Abstract
Recently, there has been a growing interest to apply bioprinting techniques to stem cell research. Several bioprinting methods have been developed utilizing acoustics, piezoelectricity, and lasers to deposit living cells onto receiving substrates. Using these technologies, spatially defined gradients of immobilized proteins can be engineered to direct stem cell differentiation into multiple subpopulations of different lineages. Stem cells can also be patterned in a high-throughput manner onto flexible implementation patches for tissue regeneration or onto substrates with the goal of accessing encapsulated stem cell of interest for genomic analysis. Here, we review recent achievements with bioprinting technologies in stem cell research, and identify future challenges and potential applications including tissue engineering and regenerative medicine, wound healing, and genomics.
Bioprinting
Manipulation of picoliter to nanoliter droplets has been a challenge for several applications including biochemical surface patterning, tissue engineering, and direct placement of cells and biomaterials for wound dressing applications [1–4]. In this regard, ejecting droplets via an actuator has emerged as a valuable technological advance addressing the issue of precise manipulation and deposition. Bioprinting is defined as the use of printing technology to deposit living cells, extracellular matrix components, biochemical factors, protein, drugs, and biomaterials on a receiving solid or gel substrate or liquid reservoir [5–8]. In recent years, there has been a growing interest in bioprinting due to its various advantages over existing patterning methods such as photolithography, soft-lithography, and stamping. Among these advantages are: (i) simple to use, (ii) enabling researchers to generate geometrically well-defined scaffolds in a rapid and inexpensive manner using polymers or blends of polymers and other cell inductive particles providing support and induction for seeded cells [9], (iii) allowing high-throughput generation of replicas of spatially and temporally well-controlled complex constructs [10], and (iv) providing three-dimensional (3-D) complexity by multilayer printing [6, 7, 10]. By using conventional single-step lithography and stamp printing methods, building 3-D constructs at high-throughput is challenging as fabricated 2-D layers have to be merged. Existing co-culture approaches either lack high-throughput (e.g., conventional multi-well plate co-culture) or require complex fabrication steps and peripheral systems (e.g., microfluidic co-culture). Emerging methods to pattern and assemble microscale hydrogels fabricated by photolithography are promising in terms of mimicking the complexity of the native microenvironment [11–15].
Although bioprinting is a young field, it has experienced a rapid growth despite the initial challenges that an emerging field experiences. First, biological challenges for bioprinting have been cell viability and long-term functionality post-printing. Concerns regarding potential apoptotic effects after and during bioprinting have been raised by the potential future end-users of this technology such as biologists, who consider bioprinting as an enabling technology for various applications. These biological requirements have set standards on the technology, leading to the development of several bioprinting technologies focusing on cytocompatibility issues maintaining a control over cell positioning in 2-D and 3-D microenvironments. Among these bioprinting technologies are inkjet based printing [16, 17], laser printing [18–22], acoustic cell encapsulation [5, 23], and valve-based printing [7, 24–26].
Recently, there has been a growing interest in the use of bioprinting for applications in biology and medicine. An emerging area is integration of bioprinting technologies with stem cell research. Microenvironments with spatially controlled gradients of immobilized macromolecules have been engineered to direct stem cell fate [27–33]. Stem cells such as embryonic stem cells (ESCs), human bone marrow stem cells, and adipose-derived stem cells (ASCs) have been also directly bioprinted onto substrates [34–37]. This review article highlights recent achievements with bioprinting technologies in stem cell research, and discusses challenges and future potential directions.
Bioprinting Technologies
Several bioprinting methods have been developed to deposit cells including acoustic [5, 23], inkjet [16, 17], valve-based [7, 24–26], and laser printing technologies [18–22] (Box 1). Initially, commercially available desktop inkjet printers have been modified and used as cell printers [38]. In these systems, cell suspensions are placed in a printer cartridge and a computer controls the printing pattern. Another technique to generate cell encapsulating hydrogel droplets is the valve-based droplet ejection method [24, 25]. In this method, cell encapsulating hydrogel droplets are ejected onto a surface drop-on-demand. Size and number of cells in a single droplet and amount of droplets are controlled by the valve opening duration and actuation frequency [26].
Box 1. Bioprinting technologies.
Table I.
Comparison of commonly used bioprinting technologies based on performance.
| Performance Metric | Valve-based Bioprinting [7, 24–26] | Laser – induced Bioprinting (LIFT, BioLP, MAPLE) [19, 21, 40–44] | Laser-guided Bioprinting [18, 39] | Inkjet Bioprinting [16, 17] | Acoustic Bioprinting [5, 23] |
|---|---|---|---|---|---|
| Throughput | Medium | Medium | Low | High | High |
| Droplet Size | 100 μm – 1 mm | >20 μm | >10 μm | 100–300 μm | 10–500 μm |
| Complexity | High | High | High | High | High |
| Spatial Resolution | Medium | Medium | High | Medium | Medium |
| Single Cell Control | Medium | Medium | High | Low | High |
In laser-guided direct writing method, photons from a laser beam trap and guide cells by exploiting the differences in refractive indexes of cells and cell media [18, 39]. Exposure of excessive thermal energy via laser light onto the cell biolayer and overheating of the cell can be one of the challenges. However, to overcome these challenges, near-infrared wavelengths (700–1,000 nm) have been used [39]. Photons in the near-infrared lack the energy to generate free radicals and are not absorbed by DNA. Hence, laser-guided direct printing is described to be unlikely to cause mutations or trigger apoptosis, although wavelength optimization remains an open issue [39]. As an alternative to laser-guided direct printing, a cell encapsulating donor film is previously deposited onto a substrate, and placed in parallel facing the receiving substrate (Box 1) [19, 21, 40–44]. Patterning is performed on the receiving substrate that is usually fixed on a computerized stage and coated with a cell culture medium or a biopolymer layer for cellular adhesion. Among these methods are laser-induced forward transfer (LIFT), absorbing film-assisted laser-induced forward transfer (AFA-LIFT), biological laser processing (BioLP), and matrix-assisted pulsed laser evaporation direct writing (MAPLE DW) [45]. The heat transfer from the laser pulse to the cell encapsulating donor film leads to the transfer of material from the donor film to the receiving substrate.
