Abstract
Although widely distributed in Nature, only two γ class carbonic anhydrases are reported besides the founding member (Cam). Although roles for active-site residues important for catalysis have been identified in Cam, second shell residues have not been investigated. Two residues (Trp19 and Tyr200), positioned distant from the catalytic metal, were investigated by structural and kinetic analyses of replacement variants. Steady-state kcat/Km and kcat values decreased 3- to 10-fold for the Trp19 variants whereas the Y200 variants showed up to a 5-fold increase in kcat. Rate constants for proton transfer decreased up to 10-fold for the Trp19 variants, and an increase of ~2-fold for Y200F. The pKa values for the proton donor decreased 1–2 pH units for Trp19 and Y200 variants. The variant structures revealed a loop composed of residues 62–64 that occupies a different conformation than previously reported. The results show that, although Trp19 and Y200 are non-essential, they contribute to an extended active-site structure distant from the catalytic metal that fine tunes catalysis. Trp19 is important for both CO2/bicarbonate interconversion, and the proton transfer step of catalysis.
Introduction
Carbonic anhydrases (CAs)1 are metalloenzymes that catalyze the reversible hydration of CO2 (CO2 + H2O = HCO3− + H+) and are found in all three domains of life (Eukarya, Bacteria and Archaea) underscoring the fundamental importance of this enzyme. Five independently evolved classes (α, β, γ, δ, ζ) of CA have been reported, among which there is little or no significant sequence or structural identity [1]. Despite this divergence, the kinetic mechanism is similar for those classes which have been investigated. The overall reaction occurs in two mechanistically distinct half-reactions (Eq. 1 and 2) where M refers to metal, E refers to enzyme and B refers to buffer.
| (Eq. 1a) |
| (Eq. 1b) |
| (Eq. 2a) |
| (Eq. 2b) |
The first half-reaction is a nucleophilic attack of metal-bound hydroxide on CO2 yielding bicarbonate which is reflected in the steady-state parameter kcat/Km. The second half-reaction is the rate-determining proton transfer from metal-bound water to buffer reflected in the steady-state parameter kcat. The proton extracted from the metal-bound water is transferred to buffer acting as the second substrate in a ping-pong mechanism. Remarkably, only four CA's from the domain Archaea have been isolated and characterized, one from the β class [2] and three γ class enzymes [3–5]. Cam from Methanosarcina thermophila is the founding member of the γ class. Cam belongs to a superfamily of proteins, comprised mainly of acyltransferases, that share a distinctive left-handed parallel β-helix fold predicted by a unique sequence motif [6, 7]. Cam overproduced in Escherichia coli and purified aerobically contains zinc in the active site [8]. However, overproduction in the closely related species Methanosarcina acetivorans yields an enzyme with 3-fold greater CA activity and containing iron in the active site, a result establishing iron as the physiologically relevant metal for Cam or any CA [9]. Kinetic analyses of Cam variants have identified Gln75, Asn73 and Asn202 involved in a hydrogen bond network required for the first half-reaction (Eq.1), and Glu62 and Glu84 required for the second half-reaction (Eq. 2) [10].
In addition to the domain Archaea, sequence-based data searches have revealed Cam homologs in plants and green algae from the domain Eukarya, and proteobacteria and cyanobacteria from the domain Bacteria [3, 11]. The three metal-coordinating His residues present in Cam appear to occur in nearly all of these sequences, indicating metal-binding competence. Alignment of these sequences suggests that there are two distinct sub-classes of the γ class represented by Cam and CamH, based on the presence (Cam) or absence (CamH) of an acidic loop that lies directly adjacent to the active site [3]. Interestingly, previous biochemical studies suggest that this acidic loop in Cam and Glu62 (not present in the CamH sub-class) are important for catalytic activity [12]. The lack of these two features in the CamH sub-class may indicate a different function, although two enzymes from this sub-class have been shown to have CA activity [3, 5].
The current biochemical understanding of the catalytic mechanism of CA's is based largely on α and β class enzymes with sequence identity and active-sites widely divergent from the independently evolved γ class. Although database sequences indicate γ class Cam and CamH homologs are widely distributed in diverse species across all three taxonomic domains of life, the biochemical mechanism of catalysis has only been investigated with Cam. Several active-site residues in Cam have been identified that are important for catalysis and proton transfer [10, 13, 14]. Although a role in catalysis and proton transfer for residues distant from active-site metal of the αCA is well documented, this level of understanding is incomplete for the γ class. Herein we report an investigation of Trp19 and Tyr200 located distant from the active-site metal of Cam. Trp19 in Cam is positioned adjacent to the proton shuttle residue Glu84 [15] and the side chain of Tyr200 is directed towards the active-site cavity and apparently interacting with solvent (Fig. 1). Based on structural and kinetic analyses of replacement variants, we conclude that these residues constitute an extended active-site structure important for fine tuning catalysis and integrity of conformation near the surface of the enzyme.
