Summary
Hepatic glucose production (HGP) maintains blood glucose levels during fasting but can also exacerbate diabetic hyperglycemia. HGP is dynamically controlled by a signaling/transcriptional network that regulates the expression/activity of gluconeogenic enzymes. A key mediator of gluconeogenic gene transcription is PGC-1α. PGC-1α’s activation of gluconeogenic gene expression is dependent upon its acetylation state, which is controlled by the acetyltransferase GCN5 and the deacetylase Sirt1. Nevertheless, whether other chromatin modifiers—particularly other sirtuins—can modulate PGC-1α acetylation is currently unknown. Herein we report that Sirt6 strongly controls PGC-1α acetylation. Surprisingly, Sirt6 induces PGC-1α acetylation and suppresses HGP. Sirt6 depletion decreases PGC-1α acetylation and promotes HGP. These acetylation effects are GCN5 dependent: Sirt6 interacts with and modifies GCN5, enhancing GCN5’s activity. Leprdb/Leprdb mice, an obese/diabetic animal model, exhibit reduced Sirt6 levels; ectopic re-expression suppresses gluconeogenic genes and normalizes glycemia. Activation of hepatic Sirt6 may therefore be therapeutically useful for treating insulin-resistant diabetes.
Introduction
In healthy individuals, hepatic glucose production (HGP) forms an integral part of the response to maintain blood glucose levels during extended periods of food deprivation or periods of high glucose demand. HGP comprises the processes of glycogenolysis and gluconeogenesis--both of which are regulated by nutrient and hormonal signaling pathways. A greater understanding of the pathways regulating the gluconeogenic component of HGP, however, has received much interest because of its contribution to the hyperglycemia observed in insulin resistant Type 2 diabetes (Magnusson et al., 1992).
One regulator of gluconeogenesis is the transcriptional co-activator peroxisome proliferator-activated receptor γ coactivator 1-α (PGC-1α). PGC-1α potently stimulates hepatic gluconeogenesis in part by increasing the expression of gluconeogenic enzymes. PGC-1α contributes to the activation of the fasting gluconeogenic program by co-activation of FoxO1 (Puigserver et al., 2003) and HNF4 (Rhee et al., 2003; Yoon et al., 2001). Because PGC-1α can promote gluconeogenesis, identification of factors that control PGC-1α activity could aid in the discovery of new regulatory components that converge upon gluconeogenesis. It is known that PGC-1α activity is controlled primarily through transcriptional regulation of total protein levels (Yoon et al., 2001), protein stability (Olson et al., 2008), as well as through post-translational modification of the mature protein (Li et al., 2007; Rodgers et al., 2010; Rodgers et al., 2005; Teyssier et al., 2005). Of these, PGC-1α acetylation has been the most extensively characterized under different nutrient and energetic states (Canto et al., 2009; Rodgers et al., 2005).
PGC-1α contains at least 13 acetylation sites (Rodgers et al., 2005). Acetylation of these residues is strongly associated with a repression of PGC-1α’s activity. General Control Non-repressed Protein 5 (GCN5, aka KAT2A) catalyzes PGC-1α acetylation, which coincides with PGC-1α’s relocalization away from the promoters of its target genes and into nuclear foci of unknown identity (Lerin et al., 2006). Furthermore, GCN5 can efficiently repress PGC-1α target genes in both hepatocytes and myotubes (Gerhart-Hines et al., 2007; Lerin et al., 2006).
Deacetylation of PGC-1α occurs primarily by the NAD+-dependent deacetylase, silent mating type information regulation 2 homolog 1 (Sirt1) (Nemoto et al., 2005; Rodgers et al., 2005). Induction of Sirt1 activity leads to PGC-1α deacetylation and activation of its respective gene targets; reductions in Sirt1 activity is accompanied by an increase in PGC-1α acetylation and a decrease in target expression (Canto et al., 2009; Gerhart-Hines et al., 2007; Rodgers et al., 2005).
Although Sirt1 is considered to be the principal PGC-1α deacetylase in vivo, there has been no systematic examination of the participation of other nuclear sirtuins in regulating PGC-1α acetylation. We therefore examined the effects of Sirt2, Sirt6, and Sirt7 (the other major nuclear sirtuins) on PGC-1α acetylation. Surprisingly, Sirt6 uniquely induced an increase in the acetylation of PGC-1α through the direct modification and activation of GCN5. This was associated with an inhibition of gluconeogenic targets both in cell culture and in whole animals. Most notably, hepatic Sirt6 was reduced in obese/diabetic mice and expression of Sirt6 normalized blood glucose levels. Overall, these findings point to a novel substrate for Sirt6, namely GCN5, and have uncovered a new sirtuin-mediated pathway for the control of PGC-1α activity. They also suggest that therapies which specifically activate hepatic Sirt6 may be useful in suppressing the chronically active hepatic gluconeogenesis commonly found in insulin-resistant diabetes.
Results
Sirt6 Is the Only Sirtuin that Increases PGC-1α Acetylation
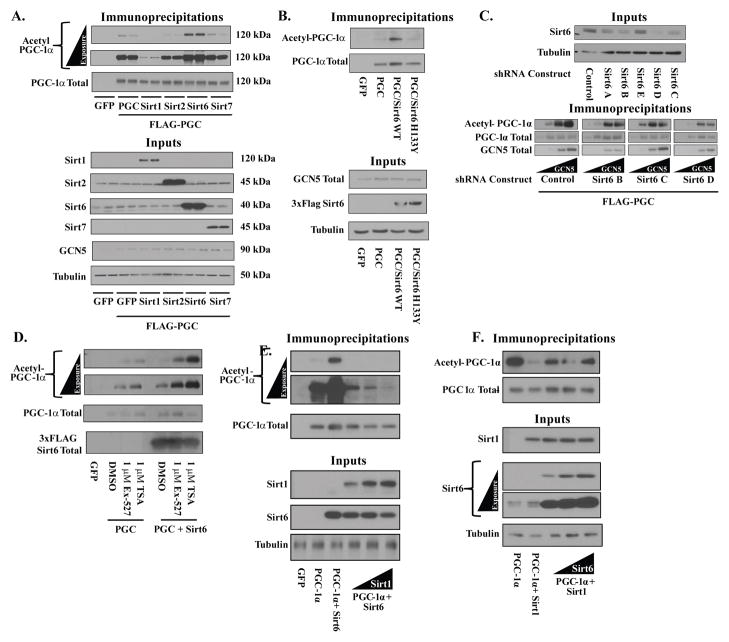
In evaluating the effects of sirtuin proteins on PGC-1α acetylation, we limited ourselves to Sirt1, Sirt2, Sirt6, and Sirt7. Like PGC-1α, these sirtuin paralogs are present within the nucleus and thus could directly affect PGC-1α’s acetylation. When ectopically expressed in the human osteosarcoma cell line U-2 OS, Sirt1 produced a significant decrease in PGC-1α acetylation (Figure 1A)—an effect that is consistent with what has been widely published. Sirt2 and Sirt7 produced no discernable change in PGC-1α acetylation—with the caveat that the degree of overexpression that was obtainable with Sirt7 was far less than with the other nuclear sirtuins and thus precludes definitely ruling out any effect on PGC-1α acetylation. Sirt6, surprisingly, dramatically increased PGC-1α acetylation. To examine whether this effect was dependent upon Sirt6’s enzymatic activity, a catalytically incompetent mutant allele of Sirt6 (H133Y) was employed. H133Y failed to elicit the effects of WT Sirt6, suggesting that the Sirt6’s effects on PGC-1α acetylation is tied to Sirt6’s enzymatic activity (Figure 1B). These results were not unique to the U-2 OS cell line, as similar results were observed in HEK293A (Figure S1A) cultures.
Figure 1. Sirt6 modulates PGC-1α acetylation in cultured cells.
