isogenic strains with the wild-type REV3 gene (Table 2); this difference was statistically significant (p=0.001, Fisher exact test). This result suggests that point mutations have a stronger dependence on Rev3p than deletions, although loss of Rev3p reduces both types of mutations.
4. Discussion
Our analysis reveals a number of important features of tract-stimulated mutagenesis in yeast. First, (GAA•TTC)230 tracts in both the GAA and TTC orientations substantially (>10-fold) elevate the frequency of large deletion mutations and point mutations in a reporter gene located about 1 kb from the tract; (GAA•TTC)20 tracts do not elevate the rate of mutations. Second, both the point mutations and the large deletions are likely to be initiated by a tract-associated DNA lesion since the large deletions always include part or all of the tract and since >80% of the point mutations were associated with a tract of reduced size. Third, sequence analysis of the large deletions indicates that these deletions were formed by non-homologous end-joining. Fourth, there was a higher frequency of double mutations in ura3 mutants associated with the long tracts than the short tracts. Fifth, the tract-associated mutations require the activity of Rev3p, part of the error-prone DNA polymerase zeta.
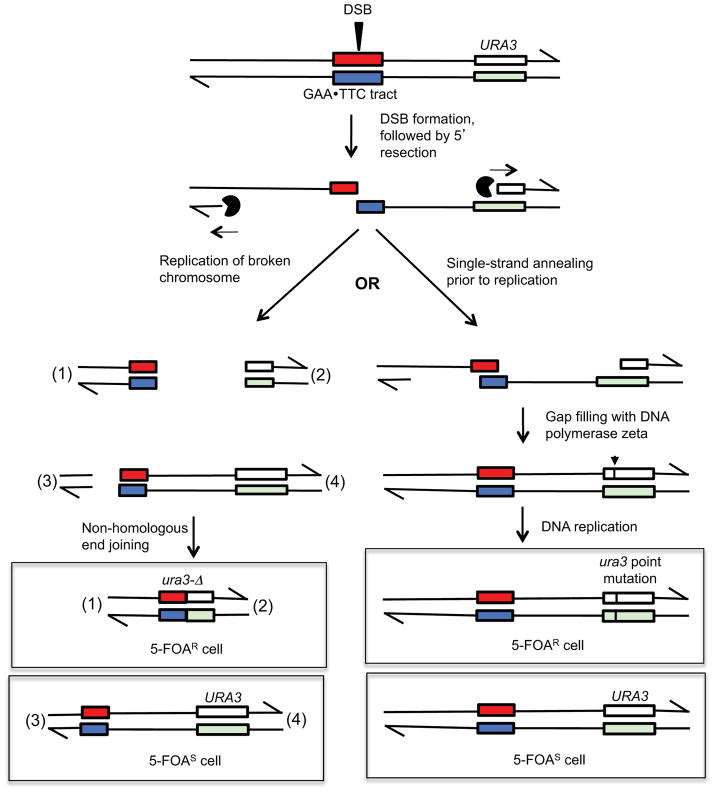
Our model to explain these observations is in Fig. 7. We suggest that the tract-associated mutations are initiated by a DSB formed within the tract in unreplicated DNA. Previously, we showed that stalled replication forks were observed in strains with tracts in one orientation (defined as GAA), but not the other (defined as TTC) [11]; this pattern indicated that the tract was replicated by replication forks emanating from ARS510, located centromere-proximal to the tract. Since we see no strong orientation-dependence of the tract on URA3 mutagenesis and since we find a tract-associated DSB in stationary phase cells [11], the event that initiates mutagenesis is likely to occur in unreplicated DNA rather than a DSB at a stalled replication fork.
Figure 7.
Alternative fates of a GAA•TTC tract-associated DSB results in deletions (left side of the figure) or point mutations (right side of the figure) of the URA3 reporter gene. We suggest that the initiating event in mutagenesis is a DSB in G1- or G0-phase cells. The broken ends are processed 5’ to 3’ with the processing complex indicated by a black circle. In this figure, processing extends into the URA3 reporter gene.
One possible fate of the broken ends is to be replicated (left side of figure) resulting in two broken chromatids. If the broken ends are rejoined by NHEJ and segregated into daughter cells, one daughter would have an ura3 gene whereas the other daughter would have a deletion of sequences that are centromere-distal to the tract. Only the strain with the ura3 deletion would be 5-FOAR.
