Abstract
Elimination of aberrantly folded polypeptides from the endoplasmic reticulum (ER) by the ER-associated degradation (ERAD) system promotes cell survival under stress conditions. This quality control mechanism requires movement of misfolded proteins across the ER membrane for targeting to the cytosolic proteasome, a process facilitated by a ‘holdase’ complex, consisting of Bag6 and the cofactors Ubl4A, Trc35. This multiprotein complex also participates in several other protein quality control processes. Here we report SGTA as a new component of the Bag6 system, which cooperates with Bag6 to channel dislocated ERAD substrates that are prone to aggregation. Using NMR spectroscopy and biochemical assays, we demonstrate that SGTA contains a non-canonical ubiquitin-like-binding domain (UBLD) that interacts specifically with an unconventional UBL in Ubl4A at least in part via electrostatics. This interaction helps recruit SGTA to Bag6, enhances substrate loading to Bag6, and thus prevents the formation of non-degradable protein aggregates in ERAD.
Keywords: ER-associated protein degradation/ERAD/retrotranslocation, ubiquitin-like domain/UBL, Bag6/Bat3, Ubl4A, SGTA, ubiquitin
Ubiquitin-like proteins/domains (UBLs) are a family of structurally related polypeptides, 45-80 amino acids in length. These proteins share striking structural similarities with ubiquitin (van der Veen and Ploegh, 2012). Some UBL proteins can be conjugated to substrates analogously to ubiquitin (Kerscher et al., 2006). These ubiquitin-like modifiers are often referred to as type I UBLs. Other UBLs are present in polypeptides where they serve as functional domains. These are termed type II UBL domains (Jentsch and Pyrowolakis, 2000). In humans, there are approximately 50 proteins bearing type II UBL domains. These proteins perform a variety of essential cellular functions, serving as proteasome adaptors, ubiquitin ligases (E3), co-chaperones, deubiquitinating enzymes, and signaling regulators (vander Veen and Ploegh, 2012, Hoeller et al., 2006). Despite structural similarities, type I UBL domains often have intrinsic differences that allow each of them to communicate with a unique downstream partner. By contrast, the conventional view on type II UBL domains is that they resemble each other more than they differ, as many of them can interact with ubiquitin binding domains (UBDs) such as UBA (Ubiquitin Associated), UIM (Ubiquitin Interacting Motif), and CUE (Coupling of Ubiquitin to ER degradation)(Hicke et al., 2005, Husnjak and Dikic, 2012).
An important function of ubiquitin and UBL domains is to regulate proteasome dependent turnover of misfolded proteins of the endoplasmic reticulum (ER) by the ER-associated degradation system (ERAD) (Tsai et al., 2002, Vembar and Brodsky, 2008, Meusser et al., 2005). This evolutionarily conserved protein quality control process requires a coordinated effort from a large number of proteins, making up a complex machinery (Smith et al., 2011, Liu and Ye, 2011). ER chaperones and lectins such as BiP, Os9, EDEM and PDI, recognize misfolded proteins in the lumen and target them to distinct ubiquitin ligase-containing membrane complexes for retrotranslocation into the cytosol (Bhamidipati et al., 2005, Carvalho et al., 2006, Denic et al., 2006, Oda et al., 2003, Molinari et al., 2003, Kim et al., 2005, Christianson et al., 2008, Gauss et al., 2006, Tsai et al., 2001). Substrates undergoing retrotranslocation are then ubiquitinated by these ligases (Mehnert et al., 2011), and subsequently dislocated into the cytosol by the p97-Ufd1-Npl4 ATPase complex for degradation by the proteasome (Rabinovich et al., 2002, Bays et al., 2001, Ye et al., 2001, Jarosch et al., 2002).
One of the best characterized ERAD-specific ubiquitin ligases in mammalian cells is gp78. gp78 is a multi-spanning transmembrane ubiquitin ligase (E3) that is homologous to Hrd1, another ERAD-dedicated E3 proposed to form a retrotranslocon (Fang et al., 2001, Carvalho et al., 2010). As a master retrotranslocation regulator, gp78 uses a G2BR (Ube2g2 Binding Region) and a VIM (VCP-interacting motif) domain to interact with its cognate conjugating enzyme (E2) Ube2g2 and the dislocation-driving p97 ATPase, respectively (Li et al., 2009, Das et al., 2009, Ye et al., 2005, Ballar et al., 2006, Christianson et al., 2011). In addition, gp78 also carries a ubiquitin-binding CUE domain (Chen et al., 2006), which binds a UBL domain in the recently identified chaperone Bag6. We previously showed that Bag6 uses an unusual ‘holdase’ activity to maintain retrotranslocated polypeptides in soluble state, facilitating their turnover (Wang et al., 2011b).
In this report, we demonstrate that the Bag6 complex contains an unusual UBL domain in its cofactor Ubl4A, which does not interact with canonical UBDs. Instead, our biochemical and structural analyses support the notion that the UBL domain in Ubl4A represents a unique class of UBL domains. Mass spectrometry studies identify a previously unknown ERAD mediator that specifically binds the Ubl4A UBL via a mode distinct from the conventional UBL-UBD interactions. Our data show that distinct means of UBL recognitions are used in the cell to integrate various ERAD components into a functional network for protein turnover.
Results
The Bag6 complex contains a canonical and a non-canonical UBL domain
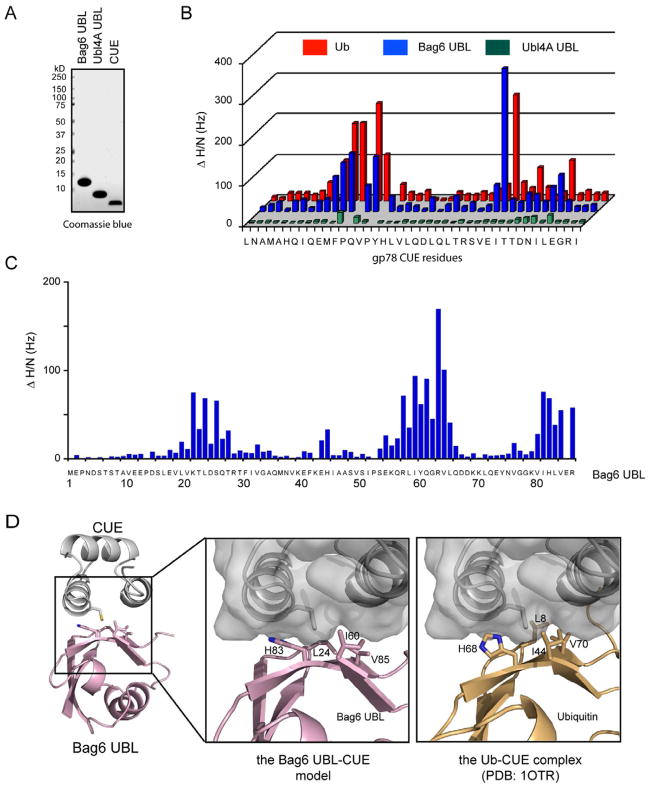
Bag6 contains a UBL domain that binds the CUE domain of gp78. It is noteworthy that another integral component of the Bag6 complex, Ubl4A also contains a UBL domain that shares significant sequence similarity with the Bag6 UBL, but it is unclear whether it can interact with CUE. We therefore characterized the interaction of purified BAG6 UBL and Ubl4A UBL with the gp78 CUE domain (Fig. 1A) using solution NMR spectroscopy. We first analyzed chemical shift perturbation of CUE by comparing the chemical shifts of free CUE (obtained from 15N/13C uniformly labeled CUE) to that of CUE in complex with BAG6 UBL (15N/13C labeled CUE plus unlabeled BAG6 UBL). We used ubiquitin as a positive control because the CUE domain was previously shown to be a ubiquitin-binding motif (Chen et al., 2006). The NMR data indicate that the chemical shift perturbation patterns of CUE upon binding BAG6 UBL and ubiquitin are almost identical (Fig. 1B; Supplementary Fig. 1A), suggesting that BAG6 UBL binds the CUE domain in a similar manner as ubiquitin. We then performed the reciprocal experiment by analyzing the chemical shift perturbation of BAG6 UBL by CUE. We observed three clusters of residues, containing residues 19-27, 54-66 and 80-86, which display significant chemical shift perturbation. Among these residues, Ile60, a residue equivalent to Ile44 of ubiquitin, and His83, Tyr61, Val65, Val85, Leu24 (Fig. 1C; Supplementary Fig. 1B) form a hydrophobic patch similar to that in the CUE binding site on ubiquitin (Kang et al., 2003, Prag et al., 2003). Based on our NMR results and previously published structures (PDB: 1OTR), we built a model of the BAG6 UBL-CUE complex. As expected, the model is highly homologous to the CUE-ubiquitin complex (Fig. 1D). From these results, we conclude that BAG6 UBL is a canonical UBL that is recognized by UBDs in a similar manner as ubiquitin. By contrast, Ubl4A UBL did not significantly alter the NMR spectra of CUE (Fig. 1B, Supplementary Fig. 1A). We concluded that the Ubl4A UBL is a distinct type II UBL unrecognizable by UBDs.