(A) Effects of ectopic nuclear sirtuin expression on PGC-1α acetylation. FLAG-HA-PGC-1α was immunoprecipitated from U-2 OS cells transfected with Sirt1, Sirt2, Sirt6, Sirt7 or GFP control and blotted for total acetylation. Molecular weights of detected proteins are indicated. (B) Mutation of catalytically essential residue H133 blocks Sirt6-induced PGC-1α acetylation. (C) Knockdown of endogenous Sirt6 reduces PGC-1α acetylation levels. (Top) Sirt6 expression in U-2 OS lines stably expressing either one control shRNA or one of five different Sirt6 shRNAs (Bottom) PGC-1α acetylation from three of the lowest Sirt6 expressing lines co-transfected with PGC-1α and increasing amounts of GCN5. (D) 16 h inhibition of Sirt1 or Class I/II HDACs fails to block Sirt6-induced PGC-1α acetylation. (E) PGC-1α acetylation from U-2 OS cells transfected with a fixed concentration of Sirt6 and increasing amounts of Sirt1. (F) PGC-1α acetylation from U-2 OS cells transfected with a fixed amount of Sirt1 and increasing amounts of Sirt6. See Figure S1.
To complement the gain-of-function effects of Sirt6 in U-2 OS cells, endogenous Sirt6 was knocked down through the use of lentiviruses expressing Sirt6 shRNAs. Accordingly, knockdown of Sirt6 was associated with a decrease in the acetylation of PGC-1α—even with the ectopic expression of GCN5 (Figure 1C). Pharmacological inhibition of Sirt1 with EX-527 did not prevent the effects of Sirt6 on PGC-1α acetylation, suggesting that Sirt6 does not affect Sirt1 activity (Figure 1D). Total cellular levels of NAD+ were also not significantly altered by changes in Sirt6 levels (Figure S1B). Inhibition of Class I/II histone deacetylases with trichostatin A, which increased PGC-1α acetylation, also failed to prevent a further increase in Sirt6-mediated PGC-1α acetylation (Figure 1D). Interestingly, although Sirt6’s effects on PGC-1α did not require Sirt1, the balance of activities between the two proteins has a large effect on steady-state PGC-1α acetylation. Increasing Sirt1 expression in U-2 OS cells overcomes the increase in acetylation induced by Sirt6 (Figure 1E). On the other hand, elevating Sirt6 activity attenuates Sirt1-induced deacetylation of PGC-1α (Figure 1F).
Collectively, these results ruled out an indirect effect of Sirt6 on the rate of PGC-1α deacetylation. We therefore focused upon whether Sirt6’s effects on PGC-1α were being directed through GCN5.
Regulation of PGC-1α Acetylation by Sirt6 Depends Upon GCN5
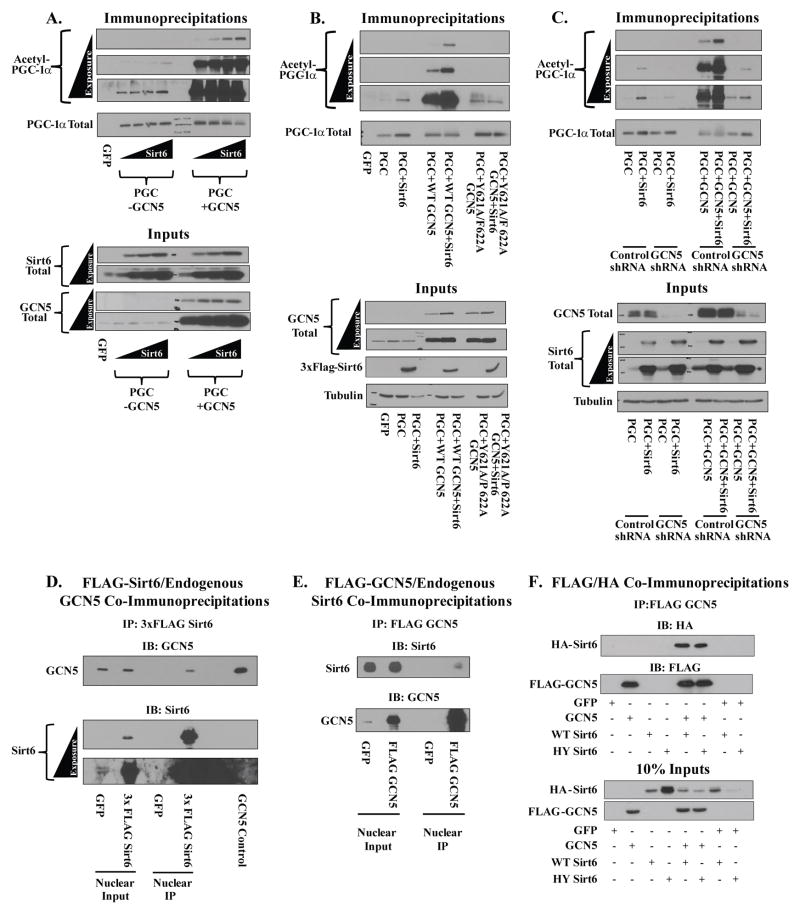
Although Sirt6 was able to induce PGC-1α acetylation in the absence of ectopically expressed GCN5, the effect was significantly magnified by elevating GCN5 expression (Figure 2A). This effect was dependent upon the catalytic activity of GCN5, as a mutant (Y621A/F622A) previously shown to have markedly impaired catalytic activity (Lerin et al., 2006) failed to amplify the Sirt6-induced acetylation of PGC-1α (Figure 2B).
Figure 2. Changing cellular GCN5 levels alters Sirt6’s ability to change PGC-1α acetylation.
(A) Effects of GCN5 on Sirt6-mediated changes in PGC-1α acetylation. U-2 OS cultures were transfected with PGC-1α and increasing concentrations of Sirt6 with or without expression of WT GCN5. (B) Catalytically impaired GCN5 mutant, Y621A/F622A, fails to augment the Sirt6-mediated increase in PGC-1α acetylation (C) GCN5 knockdown diminishes the effect of Sirt6 on PGC-1α acetylation. (D) Co-IP of endogenous GCN5 with 3xFLAG Sirt6 from the nuclear fraction of U-2 OS cells. (E) Co-IP of endogenous Sirt6 with FLAG GCN5 from the nuclear fraction of U-2 OS cultures. (F) Co-IP of WT and HY Sirt6 with GCN5 from the nuclear fractions of U-2 OS cells.
Additional support for the GCN5 involvement was garnered by knockdown of GCN5. Transient transfection of a GCN5 shRNA blunted the ability of Sirt6 to increase PGC-1α acetylation relative to a control shRNA (Figure 2C). The GCN5 shRNA, which reduced levels of ectopically expressed GCN5, also abrogated the effect of increased GCN5 expression on Sirt6.
To identify whether GCN5 and Sirt6 physically interact, co-immunoprecipitation studies were performed. Immunoprecipitation of FLAG Sirt6 from the nuclear fraction of U-2 OS revealed endogenous GCN5 bound to Sirt6 (Figure 2D). Conversely, immunoprecipitation of FLAG GCN5 showed specific binding of endogenous Sirt6 protein (Figure 2E). Both the WT and H133Y Sirt6 alleles efficiently interacted with GCN5 (Figure 2F)—suggesting that the inability of the H133Y allele to increase PGC-1α acetylation is not due to changes in GCN5 binding. Because of the binding data, we extended our investigation to see if Sirt6 was directly modulating GCN5 activity by a post-translational modification such as acetylation.
Sirt6 Increases GCN5 Activity and Induces Both Selective Deacetylation and Phosphorylation of GCN5
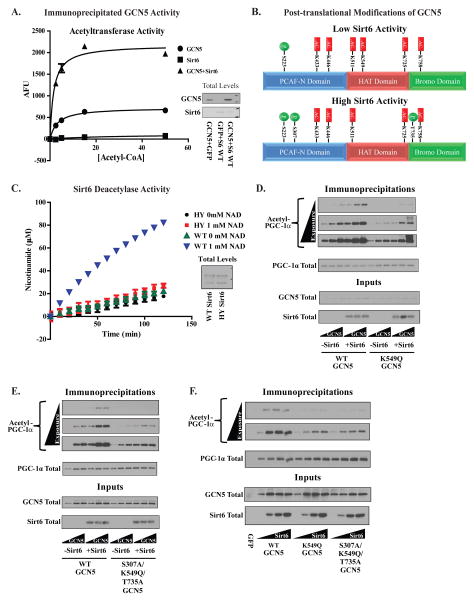
If Sirt6 were indeed modulating the activity of GCN5, then it is possible that this activity difference could be measured in vitro. GCN5’s acetyltransferase activity was therefore measured from protein immunoprecipitated from the nuclear extracts of U-2 OS in the presence or absence of ectopically expressed Sirt6. Co-transfection of Sirt6 with GCN5 produced a significant increase in the in vitro acetyltransferase activity of GCN5 (Figure 3A and Figure S2A). To investigate possible causes for this change in activity, we performed mass spectrometric analysis on GCN5 protein immunoprecipitated from U-2 OS in the presence/absence of Sirt6 (Figure 3B). Under conditions of elevated Sirt6 expression, GCN5 acetylation at residue K549 disappeared whereas two new phosphorylation sites (S307 and T735) appeared on the protein. Of these modifications, the loss of acetylation at K549 is consistent with the possibility that Sirt6 is targeting GCN5 for deacetylation, a known catalytic activity of Sirt6 (Michishita et al., 2008).