Figure 1.
Comparison of active-site structures of (top) Cam and (bottom) HCA II showing the positions of Tyr and Trp residues in the active site cavity. Residues are labeled and shown as stick models with the zinc atom depicted as a gray sphere. The figure was made using PyMOL (DeLano Scientific).
Materials and methods
Enzymes
Site-directed mutagenesis of the Cam gene-containing plasmid as described previously [10] was performed to generate replacement variants at residues 19 and 200. The variants were over-expressed in E. coli and purified as previously described [10]. In brief, the cells were harvested by centrifugation and the pellet suspended in 50 mM MOPS (pH 7.5) supplemented with 0.5 mM zinc sulfate. The cell suspension was passed twice through a French pressure cell at 6.9 × 103 kPa. DNase I and RNase A were added after the first pass to reduce lysate viscosity. The lysate was clarified by centrifugation at 20,000 × g and the protein was purified as follows. First, the supernatant was applied to a Q-Sepharose column (GE Healthcare) and the protein was eluted by a linear gradient of 0.0 to 1.0 M NaCl with elution occurring at about 0.5 M NaCl. Further purification was accomplished by adding ammonium sulfate to the fractions (1.5 M final) and applying samples to a Phenyl-Sepharose column (GE Healthcare) equilibrated with 100 mM MOPS (pH 7.5) and 1.5 M ammonium sulfate. The protein was eluted with a 1.5 to 0.0 M decreasing ammonium sulfate gradient in a peak centered at 0.75 M. Protein purity was assessed by SDS-PAGE analysis and the protein concentration was determined by the Bradford assay with bovine serum albumin as the standard.
Crystallization and Data Collection
All variants were crystallized using the hanging-drop vapor diffusion technique with a reservoir solution of 5% PEG 8000 and 250 mM ammonium sulfate. After sitting overnight, a heavy amount of precipitation was observed. Despite this, crystals were observed after approximately 4 weeks, with a size of ~1 mm3. Two of the variants, Y200S and Y200F crystallized, however there was no observed diffraction, despite multiple attempts.
Diffraction data were collected in-house at room temperature using an R-AXIS IV++ image plate system with Osmic mirrors and a Rigaku RU-H3R Cu rotating anode operating at 50 kV and 100 mA as described previously [16]. The crystal-detector distance was set at 100 mm with an oscillation angle of 1° per image. A total of 90° of data were collected for each crystal. The data were then indexed, integrated, and scaled using HKL2000 [17]. All four variants (W19A, W19F, W19N, and Y200A Cam) were solved to comparable resolution (1.6–1.8 Å) in the space group P 213 with isomorphous unit cells (a ~83.5 Å). The Rsym for each variant were: 4.6% for W19A, 6.4% for W19F, 3.4% for W19N, and 5.7% for Y200A (Table 1). The high symmetry of the unit cell allowed for nearly 100% completeness in all cases with high redundancy, though only ~50 images were used for each structure, due to radiation decay. Complete data processing statistics are presented in Table 1.
Table 1.
Data processing and refinement statistics for the structures of the Cam mutants.
| W19A | W19F | W19N | Y200A | |
|---|---|---|---|---|
| PDB accession | 3otz | 3ou9 | 3oup | 3ow5 |
| Space group | P 213 | P 213 | P 213 | P 213 |
| Cell dimensions (a) (Å) | 83.49 | 83.62 | 83.58 | 83.50 |
| Resolution (Å) | 20–1.6 | 20–1.8 | 50–1.65 | 25–1.8 |
| (1.66–1.6) a | (1.86–1.8) | (1.71–1.65) | (1.86–1.8) | |
| Rsym b (%) | 4.6 (46.0) | 6.4 (48.4) | 3.4 (24.9) | 5.7 (34.9) |
| I/(σ)I | 34.2 (3.6) | 21.8 (2.8) | 47.0 (4.7) | 26.5 (4.3) |
| Completeness | 99.8 (100.0) | 99.9 (99.9) | 99.6 (99.1) | 100.0 (100.0) |
| Redundancy | 6.8 (6.5) | 5.6 (5.4) | 7.2 (3.5) | 6.5 (6.2) |
| Rfactor c/Rfree d (%) | 14.0/15.5 | 13.5 /16.1 | 13.7 /16.8 | 12.9 /15.2 |
| No. atoms | ||||
| Protein | 1614 | 1603 | 1617 | 1562 |
| Water | 52 | 49 | 63 | 67 |
| B-factors | ||||
| Protein (main/side) | 23.6/32.3 | 25.5/33.7 | 24.3 /32.2 | 20.2/28.4 |
| Water | 35.1 | 29.2 | 35.6 | 32.7 |
| RMSD (bond/angle) | 0.012/1.4 | 0.013/1.4 | 0.013/1.5 | 0.019/1.4 |
| Ramachandran plot (%) | ||||
| Allowed | 89.5 | 90.7 | 90.1 | 90.7 |
| Additionally allowed | 9.9 | 8.7 | 9.3 | 8.7 |
| Disallowed e | 0.6 | 0.6 | 0.6 | 0.6 |
Values in parentheses are for the highest resolution shell.