Figure 1. Recognition of Bag6 UBL by the CUE domain, also see Figure S1.
A, Purified proteins used in the NMR studies.
B, NMR chemical shift perturbation analyses show that BAG6 UBL binds the gp78 CUE domain in a similar manner as ubiquitin, whereas Ubl4A UBL only interacts with CUE weakly. The NMR spectra of 15N/13C labeled CUE (400 μM) were determined in the presence or absence of the indicated proteins (800 μM). Shown is the square root summary of the chemical shift differences (ΔHz) in both nitrogen and proton dimensions as a function of protein sequence.
C, The chemical shift perturbations of BAG6 UBL by CUE. As in B, except that the BAG6 UBL is labeled whereas CUE is unlabeled.
D, A structural model of the BAG6 UBL-CUE complex. The model was obtained by aligning the BAG6 UBL structure (PDB, 1WX9, RIKEN Structural Genomics/Proteomics Initiative) to ubiquitin in the previously determined ubiquitin-CUE complex structure (PDB, 1OTR) (Kang et al., 2003). The enlarged images highlight the residues involved in CUE binding in BAG6 UBL and ubiquitin.
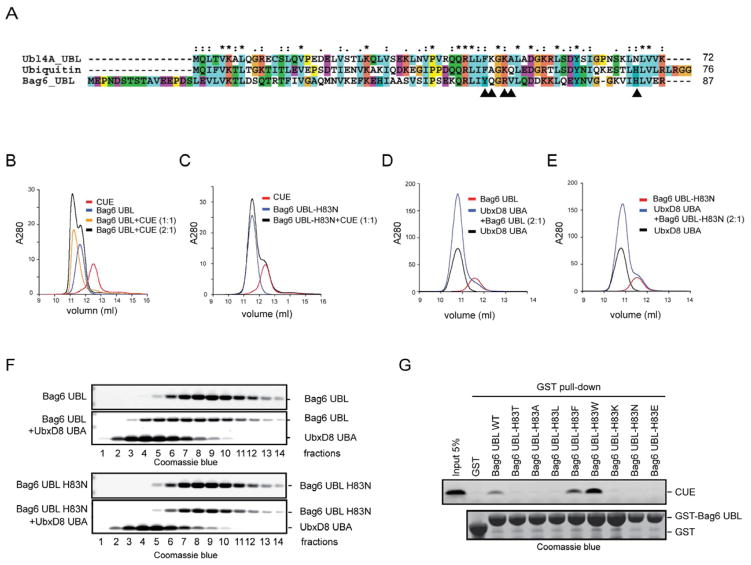
To understand why Ubl4A UBL cannot be recognized by CUE, we compared the protein sequence of Ubl4A UBL to that of BAG6 UBL. We focused on residues in BAG6 UBL that showed significant chemical shift perturbation when the CUE domain was present. Many of these residues are conserved between BAG6 UBL and Ubl4A UBL, but a few variations were noticed (Fig. 2A). We investigated the contribution of five variations to the UBD binding specificity by converting these amino acids in BAG6 UBL to the corresponding ones in Ubl4A UBL either individually or in combination. Size exclusion chromatography analyses showed that four BAG6 UBL variants (Y61F, Q62K, Q62A, R64K/V65A) bound CUE similarly to wild type BAG6 UBL (Xu, Y., unpublished results). However, a single amino acid substitution that changed His83 to Asn completely abolished the interaction of BAG6 UBL with CUE (Figs. 2B, C). This BAG6 UBL mutant also failed to bind the UBA domain of the gp78-interacting partner UbxD8 (Figs. 2D, E, F), suggesting that His83 is required for binding UBDs.
Figure 2. His83 in BAG6 UBL is required for UBD binding, also see Figure S2.
A, Sequence alignment of BAG6 UBL, Ubl4A UBL and ubiquitin (Ub). Arrowheads indicate non-conserved BAG6 UBL residues whose 1H/15N chemical shifts are significantly affected by CUE binding.
B, C, Size exclusion chromatography analyses of the interactions of BAG6 UBL, BAG6 UBL H83N with gp78 CUE. gp78 CUE (40 μM) was mixed with Bag6 UBL as indicated and incubated on ice for 30 min before analysis by a size exclusion chromatography at 4 °C.
D, E, As in B, C, except that the UBA domain from UbxD8 (40 μM) was used.
F, The peak fractions in D and E were analyzed by SDS-PAGE electrophoresis and Coomassie blue staining.
G, Histidine and strong hydrophobic residues at position 83 of Bag6 UBL support CUE binding. Shown is a GST pull-down experiment using the indicated proteins.
Many type II UBL domains contain histidine or a hydrophobic amino acid at the position equivalent to His83 of BAG6 UBL, but others have either a polar or charged residue at this position (Supplementary Fig. 2A). In either the CUE-ubiquitin or the CUE-BAG6 UBL complex, the histidine residue is in proximity to several hydrophobic residues, but its imidazole ring is oriented away from the hydrophobic UBD binding pocket (Fig. 1D)(Kang et al., 2003, Prag et al., 2003). When this residue is converted to Asn, computational modeling showed that the polar side chain of Asn can adopt many rotamers that frequently protrude into the UBD binding pocket, disrupting the hydrophobic binding site (Supplementary Fig. 2B). We therefore presumed that the side chain of the residue equivalent to His83 in BAG6 UBL might have a significant impact on whether or not a type II UBL domain could be recognized by a canonical UBD. To test this idea, we mutated His83 in BAG6 UBL to a variety of residues including alanine (A), the polar residue threonine (T), hydrophobic residues (L, F, W), or charged residues (K, E). Each mutant was purified as a GST-tagged protein from E coli. and tested for binding CUE using a GST pull-down assay. The results showed that the interaction of BAG6 UBL with CUE was maintained when His83 was substituted to a strong hydrophobic residue such as F or W. In fact, the H83W substitution consistently increased the affinity of BAG6 UBL to CUE (Fig. 2G). By contrast, substitution of His83 to either charged, polar, or even less hydrophobic residues such as leucine reduced the interaction of BAG6 UBL with CUE. Thus, a His or strong hydrophobic residue is required at this position for BAG6 UBL to be recognized by a UBD.
To further corroborate our model, we tested two other type II UBLs that have a charged residue at the position equivalent to His83. Indeed, neither GST-ZFAN4 UBL nor GST-FUBI bound CUE, whereas GST-ubiquitin or GST-Bag6 UBL could pull down CUE under the same condition (Supplementary Figs. 2C, D). Together, our results suggest the existence of a class of type II UBLs that do not bind canonical UBDs. These UBLs contain unique features including a charged or polar residue near the UBD binding site, which distinguish them from canonical UBLs that bind UBDs.