A picoliter droplet based cell encapsulation technology via acoustics has been developed [5, 23, 46]. Acoustic droplet formation rate can reach up to 100,000 droplets per second [5] with high cell viability. Among the advantages of this technology over other printing approaches are: (i) a nozzle is not required for droplet generation since droplets are created from an open liquid reservoir, which circumvents complications that may be related to shear or clogging, (ii) acoustic waves do not harm cells due to low power droplet generation with only a few microseconds of pulse duration, (iii) acoustic ejectors can be combined in an addressable array format as multiple ejectors [46]. This would allow to enhance the rate of printing and to deposit multiple cell and ECM types. These ejectors could potentially print several biomaterials such as living cells, extracellular matrix proteins, nutrients, therapeutic drugs, and growth factors (GFs) simultaneously from the same platform by integrating microfluidic systems into these ejectors [46]. To obtain reproducible results for the deposition of cell encapsulating droplets, bioprinting spatial precision should be comparable to the cell size, e.g. ~10–20 μm in suspension [5]. The acoustic technologies have been shown to eject droplets in a wide range of sizes from 3 micrometers in diameter to several hundred micrometers indicating the versatility of the technology.
Cell viability
To evaluate the limitations of bioprinting in engineering of microenvironments for stem cells, cell viability and functionality post-bioprinting have to be confirmed for various printing technologies. In bioprinting, viability of bioprinted cells was reported [47, 48]. For instance, a thermal inkjet printer was used to print hamster ovary cells and rat primary embryonic motor neurons [48]. During the preparation of cell suspension (phosphate buffered saline), on average 15% of the cells were lysed, most probably due to osmotic effects in the solution [48]. On the other hand, 3% of the cells were lysed during printing, which can be partially attributed to 4–10°C temperature increase in the ejector reservoir and sensitivity of mammalian cells to heat and mechanical stress. Additionally, primary rat embryonic hippocampal and cortical neurons were printed with 74% viability [47]. Immunostaining with dendritic marker anti-MAP2 and axonal filament anti-neurofilament NF 150 on day 15 post-bioprinting showed that the bioprinted neurons maintained their basic cellular phenotypes and functionality. Electrophysiology of cells was also evaluated and it has been shown that voltage-gated potassium and sodium channels were developed successfully, which is a requirement for differentiation of healthy neurons. Currently, several bioprinting technologies have attained viabilities exceeding 90% compared to the controls of non-printed cell cultures indicating the broad future applicability of bioprinting to various cell types ranging from neurons to stem cells. Recently, a valve-based bioprinting technology was utilized for high-throughput single cell isolation and patterning [26] using mouse embryonic stem cells. Stem cell viabilities were reported before and after the bioprinting process as 97% and 94%, respectively. Results showed that functional genomic information of stem cells (e.g. mRNA expression levels) was maintained during bioprinting.
Directing stem cell fate with patterned biomolecules
In this section, we will discuss use of bioprinting technologies to bioengineer cellular microenvironments with well-defined spatial patterns of immobilized proteins to direct stem cell fate. Stem cells can demonstrate differentiation into several subgroups of different lineages in parallel with bioprinted macromolecules. Direct write of stem cells will be discussed in the next section. Applications, advantages over existing methods, and limitations of both stem cell and biomolecule printing are given in Table 1.
Table 1.
Applications, advantages, and limitations of printing stem cells and biomolecules
| Printing Stem Cells | Refs | Printing Biomolecules | Refs |
|---|---|---|---|
| Applications | |||
| Single stem cell genomics | [26] | Protein and DNA arrays | [31] |
| Patches for wound healing | [22] | Tissue engineering applications (e.g., guiding and maximizing stem cell growth in a wound site) | [32] |
| Ex vivo generation of tissue replacement | [36] | ||
| Advantages over current methodsa | |||
| Programmable | [1, 5, 7] | Programmable (no custom stamps are required) | [1, 7, 31] |
| Inexpensive | [1, 5, 7] | Inexpensive | [1, 7, 31] |
| 3-D complexity | [1, 5, 7] | Noncontact (reduces the risk of cross-contamination originating from surface) | [31] |
| High-throughput | [1, 5, 7] | No requirement for any modifications to the proteins or substrates | [31] |
| Limitations | |||
| Cytocompatibile | [1, 3] | The resolution is lower compared with the state of-the-art protein array technologies (e.g., micro-contact printing, resolution is less than 100 nm) | [31] |
| Viscosity | [3, 54] | The number of available binding sites on the receiving substrate | [31] |
| Cytocompatibile | [1, 3] | ||
| Viscosity | [30] | ||
Printing stem cells is advantageous over centrifugal or molding techniques; Printing biomolecules is advantageous over open spotting, soft lithography, photolithography.
Bioprinting has recently been used to bioengineer microenvironments with spatially defined gradients of immobilized GFs. Biologically active bioprinted macromolecules influence the differentiation and multipotency of neural stem cells (NSCs)[30]. For example, a modified thermal inkjet printer (Canon BJC-2100) was used to print fibroblast growth factor-2 (FGF2), ciliary neurotrophic factor (CNTF) and foetal bovine serum (FBS) on a poly-acrylamide-based hydrogel [30]. Then, bioprinted macromolecules were seeded with NSCs. In culture, NSCs remained undifferentiated in the presence of FGF2. The presence of CNTF resulted in NSC differentiation into astrocytes expressing glial fibrillary acidic protein (GFAP) [49, 50]. Control on stem cell fate was achieved by patterning adjacent areas on the surface with FGF2 and CNTF. Increasing levels of CNTF were also printed and results showed linear increase of GFAP-positive cells correlated with the gradient of CNTF. NSCs cultured on regions printed with FBS demonstrated differentiation into smooth muscle cells [51]. The differentiation was only observed in FBS printed areas. These results are consistent with known effects of CNTF and FBS on NSCs[ 49–51].
A piezoelectric printing system was used to print bone morphogenic protein-2 (BMP2) onto fibrin-coated glass slides [31]. Muscle derived stem cells (MDSCs) isolated from adult mice were cultured on areas printed with BMP2. Protein activity was shown by fluorescence tests and activity linked to printed protein gradient. Similar to the NSC studies [30], differentiation of MSDCs was successfully guided through printed protein gradients immobilized to fibrin. However, the patterning approach presented in [31], also demonstrated differentiation of MDSC into two separate subpopulations with osteogenic and myogenic lineage in coordination with printed proteins on the same chip. MSDCs cultured on areas printed with BMP-2 under myogenic conditions differentiated towards osteogenic lineage. Simultaneously, MSDCs cultured on non-printed regions displayed a differentiation towards myogenic lineage. Formation of multinucleated myotubes on non-printed regions was shown by time-lapse microscopy and immunocytochemical staining. This result confirmed the differentiation toward the myogenic lineage. On the other hand, myotubes were not observed on printed regions and osteogenic lineage was evidenced by alkaline phosphatase (ALP) activity [31].
Recently, a piezoelectric inkjet-based bioprinting system [33] was used to print heparin-binding epidermal growth factor (EGF)-like macromolecules [32]. The effects of patterned exogenous GF gradients on proliferation and migration responses of mesenchymal stem cells were investigated in vitro [32]. Increasing, decreasing, and uniform levels of EGF were printed on fibrin-coated substrates. Stem cell outgrowth from a cell population source was likely mainly driven by cell-to-cell confinement gradient originating at the cell starting lines, i.e. initial locations where the cells were printed [32]. On the other hand, it was also reported that GF gradients could effectively correlate with individual cell migration [30, 31]. Despite the gradient differences among the EGF patterns, comparable collective cell migration was reported [32] (Figure 1).