Figure 3. Sirt6 increases GCN5’s acetyltransferase activity and alters the profile of post-translational modifications on GCN5 protein.
(A) In vitro acetyltransferase activities of GCN5 immunoprecipitated from the nuclear fraction of U-2 OS cells with and without Sirt6. (B) Post-translational modifications mapped by mass spectrometry on GCN5 immunoprecipitated from U-2 OS transfected with empty vector (top panel) or WT Sirt6 (bottom panel). Phosphorylated residues are indicated in green and acetylated residues in red. 49% sequence coverage was obtained in these experiments (C) Measurement of the in vitro deacetylation of a peptide containing acetyl-K549 using nicotinamide production as an indicator of deacetylase activity. WT Sirt6 and H133Y Sirt6 were immunoprecipitated from U-2 OS infected with adenoviral constructs. (D) A test of WT and K549Q GCN5’s effects on PGC-1α acetylation in the presence/absence of Sirt6 expression. Increasing concentrations of GCN5 construct were transfected along with a fixed concentration of Sirt6 and PGC-1α. (E) Evaluation of WT and S307A/K549Q/T735A GCN5’s effects on PGC-1α acetylation in the presence/absence of Sirt6. Experimental setup was identical to that shown in Figure (D). (F) Comparison of sensitivities of WT, K549Q and S307A/K549Q/T735A GCN5 to increasing concentrations of Sirt6 using acetylated PGC-1α. GCN5 and PGC-1α were transfected into U-2 OS cells at a fixed concentration along with Sirt6 at increasing concentrations. Data are means±S.E.M. See Figures S2,3.
As a product of its NAD+-dependent deacetylase reaction, Sirt6 produces free nicotinamide. We therefore first measured nicotinamide production as a means for detecting whether Sirt6 could use acetylated-GCN5 as a substrate. For this reaction, a 12-mer peptide was synthesized containing acetyl-K549 and its flanking residues from the human GCN5 sequence. Using this peptide, significant amounts of nicotinamide production were measured with WT Sirt6 in the presence of NAD+ (Figure 3C); neither the H133Y mutant in the presence or absence of NAD+ nor WT Sirt6 in the absence NAD+ generated nicotinamide. Mass spectrometry analysis of the acetyl-K549 peptide incubated with WT Sirt6 and either 0 or 1 mM NAD+ also showed that deacetylated peptide was formed only in the presence of NAD+ (Figure S2B,C).
Next, residues found to be modified on GCN5 by Sirt6 were mutated to evaluate how much they contributed to the effects on GCN5/PGC-1α acetylation. To mimic the acetylated state of GCN5, a glutamine residue was used in lieu of K549 whereas an alanine residue was used to mimic the unphosphorylated S307 and T735. The K549Q (Figure 3 D) mutant of GCN5 showed a reduced ability to increase PGC-1α acetylation in cells when Sirt6 was transfected. The T735A and S307A point mutants, however, had less pronounced effects (Figures S3A,B). Although these data indicate that residue K549 is the most important for Sirt6’s effect, a triple GCN5 mutant comprising S307A/K549Q/T735A did show a tendency towards lower levels of induction of PGC-1α acetylation than any of the individual point mutants (Figure 3E). It was also noted that the point mutants showed far less sensitivity to the absolute amount of Sirt6 expressed in cells relative to WT GCN5 (Figure 3F).
The crystal structure of the catalytic core of human GCN5 is known (Schuetz et al., 2007). Within this structure, K549 is deacetylated and forms a hydrogen bond network with M567 and Y601 (Figure S3C); acetylation of K549 would presumably disrupt this hydrogen bond network. Perhaps by deacetylating K549 and promoting hydrogen bond formation, Sirt6 causes GCN5 to undergo a structural change that facilitates acetyltransferase activity. Supporting this idea, M567A and Y601F mutants showed deficits in responding to Sirt6 that were akin to that seen in the K549Q mutant (Figure S3D, E).
In aggregate, our data indicate that Sirt6 is able to directly bind to GCN5, deacetylate it at K549, as well as induce changes in the phosphorylation of the protein which ultimately yield an increase in GCN5 activity and an increase in PGC-1α acetylation. We next sought to evaluate if this had a physiological effect on PGC-1α activity.
Sirt6-induced Changes in PGC-1α Acetylation Affect the Gluconeogenic Program in Primary Hepatocytes
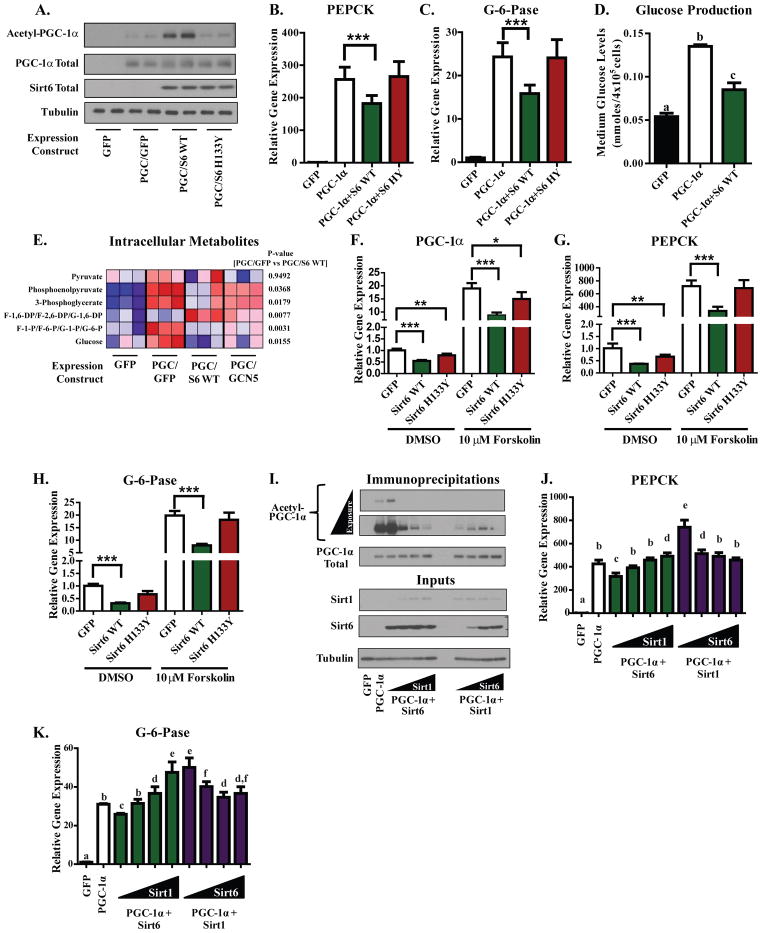
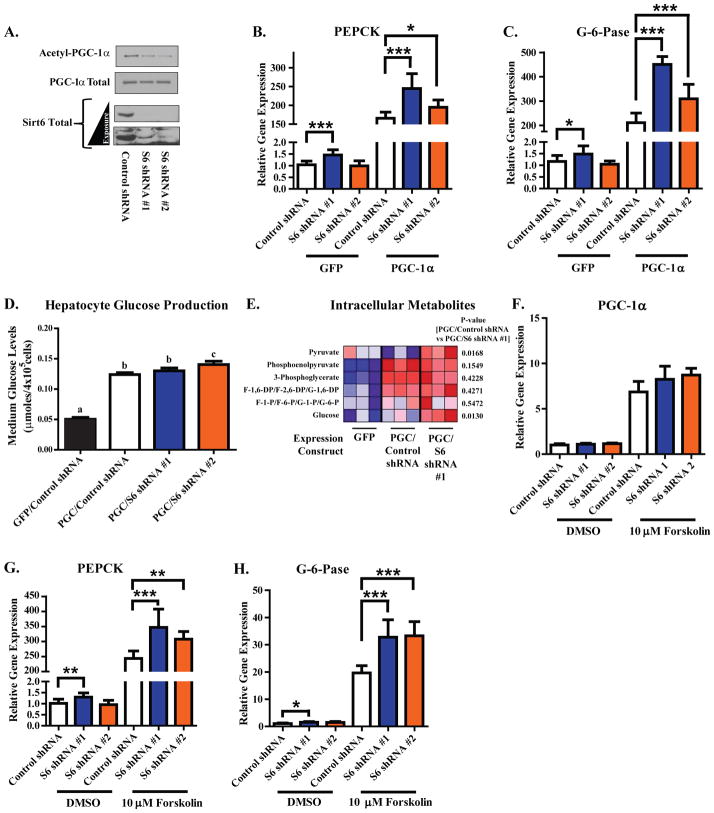
Because the gluconeogenic genes phosphoenolpyruvate carboxykinase (PEPCK) and glucose-6-phosphatase (G-6-Pase) are well described targets of PGC-1α coactivation, we tested whether Sirt6 was able to modulate their expression. Adenoviral-mediated expression of WT Sirt6—but not H133Y Sirt6—caused a significant increase in the acetylation of PGC-1α in mouse primary hepatocytes (Figure 4A). WT Sirt6 also caused a significant suppression in the PGC-1α-induced expression of PEPCK (Figure 4B) and G-6-Pase (Figure 4C) whereas the H133Y allele did not. Accompanying the Sirt6-mediated reduction in gluconeogenic gene expression was a decrease in the hepatocyte’s capacity for glucose production (Figure 4D). Indeed, Sirt6 significantly reduced many of the gluconeogenic pathway intermediates in a pattern resembling the effects of GCN5 (Figure 4E; Figure S4A). Although we