In the alternative pathway, the broken ends, following processing, reanneal within the GAA•TTC tract. We suggest that the resulting single-stranded gap is filled in by the error-prone DNA polymerase zeta. This error-prone synthesis could result in a mutation inserted within URA3 in one strand of the duplex (shown by the arrow and horizontal line). Replicative synthesis would result in one daughter cell with an ura3 point mutation and daughter with the wild-type URA3 gene.
Several other points concerning the initiating DNA lesion are relevant. First, the observation that all large URA3 deletions include all or part of the tract is a strong argument that the initiating DNA lesion occurs within the tract. Second, we do not know the proteins that are responsible for creating the orientation-independent G0/G1 tract-associated DNA lesions. The replication- and orientation-dependent DSBs observed at the long (GAA•TTC) tract in a strain in a different genetic background requires most of the proteins of the DNA mismatch repair system [13]. One possibility is that formation of H-DNA by the long (GAA•TTC) tract in non-dividing cells, triggered by negative supercoiling [35], exposes a large single-stranded region of DNA that is acted on by cellular endonucleases. Another possibly relevant observation is that Lin et al. [36] showed that R loops formed during the transcription of CTG•CAG tracts enhanced the instability of the tracts. Although we have not directly examined the transcription of the GAA•TTC tracts in our study, there is no promoter located upstream of the tracts.
Following tract-associated DSB formation, we suggest that broken chromosome ends are processed by 5’ to 3’ resection, as observed for HO-induced broken ends [37]. This process involves the Mre11p-Rad50p-Xrs2p complex and Sae2p at an early step, and Exo1p and/or Sgs1p-Top3p-Rmi1p and Dna2p at a later step. Since resection is inefficient in haploid cells in G0/G1 cells [38,39], it is possible that this resection occurs as the cells exit G1 and enter S.
We suggest that processed broken chromosome has two alternative fates, leading to the two different types of mutations. On the left side of Fig. 7, we show replication of the broken DNA molecule to yield two broken chromatids. Fusion of the resulting DNA molecules by non-homologous end-joining would lead to daughter cells with deletions.
In one cell, the deletion would include URA3 and, therefore, be selectable in medium containing 5-FOA. The other daughter cell would contain an unselectable deletion. In this model, the asymmetry of the deletions relative to the tracts is a consequence of the 5-FOA selection process rather than asymmetric processing of the DSB. In considering this model, it is important to point out that yeast cells have inefficient repair of DSBs by homologous recombination in G1 as a consequence of a number of connected factors: inefficient processing of broken ends as discussed above, weak activation of checkpoint proteins [40], and poor recruitment of Rad52p to broken ends [41]. In addition, there is strong genetic evidence that yeast chromosomes broken in G1 by treatment with gamma rays can be replicated to yield two broken chromatids [31]. Another explanation for the deletions is that they reflect nucleolytic processing of both DNA strands to yield a double-stranded DNA gap. Non-homologous end-joining could then re-join the broken ends to yield the deletion. Zierhut and Diffley [42] have recently obtained evidence for double-stranded DNA processing of HO-produced broken ends.
The alternative fate of the processed DSB, which results in point mutations in the URA3 reporter gene, is shown on the right side of Fig. 7. Before replication of the broken chromosome, the processed ends undergo single-strand annealing within the repetitive sequences. This mechanism is consistent with the observation that most of the strains with ura3 point mutations have a GAA•TTC tract that is shorter than the original size. The resulting gaps could be filled in by the error-prone DNA polymerase zeta, thus resulting in point mutations within URA3. This model is similar to those proposed by Yang et al. [21] and by others (N. Saini, Y. Zhang, Y. Nishida, and K. Lobachev, personal communication) to explain the hypermutability of single-stranded DNA formed at DSBs.
There are two alternative explanations of the Rev3p dependence of the DSB-associated mutations. First, it is possible that the single-stranded DNA resulting from processing of tract-associated DSB has a high frequency of damaged bases, and DNA polymerase zeta introduces mutations during the replication of this damaged template [21]; Yang et al. [21] showed that exogenous DNA damage in single-stranded DNA was repaired by error-prone synthesis mediated by Rev3p. Alternatively, the single-stranded gaps could be filled-in by error-prone DNA synthesis even in the absence of base damage. Consistent with this possibility, Hirano and Sugimoto [43] showed that DNA polymerase zeta is recruited to broken ends by a Mec1p-dependent mechanism. Finally, it should be noted that the observed frequency of mutations associated with the long (GAA•TTC) tracts (about 2 x 10−6/division) is much less than the frequency of DSBs associated with the long tracts (about 10−2/division [11]). The simplest interpretation of this discrepancy is that most of the DSBs are repaired by reannealing of minimally-processed broken ends. If the single-stranded gap resulting from processing of broken ends (depicted in Fig. 7) does not extend into the flanking URA3 gene, ura3 mutations would not be stimulated.