Figure 1.
Printed heparin-binding epidermal growth factor-like growth factor patterns with uniform, increasing, and decreasing gradients compared to control with no pattern. (a) Image of the cell starting line in contact with the pattern. (b) The growth factor pattern was separated into 4 windows of identical area. The cell count in each window was performed every 24 h. (c) Comparisons of cell responses among control with no pattern, uniform pattern, high-to-low pattern, and low-to-high pattern. Cells were seeded at the origin of the patterns and imaged over time (4 days in culture). 13 overprints were performed for uniform pattern. Lower and upper limit were set to 1 and 25 overprints for increasing and decreasing gradients of patterns. Reproduced with permission[32]. Copyright Year, Publisher.
The recent studies on stem cell fate with patterned proteins [30–33] present promising results for further stem cell studies. Immobilization of macromolecules, e.g. GFs, onto surfaces, enables performing experiments to evaluate the role of patterns and protein gradients. These approaches may provide higher level of control over stem cell guidance, and potentially enable multilineage stem cell differentiation to renew and repair tissues. Recent results suggested that uniform level of GFs could be as effective in providing a collective motion of stem cells into wound site as increasing protein concentration or pattern complexity [32]. Collective motion of stem cells was systematically tracked by an automated time lapse microscopy [32], which was validated previously [52, 53]. Additional applications of biomolecule printing using combinations of multiple growth factor gradients [27] and for multilineage stem cell differentiation [28] and control of cell alignment [29] have been recently reported. Such perspective enabled by bioprinting technologies through patterning can help to enhance our understanding to develop better microenvironments for stem cell fate.
Microenvironments for stem cell and protein studies could be diversified and improved with new gel materials. These materials should be compatible with bioprinting ejectors, and tested for a range of gel concentrations. Material properties such as viscosity, density, and surface tension may affect the stem cell fate. Several combinations of these parameters can be studied by computational models [54, 55] to evaluate cell viability after printing. Cells in vivo exist in a 3-D microenvironment. Cells cultured in two-dimensional (2-D) monolayers have displayed major changes in their gene expression as well as their functional behavior compared to cells in native tissues and in 3-D culture conditions [56]. As the 2-D systems do not effectively represent the complex tissue microenvironment for cells [56], another challenge would be constructing a 3-D biochemical microenvironment for stem cells. This could be accomplished by fabricating stem cell encapsulating microgel architectures in 3-D and potentially including 3-D patterns of biologically active molecules with a control over cell and protein densities spatially and temporarily.
Bioprinting stem cells
In this section, we will review the recent advances by utilizing bioprinting technologies to directly print embryonic stem cells (ESCs), human bone marrow stem cells, and adipose-derived stem cells (ASCs). ESCs show unlimited self-renewal capability and multi-lineage differentiation potential due to their pluripotent characteristic [57]. ESCs are hence an ideal source for tissue regeneration and replacement [58]. Prior to differentiation of ESCs into other phenotypes, 3-D spheroid aggregates of ESCs, i.e. embryoid bodies (EBs), are formed. EBs demonstrate early-stage embryogenesis features. For instance, germinal epithelium (three primary tissue layers or germ layers: ectoderm, mesoderm, and endoderm) form both during embryogenesis in vivo and in EB culture in vitro [59]. Hence, EB constitutes the microenvironment for ESCs in vitro and also enables lineage-specific differentiation [60]. However, uniformity in size and shape of EBs is a significant factor enabling homogeneous and synchronous in vitro differentiation of ESCs [61, 62]. The relation between EB-mediated differentiation efficiency and EB size was previously demonstrated [61, 62]. Smaller EBs differentiated towards ectoderm, while larger EBs displayed differentiation towards endoderm and mesoderm [61, 62]. Smaller EBs, e.g. 100–500 μm in the lateral dimensions and 120 μm in depth, are also more likely to show cardiomyocyte differentiation [63]. In contrast, spontaneous aggregation of individual ESCs and EB formation led to inhomogeneous size distribution of EBs and unpredictability in lineage differentiation [64–66]. Hence, a higher level of control over forming uniformly sized and shaped EBs is beneficial to successfully utilize ESCs in tissue engineering and regeneration technologies.
Several techniques have been proposed to stimulate EB formation. For example, enzymatic digestion of the ESC colonies and rotary mass suspension led to inhomogeneous size distribution of EBs [67–69]. Methods utilizing surface patterning have been only able to regulate the initial EB volume [62, 70–72]. The most common method is the hanging-drop technique, i.e. ESC encapsulating droplets are ejected on a surface by manual pipetting, and followed by EB formation after turning the substrate upside down by the help of the gravitational and surface tension forces. The droplet volume and number of ESCs per droplet may vary due to manual pipetting, and may result in different size EBs. Further, such manual approaches are labor intensive and time consuming, and the repeatability of the outcomes including EB size as well as subsequent differentiation and cellular function differ among operators. To develop control over EB size and shape, non-adhesive microwell arrays with various shapes, aspect ratios, and sizes have been developed [73]. However, microwell arrays yielded disk shaped aggregates of ESCs, whereas spheroid structures were formed in suspension cultures [73] indicating different phenotypes [74]. Additionally, the shear stress exerted on ESCs during the externally stimulated aggregation procedures (e.g., rotary mass suspension [75] and centrifugation [65]) may affect the cell-cell interaction and signaling pathways [63] and harm cells [76]. Techniques for sorting nonuniformly sized EBs have also been presented [76]. However, such approaches generally apply external driving forces that might harm ESCs and influence the subsequent cell differentiation process. Recently, bioprinting [7, 8, 77] has been integrated with the conventional hanging-drop technique to form uniform-sized EBs in a reproducible manner [37] to address the challenges described above. The results demonstrated that the system generated more uniform sized EBs compared to those formed by a manual pipetting approach (Figure 2).
Figure 2.
EB formation after bioprinting method. (a-c) Images of formed EBs with droplet sizes of 1, 4, 10, and 20 μL at a cell density of 105 cells/ ml. (a) Uniform-sized droplets encapsulating ESCs were generated by bioprinting. (b) Phase contrast images of EBs formed after hanging for 24 h and c after culture for 72 h in a 96 multiwell plate. (d) Fluorescent images of GFP positive EBs at t=96 h stained with ethidium homodimer. (e) Images of EBs formed with printed droplet size of 10 μl at t=72 h at different cell concentrations. Reproduced with permission [37]. Copyright Year, Publisher.