Ubl4A binds SGTA through a novel mode of UBL recognition
We next wished to identify the functional partner(s) of Ubl4A UBL in the context of ERAD. To this end, we expressed FLAG-tagged Ubl4A together with His-tagged Bag6 in HEK293 cells because the association of Ubl4A with the membrane is mediated by Bag6 binding to gp78. We purified the Ubl4A-Bag6 complex from both an ER-enriched membrane fraction and a cytosolic fraction by affinity chromatography. Eluted proteins were analyzed by mass spectrometry using a shotgun approach. Among proteins identified as potential Ubl4A-Bag6 interacting proteins, a protein named SGTA was chosen for further investigation because of its abundance in the eluate and because the interaction with the Bag6-Ubl4A complex was detected in both the cytosol and membrane fractions (Supplementary Table 1). In addition, the yeast SGTA homolog Sgt2p was reported to interact with Get5p, ortholog of Ubl4A(Chartron et al., 2011, Wang et al., 2010, Chang et al., 2010, Kohl et al., 2011).
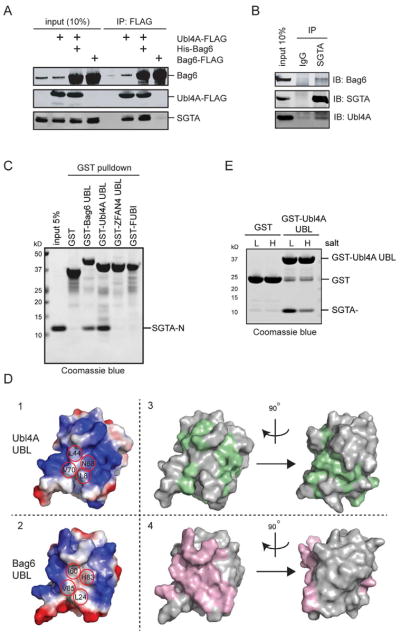
To validate the mass spectrometric results, we carried out co-immunoprecipitation and immunoblotting experiments. When FLAG-tagged Ubl4A was expressed either by itself or together with His-tagged Bag6, immunoprecipitation of Ubl4A pulled down endogenous SGTA. By contrast, overexpressed Bag6 alone did not co-precipitate with SGTA efficiently (Fig. 3A). Endogenous SGTA could also be co-precipitated with the endogenous Bag6 complex (Fig. 3B). These results suggest that SGTA interacts with the Bag6 complex in cells likely through Ubl4A.
Figure 3. SGTA binds Ubl4A UBL via electrostatics, also see Figure S3, Table S1.
A, Ubl4A interacts with SGTA. Cells expressing the indicated proteins were analyzed by immunoprecipitation and immunoblotting.
B, Endogenous interaction between SGTA and the Bag6 complex. HEK293 cell extract was subject to immunoprecipitation by the indicated antibodies.
C, SGTA interacts directly with the UBL domain of Ubl4A. Shown is a GST pull-down experiment using the indicated proteins.
D, SGTA binds Ubl4A UBL by a means that is distinct from the canonical mode of UBL recognition. Panels 1, 2, show the electrostatic surface potential of the surface around the UBD binding hydrophobic residues (red circles) of BAG6 UBL and the corresponding ones on Ubl4A UBL. Panels 3, 4 are surface views of Ubl4A UBL and BAG6 UBL, respectively, showing residues whose NMR spectra are significantly affected by their corresponding partner (green for Ubl4A UBL and pink for BAG6 UBL).
E, The interaction of Ubl4A UBL with SGTA is sensitive to salt. The indicated GST-tagged proteins were immobilized and incubated with SGTA-N either under low (L, 150 mM) or high salt conditions (H, 500 mM). The precipitated proteins were analyzed by SDS-PAGE and Coomassie blue staining.
We next determined the region in SGTA that is responsible for Ubl4A binding. We created constructs expressing various SGTA fragments. Immunoprecipitation showed that the N-terminal 80 amino acids of SGTA (SGTA-N) were both necessary and sufficient for Ubl4A binding (Supplementary Fig. 3A, B), consistent with studies in yeast (Chartron et al., 2011, Chartron et al., 2012).
To see whether the UBL domain in Ubl4A is involved in SGTA binding, we incubated GST or GST-tagged Ubl4A UBL with a whole cell extract. Indeed, immunoblotting showed that GST-Ubl4A UBL, but not GST, interacted with endogenous SGTA (Supplementary Fig. 3C). In addition, GST-Ubl4A UBL could also pull down purified recombinant SGTA N-domain (SGTA-N), demonstrating a direct interaction between these proteins (Fig. 3C). Compared to GST-Ubl4A UBL, GST-BAG6 UBL pulled down less SGTA-N, whereas GST-ubiquitin, GST-FUBI and GST-ZFAN4 UBL did not pull down SGTA-N above the background level (Fig. 3C, Supplementary Fig. 3D). Thus, SGTA is a novel UBL-binding protein that preferentially interacts with Ubl4A UBL.
To further characterize the interaction between SGTA-N and Ubl4A UBL, we uniformly labeled Ubl4A UBL with 15N/13C and obtained backbone 1H/15N chemical shift assignments of Ubl4A UBL in complex with SGTA-N. The chemical shift perturbation profile for Ubl4A UBL upon binding of SGTA was compared to the CUE-induced chemical shift perturbation of BAG6 UBL. The overall chemical shift perturbation patterns appeared similar, but a careful comparison of the two profiles showed that some Ubl4A UBL residues perturbed by SGTA were not significantly affected by CUE in BAG6 UBL and vice versa (Supplementary Figs. 3E, F). These results indicate that Ubl4A UBL employs a site similar to the UBD binding site on canonical UBL domains for binding SGTA, but the precise mode of interactions may be different. Indeed, the electrostatic surface potential of the SGTA binding site in Ubl4A UBL is significantly different from that of the canonical UBD binding surface on BAG6 UBL (Fig. 3D). In BAG6 UBL, the imidazole ring of His83 is pointed away from Leu24, Ile60 and Val85, allowing the latter to form a continuous hydrophobic binding surface. By contrast, Asn68 in Ubl4A UBL disrupts this hydrophobic surface. Importantly, Leu44 and Val70 in Ubl4A UBL (equivalent to Ile60 and Val80 in BAG6 UBL, respectively) are surrounded by positively charged residues (Fig. 3D, panel 1), and 1H/15N chemical shift perturbation shows that many of these charged residues were significantly perturbed upon binding SGTA (panel 3). Thus, electrostatic side chain contacts may contribute significantly to the Ubl4A UBL-SGTA interaction. In support of this notion, we found that the interaction of SGTA with Ubl4A was highly sensitive to salt treatment (Fig. 3E). Together, these results demonstrate a means of UBL recognition that is mediated at least in part by electrostatics, a conclusion that is accordance with a study in yeast (Chartron et al., 2012).