DSBs in yeast can also be repaired by mutagenic processes that are independent of DNA polymerase zeta [22,23]. For example, mutations induced in a URA3 gene by a shorter (GAA•TTC)100 tract located within its artificial intron are independent of Rev3p (K. Shah and S. Mirkin, personal communication). It is currently unclear whether the variation in the requirement for Rev3p in the DSB-associated mutagenesis reflects sequence context, variation in the timing of the DSB relative to the cell cycle, the nature of the DSB (blunt-ended, or with single-stranded ends), or some other factor.
5. Summary
In conclusion, we demonstrated that (GAA•TTC)230 tracts induce both deletions and point mutations in a reporter gene located about 1 kb from the tract. These mutations require the error-prone DNA polymerase zeta. The mutagenic effects of the tract do not appear to be related to DSBs forming at a stalled replication fork, since the effects are largely independent of the orientation of the tract. Our data and the data of others argue that DSB-prone sequences may influence the rate of evolution of closely-linked genes.
Supplementary Material
Table 3.
The (GAA•TTC)230 tract increases the frequency of 5-FOAR strains that have two mutations within the URA3 coding sequence1
| Strain name and tract size | # 5-FOAR strains examined | # strains with single mutation | # strains with double mutations |
|---|---|---|---|
|
| |||
| No tract2 | 207 | 198 | 9 |
| WXT10 (TTC)20 | 34 | 33 | 1 |
| WXT11 (TTC)230 | 55 | 44 | 11 |
| WXT12 (TTC)20 | 25 | 24 | 1 |
| WXT13 (TTC)230 | 57 | 47 | 10 |
5-FOAR derivatives were isolated and the ura3 genes were amplified by PCR and sequenced.
Data for the strain within no GAA•TTC tracts are from Lang and Murray [34].
Highlights.
Trinucleotide repeats (GAA•TTC) stimulate mutations in adjacent sequences. Both point mutations and deletions are induced. An error-prone DNA polymerase is required for the generation of point mutations. This mechanism will be a factor in genome evolution.
Acknowledgments
We thank Dr. K. S. Lobachev, H.-M. Kim, and V. Narayanan for strains used in the study, K. Lobachev, D. Gordenin, and S. Mirkin for useful discussions and communication of unpublished results. We thank the other members of the Petes Lab for their suggestions during the course of these experiments. The research was supported by NIH grants GM24110, and GM52319 to T.D.P.
Footnotes
Conflict of interest
None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sutherland GR, Richards RI. Simple tandem DNA repeats and human genetic disease. Proc Natl Acad Sci USA. 1995;92:3636–3641. doi: 10.1073/pnas.92.9.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campuzano V, Montermini L, Molto MD, Pianese L, Cossee M, et al. Friedreich’s ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- 3.Pearson CD, Nichol Edamura K, Cleary JD. Repeat instability: mechanisms of dynamic mutations. Nat Rev Genet. 2005;6:729–742. doi: 10.1038/nrg1689. [DOI] [PubMed] [Google Scholar]
- 4.Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- 5.Lopez Castel A, Cleary JD, Pearson CE. Repeat instability as the basis for human diseases and as a potential target for therapy. Nat Rev Mol Cell Biol. 2010;11:165–170. doi: 10.1038/nrm2854. [DOI] [PubMed] [Google Scholar]
- 6.Cherng N, Shishkin AA, Schlager LI, Tuck RH, Sloan R, et al. Expansions, contractions, and fragility of the spinocerebellar ataxia type 10 pentanucleotide repeat in yeast. Proc Natl Acad Sci USA. 2011;108:2843–2848. doi: 10.1073/pnas.1009409108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lenzmeier BA, Freudenreich CH. Trinucleotide repeat instability: a hairpin curve at the crossroads of replication, recombination, and repair. Cytogenetics and Genome Res. 2003;100:7–24. doi: 10.1159/000072836. [DOI] [PubMed] [Google Scholar]
- 8.Kovtun IV, McMurray CT. Features of trinucleotide repeat instability in vivo. Cell Res. 2008;18:198–213. doi: 10.1038/cr.2008.5. [DOI] [PubMed] [Google Scholar]