initially used overexpressed PGC-1α to evaluate the effects of Sirt6, it is known that endogenous hepatic PGC-1α expression is induced upon fasting through the CREB pathway and this can be mimicked in primary hepatocytes by forskolin treatment (Yoon et al., 2001). Increased expression of WT Sirt6 blunted forskolin-induced increases in the expression of PGC-1α, PEPCK, and G-6-Pase (Figures 4F–H). This effect was dependent upon Sirt6 catalytic activity as the H133Y allele showed reduced repressive effects. Another forskolin-inducible gene, NOR-1, was not significantly affected by Sirt6 expression (Figure S4B). The ability of Sirt6 to blunt forskolin-mediated induction of endogenous PGC-1α expression could be due to an auto-regulatory mechanism of control that has been previously observed for PGC-1α (Handschin et al., 2003).
Figure 4. Sirt6 elevates PGC-1α acetylation in murine primary hepatocyte cultures and suppresses the gluconeogenic program in direct opposition to Sirt1.
(A) Measurement of PGC-1α acetylation in murine primary hepatocyte cultures expressing GFP, WT Sirt6, or H133Y Sirt6. (B,C) Assessment of PEPCK (B) and G-6-Pase (C) mRNA levels in murine primary hepatocyte cultures expressing the indicated constructs. In Figures (A–C), data are a composite of three separate experiments. (D) PGC-1α-induced glucose production in primary hepatocytes co-expressing either GFP or Sirt6. Data are an average of two independent experiments. Columns with different letters above them are statistically significant. (E) Heat map depicting LC-MS analysis of intracellular metabolites from primary hepatocytes expressing the indicated constructs. (F–H) Assessment of PGC-1α (F), PEPCK (G), and G-6-Pase (H) mRNA levels in primary hepatoctyes infected with GFP, WT Sirt6, or H133Y Sirt6 expression constructs and treated for 1.5 h with DMSO vehicle or 10 μM forskolin. In Figures (F–H), data are pooled from three independent experiments. (I) Competitive effects of Sirt6 and Sirt1 on PGC-1α acetylation in primary hepatocytes. Cultures were infected with a fixed dose of one sirtuin while the amount of the other was progressively increased (total viral infection was equal across all treatments). (J,K) Gluconeogenic gene expression from the experimental setup in (I). Data are from two independent experiments. Columns with different letters above them are statistically significant (p<0.05). All data expressed as means±S.E.M. and analyzed by one-way ANOVA with a Tukey’s post-test. See Figure S4.
In primary hepatocytes, Sirt6 and Sirt1 had opposite effects on PGC-1α acetylation and biological activity. Increasing Sirt1 activity reversed the increase in PGC-1α acetylation caused by ectopic expression of Sirt6 whereas increasing Sirt6 activity partially reversed Sirt1-mediated deacetylation of PGC-1α (Figure 4I). Elevating Sirt1 activity reversed suppressions in PEPCK and G-6-Pase expression that were caused by Sirt6 whereas elevating Sirt6 activity reduced Sirt1-mediated increases in PEPCK and G-6-Pase expression (Figures 4J,K).
Two separate adenoviral shRNAs contructs were used to efficiently knockdown murine Sirt6 at the level of protein (Figure 5A) and mRNA (Figure S4C). Decreasing Sirt6 in hepatocytes caused a decrease in the acetylation of PGC-1α protein (Figure 5A) and a significant increase in PEPCK (Figure 5B) and G-6-Pase (Figure 5C). This increase in gene expression was also associated with a modest elevation in the capacity for de novo glucose synthesis (Figure 5D) and an increase in intracellular intermediates of the gluconeogenic pathway (Figure 5E). Whereas forskolin-induced expression of PGC-1α was not significantly elevated by Sirt6 knockdown (Figure 5F), the induction of both PEPCK (Figure 5G) and G-6-Pase (Figure 5H) were enhanced. Induction of NOR-1 by forskolin was not significantly affected by Sirt6 expression (Figure S4D)—suggesting that Sirt6 was not affecting the cAMP/PKA transcriptional response.
Figure 5. Reduction of endogenous Sirt6 decreases PGC-1α acetylation and enhances the gluconeogenic program of murine primary hepatocytes.
(A) Assessment of PGC-1α acetylation and Sirt6 protein levels in murine primary hepatoctyes infected with the indicated adenoviral shRNA-expression constructs. (B,C) qRT-PCR measurement of PEPCK (B) and G-6-Pase (C) mRNA levels in primary hepatoctyes infected with GFP or PGC-1α and the indicated shRNA constructs. Data in Figures (A–C) are from three independent experiments. (D) Glucose production in primary hepatocytes infected with either GFP or PGC-1α and the indicated shRNA constructs. Data are pooled from two independent experiments. Columns with different letters above them are statistically significant (p<0.05). (E) Heat map depicting LC-MS analysis of intracellular metabolites from primary hepatocytes expressing the indicated constructs. (F–H) Measurement of PGC-1α (F), PEPCK (G), and G-6-Pase (H) mRNA levels in primary hepatoctyes infected with the indicated shRNA expression constructs and treated for 1.5 h with DMSO vehicle or 10 μM forskolin. In Figures (F–H), data are pooled from three independent experiments. All data expressed as means±S.E.M. *-p<0.05; **-p<0.01; ***-p<0.001 by one-way ANOVA with a Tukey’s post-test. See Figure S4.