Ubl4A enhances the association of SGTA with Bag6
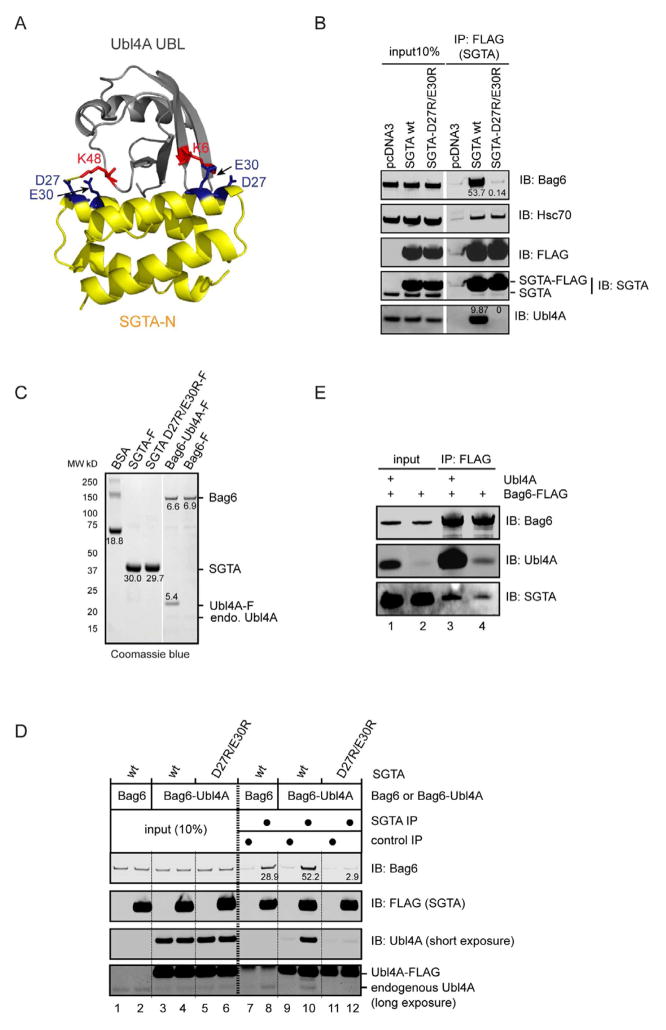
To test whether Ubl4A helps recruit SGTA to Bag6, we took two approaches. First, we generated a structural model of the human SGTA N-domain in complex with Ubl4A UBL by aligning the SGTA-N and Ubl4A UBL structures with the yeast homologous complex (Chartron et al., 2012). The model indicates that the side chains of two highly conserved acidic residues in SGTA (Asp27 and Glu30) make contacts with Lys48 and Lys6 in Ubl4A, respectively, two residues showing significant chemical shift perturbation in our NMR study (Fig. 4A). We expressed either wild type SGTA or a SGTA mutant bearing D27R and E30R substitutions in cells. Immunoprecipitation showed that compared to wild type SGTA, the D27R/E30R mutant was completely inactive in binding Ubl4A, but its interaction with Hsc70 was maintained (Fig. 4B). The results suggest that the mutations specifically affect the binding site for Ubl4A. Consistent with the notion that Ubl4A serves a link between SGTA and Bag6, Bag6 was not co-precipitated with the SGTA mutant defective in Ubl4A binding. Next, we reconstituted the Ubl4A dependent interaction of SGTA with Bag6 in vitro using purified SGTA and Bag6 or a Bag6-Ubl4A complex. We purified these proteins from HEK293 cells under a high salt condition (see experimental procedure). Nonetheless, the Bag6 sample contained some endogenous Ubl4A. As expected, when Ubl4A was co-expressed with Bag6, the purified complex contained Bag6 and Ubl4A in stoichiometric ratio (Fig. 4C). Co-immunoprecipitation experiments demonstrate that purified Bag6 only moderately co-precipitated SGTA (Fig. 4D, lane 8). This association is probably mediated by the week affinity between the Bag6 UBL and SGTA N-domain (Winnefeld et al., 2006). The small amount of endogenous Ubl4A present in the sample may also contribute to this interaction. Importantly, the interaction of Bag6 with SGTA, but not SGTA D27R/E30R was significantly enhanced when stoichiometric amount of Ubl4A was present (lane 10 vs. lanes 8 and 12). Likewise, ectopically expressed Bag6 did not interact significantly with endogenous SGTA in cells (Fig. 3A, Fig. 4E, lane 4), but co-expression of Ubl4A enhanced the interaction (Fig. 4E, lane 3 vs. 4). We conclude from these experiments that Ubl4A can serve as a matchmaker to enhance SGTA binding to Bag6.
Figure 4. Ubl4A helps recruit SGTA to Bag6.
A, A model of the Ubl4A UBL-SGTA N-domain complex. The structures of Ubl4A UBL (PDB, 2DZI, RIKEN Structural Genomics/Proteomics Initiative) and SGTA-N (PDB, 4GOD) were aligned to the corresponding domain in the homologous yeast Get5p UBL-Sgt2p N complex (PDB, 2LXC).
B, Two acidic residues on SGTA-N are essential for interaction with Ubl4A and Bag6. HEK293 cells transfected with plasmids expressing the indicated FLAG-tagged proteins were lysed in the NP40 lysis buffer. Cell extracts were subject to immunoprecipitation with the FLAG antibody and the precipitated materials were analyzed by immunoblotting.
C, Purified proteins for in vitro binding studies. A known amount of BSA was used to estimate protein concentrations. The numbers indicate the protein levels.
D, Ubl4A promotes SGTA binding Bag6 in vitro. Purified SGTA or the D27R/E30R mutant was incubated with either Bag6 or the Bag6-Ubl4A complex (Bag6-Ubl4A). After incubation, the samples were subject to immunoprecipitation and immunoblotting analyses. The numbers indicate the amount of Bag6 co-precipitated with SGTA.
E, Ubl4A promotes the interaction of SGTA with Bag6 in cells. HEK293 cells expressing the indicated proteins were lysed. The cell extracts were subject to immunoprecipitation by FLAG antibodies.
Depletion of SGTA impairs ERAD and induces UPR
The Bag6 complex was recently established as key regulator in membrane targeting of tail-anchored (TA) proteins (Mariappan et al., 2010). Importantly, the same complex also plays pivotal roles in several aspects of protein quality control (Wang et al., 2011b, Hessa et al., 2011, Minami et al., 2010). Notably, yeast does not contain a Bag6 homologue, but it contains a SGTA ortholog named Sgt2p, which binds Get5p, the ortholog of Ubl4A. In yeast, Sgt2p appears to serve as a functional ‘substituent’ for Bag6 in TA protein biogenesis, but it is unclear whether Sgt2p or SGTA has the capacity to regulate any proteasomal degradation processes.
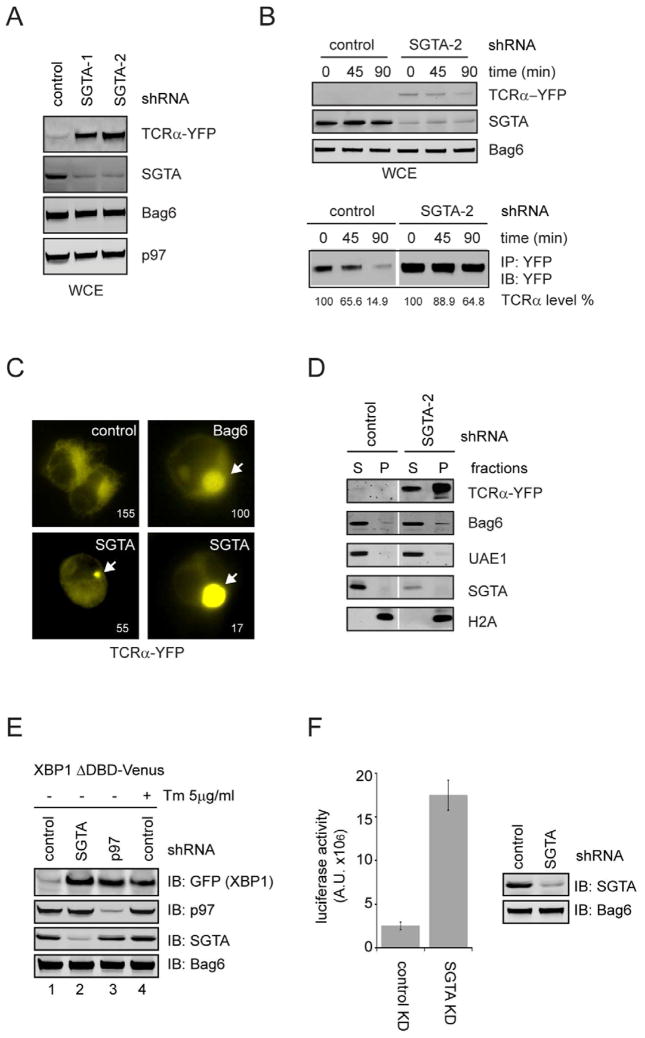
We therefore tested whether SGTA can function in ERAD. We first used two different SGTA specific shRNAs to deplete SGTA in a cell line stably expressing the model ERAD substrate TCRα-YFP because the degradation of this substrate requires both gp78 and the Bag6 complex (Wang et al., 2011b). Immunoblotting showed that depletion of SGTA by >80% increased the steady state level of TCRα-YFP by at least 5-fold (Fig. 5A). Cycloheximide chase experiments showed that the turnover of TCRα-YFP was significantly inhibited by SGTA depletion, demonstrating that SGTA is functionally required for the turnover of this ERAD substrate (Fig. 5B). SGTA depletion also consistently caused accumulation of another Bag6-dependent ERAD substrate CD4 in Vpu-expressing cells (Supplementary Fig. S4). Intriguingly, fluorescence microscopy revealed that in SGTA knock-down cells, TCRα-YFP often accumulated in aggresome-like structures (Fig. 5C), a phenotype similarly observed in Bag6-depleted cells (Wang et al., 2011b). Consistently, a significant fraction of TCRα-YFP in SGTA knock-down cells could not be extracted by the non-ionic detergent NP40 due to aggregation (Fig. 5D). Collectively, these results suggest that SGTA may cooperate with Bag6 to maintain the solubility of retrotranslocated ERAD substrates and therefore promote their turnover.