- 9.Pollard LM, Sharma R, Gomez M, Shah S, Delatycki MB, et al. Replication-mediated instability of the GAA triplet repeat mutation in Friedreich ataxia. Nucleic Acids Res. 2004;32:5962–5971. doi: 10.1093/nar/gkh933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krasilnikova MM, Mirkin SM. Replication stalling at Friedreich’s ataxia (GAA)n repeats in vivo. Mol Cell Biol. 2004;24:2286–2295. doi: 10.1128/MCB.24.6.2286-2295.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang W, Dominska M, Greenwell PW, Harvanek Z, Lobachev KS, et al. The triplet repeats associated with Friedreich’s ataxia [(GAA)n • (TTC)n] strongly stimulate mitotic crossovers in Saccharomyces cerevisiae. PLoS Genet. 2011;7:e1001270. doi: 10.1371/journal.pgen.1001270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Napierala M, Dere R, Vetcher A, Wells RD. Structure-dependent recombination hot spot activity of GAA.TTC sequences from intron 1 of the Friedreich’s ataxia gene. J Biol Chem. 2004;279:6444–6454. doi: 10.1074/jbc.M309596200. [DOI] [PubMed] [Google Scholar]
- 13.Kim HM, Narayanan V, Mieczkowski PA, Petes TD, Krasilnikova MM, et al. Chromosome fragility at GAA tracts in yeast depends on repeat orientation and requires mismatch repair. EMBO J. 2008;27:2896–2906. doi: 10.1038/emboj.2008.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang G, Vasquez KM. Naturally occurring H-DNA-forming sequences are mutagenic in mammalian cells. Proc Natl Acad Sci USA. 2004;101:13448–13453. doi: 10.1073/pnas.0405116101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wojciechowska M, Napierala M, Larson JE, Wells RD. Non-B DNA conformations formed by long repeating tracts of myotonic dystrophy type 1, myotonic dystrophy type 2, and Friedreich’s ataxia genes, not the sequences per se, promote mutagenesis in flanking regions. J Biol Chem. 2006;281:24531–24543. doi: 10.1074/jbc.M603888200. [DOI] [PubMed] [Google Scholar]
- 16.Bidichandani SI, Purandare SM, Taylor EE, Gumin G, Machkhas H, et al. Somatic sequence variation at the Friedreich ataxia locus includes complete contraction of the expanded GAA triplet repeat, significant length variation in serially passaged lymphoblasts and enhanced mutagenesis in the flanking sequence. Hum Mol Genet. 1999;8:2425–2436. doi: 10.1093/hmg/8.13.2425. [DOI] [PubMed] [Google Scholar]
- 17.Wang G, Seidman MM, Glazer PM. Mutagenesis in mammalian cells induced by triple helix formation and transcription-coupled repair. Science. 1996;271:802–805. doi: 10.1126/science.271.5250.802. [DOI] [PubMed] [Google Scholar]
- 18.Shishkin AA, Voineagu I, Matera R, Cherng N, Chernet BT, et al. Large-scale expansions of Friedreich’s ataxia GAA repeats in yeast. Mol Cell. 2009;35:82–92. doi: 10.1016/j.molcel.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ponder RG, Fonville NC, Rosenberg SM. A switch from high-fidelity to error-prone DNA double-strand break repair underlies stress-induced mutation. Mol Cell. 2005;19:791–804. doi: 10.1016/j.molcel.2005.07.025. [DOI] [PubMed] [Google Scholar]
- 20.Strathern JN, Shafer BK, McGill CB. DNA synthesis errors associated with double-strand-break repair. Genetics. 1995;140:965–972. doi: 10.1093/genetics/140.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Y, Sterling J, Storici F, Resnick MA, Gordenin DA. Hypermutability of damaged single-strand DNA formed at double-strand breaks and uncapped telomeres in yeast Saccharomyces cerevisiae. PLoS Genet. 2008;4:e1000264. doi: 10.1371/journal.pgen.1000264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hicks WM, Kim M, Haber JE. Increased mutagenesis and unique mutation signature associated with mitotic gene conversion. Science. 2010;329:82–85. doi: 10.1126/science.1191125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deem A, Keszthely A, Blackgrove T, Vayl A, Coffey B, et al. Break-induced replication is highly inaccurate. PLoS Biol. 2011;9:e1000594. doi: 10.1371/journal.pbio.1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holbeck SL, Strathern JN. A role for REV3 in mutagenesis during double-strand break repair in Saccharomyces cerevisiae. Genetics. 1997;147:1017–1024. doi: 10.1093/genetics/147.