Rsym = (Σ|I − <I>|/ Σ<I>) × 100.
Rcryst = (Σ‖Fo| − |Fc‖/ Σ|Fo|) × 100.
Rfree is calculated the same as Rcryst, except with 5% of the data omitted from refinement.
The orientation of Met64 is disallowed, however the electron density for this residue clearly shows its position.
All structures were refined using the program Phenix [18]. Structures were refined until the Rfactor reached convergence, with a final Rfactor and Rfree of approximately 13 and 17%, respectively for all four structures. One residue, Met65 (chain A), refined to a position with unfavorable φ and ψ angles. However, as the electron density was well defined for this residueit was left in this strained conformation. Final refinement statistics are given in Table 1.
Steady-state kinetics
Carbon dioxide hydration activity was assayed by the pH indicator method [19] with a model SF-2001 KinTek spectrophotometer (KinTek Corp., Austin, TX). Enzyme monomer concentrations ranged from 400 nM to 1 µM. Buffer-indicator dye pairs used were MES and chlorophenol red (at pH 5.7–6.9) measured at a wavelength of 574 nm, MOPS or imidazole and 4-nitrophenol (at pH 6.5–7.7) measured at a wavelength of 400 nm, and TAPS and m-cresol purple (at pH 7.7–9.1) measured at a wavelength of 578 nm. Buffer concentrations were 50 mM, the total ionic strength was adjusted to 50 mM with Na2SO4, and final pH indicator concentrations were 50 µM. Saturated solutions of CO2 (32.9 mM in H2O) were prepared by bubbling CO2 gas into deionized water at 25°C. The final CO2 concentrations after mixing were varied from 7.9 to 24.7 mM. The initial linear regions, 5–10% of the total absorbance changes, were used to calculate initial velocities for kinetic analysis, using the average of 10–15 reaction traces per experiment. The initial rate data were fit to the Michaelis-Menten equation to obtain experimental values for kcat and Km.
Oxygen-18 exchange
This method is based on the measurement by mass spectrometry of the exchange of 18O between CO2 and water catalyzed by carbonic anhydrase [20]. The method is covered in detail previously [20–22] and described briefly here. Using an Extrel EXM-200 mass spectrometer and a membrane inlet probe, we determine two rate constants for the catalysis observed at chemical equilibrium. The first is kcatex /KeffCO2 which is equivalent in theory to kcat/Km for hydration of CO2. Here, kcatex is the maximal rate of interconversion of CO2 and bicarbonate at chemical equilibrium, and KeffCO2 is an effective binding constant of substrate to all species of enzyme (20, 22). The second rate constant is RH2O/[E] which is a rate constant for the dissociation of 18O–labeled water from the active site. This constant is determined in rate by proton transfer to the zinc-bound hydroxide; its bell shaped pH profile allows estimates of the rate constant for proton transfer as well as the pKa of the proton donor and the zinc-bound solvent molecule [20, 23].
Results
Crystallography
Overall, the structures of four variants (Y200A, W19A, W19F, W19N) were very similar to that of wild-type Cam (PDB code: 1QRG, [15]). The residues of the active-site region in each of these variants are closely superimposable with wild-type, as shown for Y200A Cam and W19F Cam in Figures 2 and 3. This includes the dual conformations observed for the side-chains of Glu62 (Figs. 2 and 3); the electron densities for Glu84 were relatively weak. The largest difference between the structures of the variants and wild-type was the dual conformation of a loop composed of resides Glu62, Gly63, and Met64 (Figs. 4–6). This conformation was observed in three of the four variants (not the Y200A) and has not been observed in any previously determined Cam structure currently available in the PDB. To ensure a lack of model bias in the electron density maps, the loop was removed prior to refinement. The FO-FC density map clearly shows the position of the loop, indicating that this is the correct conformation despite the unfavorable nature of the φ and ψ angles of Met65 (Fig. 5).
Figure 2.
The superposition of amino acids in the active site regions of (yellow) Y200A Cam; and (pink) wild-type Cam. Residues are labeled and shown as stick models with the zinc atom depicted as a gray sphere. This figure was generated and rendered with Pymol (Delano Scientific).
Figure 3.