Figure 5. SGTA is required for degradation of misfolded ER proteins, also see Figure S4.
A, Depletion of SGTA causes accumulation of the model ERAD substrate TCRα. Whole cell extracts from TCRα-YFP-expressing cells transfected with the indicated shRNA constructs were analyzed by immunoblotting.
B, Cycloheximide chase analysis of TCRα turnover in control and SGTA knockdown cells. Because the level of TCRα in control cells is too low to allow an accurate estimation of the degradation kinetics, we also performed immunoprecipitation and quantitative immunoblotting using cell extracts from the SGTA knockdown and control cells (bottom panel).
C, Cells expressing TCRα-YFP together with the indicated shRNA constructs were imaged by a fluorescence microscope. Two examples of SGTA knockdown cells bearing TCRα aggregates of different sizes are shown. The numbers indicate the exposure time in millisecond.
D, TCRα accumulates in SGTA knockdown cells in NP40 insoluble fractions. Cells transfected with a TCRα-YFP-expressing plasmid together with the indicated shRNAs were extracted first by the NP40 lysis buffer to obtain soluble extracts (S). The NP40 insoluble fractions (P) were subject to further extraction by the SDS-containing Laemmli buffer prior to immunoblotting.
E, F, SGTA depletion induces ER stress. E, Cells transfected with the indicated knockdown constructs together with the XBP1-Venus reporter were lysed. Whole cell extracts were analyzed by immunoblotting. Where indicated, cells were treated with Tunicamycin (Tm) for 9h. F, HEK293 cells were transfected with the indicated knockdown (KD) constructs together with the pGRP-luciferase reporter. A fraction of the cell extracts were subject to immunoblotting. The remaining samples were used to determine the luciferase activities. Shown is the quantification of 3 independent experiments (error bars, standard deviation, n=3).
To test whether SGTA has a broad role in ERAD, we asked whether depletion of SGTA elicits the unfolded protein response (UPR), a stress response induced when misfolded proteins accumulate in the ER. If SGTA is a general regulator of ERAD, its deficiency should cause accumulation of misfolded ER proteins and induce ER stress. We first used an EGFP-tagged XBP1 (XBP1 ΔDBD Venus) as a reporter (Iwawaki et al., 2004). The splicing of XBP1 upon ER stress induction (e.g. in cells depleted of p97 or tunicamycin-treated cells) activates the expression of EGFP, which was also detected in cells depleted of SGTA (Fig. 5E). UPR induction in SGTA knockdown cells could also be verified using a BiP promoter-controlled luciferase reporter [GRP78 (−132)-Luc] (Fig. 5F)(Yoshida et al., 2001). These results are consistent with the proposed function of SGTA in ERAD regulation. However, the data cannot rule out the possibility that SGTA may also use another mechanism independent of ERAD to regulate ER homeostasis because UPR induction was similarly observed in yeast strain deficient in Sgt2 (Jonikas et al., 2009), yet no evidence suggests that Sgt2p has a role in ERAD regulation in S. cerevisiae.
SGTA promotes substrate binding by Bag6 to facilitate ERAD
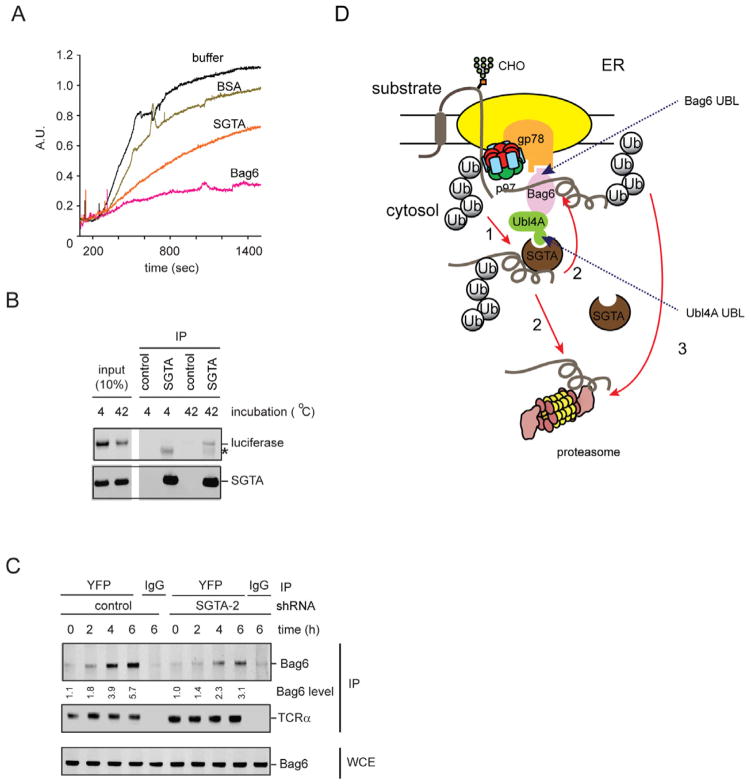
Bag6 has a chaperone-like activity that binds proteins bearing exposed hydrophobic surfaces to inhibit their aggregation (Wang et al., 2011b, Mariappan et al., 2010). To capture retrotranslocated ERAD substrates in cells, Bag6 needs to compete with many abundant cytosolic chaperones that have similar activities. We hypothesized that SGTA may be a substrate recruiting co-factor that improves the efficiency of substrate binding by Bag6 in the complex cellular environment. We therefore tested whether SGTA itself could bind proteins with exposed hydrophobic patches. We used luciferase as a model substrate because it could be readily denatured by heat, exposing a stretch of hydrophobic residues. As shown previously, purified Bag6 can capture heat-denatured luciferase and keep it in an unfolded yet soluble form (Wang et al., 2011b). Under the same condition, purified SGTA was significantly less effective than Bag6 in suppressing luciferase aggregation, as demonstrated by both light scattering and sedimentation assays (Fig. 6A; Supplementary Fig. S5A). Nonetheless, an interaction between SGTA and the denatured luciferase, but not native luciferase could be detected by co-immunoprecipitation (Fig. 6B). Importantly, luciferase binding is not dependent on the association of SGTA with Hsc70 because mutations in the predicted Hsc70 binding site abolished Hsc70 binding (data not shown) but did not affect SGTA binding to luciferase (Supplementary Fig. S5B). These results indicate that SGTA has a chaperone-like activity distinct from that of Bag6: while Bag6 can bind and hold its substrates, SGTA might bind substrates in a more transient and dynamic fashion.
Figure 6. SGTA assists Bag6 in chaperoning ERAD substrates, also see Figure S5.
A, SGTA weakly suppresses luciferase aggregation in vitro. Light scattering analysis of luciferase aggregation in the presence of the purified proteins at 42 °C. The molar ratio of luciferase to the chaperones or BSA is 1:3.
B, SGTA preferentially binds unfolded luciferase. SGTA was incubated with luciferase at 42 °C or 4 °C for 10 min. The soluble fractions were subject to immunoprecipitation with FLAG or control agarose beads. The asterisk indicates IgG.
C, SGTA facilitates substrate binding by Bag6. TCRα-YFP-expressing cells transfected with the indicated shRNA constructs were treated with MG132 for the indicated periods of time. The interaction of Bag6 with TCRα was analyzed by co-immunoprecipitation. WCE, whole cell extract.
D, A model shows the roles of the two UBL domains of the Bag6 complex in ERAD. The red arrows indicate the proposed routes for targeting the ERAD substrates to the proteasome.