Laser-induced forward transfer (LIFT) was also used to print human mesenchymal stem cells (hMSC) and skin cell lines (fibroblasts/keratinocytes) [36]. To assess the effects of bioprinting, survival rates of cells, their proliferation, and DNA injuries and alterations of cell surface markers were evaluated. The cells demonstrated cell viability after LIFT as 90% (hMSC) and 98% (skin cells). The skin cell lines and the hMSCs preserved their proliferation capacity after bioprinting, and showed no significant difference in apoptosis or DNA fragmentation compared to controls. Furthermore, the hMSC retained their phenotype as validated by fluorescence activated cell sorting (FACS) analysis. Recently, same printing technology (i.e. LIFT) was used to generate grafts consisting of MSCs [35]. Results showed that bioprinted MSC grafts were able differentiate towards cartilage and bone, which was validated by evaluating the ALP activity and the amount of accumulated calcium over several weeks. It was also shown that LIFT could yield sufficiently high cell densities for chondrogenic differentiation. Further, pre-differentiated MSCs survived the entire bioprinting process and maintained their functionality (Figure 3A–F) [35]. In another work, human adipose-derived stem cells (ASCs) and endothelial colony-forming cells (ECFCs) were printed using LIFT [34]. For separately printed ASCs and ECFCs regions, cell-cell interactions in vascular endothelial growth factor-free medium were reported. It was also shown that direct cell-cell interactions activated vascular-like network formations. Recently, LIFT was used to print hMSCs and human umbilical vein endothelial cells (HUVEC) onto a cardiac patch for cardiac regeneration [22] (Figure 3G&H). Cells were seeded in a pre-specified pattern on a polyester urethane urea (PEUU) cardiac patch. As a control, cells were randomly and equally seeded onto patches (without LIFT). Cardiac patches were cultivated in vitro or transplanted in vivo to the infarcted zone of rat hearts after left anterior descending (LAD)-ligation. Cardiac performance was evaluated by left ventricular catheterization 8 weeks post infarction. It was demonstrated that bioprinted cell pattern changed growth characteristics of co-cultured hMSC and HUVEC leading to improved vessel formation. Significant functional progress of infarcted hearts was shown after transplantation of a bioprinted patch. It was also demonstrated that capillary density was improved and human cells integrated into the functionally connected vessels of murine vascular system.
Figure 3.
Bioprinted patch implementation. (a) Microscopic image of the mesenchymal stem cell (MSC) pattern directly after laser induced forward transfer (LIFT) printing. (b) fluorescence live/dead image of the printed cells after 6 days under chondrogenic culture conditions. (c) Image of MSCs, predifferentiated in osteogenic lineage for 7 days, printed in two chessboard patterns and cultivated for 17 days under osteogenic conditions. (d) Transmission image, (e) fluorescence images of nuclei stained with Hoechst 33342 (blue staining), and (f) membranes stained with DIL (red staining) of the chessboard section. Scale bar is 1 mm. (g) Arrangement of transferred cells by LIFT observed after 24 h: Human MSC were prestained with PKH26 and patches were stained with polyclonal goat anti-Pecam1 24 h after LIFT to separate grid patterned HUVEC. (h) Patch implantation in vivo: After LAD-ligation rats received the cardiac patch sutured onto the area of blanched myocardium. Reproduced with permission. Copyright Year, Publisher.
A recent study utilized a gelatin-based laser direct write (LDW) method to deposit mouse ESCs into defined arrays of spots [41]. The results demonstrated that ESCs retained the canonical stem cell marker expression of Oct4 in nuclei 3 hours post-printing (Figure 4). Results showed that marker proteins of nestin (ectoderm), Myf-5 (mesoderm) and PDX-1 (endoderm), were expressed after 7 days of cultivation, which confirmed the differentiation of mouse ESCs after bioprinting. In a recent study [78], a donor surface composed of hydrogel microbeads was used as an alternative to a continuous film during laser-assisted bioprinting. Stem cell-encapsulating hydrogel beads were targeted from the donor surface and transferred onto a receiving substrate [78].
Figure 4.
Cell growth profiles after laser printing on (a) day 0, (b) day 1, (c) day 4, and (d) day 7. EB formation after 7 day culture (scale bar is 500 μm). Fluorescence images of mouse ESCs expressing (e) pluripotency marker, Oct4, and (f) DAPI stain to reveal the cell nuclei. (g) and (h) merged images to show the expression of Oct4 in nuclei (scale bar is 100 μm). The marker proteins of (i and l) nestin (ectoderm), (j and m) Myf-5 (mesoderm) and (k and n) PDX-1 (endoderm), were expressed after 7 days of culture, which confirmed the differentiation potential of mouse ESCs after printing (scale bar is 100 μm for i-k, and 50 μm for l-n). Reproduced with permission[41]. Copyright Year, Publisher.
Stem cell genomics
Advances in single cell level functional genomic studies enables a better understanding of stem cells and their characterization [79]. As new regenerative therapies [80] emerge, the existence of tissue-specific stem cells in adult organs has been broadly studied in bone marrow, heart, lung, muscle, skin, pancreas, and as well as the nervous system. However, insufficient cell markers and low viability of the purified cells have negatively affected the characterization of differentiated progeny [81]. Efficient cell isolation and handling methods integrated with in vivo bioprinting technologies can be useful for single cell transplantation methods [82].
A simple, high-throughput system was developed for single cell isolation with direct access to patterned cells [26]. The technique was based on a drop-on-demand cell patterning method that applies simple random sampling (Figure 5). The platform deposited an array of ~10k droplets that encapsulated single cells from a heterogeneous cell population within seconds, where imaging platforms can be used to track the patterned droplets within a few seconds demonstrating target cell positions [26]. As the droplets were deposited onto a glass surface, each droplet encapsulating cells of interest, could then be reached directly. Immunostaining was utilized before patterning to separate target cells from non-target cells of the heterogeneous population. Based on the optical images, each patterned droplet was classified as droplet encapsulating (i) a single target cell, (ii) no cells (empty droplet), (iii) a non-target cell, or (iv) multiple cells (either the target, non-target cell type or both cell types). RNA extraction was subsequently performed for genomic analysis of bioprinted stem cells. In a genome-wide analysis, using DNA microarrays for the printed and control stem cells, stem cell-related markers were employed to assess whether the RNA obtained from the printed group provided useful biological information. 1000 genes with the highest expression levels were measured for printed and non-printed stem cells. The results presented that 11 stem cell markers (including Oct4, Notch1 and Kit) were observed in both groups. Further, printed stem cells demonstrated more than 90% viability following 1.7 minutes of bioprinting and patterning process creating a 50×50 array of cell encapsulating droplets. This approach enables high-throughput processing for stem cell isolation [26]. Such methods can significantly contribute to stem cell studies by reducing time required for isolation and by automating some of the laboratory procedures such as EB cultures with higher spatial and temporal precision over the deposited cells.