The superposition of amino acids in the active site regions of (yellow) W19F Cam; and (pink) wild-type Cam. Residues are labeled and shown as stick models with the zinc atom depicted as a gray sphere. This figure was generated and rendered with Pymol (Delano Scientific).
Figure 4.
The structure of a Cam monomer shows a left-handed β-helix topology with several N-terminal (blue) surface loops and a C-terminal (red) α-helix. The acidic loop that differentiates the Cam and CamH classes is identified in green and the loop 63-64-65 loop is indicated by a red arrowhead.
Figure 6.
The electron density for the 62-63-64 loop clearly defines its orientation in the structure of W19N Cam. Shown as blue mesh is FO-FC (3σ contour) electron density calculated with the loop removed. The side chains are built into this density (green sticks) and are in a novel conformation as compared to wild-type (magenta backbone) (PDB ID 1qrg, [15]). Also shown is Pro65, a residue which remains more or less unchanged between the mutant and wild-type structures.
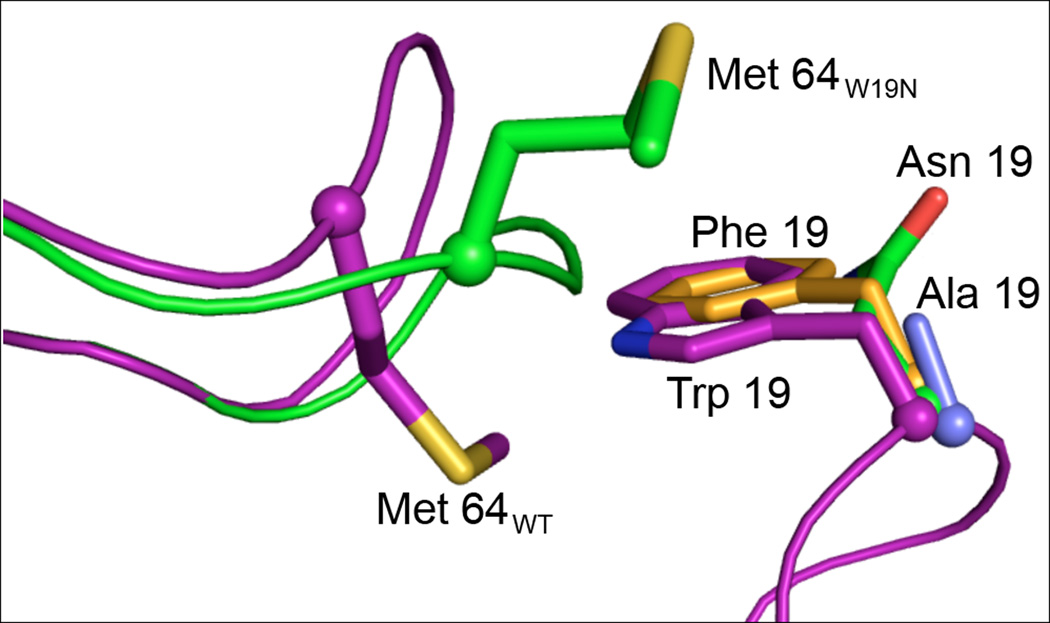
Figure 5.
The 63-64-65 loop in the mutants of Cam occupies a different conformation than that observed in all reported Cam structures. The orientation of the side chain of Met64 is clearly different in the two structures (magenta = wild-type, green = mutants) with a maximal distance of 11 Å between identical atoms. The loop of W19N is shown here, however the conformation of the loop is the same for all mutants, with W19F displaying both conformations. Side chains for the W19 mutants are also shown here.
There was a large electron density peak observed adjacent to the zinc ion in the electron density maps for all four variants, indicating an electron rich entity was coordinated to the metal. Two previous structures of Zn-containing and Co-containing Cam have been solved with a sulfate molecule in this exact position (3). In the four variant structures reported here, this density was modeled as a water in W19N and W19A, a bicarbonate in W19F, and with a dual occupancy of both a bicarbonate and sulfate in Y200A. These were modeled so that there were no large, negative FO-FC density peaks. Additionally a metal was found at the 3-fold symmetry axis in Y200A Cam. This metal was modeled as an iron, which left no residual FO-FC electron density (negative or positive). The iron is coordinated by three methionines, Met55, one from each of the three monomers. It should be noted that the analysis of this density is convoluted due to its overlap with the 3-fold crystallographic symmetry axis.
Catalysis
The steady-state parameter kcat/Km for the Trp19 and Tyr200 variants (Table 2) indicated that these two residues are not essential for the CO2 hydration step, which is expected since the side chains are 10 to 12 Å from the catalytic metal. The constant kcat/Km for hydration of CO2 for each variant at pH 8.8, near the expected maximum, was within 3-fold of the wild-type value with the exception of W19F in which kcat/Km was 5-fold smaller (Table 2). The values of the turnover number kcat were decreased by about 3- to 6-fold for the Trp19 variants, and enhanced up to 5-fold for the Tyr200 variants (Table 2). The results indicate that, although not essential, Trp19 is important for optimal activity of Cam.