Our in vitro experiments so far support the notion that SGTA may act as a cofactor to enhance substrate binding by Bag6. This would explain the substrate aggregation phenotype observed in cells lacking either SGTA or Bag6. We previously showed that a Bag6-containg complex carrying dislocated TCRα could be detected by immunoprecipitation upon inhibition of the proteasome. Using this assay, we found that depletion of SGTA by expressing a SGTA specific shRNA reproducibly reduced the level of Bag6-associated TCRα, even though the total level of TCRα was significantly increased by SGTA knockdown (Fig. 6C). We concluded that SGTA may assist Bag6 in capturing retrotranslocated ERAD substrates, promoting their turnover.
Discussion
Many type II UBL domains in cells can be recognized by UBDs in a manner similar to ubiquitin (van der Veen and Ploegh, 2012, Mueller and Feigon, 2003). Consistent with this notion, we establish the UBL domain in Bag6 as a canonical type II UBL that is recognized by UBDs in a similar mode to ubiquitin. However, our study also reveals the existence of a class of type II UBL that cannot be recognized by UBDs. We propose to refer to the canonical UBLs as type IIa UBLs and the non-conventional ones as type IIb UBLs. A representative of the type IIb class is found in Ubl4A, which does not bind either CUE or UBA domains.
We identify SGTA as a functional partner of Ubl4A UBL. As expected, SGTA does not contain any previously known UBDs, and the interaction of SGTA with Ubl4A UBL does not resemble UBD binding to ubiquitin. Instead, SGTA utilizes its N-terminal domain to form a dimer, which uses symmetric electrostatics to interact with the Ubl4A UBL. a feature that is conserved in the yeast system (Chartron et al., 2012). Notably, Ubl4A has an unusually high pI of 8.7, whereas the pI of SGTA is 4.7, suggesting that these proteins carry opposite charges at the physiological pH. This provides a plausible explanation for the observed electrostatic interactions. Interestingly, notwithstanding the distinct UBL recognition mechanisms, SGTA binds Ubl4A UBL to a site overlapping with the UBD binding surface on canonical UBLs. Thus, evolution seems to have reengineered the UBD binding surface on Ubl4A UBL to accommodate a unique functional partner. This theme may be reiterated by other type IIb UBL domains. In this regard, it would be interesting to identify the functional partners of other type IIb UBL domains in the cell.
We showed previously that the Bag6 complex interacts with gp78, a key component of a multi-subunit complex required for degradation of many misfolded ER proteins by ERAD. Intriguingly, many proteins in the gp78 pathway have either a type II UBL domain or a UBD. Specifically, gp78 itself has a CUE domain and two of its interactors, UbxD8 and UBAC2, each contain a UBA domain (Christianson et al., 2011). The p97 complex also contains several ubiquitin-binding domains (Ye et al., 2003). Proteins bearing a UBL domain in the gp78 complex include HERP and TMUB1 (Okuda-Shimizu and Hendershot, 2007, Jo et al., 2011). In addition, the Bag6 complex contains two UBL domains, one in Bag6 and the other in Ubl4A(Wang et al., 2011b). It was originally thought that the ERAD machineries might use UBDs to capture misfolded proteins undergoing retrotranslocation and ubiquitination. However, the frequent presence of UBL domains in the ERAD machineries suggests an alternative model in which the UBDs in the ERAD network may use these UBLs as ‘connecting bolts’ to facilitate protein-protein interactions between distinct ERAD sub-complexes. Indeed, we demonstrate that the two UBL domains in the Bag6 complex can serve an adaptor function that links this ‘holdase’ to different ERAD machineries. The UBL domain in Bag6 binds the CUE domain in the gp78 ligase complex, whereas the Ubl4A uses a non-canonical UBL to recruit SGTA. These observations establish a UBL-dependent protein network essential for ER protein quality control (Fig. 6D).
The Bag6-Ubl4A-Trc35 complex can function as a chaperone holdase to channel retrotranslocated ERAD substrates to the proteasome for degradation while maintaining their solubility (Wang et al., 2011b). Intriguingly, this holdase activity can be used for chaperoning newly synthesized tailed-anchored (TA) proteins to the ER membrane or for degradation of mislocalized membrane proteins (Mariappan et al., 2010, Leznicki et al., 2010, Hessa et al., 2011). Our studies implicate SGTA as another critical component of the Bag6 system in ERAD. Given the broad participation of Bag6 in various protein quality control processes, SGTA may also function in other protein degradation pathways. SGTA is homologous to Sgt2p in yeast. It was shown previously that Get5p, the ortholog of Ubl4A can interact and function with Sgt2p as well as a downstream ATPase named Get3p, resulting in the handoff of TA proteins from Sgt2p to Get3p (Wang et al., 2010, Chartron et al., 2011, Chang et al., 2011, Wang et al., 2011a). Our results suggest that in mammalian cells, Ubl4A may promote the interaction of SGTA with Bag6 to form a similar chaperone axis that facilitates substrate transfer from SGTA to Bag6 (Fig. 6D). We propose that SGTA may serve as the initial ‘interrogator’ when misfolded substrates are emerging from an ER ‘retrotranslocon’. SGTA may identify aggregation-prone substrates and transfer them to Bag6 for further shielding until degradation occurs. It is also possible that SGTA may function with other downstream chaperones/effectors to promote substrate targeting to the proteasome. These chaperoning cascades would effectively protect cells against protein aggregation by ensuring that all aberrantly folded ER proteins are efficiently ‘shepherded’ to the proteasome for degradation.
Experimental procedures
Immunoblotting, immunoprecipitation and pulldown
Cells were lysed in the NP40 lysis buffer containing 50 mM Tris-HCl pH 7.4, 150 mM sodium chloride, 2 mM magnesium chloride, 0.5% NP40, and a protease inhibitor cocktail. Cell extracts were subject to centrifugation to remove insoluble materials. For most experiments, the supernatant fractions were analyzed. Where indicated in the figure legends, the NP40 insoluble pellet fractions were resolubilized by the Laemmli buffer for immunoblotting. Immunoblotting was performed according to the standard protocol. Fluorescence-labeled secondary antibodies (Rockland) were used for detection. The fluorescent bands were imaged and quantified on a LI-COR Odyssey infrared imager using the software provided by the manufacturer. For immunoprecipitation, the whole cell extract was incubated with FLAG-agarose beads (Sigma) or protein A-Sepharose CL-4B (GE Healthcare) bound with antibodies against specific proteins. After incubating, the beads were washed two times by NP40 wash buffer containing 50 mM Tris-HCl pH 7.4, 150 mM sodium chloride, 2 mM magnesium chloride, 0.1% NP40. The proteins on beads were assayed by immunoblotting.
For in vitro pulldown, the GST beads bound GST tagged bait proteins were incubated with prey protein, the pulled down materials were analyzed by SDS-PAGE. To assay the effect of salt concentration on Ubl4A UBL-SGTA-N interaction, GST-Ubl4A UBL was bound to GST beads, incubated with SGTA-N in PBS or PBS plus salt (400 mM potassium acetate). The beads were washed once with the corresponding binding buffer and assayed by SDS-PAGE.
Analytical size exclusion chromatography
Size exclusion chromatography was performed as follows. To study UBL-UBD interactions, protein or proteins mixtures at indicated concentration were incubated on ice for 3 min and then loaded onto a Superdex75 10/300 GL size exclusion column (GE Healthcare) pre-equilibrated with a buffer containing 20 mM Tris-HCl pH7.5, 10 mM 2-ME). The protein(s) was resolved at a flow rate of 0.5 ml/min on an AKTA (GE Healthcare) automated liquid chromatography system and assayed by SDS-PAGE. To analyze purified Bag6, the protein was applied on a Superdex 200 10/300 GL size exclusion column (GE Healthcare) pre-equilibrated with the PB buffer and resolved at a flow rate of 0.4 ml/min on an AKTA (GE Healthcare) automated liquid chromatography system. To analyze endogenous Bag6, 293T cells were collected from an 80% confluent 15 cm dish and lysed in 600 ml NP40 lysis buffer containing a protease inhibitor cocktail. The whole cell extract (WCE) was filtered through a 0.22 mm filter and applied onto a Superose 6 10/300 GL size exclusion column (GE Healthcare) pre-equilibrated with the PB buffer. Fractions of 0.4 ml were collected and analyzed by immunoblotting.