Modulating Hepatic Sirt6 Levels in Fasted, Lean, Non-diabetic Mice Changes Gluconeogenic Gene Expression and Blood Glucose Levels
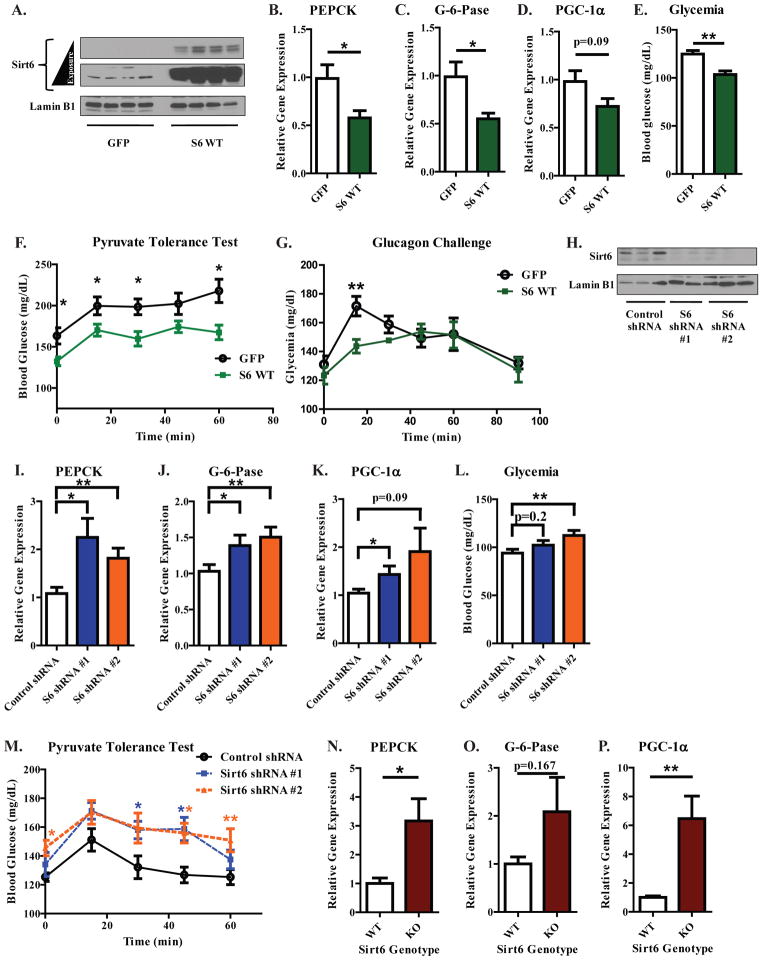
To confirm whether the effects of Sirt6 observed in primary murine hepatocytes also occurred within the context of the whole animal, hepatic Sirt6 levels were altered by tail vein administered injections of adenoviral expression constructs—which are targeted primarily to the liver (Shayakhmetov et al., 2004)—into fasted mice, a condition known to induce PGC-1α (Rodgers and Puigserver, 2007). Hepatic Sirt6 protein levels were significantly elevated in C57BL/6 mice following tail vein injections of Sirt6 expression constructs (Figure 6A). With Sirt6 overexpression, PEPCK and G-6-Pase were significantly repressed (Figures 6B,C), PGC-1α levels exhibited a declining trend (Figure 6D), and blood glucose levels were significantly reduced (Figure 6E). Overall, a similar pattern was observed in BALB/c mice (Figures S5A–E). In C57BL/6 mice, increased expression of Sirt6 reduced hepatic glucose output, as assessed by a pyruvate tolerance test (Figure 6F). In addition, Sirt6 attenuated hepatic glucose output in mice injected with glucagon—an important hormonal mediator of the fasting gluconeogenic program (Figure 6G).
Figure 6. Hepatic Sirt6 levels regulate the gluconeogenic transcriptional response of fasted C57BL/6 mice.
(A) Hepatic Sirt6 protein levels from mice injected with either GFP or 3xFLAG WT Sirt6 adenoviruses. Mice were fed ad libitum for 4 days after injection and then fasted for 16 h before being sacrificed. (B–D) Hepatic gene expression data for PEPCK (B), G-6-Pase (C), and PGC-1α (D). (E) Blood glucose levels from mice infected with the indicated constructs (A). Figures (A–E) were compiled from two independent experiments (N=8–9 mice per treatment group) and analyzed by a two-tailed unpaired t-test. (F) Effect of hepatic Sirt6 over-expression on a pyruvate tolerance test (N=8 per group, analyzed by a two-tailed unpaired t-test). (G) Effect of hepatic Sirt6 over-expression on glucagon-induced changes in glycemia (N=8 per treatment group, analyzed by a two-tailed unpaired t-test). (H) Hepatic Sirt6 protein levels from mice receiving a tail-vein injection of the indicated adenoviral shRNA constructs. Mice were fed ad libitum for 3 days after injection and then fasted for 15 h before being sacrificed. (I–K) Hepatic PEPCK (I), G-6-Pase (J), and PGC-1α (K) mRNA levels from mice treated as in (H). (L) Blood glucose levels from mice treated as in Figure (H). Figures (H–L) were compiled from two separate experiments (N=13–14 mice per treatment group; analyzed by one-way ANOVA with Dunnet’s post-test). (M) Effect of hepatic Sirt6 knockdown on a pyruvate tolerance test (N=8 per treatment group; analyzed by one-way ANOVA with Dunnet’s post-test). See Figure S5.
Adenoviral delivery of Sirt6 shRNAs reduced hepatic Sirt6 protein (Figure 6H) and produced a significant elevation in the mRNA of PEPCK (Figure 6I), G-6-Pase (Figure 6J), and PGC-1α (Figure 6K). Blood glucose levels were also elevated by Sirt6 knockdown (Figure 6L). Similar results were observed in BALB/c mice (Figures S5F–J). Coincident with the change in gluconeogenic gene expression, hepatic glucose output was significantly increased by hepatic Sirt6 knockdown (Figure 6M). In animals with a whole body deletion of Sirt6, the hepatic gluconeogenic genes PEPCK (Figure 6N) and G-6-Pase (Figure 6O) were also found to be elevated along with PGC-1α (Figure 6P). In sum, these data support a repressive role for Sirt6 in the regulation of hepatic gluconeogenesis in the animal.
Increased Hepatic Sirt6 Expression Suppresses the Gluconeogenic Transcriptional Program and Blood Glucose Levels in Diabetic db/db Mice
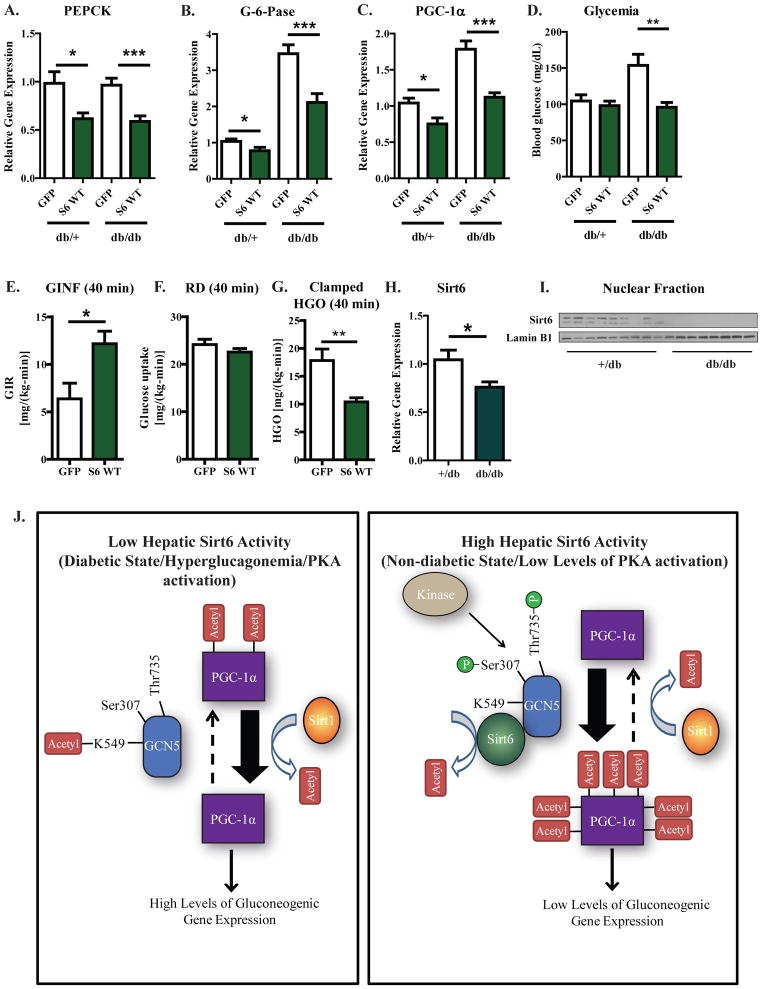
The diabetic phenotype displayed by db/db mice is characterized in part by a chronically active and insulin insensitive hepatic gluconeogenic program. Furthermore, PGC-1α activity contributes to the elevated gluconeogenesis in this diabetic mouse model (Koo et al., 2004; Yoon et al., 2001). We therefore tested whether increased expression of Sirt6 would be able to attenuate the diabetic profile of db/db mice. Adenoviral-mediated expression of Sirt6 in both +/db and db/db mice caused a significant repression in the expression of PEPCK (Figure 7A), G-6-Pase (Figure 7B), and PGC-1α (Figure 7C). Sirt6 expression significantly reduced the blood glucose levels of fasted db/db mice to levels that were comparable to +/db (Figure 7D). Increased Sirt6 levels did not appear to alter insulin/AKT pathway signaling in the livers of db/db mice as no significant changes in AKT phosphorylation were detected (Figure S6A). To confirm that changes in the blood glucose of db/db mice were due to reduced hepatic glucose production, hyperinsulinemic-euglycemic clamp studies were performed (Figures 7E–G; Figure S6B; Table S1). In db/db mice expressing Sirt6, the glucose infusion rate required to maintain euglycemia was significantly greater compared with control mice (Figure 7E; Figure S6B). This was not due to a changes in insulin-stimulated peripheral glucose uptake (Figure 7F), but rather due to reduced rates of hepatic glucose production.