Figure 5.
Comparison of methods for total RNA expression analysis: conventional method vs. a hybrid bioprinting method. (a) Conventional single cell isolation method. 1) Heterogeneous sample was collected from a specific tissue, macro-dissection, 2) cells were stained with a specific antibody, 3) target cells were collected with conventional FACS, 4) multiple dilution steps created a small population of target cells, 5) total RNA gene expression analysis with microarray. (b) Drop-on-demand total RNA analysis utilizing bioprinting method. 1) Heterogeneous sample was collected, 2) cells were stained with antibody and patterned with cell-encapsulating droplets, 3) specific homogeneous samples containing target cells were generated by a cell droplet patterning platform; homogeneous samples were identified by an automated imaging system, 4) total RNA gene expression analysis with microarray. Reproduced with permission [26]. Copyright Year, Publisher.
Outlook
Bioprinting technologies enable powerful methods to address the challenges related to multiple potential applications in stem cell research. These technologies offer viable approaches to fabricate protein- and enzyme-based platforms. Recently, significant progress has been made to find the biocompatible and minimum-damage printing conditions for cells and biomacromolecules. Furthermore, spatially controlled multi-lineage differentiation of stem cells was shown [30–32] using patterned and immobilized GFs [30–32]. Bioprinting methods may offer a strategy to bioengineer replacement tissues using stem cells as a source to regenerate multiple tissues through precise patterning of immobilized bioactive molecules. These approaches may be useful to enhance our understanding of cell behavior on immobilized biological patterns and could have potential broad applications in regenerative medicine and tissue engineering.
There is still an unmet need to understand underlying mechanisms of cellular damage during bioprinting, although there has been efforts that merge probabilistic models with experimental studies to enhance our understanding of how cell get encapsulated in droplets during bioprinting [26, 54, 55, 83, 84]. The future success and broad applicability of bioprinting technologies will benefit from evaluation of cell function during and after bioprinting. Additionally, mathematical and computational studies could advance our understanding of bioprinting processes and technologies such as the mechanical effects during printing as well as probabilistic understanding of cell encapsulation in droplets [54, 55, 83]. These studies will enable further steps towards validating the cyto- and bio-compatibility of various bioprinting technologies. Additionally, biomaterials are significant in the integration of cell delivery with 3-D scaffold fabrication. Various hydrogels such as photo and temperature crosslinkable gels have been used for bioprinting such as PEG, GELMA, agarose, and alginate [7, 11, 12, 15, 26, 84–88]. Biomaterial development for bioprinting needs to take biological variables such as cytotoxicity into consideration, thus limiting the available chemistries and temperature ranges, and the rheological range of fluids that are compatible with various bioprinting technologies.
The ability manipulate cells in microdroplets has enabled biopreservation of cells in nanoliter droplets creating a new biopreservation approach, i.e., cryoprinting [2, 25, 89, 90]. The advantages of nanoliter droplet cryopreservation include reduced cryoprotectant agent (CPA) concentration and toxicity and enhanced heat transfer rates during vitrification [90]. Bioprinting technologies have enabled biopreservation of multiple cell types including mouse oocytes [89], embryonic stem cells [37], cardiomyocytes [24], blood cells [2]. The CPAs used for stem cell preservation such as DMSO, glycol are toxic and they are used at high concentrations, which is shown to have adverse effects on stem cell functionality and differentiation after thawing [91, 92]. These nanoliter droplet vitrification technologies create a new direction for biopreservation of stem cells with potentially better functional outcomes and reduced CPA toxicity [2, 89, 93].
Bioprinting is an emerging field with a lot of future potential, as this method can be integrated with conventional laboratory techniques, and create high-throughput, efficient, and scalable alternatives to existing methods. Such techniques provide effective tools for various applications in broad fields of study such as tissue engineering and regenerative medicine as well as in cancer studies [77]. Bioprinting methods provide a new avenue for stem cell patterning and differentiation, with precision and control over the cellular microenvironment. Beyond the in vitro laboratory approaches, the future of bioprinting is likely to have applications for in vivo. Further advances in the bioprinting field may lead to applications in the operating room in clinical settings in the future creating novel biomaterials directly printed in vivo.
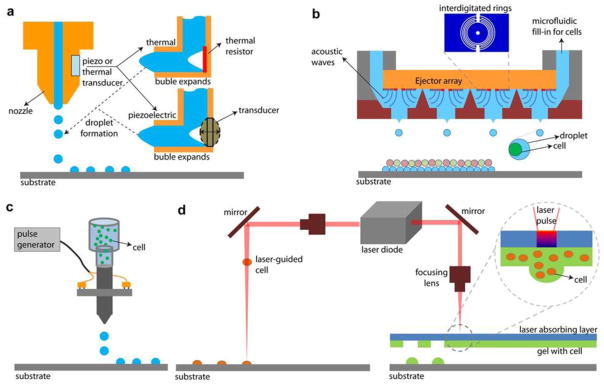
Box 1, Figure I.
Sketch of bioprinting technologies. (a) Thermal and piezoelectric ink-jet printing. Two major methods to jet the bioink are demonstrated. The thermal technique heats a resistor and expands an air bubble. The piezoelectric technique charges crystals that expand. (b) Setup for acoustic pico-liter droplet generation. Droplets can be deposited drop-on-demand with predetermined separation and locations. Periodically spaced interdigitated gold rings of an acoustic picoliter droplet ejector are demonstrated. The wavelength of the acoustic wave (low ‘f’) is much larger than the cell size resulting in harmless ejection of cells [5]. (c) Sketch of the valve-based printing setup [7, 24–26]. (d) Sketch of the laser printing setup. (left) Laser-guided direct cell printing [18, 39]. The laser is focused into a cell suspension and the force due to difference in refractive indexes moves the cells onto an acceptor substrate (right) The cell-hydrogel compound is propelled forward as a jet by the pressure of a laser-induced vapor bubble.
Highlights.
There is a growing number of applications of bioprinting in stem cell research.
Bioprinting offers a strategy to engineer a precise microenvironment for stem cells.
Stem cells can be printed in a highthroughput manner onto regenerative patches.
Bioprinting provides a new avenue for stem cell research.