NMR 1H/15N chemical shift perturbation experiments
All NMR spectra were collected on 0.4 mM protein 15N/13C uniformly labeled proteins, or 0.4 mM 15N/13C-labeled protein mixed with 0.8 mM unlabeled protein dissolved in 25 mM sodium phosphate at pH 7.4. Sequence-specific backbone assignments for labeled free proteins and protein/protein complexes were obtained through HSQC (Grzesiek and Bax, 1992), CBCA(CO)NH and HNCACB(Clore and Gronenborn, 1991) experiments, recorded at either 25 °C for analysis of the Bag6 UBL-CUE interaction or 40 °C for Ubl4A UBL-SGTA interaction on Bruker DRX600 or DRX500 equipped with Z-gradient and cryoprobe. Spectra were processed using the program NMRPipe (Delaglio et al., 1995) and analyzed using the programs PIPP (Garrett et al., 1991). The chemical shift perturbation was determined according to ΔHz = [(ΔHz15N)2 + (ΔHz1H)2]1/2 (ΔHz1H and ΔHz15N are the observed chemical shift changes in Hz for 1H and 15N, respectively).
Gene knockdown and various ERAD assays
To knock down UbxD8 and SGTA, 0.5×106 293T cells were seeded on Day 0 and transfected with shRNA constructs using Lipofectamine 2000 on Day 1 and Day 2. 72h post the first transfection, cells were harvested for various assays. For cycloheximide chase experiments, 3.0×106 cells were harvested and incubated in 1.8 ml DMEM medium containing 50 μg/ml cycloheximide. Cells were then incubated at 37 °C for different time periods. At each time point, 1×106 cells were collected. Cell extracts were prepared in the NP40 lysis buffer. TCRα-YFP was either analyzed directly by immunoblotting or was first immunoprecipitated using a GFP antibody followed by immunoblotting. To analyze the in vivo aggregation of ERAD substrates, cells transfected with the TCRα-YFP-expressing plasmid together with SGTA shRNA knockdown construct were first solubilized in the NP40 lysis buffer. The detergent insoluble fractions were further solubilized by the Laemmli buffer.
Luciferase aggregation and refolding assay
To assay luciferase aggregation, luciferase (80 nM) was incubated with the indicated amount of BSA, Bag6, or SGTA at 42°C. The scattered light at 330 nm was measured by Amico-Bowman Series 2 Spectrofluorometer. Luciferase incubated in the absence of chaperones or in the presence of bovine albumin (BSA) was assayed as controls. For the luciferase refolding assay, luciferase (500 nM) alone or with Bag6 (1 μM), or SGTA(1 μM) was heat inactivated (42°C, 20 min) in buffer A (20 mM HEPES pH7.3, 5 mM Magnesium acetate, 50 mM Potassium chloride, 1 mM DTT). The total volume was 28 μl. The mixture was then divided into two equal portions. One portion was further incubated with 6 μl buffer B (buffer A plus 5 mM ATP) whereas the other portion was incubated with buffer C (buffer B plus a chaperone mixture containing 2 μM HSP70, 2.4 μM HOP, 3.3 μM Hdj). The mixtures were incubated at 25°C for 30 min. The luciferase activity was assayed using the Luciferase Reporter Gene Assay-high sensitivity kit (Roche) according to the protocol provided by the manufacturer.
Supplementary Material
Highlights.
The two UBLs in the Bag6 complex have distinct features that specify their interactors
The Ubl4A UBL interacts with SGTA to enhance its binding to Bag6
The Ubl4A UBL binds SGTA-N via an unconventional means of UBL recognition
SGTA assists Bag6 in maintaining the solubility of ERAD substrates and promoting ERAD
Acknowledgments
We thank R. Hegde (MRC, UK) for the SGTA antibody, J. W. Chartron and W. Clemons (Caltech) for sharing unpublished information and for building the human SGTA N-domain-Ubl4A UBL model, Fred Dyda, Marius Clore, and Martin Gellert (NIDDK) for critical reading of the manuscript. The research is supported by the Intramural Research Program of the NIDDK, and by a grant from the NIH Intramural AIDS Targeted Antiviral Program (IATAP) to Y.Y., and by National Institutes of Health grant RO1GM74830-06A1 to L.H.
Footnotes
Author contributions
M-L. C. designed and conducted the NMR experiments. Y-Y.Y. and L. H. performed the mass spectrometry analyses. Y.X. and Y-H.Y. contributed to the design of all other experiments. All authors contributed to the interpretation and conclusions of the experiments.
Competing financial interests
The authors declare no competing financial interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- BALLAR P, et al. The role of a novel p97/valosin-containing protein-interacting motif of gp78 in endoplasmic reticulum-associated degradation. J Biol Chem. 2006;281:35359–35368. doi: 10.1074/jbc.M603355200. [DOI] [PubMed] [Google Scholar]
- BAYS NW, et al. HRD4/NPL4 is required for the proteasomal processing of ubiquitinated ER proteins. Mol Biol Cell. 2001;12:4114–4128. doi: 10.1091/mbc.12.12.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BHAMIDIPATI A, et al. Exploration of the topological requirements of ERAD identifies Yos9p as a lectin sensor of misfolded glycoproteins in the ER lumen. Mol Cell. 2005;19:741–751. doi: 10.1016/j.molcel.2005.07.027. [DOI] [PubMed] [Google Scholar]
- CARVALHO P, et al. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- CARVALHO P, et al. Retrotranslocation of a misfolded luminal ER protein by the ubiquitin-ligase Hrd1p. Cell. 2010;143:579–591. doi: 10.1016/j.cell.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHANG YW, et al. Crystal structure of Get4-Get5 complex and its interactions with Sgt2, Get3, and Ydj1. J Biol Chem. 2010;285:9962–9970. doi: 10.1074/jbc.M109.087098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHANG YW, et al. Crystal structure of Get4-Get5 complex and its interactions with Sgt2, Get3, and Ydj1. J Biol Chem. 2011;285:9962–9970. doi: 10.1074/jbc.M109.087098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHARTRON JW, et al. A structural model of the Sgt2 protein and its interactions with chaperones and the Get4/Get5 complex. J Biol Chem. 2011;286:34325–34334. doi: 10.1074/jbc.M111.277798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHARTRON JW, et al. Structures of the Sgt2/SGTA dimerization domain with the Get5/Ubl4a UBL domain reveal a novel interaction that forms a conserved dynamic interface. Cell Reports. 2012 doi: 10.1016/j.celrep.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHEN B, et al. The activity of a human endoplasmic reticulum-associated degradation E3, gp78, requires its Cue domain, RING finger, and an E2-binding site. Proc Natl Acad Sci U S A. 2006;103:341–346. doi: 10.1073/pnas.0506618103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHRISTIANSON JC, et al. Defining human ERAD networks through an integrative mapping strategy. Nat Cell Biol. 2011;14:93–105. doi: 10.1038/ncb2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHRISTIANSON JC, et al. OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat Cell Biol. 2008;10:272–282. doi: 10.1038/ncb1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLORE GM, GRONENBORN AM. Structures of larger proteins in solution: three- and four-dimensional heteronuclear NMR spectroscopy. Science. 1991;252:1390–1399. doi: 10.1126/science.2047852. [DOI] [PubMed] [Google Scholar]
- DAS R, et al. Allosteric activation of E2-RING finger-mediated ubiquitylation by a structurally defined specific E2-binding region of gp78. Mol Cell. 2009;34:674–685. doi: 10.1016/j.molcel.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DELAGLIO F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- DENIC V, et al. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 2006;126:349–359. doi: 10.1016/j.cell.2006.05.045. [DOI] [PubMed] [Google Scholar]
- FANG S, et al. The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proc Natl Acad Sci U S A. 2001;98:14422–14427. doi: 10.1073/pnas.251401598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GARRETT DS, et al. A common sense approach to peak picking in two-, three-, and four-dimensional spectra using automatic computer analysis of contour diagrams. 1991. J Magn Reson. 1991;213:357–363. doi: 10.1016/j.jmr.2011.09.007. [DOI] [PubMed] [Google Scholar]