Table 2.
Steady state constants for the catalysis of the hydration of CO2 catalyzed by Cam variants.
| pH 7.5 | pH 8.8 | |||
|---|---|---|---|---|
| Variant |
kcat (ms−1) |
kcat/Km (µM−1 s−1) |
kcat (ms−1) |
kcat/Km (µM−1 s−1) |
| Wild-type Zn-Cam |
32.4 ± 2.2 | 2.0 ± 0.4 | 29.4 ± 1.7 | 3.1 ± 0.6 |
| W19A | 7.9 ± 0.2 | 0.79 ± 0.06 | 8.5 ± 0.2 | 1.4 ± 0.2 |
| W19N | 5.8 ± 0.1 | 0.63 ± 0.02 | 5.2 ± 0.2 | 1.3 ± 0.3 |
| W19F | 6.5 ± 0.1 | 0.40 ± 0.02 | 9.5 ± 0.2 | 0.60 ± 0.02 |
| Y200A | 95.0 ± 3.9 | 7.0 ± 0.9 | 88.5 ± 2.7 | 10.3 ± 1.2 |
| Y200S | 79.3 ± 5.0 | 5.9 ± 1.1 | 68.8 ± 8.5 | 10.2 ± 0.5 |
| Y200F | 120.1 ± 2.7 | 7.1 ± 0.5 | 153.6 ± 5.8 | 7.9 ± 0.8 |
Catalysis was also determined by the exchange of 18O between CO2 and water at chemical equilibrium as measured by mass spectrometry [20]. The maximal values of kcatex /KeffCO2 for selected variants (Figs. 7 and 8) (Table 3) were in reasonable agreement with kcat/Km measured at steady-state (Table 2). As described previously for wild-type Cam [24], the pH profiles of kcatex /KeffCO2 for the variants were dependent on two ionizations with values of pKa near 5.7 and 8.1, the same values observed for wild-type (Figs. 7 and 8) (Table 3, column 3). The smaller pKa has been assigned to protolysis of the zinc-bound water and the larger value to a second, perturbing ionization influencing the first [24].
Figure 7.
(top) The pH profile for kcatex /KeffCO2; and (bottom) the pH profile for RH2O/[E] measured for (●) Y200A Cam; (Δ) Y200F Cam; and (■) wild-type Cam by 18O exchange at 25 °C. The total concentration of all species of CO2 was 25 mM and the ionic strength of solution was maintained at 0.2 M by addition of Na2SO4. No buffers were used. The solid lines for kcatex /KeffCO2 are least squares fits to two ionization, and solid lines for RH2O/[E] are fits to eq 6 of [21] with parameters given in Table 3.
Figure 8.
(top) The pH profile for kcatex /KeffS; and (bottom) the pH profile for RH2O/[E] measured for (●) W19F Cam; (◊) W19N Cam; and (■) wild-type Cam by 18O exchange at 25 °C. The total concentration of all species of CO2 was 25 mM and the ionic strength of solution was maintained at 0.2 M by addition of Na2SO4. No buffers were used. The solid lines for kcatex /KeffCO2 are least squares fits to two ionization, and solid lines for RH2O/[E] are fits to eq 6 of [21] with parameters given in Table 3.
Table 3.
Maximal values of rate constants for the hydration of CO2 and dehydration of bicarbonate catalyzed by variants of cam, and related kinetic pKa values.a
| Variant | kcatexch/KeffCO2 (µM−1 s−1) |
pKa b | kB (ms−1) |
pKZnH2O c | pK donor c |
|---|---|---|---|---|---|
| Wild-type | 14 ± 1 | 5.7, 8.1 | 65 ± 7 | 6.0 | 8.6 |
| W19N | 1.2 ± 0.1 | 5.3, 8.4 | 4.9 ± 0.5 | 5.7 | 7.8 |
| W19F | 0.9 ± 0.1 | 5.7, 8.2 | 13 ± 4 | 6.0 | 7.2 |
| Y200A | 4.0 ± 0.4 | 5.2, 7.8 | 30 ± 3 | 5.7 | 8.3 |
| Y200F | 3.7 ± 0.4 | 5.6, 8.1 | 150 ± 60 | 6.5 | 6.8 |
The standard errors are 0.3 pK units or less. These data were obtained from values of kcatexch/KeffCO2 using a fit to the following: (k/K)obs = (k/K)1/(1 + 10pK1-pH) + (k/K)2/(1 + 10pK2-pH).