Acknowledgments
We acknowledge that this material is based in part upon work supported by the National Science Foundation under NSF CAREER Award Number 1150733, NIH R01EB015776 and NIH R21HL095960, and R21 HL112114. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Geckil H, et al. Engineering hydrogels as extracellular matrix mimics. Nanomedicine. 2010;5:469–484. doi: 10.2217/nnm.10.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Samot J, et al. Blood banking in living droplets. PLoS One. 2011;6:e17530. doi: 10.1371/journal.pone.0017530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Derby B. Bioprinting: inkjet printing proteins and hybrid cell-containing materials and structures. Journal of Materials Chemistry. 2008;18:5717–5721. [Google Scholar]
- 4.Lee WG, et al. Microscale electroporation: challenges and perspectives for clinical applications. Integr Biol (Camb) 2009;1:242–251. doi: 10.1039/b819201d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Demirci U, Montesano G. Single Cell Epitaxy by Acoustic Picoliter Droplets. Lab Chip. 2007;7:1139–1145. doi: 10.1039/b704965j. [DOI] [PubMed] [Google Scholar]
- 6.Mironov V, et al. Organ printing: promises and challenges. Regenerative medicine. 2008;3:93–103. doi: 10.2217/17460751.3.1.93. [DOI] [PubMed] [Google Scholar]
- 7.Moon S, et al. Layer by layer three-dimensional tissue epitaxy by cell-laden hydrogel droplets. Tissue Eng Part C Methods. 2010;16:157–166. doi: 10.1089/ten.TEC.2009.0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu F, et al. A droplet-based building block approach for bladder smooth muscle cell (SMC) proliferation. Biofabrication. 2010;2:9. doi: 10.1088/1758-5082/2/1/014105. [DOI] [PubMed] [Google Scholar]
- 9.Hamid Q, et al. Fabrication of three-dimensional scaffolds using precision extrusion deposition with an assisted cooling device. Biofabrication. 2011;3:034109. doi: 10.1088/1758-5082/3/3/034109. [DOI] [PubMed] [Google Scholar]
- 10.Mironov V, et al. Organ printing: computer-aided jet-based 3D tissue engineering. Trends in biotechnology. 2003;21:157–161. doi: 10.1016/S0167-7799(03)00033-7. [DOI] [PubMed] [Google Scholar]
- 11.Gurkan U, et al. Emerging Technologies for Assembly of Microscale Hydrogels. Advanced Healtcare Materials. 2012;1:149–158. doi: 10.1002/adhm.201200011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu F, et al. Three-dimensional magnetic assembly of microscale hydrogels. Adv Mater. 2011;23:4254–4260. doi: 10.1002/adma.201101962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gurkan UA, et al. Simple precision creation of digitally specified, spatially heterogeneous, engineered tissue architectures. Adv Mater. 2012 doi: 10.1002/adma.201203261. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tasoglu S, et al. Paramagnetic Levitational Assembly of Hydrogels. Adv Mater. 2012 doi: 10.1002/adma.201200285. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu F, et al. Release of Magnetic Nanoparticles from Cell-Encapsulating Biodegradable Nanobiomaterials. ACS Nano. 2012;6:6640–6649. doi: 10.1021/nn300902w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boland T, et al. Application of inkjet printing to tissue engineering. Biotechnol J. 2006;1:910–917. doi: 10.1002/biot.200600081. [DOI] [PubMed] [Google Scholar]
- 17.Nakamura M, et al. Biocompatible inkjet printing technique for designed seeding of individual living cells. Tissue engineering. 2005;11:1658–1666. doi: 10.1089/ten.2005.11.1658. [DOI] [PubMed] [Google Scholar]
- 18.Odde DJ, Renn MJ. Laser-guided direct writing for applications in biotechnology. Trends in biotechnology. 1999;17:385–389. doi: 10.1016/s0167-7799(99)01355-4. [DOI] [PubMed] [Google Scholar]
- 19.Barron JA, et al. Application of laser printing to mammalian cells. Thin Solid Films. 2004;453:383. [Google Scholar]
- 20.Nahmias Y, et al. Laser-guided direct writing for three-dimensional tissue engineering. Biotechnology and Bioengineering. 2005;92:129–136. doi: 10.1002/bit.20585. [DOI] [PubMed] [Google Scholar]
- 21.Guillotin B, et al. Laser assisted bioprinting of engineered tissue with high cell density and microscale organization. Biomaterials. 2010;31:7250–7256. doi: 10.1016/j.biomaterials.2010.05.055. [DOI] [PubMed] [Google Scholar]
- 22.Gaebel R, et al. Patterning human stem cells and endothelial cells with laser printing for cardiac regeneration. Biomaterials. 2011;32:9218–9230. doi: 10.1016/j.biomaterials.2011.08.071. [DOI] [PubMed] [Google Scholar]
- 23.Fang Y, et al. Rapid Generation of Multiplexed Cell Cocultures Using Acoustic Droplet Ejection Followed by Aqueous Two-Phase Exclusion Patterning. Tissue Engineering Part C: Methods. 2012 doi: 10.1089/ten.tec.2011.0709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Demirci U, Montesano G. Cell encapsulating droplet vitrification. Lab Chip. 2007;7:1428–1433. doi: 10.1039/b705809h. [DOI] [PubMed] [Google Scholar]
- 25.Song YS, et al. Vitrification and levitation of a liquid droplet on liquid nitrogen. Proc Natl Acad Sci U S A. 2010;107:4596–4600. doi: 10.1073/pnas.0914059107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moon S, et al. Drop-on-demand single cell isolation and total RNA analysis. PLoS One. 2011;6:e17455. doi: 10.1371/journal.pone.0017455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller ED, et al. Inkjet Printing of Growth Factor Concentration Gradients and Combinatorial Arrays Immobilized on Biologically-Relevant Substrates. Combinatorial Chemistry & High Throughput Screening. 2009;12:604–618. doi: 10.2174/138620709788681907. [DOI] [PubMed] [Google Scholar]
- 28.Ker EDF, et al. Engineering spatial control of multiple differentiation fates within a stem cell population. Biomaterials. 2011;32:3413–3422. doi: 10.1016/j.biomaterials.2011.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ker EDF, et al. Bioprinting of growth factors onto aligned sub-micron fibrous scaffolds for simultaneous control of cell differentiation and alignment. Biomaterials. 2011;32:8097–8107. doi: 10.1016/j.biomaterials.2011.07.025. [DOI] [PubMed] [Google Scholar]
- 30.Ilkhanizadeh S, et al. Inkjet printing of macromolecules on hydrogels to steer neural stem cell differentiation. Biomaterials. 2007;28:3936–3943. doi: 10.1016/j.biomaterials.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 31.Phillippi JA, et al. Microenvironments engineered by inkjet bioprinting spatially direct adult stem cells toward muscle- and bone-like subpopulations. Stem Cells. 2008;26:127–134. doi: 10.1634/stemcells.2007-0520. [DOI] [PubMed] [Google Scholar]