- GAUSS R, et al. A complex of Yos9p and the HRD ligase integrates endoplasmic reticulum quality control into the degradation machinery. Nat Cell Biol. 2006;8:849–854. doi: 10.1038/ncb1445. [DOI] [PubMed] [Google Scholar]
- GRZESIEK S, BAX A. Correlating backbone amide and sidechain resonances in large proteins by multiple relayed tripe resonance NMR. J Am Chem Soc. 1992;114:6291–6293. [Google Scholar]
- HESSA T, et al. Protein targeting and degradation are coupled for elimination of mislocalized proteins. Nature. 2011;475:394–397. doi: 10.1038/nature10181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HICKE L, et al. Ubiquitin-binding domains. Nat Rev Mol Cell Biol. 2005;6:610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- HOELLER D, et al. Ubiquitin and ubiquitin-like proteins in cancer pathogenesis. Nat Rev Cancer. 2006;6:776–788. doi: 10.1038/nrc1994. [DOI] [PubMed] [Google Scholar]
- HUSNJAK K, DIKIC I. Ubiquitin-Binding Proteins: Decoders of Ubiquitin-Mediated Cellular Functions. Annu Rev Biochem. 2012 doi: 10.1146/annurev-biochem-051810-094654. [DOI] [PubMed] [Google Scholar]
- IWAWAKI T, et al. A transgenic mouse model for monitoring endoplasmic reticulum stress. Nat Med. 2004;10:98–102. doi: 10.1038/nm970. [DOI] [PubMed] [Google Scholar]
- JAROSCH E, et al. Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nat Cell Biol. 2002;4:134–139. doi: 10.1038/ncb746. [DOI] [PubMed] [Google Scholar]
- JENTSCH S, PYROWOLAKIS G. Ubiquitin and its kin: how close are the family ties? Trends Cell Biol. 2000;10:335–342. doi: 10.1016/s0962-8924(00)01785-2. [DOI] [PubMed] [Google Scholar]
- JO Y, et al. Membrane-associated ubiquitin ligase complex containing gp78 mediates sterol-accelerated degradation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase. J Biol Chem. 2011;286:15022–15031. doi: 10.1074/jbc.M110.211326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JONIKAS MC, et al. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323:1693–1697. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KANG RS, et al. Solution structure of a CUE-ubiquitin complex reveals a conserved mode of ubiquitin binding. Cell. 2003;113:621–630. doi: 10.1016/s0092-8674(03)00362-3. [DOI] [PubMed] [Google Scholar]
- KERSCHER O, et al. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- KIM W, et al. Yos9p detects and targets misfolded glycoproteins for ER-associated degradation. Mol Cell. 2005;19:753–764. doi: 10.1016/j.molcel.2005.08.010. [DOI] [PubMed] [Google Scholar]
- KOHL C, et al. Cooperative and independent activities of Sgt2 and Get5 in the targeting of tail-anchored proteins. Biol Chem. 2011;392:601–608. doi: 10.1515/BC.2011.066. [DOI] [PubMed] [Google Scholar]
- LEZNICKI P, et al. Bat3 promotes the membrane integration of tail-anchored proteins. J Cell Sci. 2010;123:2170–2178. doi: 10.1242/jcs.066738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI W, et al. Mechanistic insights into active site-associated polyubiquitination by the ubiquitin-conjugating enzyme Ube2g2. Proc Natl Acad Sci U S A. 2009;106:3722–3727. doi: 10.1073/pnas.0808564106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIU Y, YE Y. Proteostasis regulation at the endoplasmic reticulum: a new perturbation site for targeted cancer therapy. Cell Res. 2011;21:867–883. doi: 10.1038/cr.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARIAPPAN M, et al. A ribosome-associating factor chaperones tail-anchored membrane proteins. Nature. 2010;466:1120–1124. doi: 10.1038/nature09296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MEHNERT M, et al. ERAD ubiquitin ligases: multifunctional tools for protein quality control and waste disposal in the endoplasmic reticulum. Bioessays. 2011;32:905–913. doi: 10.1002/bies.201000046. [DOI] [PubMed] [Google Scholar]
- MEUSSER B, et al. ERAD: the long road to destruction. Nat Cell Biol. 2005;7:766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- MINAMI R, et al. BAG-6 is essential for selective elimination of defective proteasomal substrates. J Cell Biol. 2010;190:637–650. doi: 10.1083/jcb.200908092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MOLINARI M, et al. Role of EDEM in the release of misfolded glycoproteins from the calnexin cycle. Science. 2003;299:1397–1400. doi: 10.1126/science.1079474. [DOI] [PubMed] [Google Scholar]
- MUELLER TD, FEIGON J. Structural determinants for the binding of ubiquitin-like domains to the proteasome. Embo J. 2003;22:4634–4645. doi: 10.1093/emboj/cdg467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ODA Y, et al. EDEM as an acceptor of terminally misfolded glycoproteins released from calnexin. Science. 2003;299:1394–1397. doi: 10.1126/science.1079181. [DOI] [PubMed] [Google Scholar]
- OKUDA-SHIMIZU Y, HENDERSHOT LM. Characterization of an ERAD pathway for nonglycosylated BiP substrates, which require Herp. Mol Cell. 2007;28:544–554. doi: 10.1016/j.molcel.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PRAG G, et al. Mechanism of ubiquitin recognition by the CUE domain of Vps9p. Cell. 2003;113:609–620. doi: 10.1016/s0092-8674(03)00364-7. [DOI] [PubMed] [Google Scholar]
- RABINOVICH E, et al. AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol Cell Biol. 2002;22:626–634. doi: 10.1128/MCB.22.2.626-634.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SMITH MH, et al. Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science. 2011;334:1086–1090. doi: 10.1126/science.1209235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TSAI B, et al. Protein disulfide isomerase acts as a redox-dependent chaperone to unfold cholera toxin. Cell. 2001;104:937–948. doi: 10.1016/s0092-8674(01)00289-6. [DOI] [PubMed] [Google Scholar]
- TSAI B, et al. Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat Rev Mol Cell Biol. 2002;3:246–255. doi: 10.1038/nrm780. [DOI] [PubMed] [Google Scholar]
- VAN DER VEEN AG, PLOEGH HL. Ubiquitin-Like Proteins. Annu Rev Biochem. 2012;81:323–357. doi: 10.1146/annurev-biochem-093010-153308. [DOI] [PubMed] [Google Scholar]
- VEMBAR SS, BRODSKY JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG F, et al. A chaperone cascade sorts proteins for posttranslational membrane insertion into the endoplasmic reticulum. Mol Cell. 2010;40:159–171. doi: 10.1016/j.molcel.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG F, et al. The mechanism of tail-anchored protein insertion into the ER membrane. Mol Cell. 2011a;43:738–750. doi: 10.1016/j.molcel.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG Q, et al. A ubiquitin ligase-associated chaperone holdase maintains polypeptides in soluble States for proteasome degradation. Mol Cell. 2011b;42:758–770. doi: 10.1016/j.molcel.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WINNEFELD M, et al. Human SGT interacts with Bag-6/Bat-3/Scythe and cells with reduced levels of either protein display persistence of few misaligned chromosomes and mitotic arrest. Exp Cell Res. 2006;312:2500–2514. doi: 10.1016/j.yexcr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- YE Y, et al. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- YE Y, et al. Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J Cell Biol. 2003;162:71–84. doi: 10.1083/jcb.200302169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YE Y, et al. Inaugural Article: Recruitment of the p97 ATPase and ubiquitin ligases to the site of retrotranslocation at the endoplasmic reticulum membrane. Proc Natl Acad Sci U S A. 2005;102:14132–14138. doi: 10.1073/pnas.0505006102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YOSHIDA H, et al. Endoplasmic reticulum stress-induced formation of transcription factor complex ERSF including NF-Y (CBF) and activating transcription factors 6alpha and 6beta that activates the mammalian unfolded protein response. Mol Cell Biol. 2001;21:1239–1248. doi: 10.1128/MCB.21.4.1239-1248.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.