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rattray AJ, Shafer BK, McGill CB, Strathern JN. The roles of REV3 and RAD57 in double-strand-break-repair-induced mutagenesis of Saccharomyces cerevisiae. Genetics. 2002;162:1063–1077. doi: 10.1093/genetics/162.3.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee PS, Greenwell PW, Dominska M, Gawel M, Hamilton M, et al. A fine-structure map of spontaneous mitotic crossovers in the yeast Saccharomyces cerevisiae. PLoS Genet. 2009;5:e1000410. doi: 10.1371/journal.pgen.1000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei W, McCusker JH, Hyman RW, Jones T, Ning Y, et al. Genome sequencing and comparative analysis of Saccharomyces cerevisiae strain YJM789. Proc Natl Acad Sci USA. 2007;104:12825–12830. doi: 10.1073/pnas.0701291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guthrie C, Fink GR. Guide to Yeast Genetics and Molecular Biology. Academic Press; San Diego, CA: 1991. [Google Scholar]
- 29.Barbera MA, Petes TD. Selection and analysis of spontaneous reciprocal mitotic cross-overs in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2006;103:12819–12824. doi: 10.1073/pnas.0605778103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stone JE, Ozbirn RG, Petes TD, Jinks-Robertson S. Role of proliferating cell nuclear antigen interactions in the mismatch repair-dependent processing of mitotic and meiotic recombination intermediates in yeast. Genetics. 2008;178:1221–1236. doi: 10.1534/genetics.107.085415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee PS, Petes TD. Mitotic gene converson events induced in G1-synchronized yeast cells by gamma rays are similar to spontaneous conversion events. Proc Natl Acad Sci USA. 2010;107:7383–7388. doi: 10.1073/pnas.1001940107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boeke JD, Lacroute F, Fink GR. A positive selection for mutants lacking orotidine-5’-phosphate decarboxylase activity in yeast; 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 33.Lea DE, Coulson CA. The distribution of the numbers of mutants in bacterial populations. J Genet. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 34.Lang GI, Murray AW. Estimating the per-base-pair mutation rate in the yeast Saccharomyces cerevisiae. Genetics. 2008;178:67–82. doi: 10.1534/genetics.107.071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mirkin SM, Frank-Kamenetskii MD. H-DNA and related structures. Ann Rev Biophys Biomol Struct. 1994;23:541–576. doi: 10.1146/annurev.bb.23.060194.002545. [DOI] [PubMed] [Google Scholar]
- 36.Lin Y, Dent SY, Wilson JH, Wells RD, Napierala M. R loops stimulate genetic instability of CTG.CAG repeats. Proc Natl Acad Sci USA. 2010;107:692–697. doi: 10.1073/pnas.0909740107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mimitou EP, Symington LS. DNA end resection: many nucleases make light work. DNA Repair. 2009;8:983–995. doi: 10.1016/j.dnarep.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aylon Y, Liefshitz B, Kupiec M. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J. 2004;23:4868–4875. doi: 10.1038/sj.emboj.7600469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pellicioli A, Lee SE, Lucca C, Foiani M, Haber JE. Regulation of Saccharomyces Rad53 checkpoint kinase during adaptation from DNA damage-induced G2/M arrest. Mol Cell. 2001;7:293–300. doi: 10.1016/s1097-2765(01)00177-0. [DOI] [PubMed] [Google Scholar]
- 41.Lisby M, Rothstein R, Mortensen UH. Rad52 forms DNA repair and recombination centers during S phase. Proc Natl Acad Sci USA. 2001;98:8276–8282. doi: 10.1073/pnas.121006298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zierhut C, Diffley JFX. Break dosage, cell cycle stage and DNA replication influence DNA double strand break response. EMBO J. 2008;27:1875–1885. doi: 10.1038/emboj.2008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hirano Y, Sugimoto K. ATR homolog Mec1 controls association of DNA polymerase zeta–Rev1 complex with regions near a double-strand break. Curr Biol. 2006;16:585–590. doi: 10.1016/j.cub.2006.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.