- 32.Miller ED, et al. Spatially directed guidance of stem cell population migration by immobilized patterns of growth factors. Biomaterials. 2011;32:2775–2785. doi: 10.1016/j.biomaterials.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Campbell PG, et al. Engineered spatial patterns of FGF-2 immobilized on fibrin direct cell organization. Biomaterials. 2005;26:6762–6770. doi: 10.1016/j.biomaterials.2005.04.032. [DOI] [PubMed] [Google Scholar]
- 34.Gruene M, et al. Adipogenic differentiation of laser-printed 3D tissue grafts consisting of human adipose-derived stem cells. Biofabrication. 2011:3. doi: 10.1088/1758-5082/3/1/015005. [DOI] [PubMed] [Google Scholar]
- 35.Gruene M, et al. Laser Printing of Stem Cells for Biofabrication of Scaffold-Free Autologous Grafts. Tissue Engineering Part C-Methods. 2011;17:79–87. doi: 10.1089/ten.TEC.2010.0359. [DOI] [PubMed] [Google Scholar]
- 36.Koch L, et al. Laser Printing of Skin Cells and Human Stem Cells. Tissue Engineering Part C-Methods. 2010;16:847–854. doi: 10.1089/ten.TEC.2009.0397. [DOI] [PubMed] [Google Scholar]
- 37.Xu F, et al. Embryonic stem cell bioprinting for uniform and controlled size embryoid body formation. Biomicrofluidics. 2011;5:022207. doi: 10.1063/1.3580752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cui X, Boland T. Human microvasculature fabrication using thermal inkjet printing technology. Biomaterials. 2009;30:6221–6227. doi: 10.1016/j.biomaterials.2009.07.056. [DOI] [PubMed] [Google Scholar]
- 39.Nahmias Y, Odde DJ. Micropatterning of living cells by laser-guided direct writing: application to fabrication of hepatic-endothelial sinusoid-like structures. Nat Protoc. 2006;1:2288–2296. doi: 10.1038/nprot.2006.386. [DOI] [PubMed] [Google Scholar]
- 40.Wu PK, et al. The deposition, structure, pattern deposition, and activity of biomaterial thin-films by matrix-assisted pulsed-laser evaporation (MAPLE) and MAPLE direct write. Thin Solid Films. 2001;398:607–614. [Google Scholar]
- 41.Raof NA, et al. The maintenance of pluripotency following laser direct-write of mouse embryonic stem cells. Biomaterials. 2011;32:1802–1808. doi: 10.1016/j.biomaterials.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keriquel V, et al. In vivo bioprinting for computer- and robotic-assisted medical intervention: preliminary study in mice. Biofabrication. 2010;2:014101. doi: 10.1088/1758-5082/2/1/014101. [DOI] [PubMed] [Google Scholar]
- 43.Kattamis NT, et al. Laser direct write printing of sensitive and robust light emitting organic molecules. Applied Physics Letters. 2009;94:103306. [Google Scholar]
- 44.Colina M, et al. Laser-induced forward transfer of liquids: Study of the droplet ejection process. Journal of Applied Physics. 2006;99:084909. [Google Scholar]
- 45.Schiele NR, et al. Laser-based direct-write techniques for cell printing. Biofabrication. 2010:2. doi: 10.1088/1758-5082/2/3/032001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Demirci U. Acoustic picoliter droplets for emerging applications in semiconductor industry and biotechnology. Journal of Microelectromechanical Systems. 2006;15:957–966. [Google Scholar]
- 47.Xu T, et al. Viability and electrophysiology of neural cell structures generated by the inkjet printing method. Biomaterials. 2006;27:3580–3588. doi: 10.1016/j.biomaterials.2006.01.048. [DOI] [PubMed] [Google Scholar]
- 48.Xu T, et al. Inkjet printing of viable mammalian cells. Biomaterials. 2005;26:93–99. doi: 10.1016/j.biomaterials.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 49.Johe KK, et al. Single factors direct the differentiation of stem cells from the fetal and adult central nervous system. Genes & development. 1996;10:3129–3140. doi: 10.1101/gad.10.24.3129. [DOI] [PubMed] [Google Scholar]
- 50.Hermanson O, et al. N-CoR controls differentiation of neural stem cells into astrocytes. Nature. 2002;419:934–939. doi: 10.1038/nature01156. [DOI] [PubMed] [Google Scholar]
- 51.Oishi K, et al. Contractile responses of smooth muscle cells differentiated from rat neural stem cells. The Journal of physiology. 2002;540:139–152. doi: 10.1113/jphysiol.2001.013278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li K, et al. Cell population tracking and lineage construction with spatiotemporal context. Med Image Anal. 2008;12:546–566. doi: 10.1016/j.media.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li K, et al. Online Tracking of Migrating and Proliferating Cells Imaged with Phase-Contrast Microscopy. Proceedings of the 2006 Conference on Computer Vision and Pattern Recognition Workshop (CVPRW ‘06); 2006. pp. 65–72. [Google Scholar]
- 54.Tasoglu S, et al. Impact of a compound droplet on a flat surface: A model for single cell epitaxy. Phys Fluids. 2010:22. doi: 10.1063/1.3475527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Muradoglu M, Tasoglu S. A Front Tracking Method for Computational Modeling of Impact and Spreading of Viscous Droplets on Solid Walls. Computers & Fluids. 2010;39:615–626. [Google Scholar]
- 56.Birgersdotter A, et al. Gene expression perturbation in vitro - A growing case for three-dimensional (3D) culture systems. Seminars in Cancer Biology. 2005;15:405–412. doi: 10.1016/j.semcancer.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 57.Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 58.Xu Y, et al. A chemical approach to stem-cell biology and regenerative medicine. Nature. 2008;453:338–344. doi: 10.1038/nature07042. [DOI] [PubMed] [Google Scholar]
- 59.Itskovitz-Eldor J, et al. Differentiation of human embryonic stem cells into embryoid bodies compromising the three embryonic germ layers. Mol Med. 2000;6:88–95. [PMC free article] [PubMed] [Google Scholar]
- 60.Kurosawa H. Methods for inducing embryoid body formation: in vitro differentiation system of embryonic stem cells. Journal of bioscience and bioengineering. 2007;103:389–398. doi: 10.1263/jbb.103.389. [DOI] [PubMed] [Google Scholar]
- 61.Peerani R, et al. Niche-mediated control of human embryonic stem cell self-renewal and differentiation. The EMBO journal. 2007;26:4744–4755. doi: 10.1038/sj.emboj.7601896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park J, et al. Microfabrication-based modulation of embryonic stem cell differentiation. Lab Chip. 2007;7:1018–1028. doi: 10.1039/b704739h. [DOI] [PubMed] [Google Scholar]