Figure 7. Sirt6 suppresses the gluconeogenic program and reduces blood glucose levels in diabetic db/db mice.
(A–C) Hepatic PEPCK (A), G-6-Pase (B), and PGC-1α (C) mRNA levels from +/db and db/db mice injected with GFP or 3×FLAG WT Sirt6-expressing adenovirus. For 3–4 days after tail vein injection of virus, mice were fed ad libitum and then fasted for 16 h before being sacrificed. (D) Blood glucose levels measured from mice treated as in (A). (E–G) Hyperinsulinemic-euglycemic clamp data from db/db mice injected with GFP or 3×FLAG WT Sirt6 virus. Clamped glucose infusion rate (E), glucose uptake (F), and hepatic glucose production (G) are shown. (F) Hepatic levels of endogenous Sirt6 mRNA in GFP-infected control mice treated as in (A). (G) Hepatic levels of endogenous Sirt6 protein in GFP-infected control mice treated as in (A). In Figures (A–D), results are data pooled from two independent experiments; N=15 for GFP injected +/db mice, N=10 for Sirt6 injected +/db mice, N=13 for GFP injected db/db mice, N=12 for Sirt6 injected db/db mice. In Figures (E–G), data are from 6-GFP injected and 7-Sirt6 injected mice and analyzed by a two-tailed unpaired t-test. (J) A model for how Sirt6 is able to regulate the acetylation state of PGC-1α and the gluconeogenic program of hepatoctyes. Left—when hepatic Sirt6 activity is low, GCN5 is acetylated at K549 and residues Ser307 and Thr735 are unphosphorylated. In this state, GCN5 activity is low. This, coupled with the action of Sirt1, results in low levels of PGC-1α acetylation, high levels of PGC-1α activity, and an activation of gluconeogenic gene expression. Right—when hepatic Sirt6 activity is high, GCN5 is deacetylated at K549 and residues Ser307 and Thr735 are phosphorylated. In this modified state, GCN5 activity is enhanced and exceeds the rate of Sirt1-mediated deacetylation, resulting in high levels of PGC-1α acetylation, reduced levels of PGC-1α activity, and a decrease in gluconeogenic gene expression. See Figures S6,7 and Table S1.
In our investigations with +/db and db/db mice, we noticed that Sirt6 levels were significantly lower in the db/db cohorts both at the level of mRNA (Figure 7H) and total protein (Figure 7I). One possible contributor to Sirt6 repression in db/db mice is hyperglucagonemia and chronic hepatic PKA activation; db/db mice exhibit significantly higher levels of circulating glucagon than genetic controls and an enhanced hepatic catecholaminergic tone (Liang and Cincotta, 2001). Intraperitoneal injections of glucagon into non-diabetic BALB/c mice significantly increased blood glucose levels as well as the expression of PGC-1α, PEPCK, and G-6-Pase (Figure S7A–D). It also caused a significant repression of Sirt6 by a magnitude that was comparable with db/db mice (Figure S7E). Moreover, activation of PKA signaling by either forskolin or glucagon reduced Sirt6 mRNA and protein levels in primary hepatocyte cultures (Figure S7F,G).
These data, in conjunction with the partial rectification of the diabetic phenotype when Sirt6 levels were increased in db/db mice and the effects of Sirt6 knockdown in lean, non-diabetic mice, suggests that hepatic Sirt6 deficiency contributes to a transcriptional and metabolic phenotype that favors gluconeogenesis in the liver and hyperglycemia.
Discussion
To explain the effects of Sirt6 on PGC-1α acetylation, we propose a model (Figure 7J) wherein Sirt6 associates with GCN5 and deacetylates it. This deacetylation is accompanied by changes in GCN5’s phosphorylation state which, collectively, enhance its acetyltransferase activity—though it would appear that the principal driver of the effect of Sirt6 on GCN5 activity is the deacetylation of K549. This causes an increase in the acetylation of PGC-1α and a repression in its ability to function as a transcriptional co-activator. In hepatocytes, this repression causes a decrease in the expression of gluconeogenic genes and hepatic glucose output. In the case of primary hepatocytes, manipulation of Sirt6 expression produced significant changes in the ability of PGC-1α to potentiate gluconeogenic gene expression. Although the absolute levels of gene expression still remained high, the relative effects on metabolism were manifest in glucose production assays as well as in intracellular metabolite analyses, which confirmed that the relative change in gluconeogenic gene expression caused by Sirt6 overexpression/knockdown produced a similar magnitude change in the intracellular concentration of gluconeogenic intermediates.
From an evolutionary perspective, the nutrient-dependent control of protein acetylation through acetyltransferases and deacetylases is highly conserved and is a major mechanism for coupling metabolic activity with carbon/energy availability. The regulated acetylation of PGC-1α by GCN5 and Sirt1 is an excellent example of this: PGC-1α acetylation by GCN5 is favored under conditions of nutrient/energy abundance (Coste et al., 2008; Rodgers et al., 2005) whereas deacetylation by Sirt1 is favored under conditions of nutrient dearth/ and high energy demand (Canto et al., 2009; Rodgers et al., 2005). Our findings add an additional layer of complexity to this regulation: Sirt6 promotes PGC-1α acetylation and suppress its activation of hepatic gluconeogenesis. This raises several questions. Under what conditions does the effect of one sirtuin predominate over the other on PGC-1α? Why should Sirt1 and Sirt6 have opposing effects on PGC-1α acetylation and activity? Moreover, can any generalizations be made about the cooperatively/antagonism of Sirt1 and Sirt6 in hepatic gluconeogenesis? Regarding the first question, a change in the expression of Sirt1 and Sirt6 could alter the balance of PGC-1α acetylation. Indeed, our experiments with primary hepatocytes showed that altering the levels/activities of Sirt1 and Sirt6 can shift the balance of PGC-1α activity. In the animal, we have found conditions which perturb hepatic Sirt6 levels and alter PGC-1α/gluconeogenic activity. First, the diabetic db/db mouse has lower levels of hepatic Sirt6. Second, hepatic Sirt6 levels were suppressed upon PKA axis stimulation. This latter point is intriguing as the cAMP/PKA pathway increases Sirt1 activity (Gerhart-Hines et al., 2011; Nin et al., 2012). The repression of Sirt6 by PKA stimulation may thus represent an additional mechanism whereby the PKA pathway maintains low levels of PGC-1α acetylation and favors the gluconeogenic program in the liver.