The standard errors are 0.3 pK units or less. These data were obtained from values of the rate constant for release of 18O-labeled water from the enzyme RH2O/[E] using a fit to the following: RH2O/[E] = kB / ([1 + (Ka)His64 /[H+]][1 + [H+]/(Ka)ZnH2O]).
The 18O exchange method was used to measure the pH profiles for RH2O/[E], a rate constant that is dependent on proton transfer. These pH profiles (Figs. 7 and 8) gave estimates of rate constants for proton transfer in the dehydration direction (kB), showing a decrease as much as 10-fold for the Trp19 variants, and an increase of about 2-fold for Y200F compared with wild-type (Table 3). Additional data from the 18O method are estimates of the values of pKa for proton acceptor and donor (Table 3, columns 5 and 6). The value of pKa near 6 was assigned to the zinc-bound water based on the evidence presented earlier [24]. The second ionization, that of the proton donor, appeared decreased for each of the variants of Table 3. This suggests that the replacements made of Trp19 and Tyr 200 have lowered the pKa of the proton donor group from 8.6 for wild-type to as low as 6.8 (Table 3). This is consistent with the location of these replacements far from the zinc but closer to the proton shuttle residue Glu84 (Fig. 2). Prior studies suggest that the proton donor group in catalysis is Glu84 [12]. The high pKa of this proton transfer group is possibly a result of the concentration of negative charge in the active-site cavity due to the side chains of Glu82 and Glu84. The assignment of this high pKa is discussed by Alber et al. (24).
Discussion
The role of Trp19 and Tyr200 adjacent to active-site residues in Cam, the model γ class CA, were investigated to determine roles in catalysis. The results support that these residues constitute an extended active-site structure important for fine tuning catalysis and integrity of conformation near the surface of the enzyme.
Cam is from the domain Archaea that contains iron in the active and bears no sequence identity or overall structure to the independently-evolved zinc-containing α class HCA II from humans of the domain Eukarya. This striking divergence extends to the active-sites where catalytically important residues other than ligands to the metals are non-identical. Thus, it is remarkable that Trp19 and Tyr200 in Cam occupy locations distant from the active-site metal similar to Trp5 and Tyr7 in the independently-evolved α class HCA II (Fig. 1). This exception to the active-sites of Cam and HCA II prompts a discussion of these residues in Cam versus HCA II. In this regard it is intriguing that the replacement of Tyr200 with Phe in Cam resulted in more rapid intramolecular proton transfer, both kcat measured in the hydration direction (Table 2) and kB measured in dehydration (Table 3). By examination of crystal structures and by analogy with results for Y7F HCA II [25], we suggest enhancement is due to solvation effects in the active-site cavity. Replacement of Tyr7 with Phe in HCA II results in a 7-fold enhancement of kB for dehydration but not kcat for hydration [26, 27]; this latter result may indicate that another step, such as product dissociation, is limiting kcat for the Y7F variant of HCA II compared to Cam.
The source of enhanced proton transfer in the Y7F variant of HCA II is suggested to be associated with the structure of the ordered water in the active-site cavity of Y7F being less branched than in wild-type, with the background that proton transfer through un-branched, hydrogen-bonded water structures is expected to be more rapid than in branched structures [28, 29]. This is a feature on which we cannot comment for the structure of the Y200F variant of Cam since it has a dual occupancy of both a bicarbonate and sulfate bound at the zinc atom and the water structure is partially displaced. We were unable to crystallize the enzyme without this anion bound.
The neutron diffraction structure of HCA II crystallized at pH 9.0 shows that the side chain of Tyr7 is a phenolate anion [30]. This provides another possible explanation for the enhanced catalytic activity of these variants with the active-site Tyr replaced, since deletion of such an anion would be expected to have major electrostatic effects on catalysis. Such an explanation presumes that the side chain of Tyr200 in Cam is also a phenolate anion under the conditions of these experiments.
The steady-state kinetic data (Table 2) show that replacement of Trp19 in Cam is associated with decreases in both kcat/Km and kcat of 3- to 6-fold, and for some variants were closer to 10-fold less with the 18O exchange method of analysis (Table 3). The observation that these decreases affect kcat/Km as well as kcat suggests influences on catalysis for CO2/bicarbonate interconversion, as well as proton transfer. These results are of further interest since the value of the pKa for the zinc-bound water appears mostly unchanged in these variants. Changes in kcat/Km could be in steps such as bicarbonate departure from the enzyme. In the absence of distinct structural changes, these effects could be due to non-specific influences such as electrostatics and effects on hydrophobicity and dielectric properties of the active-site cavity. Additionally, the observation of a dual conformation of the methionine loop suggests that replacements at Trp19 and Tyr200 of Cam increase mobility of the active site. This is expected, as replacement of these residues relieves the protein of having to bury a large hydrophobic side chain. Regardless, we can conclude that both Trp19 and Tyr200 serve to fine tune catalysis, It is pertinent to comment on why nature has not evolved Cam with Phe at residue 200, since the both the values of kcat and kcat/Km are enhanced with Phe at this position (Table 2). The reason probably lies in effects not measured in this study, such as thermal stability and folding. Also, residues Trp19 and Tyr200 may interact with other components of the cell unique to M. thermophila. As an example, a series of His residues at the amino terminus of HCA II enhance binding of this enzyme to an anion exchange protein [32].