- 63.Mohr JC, et al. The microwell control of embryoid body size in order to regulate cardiac differentiation of human embryonic stem cells. Biomaterials. 2010;31:1885–1893. doi: 10.1016/j.biomaterials.2009.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reubinoff BE, et al. Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nature biotechnology. 2000;18:399–404. doi: 10.1038/74447. [DOI] [PubMed] [Google Scholar]
- 65.Burridge PW, et al. Improved human embryonic stem cell embryoid body homogeneity and cardiomyocyte differentiation from a novel V-96 plate aggregation system highlights interline variability. Stem Cells. 2007;25:929–938. doi: 10.1634/stemcells.2006-0598. [DOI] [PubMed] [Google Scholar]
- 66.Choi YY, et al. Controlled-size embryoid body formation in concave microwell arrays. Biomaterials. 2010;31:4296–4303. doi: 10.1016/j.biomaterials.2010.01.115. [DOI] [PubMed] [Google Scholar]
- 67.Rohani L, et al. Embryonic stem cell sphere: a controlled method for production of mouse embryonic stem cell aggregates for differentiation. The International journal of artificial organs. 2008;31:258–265. doi: 10.1177/039139880803100310. [DOI] [PubMed] [Google Scholar]
- 68.Youn BS, et al. Scale-up of breast cancer stem cell aggregate cultures to suspension bioreactors. Biotechnology progress. 2006;22:801–810. doi: 10.1021/bp050430z. [DOI] [PubMed] [Google Scholar]
- 69.Singla DK, et al. Human Cell Culture 6: Embryonic Stem Cells. Springer-Verlag; 2007. [Google Scholar]
- 70.Bauwens CL, et al. Control of human embryonic stem cell colony and aggregate size heterogeneity influences differentiation trajectories. Stem Cells. 2008;26:2300–2310. doi: 10.1634/stemcells.2008-0183. [DOI] [PubMed] [Google Scholar]
- 71.Ungrin MD, et al. Reproducible, ultra high-throughput formation of multicellular organization from single cell suspension-derived human embryonic stem cell aggregates. PLoS One. 2008;3:e1565. doi: 10.1371/journal.pone.0001565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gothard D, et al. Controlled embryoid body formation via surface modification and avidin-biotin cross-linking. Cytotechnology. 2009;61:135–144. doi: 10.1007/s10616-010-9255-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Karp JM, et al. Controlling size, shape and homogeneity of embryoid bodies using poly(ethylene glycol) microwells. Lab Chip. 2007;7:786–794. doi: 10.1039/b705085m. [DOI] [PubMed] [Google Scholar]
- 74.Nelson CM, et al. Emergent patterns of growth controlled by multicellular form and mechanics. Proc Natl Acad Sci U S A. 2005;102:11594–11599. doi: 10.1073/pnas.0502575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ng ES, et al. Forced aggregation of defined numbers of human embryonic stem cells into embryoid bodies fosters robust, reproducible hematopoietic differentiation. Blood. 2005;106:1601–1603. doi: 10.1182/blood-2005-03-0987. [DOI] [PubMed] [Google Scholar]
- 76.Lillehoj PB, et al. Continuous sorting of heterogeneous-sized embryoid bodies. Lab Chip. 2010;10:1678–1682. doi: 10.1039/c000163e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xu F, et al. A three-dimensional in vitro ovarian cancer coculture model using a high-throughput cell patterning platform. Biotechnology journal. 2011;6:204–212. doi: 10.1002/biot.201000340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Phamduy TB, et al. Laser direct-write of single microbeads into spatially-ordered patterns. Biofabrication. 2012:4. doi: 10.1088/1758-5082/4/2/025006. [DOI] [PubMed] [Google Scholar]
- 79.Kurimoto K, et al. Global single-cell cDNA amplification to provide a template for representative high-density oligonucleotide microarray analysis. Nat Protocols. 2007;2:739–752. doi: 10.1038/nprot.2007.79. [DOI] [PubMed] [Google Scholar]
- 80.Bianco P, Robey PG. Stem cells in tissue engineering. Nature. 2001;414:118–121. doi: 10.1038/35102181. [DOI] [PubMed] [Google Scholar]
- 81.Fabbri R, et al. Human oocyte cryopreservation: new perspectives regarding oocyte survival. Human Reproduction. 2001;16:411–416. doi: 10.1093/humrep/16.3.411. [DOI] [PubMed] [Google Scholar]
- 82.Leong KG, et al. Generation of a prostate from a single adult stem cell. Nature. 2008;456:804–808. doi: 10.1038/nature07427. [DOI] [PubMed] [Google Scholar]
- 83.Ceyhan E, et al. Prediction and Control of Number of Cells in Microdroplets by Stochastic Modeling. Lab on a Chip. 2012 doi: 10.1039/C1032LC40523G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Moon S, et al. Statistical modeling of single target cell encapsulation. PLoS One. 2011;6:e21580. doi: 10.1371/journal.pone.0021580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jeon O, et al. Photocrosslinked alginate hydrogels with tunable biodegradation rates and mechanical properties. Biomaterials. 2009;30:2724–2734. doi: 10.1016/j.biomaterials.2009.01.034. [DOI] [PubMed] [Google Scholar]
- 86.Xu F, et al. Living Bacterial Sacrificial Porogens to Engineer Decellularized Porous Scaffolds. PLoS ONE. 2011;6:12. doi: 10.1371/journal.pone.0019344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xu F, et al. The assembly of cell-encapsulating microscale hydrogels using acoustic waves. Biomaterials. 2011;32:7847–7855. doi: 10.1016/j.biomaterials.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Song YS, et al. Engineered 3D tissue models for cell-laden microfluidic channels. Anal Bioanal Chem. 2009;395:185–193. doi: 10.1007/s00216-009-2935-1. [DOI] [PubMed] [Google Scholar]
- 89.Zhang X, et al. Nanoliter droplet vitrification for oocyte cryopreservation. Nanomedicine. 2011;7:553–564. doi: 10.2217/nnm.11.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang X, et al. Emerging technologies in medical applications of minimum volume vitrification. Nanomedicine (Lond) 2011;6:1115–1129. doi: 10.2217/nnm.11.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Benekli M, et al. Severe respiratory depression after dimethylsulphoxide-containing autologous stem cell infusion in a patient with AL amyloidosis. Bone Marrow Transplant. 2000;25:1299–1301. doi: 10.1038/sj.bmt.1702452. [DOI] [PubMed] [Google Scholar]
- 92.Higman MA, et al. Reversible leukoencephalopathy associated with re-infusion of DMSO preserved stem cells. Bone Marrow Transplant. 2000;26:797–800. doi: 10.1038/sj.bmt.1702589. [DOI] [PubMed] [Google Scholar]
- 93.Song YS, et al. Microfluidics for cryopreservation. Lab on a Chip. 2009;9:1874–1881. doi: 10.1039/b823062e. [DOI] [PMC free article] [PubMed] [Google Scholar]