As for question two, we are currently pursuing additional research to assess the physiological significance underpinning the opposing regulation of PGC-1α by Sirt1 and Sirt6. We are also investigating the larger physiological role of Sirt6 in the control of whole body glucose metabolism. Previous work has indicated that Sirt6 is able to suppress peripheral glucose utilization through the control of glycolysis (Zhong et al., 2010). Taken together with our findings that Sirt6 represses gluconeogenesis, this would suggest that Sirt6 serves to reduce whole body glucose turnover, a phenomenon that occurs during the early stages of fasting. In conjunction with the work showing that hepatic knockout of Sirt6 also yields higher rates of glycolysis (Kim et al., 2010), our data raise the possibility that loss of Sirt6 in the liver produces a futile cycling of glucose production and breakdown. Indeed, there are some reports indicating that hepatic futile cycling of glucose is significantly elevated in certain diabetic animal models as well as humans with Type II diabetes (Efendic et al., 1988; Kleckner et al., 1987). The significance of this deserves further investigation.
In terms of generalizing the interactions between Sirt1 and Sirt6 in the overall control of hepatic gluconeogenesis, we have observed that Sirt1 can promote PGC-1α-mediated hepatic glucose output whereas Sirt6 can inhibit it. However, others have reported that Sirt1 is able to suppress PGC-1α-independent regulatory elements of gluconeogenesis, such as CRTC2 (Liu et al., 2008), and reduce hepatic glucose output during short-term fasting. This would phenocopy the effects of Sirt6 on gluconeogenesis. Integrating these observations, the effect of each sirtuin on hepatic gluconeogenic rates is going to be dependent upon the duration of fasting and the dominant transcription factor/transcriptional co-activators contributing to the maintenance of gluconeogenesis.
Our experiments with Sirt6 in the livers of mice also suggest that liver-targeted activators of this sirtuin may be useful in controlling insulin-resistant (Type II) diabetes. One of the hallmarks of this disease state is an unrestrained production of glucose in the liver under both pre- and post-prandial conditions, which exacerbates peripheral hyperglycemia. Livers of diabetic db/db were found to contain significantly reduced levels of hepatic Sirt6 and re-expression of Sirt6 in this mouse strain suppressed gluconeogenic genes and improved circulating glucose levels. Even in lean, non-diabetic mice, a reduction in the level of Sirt6 was sufficient to increase gluconeogenic genes and raise blood glucose levels. Interestingly, our data on Sirt6’s effects on glycemia agree with a recent report showing that fasting blood glucose levels were moderately elevated in mice containing a liver-specific knockout of Sirt6—though no mechanism was proposed (Kim et al., 2010).
Finally, the regulatory control of GCN5 by Sirt6 may extend beyond metabolism and have important implications in cellular processes in which the two proteins have been previously and separately implicated, such as the DNA damage response and maintenance of genomic stability (Burgess et al., 2010; Mostoslavsky et al., 2006; Robert et al., 2011). At this point, additional work will be needed to identify the non-histone targets of GCN5 and their effects on cellular physiology.
Experimental Procedures
Animal Experiments
For mouse experiments, 7 week old male BALB/c mice were purchased from Taconic Farms, 8 week old male C57/Bl6 mice were also purchased from Taconic Farms, whereas 8 week old male BKS.Cg-Dock7m +/+ Leprdb/J heterozygous (+/db) and homozygous (db/db) mice were purchased from Jackson Laboratory. See Supplemental Procedures for additional details on the mouse procedures used in this paper. Animal experiments were conducted in accordance with the DFCI’s Institutional Animal Care and Use Committee.
Cell Culture
U-2 OS and HEK 293A cell lines were maintained in DMEM containing 10% FBS. Transfections were performed with Polyfect (Qiagen) with a fixed total quantity of DNA (1.5 μg/well of a 6 well plate).
Mouse primary hepatocytes were prepared from 6–10 week old male BALB/c mice by perfusion with liver digest medium (Invitrogen). See Supplemental Procedures for details on adenoviral infection, glucose production assays, and metabolite analysis of primary hepatocyte cultures.
Hyperinsulinemic-euglycemic clamp studies
Hyperinsulinemic-euglycemic clamps were performed as previously described with minor modifications (Jurczak et al.). Briefly, an indwelling catheter was surgically implanted in the right jugular vein seven days prior to study. Three days after surgery and four days before study, mice were infected with GFP control or Sirt6 WT adenovirus by tail vein injection. Mice were fasted overnight prior to clamps and then infused with a fixed amount of insulin (20 mU/kg/min) and a variable amount of 20% dextrose to maintain euglycemia. 3-[3H] glucose was included in the infusate to allow for the calculation of whole-body rates of glucose metabolism. Hepatic overexpression of Sirt6 was confirmed post-clamp by qRT-PCR.
Construct Generation
FLAG-HA-PGC-1α and FLAG-GCN5 pcDNA constructs were made as previously described (Lerin et al., 2006; Rodgers et al., 2005). HA-tagged human GCN5 and human Sirt6 constructs were amplified from their respective cDNAs and cloned into pcDNA 3.0. Murine 3xFLAG Sirt6 constructs were made as previously described (Zhong et al., 2010). Point mutants of GCN5 were generated by PCR-based mutagenesis.
Lentiviral shRNA constructs were generated in a pLKO.1 backbone (see Table S2 for shRNA hairpin sequences). Mature virus was produced in HEK-293T cells transfected with the pLKO.1 construct, psPAX2 and pMD.2G plasmids.
Adenoviruses were produced with the pAd-Track/pAd-Easy system. FLAG-HA-PGC-1a and FLAG-GCN5 viruses were made as previously described (Lerin et al., 2006; Rodgers et al., 2005). 3×FLAG Sirt6 adenoviruses were subcloned from pCMV7.1–3×FLAG constructs. Adenoviruses containing shRNAs were subcloned from pLKO.1 vectors with the U6 promoter into the pAD-Track/pAd-Easy system.
Immunoprecipitations and Western blot analysis
Immunoprecipitations for detection of PGC-1α acetylation were done with clarified lysates of cells harvested in RIPA buffer with 10 mM nicotinamide and 5 μM trichostatin A. Co-immunoprecipitations were done with the clarified nuclear fractions of cells reconstituted in 10 mM HEPES, pH = 7.5, 150 mM NaCl, 0.5% Igepal, 0.3% CHAPS, and 10% glycerol. See Table S2 for a list of antibodies used for Western blotting.
In vitro acetyltransferase assays
Acetyltransferase assays were conducted with FLAG-tagged GCN5 immunoprecipitated from the nuclear fraction of U-2 OS cells transfected with an empty vector control or Sirt6. Protein was eluted from FLAG antibody beads with 3×FLAG peptide and used in a fluorimetric acetyltransferase assay as per the manufacturer’s instructions (Active Motif).
In vitro deacetylase assays
Sirt6 protein was immunopurified from U-2 OS infected with 3×FLAG Sirt6 adenovirus. Measurement of nicotinamide production over time was performed as previously described (Smith et al., 2009). Assays were conducted with 8 μg Sirt6 protein and 500 μM acetylated GCN5 peptide (FDPKH[Acetyl-K]TLALIK) in the presence or absence of 1 mM NAD+. For mass spectrometry experiments, 2 μg of Sirt6 and 15 μM acetylated K549 GCN5 peptide was used.
Gene Expression Analysis
Total RNA was isolated from cells or pulverized liver using Trizol (Invitrogen). For Q-RT-PCR analysis, cDNA was synthesized from 2 μg RNA using random primers and High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Gene expression analysis was performed using a CFX384 Real-Time system (Bio-Rad) and Power Sybr Green qPCR mastermix (Applied Biosystems). All gene expression data were corrected by β-2 microglobulin and 36B4 control gene expression and normalized using the Δ ΔCT method. All primers are available upon request.
Mass Spectrometry
In-gel protein digests were analyzed by microcapillary liquid chromatography electrospray ionization tandem mass spectrometry (LC-MS/MS). Analyses were done on a LTQ Orbitrap mass spectrometer (Thermo Fisher Scientific, Germany) equipped with a Thermo Fisher Scientific nanospray source, an Agilent 1100 Series binary HPLC pump, and a Famos autosampler. For additional details, see Supplemental Procedures.
Statistics
All data are presented as means +/− standard error of the mean. t-tests and one-way analysis of variance (ANOVA) tests were conducted—along with corresponding post tests—as indicated; p<0.05 was considered significant. *-p<0.05, **-p<0.01, ***-p<0.001.