The structures of the four variants of Cam (W19A, W19F, W19N, Y200F) revealed a previously unseen structural feature (Figs. 4 and 6). The loop composed of residues 62–65 occupies a distinctly different conformation(s) than that reported in previous Cam structures. In this conformation the side chain of Met64 is shifted ~10 Å as measured at the Sδ atoms. In wild-type Cam the Met64 side chain buries itself into the core of the protein. The observed rotation of the loop seen in the variant Cam structures results in the side chain of Met64 pointing out towards the surface of the protein, almost completely solvent exposed (Figs. 5 and 6). The Sδ atom of Met64 in wild-type is 12 Å from the zinc atom in the active site, so it is unlikely that this conformational change has significant effects on the catalysis of the interconversion of CO2 and bicarbonate at zinc. Met64 is 9 Å from the side chain of the proton shuttle Glu84, and is also unlikely to have a significant effect on proton transfer. The conformation of the side chain of Glu62 is not altered by these replacements. Glu62 plays a significant role in catalysis and its replacement results in large decreases in the steady-state constants of catalysis [12].
Conclusions
Residues within and adjacent to the active site of γ class CAs bear little resemblance to the well-studied α class, a consequence of independent evolution. Although roles in catalysis are well documented for residues distant from the active-sites of enzymes, this level of understanding for the γ class of CAs is incomplete. The results presented here identify two residues which contribute to an extended active-site structure that fine tunes catalysis and influences the conformation of Cam, the model γ class CA. Although the same residues are adjacent to the active-site of the independently-evolved HCA II, further research with both enzymes is necessary to draw firm conclusions regarding similar or dissimilar functions. Finally, the results presented for the Cam extend an understanding of the γ class CAs.
Cam is the founding member of the class of carbonic anhydrases
W19 and Y200 contribute to the active-site structure that fine tunes catalysis
W19 is important for the CO2/HCO3− exchange and proton transfer steps of catalysis
Replacement of W19 and Y200 yields conformational changes near the surface
Acknowledgements
This work was supported in part by grants from the Department of Energy, Energy Biosciences Program DE-FG02-95ER20198 to J.G.F. and by the National Institutes of Health GM25154 to D.N.S and R.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations used:CA, carbonic anhydrase
References
- 1.Zimmerman SA, Ferry JG. Curr. Pharm. Des. 2008;14:716–721. doi: 10.2174/138161208783877929. [DOI] [PubMed] [Google Scholar]
- 2.Smith KS, Ferry JG. Journal of Bacteriology. 1999;181:6247–6253. doi: 10.1128/jb.181.20.6247-6253.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zimmerman SA, Tomb JF, Ferry JG. Journal of Bacteriology. 2010;192:1353–1360. doi: 10.1128/JB.01164-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jeyakanthan J, Rangarajan S, Mridula P, Kanaujia SP, Shiro Y, Kuramitsu S, Yokoyama S, Sekar K. Acta Crystallogr. D Biol. Crystallogr. 2008;64:1012–1019. doi: 10.1107/S0907444908024323. [DOI] [PubMed] [Google Scholar]
- 5.Pena KL, Castel SE, de Araujo C, Espie GS, Kimber MS. Proc. Natl. Acad. Sci. U S A. 2010;107:2455–2460. doi: 10.1073/pnas.0910866107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kisker C, Schindelin H, Alber BE, Ferry JG, Rees DC. EMBO J. 1996;15:2323–2330. [PMC free article] [PubMed] [Google Scholar]
- 7.Parisi G, Fornasari M, Echave J. Mol. Phylogenet. Evol. 2000;14:323–334. doi: 10.1006/mpev.1999.0734. [DOI] [PubMed] [Google Scholar]