Supplementary Material
Acknowledgments
We thank members of the Puigserver lab for important discussions about this project. J.E.D. was supported in part by an NRSA Kirschstein fellowship from the National Institutes of Health. Research was supported with funds from the DFCI and with grants from the American Diabetes Association, Department of Defense and NIH/NIDDK (RO1 069966) awarded to P.P. as well as with a grant from NIH (DK059635) awarded to Yale’s Mouse Metabolic Phenotyping Center/G.I.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Burgess RJ, Zhou H, Han J, Zhang Z. A role for Gcn5 in replication-coupled nucleosome assembly. Mol Cell. 2010;37:469–480. doi: 10.1016/j.molcel.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coste A, Louet JF, Lagouge M, Lerin C, Antal MC, Meziane H, Schoonjans K, Puigserver P, O’Malley BW, Auwerx J. The genetic ablation of SRC-3 protects against obesity and improves insulin sensitivity by reducing the acetylation of PGC-1{alpha} Proc Natl Acad Sci U S A. 2008;105:17187–17192. doi: 10.1073/pnas.0808207105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efendic S, Karlander S, Vranic M. Mild type II diabetes markedly increases glucose cycling in the postabsorptive state and during glucose infusion irrespective of obesity. J Clin Invest. 1988;81:1953–1961. doi: 10.1172/JCI113543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhart-Hines Z, Dominy JE, Jr, Blattler SM, Jedrychowski MP, Banks AS, Lim JH, Chim H, Gygi SP, Puigserver P. The cAMP/PKA pathway rapidly activates SIRT1 to promote fatty acid oxidation independently of changes in NAD(+) Mol Cell. 2011;44:851–863. doi: 10.1016/j.molcel.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, Alt FW, Wu Z, Puigserver P. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J. 2007;26:1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handschin C, Rhee J, Lin J, Tarr PT, Spiegelman BM. An autoregulatory loop controls peroxisome proliferator-activated receptor gamma coactivator 1alpha expression in muscle. Proc Natl Acad Sci U S A. 2003;100:7111–7116. doi: 10.1073/pnas.1232352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurczak MJ, Lee AH, Jornayvaz FR, Lee HY, Birkenfeld AL, Guigni BA, Kahn M, Samuel VT, Glimcher LH, Shulman GI. Dissociation of inositol-requiring enzyme (IRE1alpha)-mediated c-Jun N-terminal kinase activation from hepatic insulin resistance in conditional X-box-binding protein-1 (XBP1) knock-out mice. J Biol Chem. 287:2558–2567. doi: 10.1074/jbc.M111.316760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Xiao C, Wang RH, Lahusen T, Xu X, Vassilopoulos A, Vazquez-Ortiz G, Jeong WI, Park O, Ki SH, et al. Hepatic-specific disruption of SIRT6 in mice results in fatty liver formation due to enhanced glycolysis and triglyceride synthesis. Cell Metab. 2010;12:224–236. doi: 10.1016/j.cmet.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner NW, Kizaki Z, Thurman RG. Potential intercellular futile cycling of carbohydrates in diabetes. Biochem J. 1987;246:417–423. doi: 10.1042/bj2460417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo SH, Satoh H, Herzig S, Lee CH, Hedrick S, Kulkarni R, Evans RM, Olefsky J, Montminy M. PGC-1 promotes insulin resistance in liver through PPAR-alpha-dependent induction of TRB-3. Nat Med. 2004;10:530–534. doi: 10.1038/nm1044. [DOI] [PubMed] [Google Scholar]
- Lerin C, Rodgers JT, Kalume DE, Kim SH, Pandey A, Puigserver P. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC- 1alpha. Cell Metab. 2006;3:429–438. doi: 10.1016/j.cmet.2006.04.013. [DOI] [PubMed] [Google Scholar]
- Li X, Monks B, Ge Q, Birnbaum MJ. Akt/PKB regulates hepatic metabolism by directly inhibiting PGC-1alpha transcription coactivator. Nature. 2007;447:1012–1016. doi: 10.1038/nature05861. [DOI] [PubMed] [Google Scholar]
- Liang Y, Cincotta AH. Increased responsiveness to the hyperglycemic, hyperglucagonemic and hyperinsulinemic effects of circulating norepinephrine in ob/ob mice. Int J Obes Relat Metab Disord. 2001;25:698–704. doi: 10.1038/sj.ijo.0801614. [DOI] [PubMed] [Google Scholar]
- Liu Y, Dentin R, Chen D, Hedrick S, Ravnskjaer K, Schenk S, Milne J, Meyers DJ, Cole P, Yates J, 3rd, et al. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature. 2008;456:269–273. doi: 10.1038/nature07349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson I, Rothman DL, Katz LD, Shulman RG, Shulman GI. Increased rate of gluconeogenesis in type II diabetes mellitus. A 13C nuclear magnetic resonance study. J Clin Invest. 1992;90:1323–1327. doi: 10.1172/JCI115997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- Nemoto S, Fergusson MM, Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha} J Biol Chem. 2005;280:16456–16460. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- Nin V, Escande C, Chini CC, Giri S, Camacho-Pereira J, Matalonga J, Lou Z, Chini EN. Role of deleted in breast cancer 1 (DBC1) in SIRT1 activation induced by protein kinase A and AMP activated protein kinase. J Biol Chem. 2012 doi: 10.1074/jbc.M112.365874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson BL, Hock MB, Ekholm-Reed S, Wohlschlegel JA, Dev KK, Kralli A, Reed SI. SCFCdc4 acts antagonistically to the PGC-1alpha transcriptional coactivator by targeting it for ubiquitin-mediated proteolysis. Genes Dev. 2008;22:252–264. doi: 10.1101/gad.1624208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver P, Rhee J, Donovan J, Walkey CJ, Yoon JC, Oriente F, Kitamura Y, Altomonte J, Dong H, Accili D, et al. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature. 2003;423:550–555. doi: 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
- Rhee J, Inoue Y, Yoon JC, Puigserver P, Fan M, Gonzalez FJ, Spiegelman BM. Regulation of hepatic fasting response by PPARgamma coactivator-1alpha (PGC-1): requirement for hepatocyte nuclear factor 4alpha in gluconeogenesis. Proc Natl Acad Sci U S A. 2003;100:4012–4017. doi: 10.1073/pnas.0730870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert T, Vanoli F, Chiolo I, Shubassi G, Bernstein KA, Rothstein R, Botrugno OA, Parazzoli D, Oldani A, Minucci S, et al. HDACs link the DNA damage response, processing of double-strand breaks and autophagy. Nature. 2011;471:74–79. doi: 10.1038/nature09803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JT, Haas W, Gygi SP, Puigserver P. Cdc2-like kinase 2 is an insulin-regulated suppressor of hepatic gluconeogenesis. Cell Metab. 2010;11:23–34. doi: 10.1016/j.cmet.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- Rodgers JT, Puigserver P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc Natl Acad Sci U S A. 2007;104:12861–12866. doi: 10.1073/pnas.0702509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetz A, Bernstein G, Dong A, Antoshenko T, Wu H, Loppnau P, Bochkarev A, Plotnikov AN. Crystal structure of a binary complex between human GCN5 histone acetyltransferase domain and acetyl coenzyme A. Proteins. 2007;68:403–407. doi: 10.1002/prot.21407. [DOI] [PubMed] [Google Scholar]
- Shayakhmetov DM, Li ZY, Ni S, Lieber A. Analysis of adenovirus sequestration in the liver, transduction of hepatic cells, and innate toxicity after injection of fiber-modified vectors. J Virol. 2004;78:5368–5381. doi: 10.1128/JVI.78.10.5368-5381.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BC, Hallows WC, Denu JM. A continuous microplate assay for sirtuins and nicotinamide-producing enzymes. Anal Biochem. 2009;394:101–109. doi: 10.1016/j.ab.2009.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teyssier C, Ma H, Emter R, Kralli A, Stallcup MR. Activation of nuclear receptor coactivator PGC-1alpha by arginine methylation. Genes Dev. 2005;19:1466–1473. doi: 10.1101/gad.1295005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JC, Puigserver P, Chen G, Donovan J, Wu Z, Rhee J, Adelmant G, Stafford J, Kahn CR, Granner DK, et al. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001;413:131–138. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- Zhong L, D’Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, Guimaraes A, Marinelli B, Wikstrom JD, Nir T, et al. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell. 2010;140:280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.