- 8.Alber BE, Ferry JG. Proc. Natl. Acad. Sci. USA. 1994;91:6909–6913. doi: 10.1073/pnas.91.15.6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.MacAuley SR, Zimmerman SA, Apolinario EE, Evilia C, Hou Y, Ferry JG, Sowers KR. Biochemistry. 2009;48:817–819. doi: 10.1021/bi802246s. [DOI] [PubMed] [Google Scholar]
- 10.Zimmerman SA, Ferry JG. Biochemistry. 2006;45:5149–5157. doi: 10.1021/bi052507y. [DOI] [PubMed] [Google Scholar]
- 11.Parisi G, Perales M, Fornasari MS, Colaneri A, Gonzalez-Schain N, Gomez-Casati D, Zimmermann S, Brennicke A, Araya A, Ferry JG, Echave J, Zabaleta E. Plant Molecular Biology. 2004;55:193–207. doi: 10.1007/s11103-004-0149-7. [DOI] [PubMed] [Google Scholar]
- 12.Tripp BC, Ferry JG. Biochemistry. 2000;39:9232–9240. doi: 10.1021/bi0001877. [DOI] [PubMed] [Google Scholar]
- 13.Tripp BC, Ferry JG. Biochemistry. 2000;39:9232–9240. doi: 10.1021/bi0001877. [DOI] [PubMed] [Google Scholar]
- 14.Tripp BC, Tu C, Ferry JG. Biochemistry. 2002;41:669–678. doi: 10.1021/bi010768b. [DOI] [PubMed] [Google Scholar]
- 15.Iverson TM, Alber BE, Kisker C, Ferry JG, Rees DC. Biochemistry. 2000;39:9222–9231. doi: 10.1021/bi000204s. [DOI] [PubMed] [Google Scholar]
- 16.Domsic JF, Williams W, Fisher SZ, Tu C, Agbandje-McKenna M, Silverman DN, McKenna R. Biochemistry. 2010;49:6394–6399. doi: 10.1021/bi1007645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 18.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallographica Section D-Biological Crystallography. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 19.Khalifah RG. J. Biol. Chem. 1971;246:2561–2573. [PubMed] [Google Scholar]
- 20.Silverman DN. Methods Enzymol. 1982;87:732–752. doi: 10.1016/s0076-6879(82)87037-7. [DOI] [PubMed] [Google Scholar]
- 21.Zheng JY, Avvaru BS, Tu C, McKenna R, Silverman DN. Biochemistry. 2008;47:12028–12036. doi: 10.1021/bi801473w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simonsson I, Jonsson BH, Lindskog S. European Journal of Biochemistry. 1979;93:409–417. doi: 10.1111/j.1432-1033.1979.tb12837.x. [DOI] [PubMed] [Google Scholar]
- 23.Silverman DN, Tu C, Chen X, Tanhauser SM, Kresge AJ, Laipis PJ. Biochemistry. 1993;32:10757–10762. doi: 10.1021/bi00091a029. [DOI] [PubMed] [Google Scholar]
- 24.Alber BE, Colangelo CM, Dong J, Stalhandske CM, Baird TT, Tu C, Fierke CA, Silverman DN, Scott RA, Ferry JG. Biochemistry. 1999;38:13119–13128. doi: 10.1021/bi9828876. [DOI] [PubMed] [Google Scholar]
- 25.Fisher SZ, Tu C, Bhatt D, Govindasamy L, Agbandje-McKenna M, McKenna R, Silverman DN. Biochemistry. 2007;46:3803–3813. doi: 10.1021/bi602620k. [DOI] [PubMed] [Google Scholar]
- 26.Fisher SZ, Tu CK, Bhatt D, Govindasamy L, Agbandje-McKenna M, McKenna R, Silverman DN. Biochemistry. 2007;42:3803–3813. doi: 10.1021/bi602620k. [DOI] [PubMed] [Google Scholar]
- 27.Liang ZW, Xue YF, Behravan G, Jonsson BH, Lindskog S. European Journal of Biochemistry. 1993;211:821–827. doi: 10.1111/j.1432-1033.1993.tb17614.x. [DOI] [PubMed] [Google Scholar]
- 28.Cui Q, Karplus M. Journal of Physical Chemistry B. 2003;107:1071–1078. [Google Scholar]
- 29.Maupin CM, Saunders MG, Thorpe IF, McKenna R, Silverman DN, Voth GA. Journal of the American Chemical Society. 2008;130:11399–11408. doi: 10.1021/ja802264j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fisher SZ, Kovalevsky AY, Domsic JF, Mustyakimov M, McKenna R, Silverman DN, Langan PA. Biochemistry. 2010;49:415–421. doi: 10.1021/bi901995n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.An H, Tu C, Duda D, Montanez-Clemente I, Math K, Laipis PJ, McKenna R, Silverman DN. Biochemistry. 2002;41:3235–3242. doi: 10.1021/bi0120695. [DOI] [PubMed] [Google Scholar]
- 32.Sterling D, Reithmeier RAF, Casey JR. Journal of Biological Chemistry. 2001;276:47886–47894. doi: 10.1074/jbc.M105959200. [DOI] [PubMed] [Google Scholar]