SUMMARY
Many genes are regulated at the level of a Pol II that is recruited to a nucleosome-free region upstream of the +1 nucleosome. How the Mediator coactivator complex, which functions at multiple steps, affects transcription through the promoter proximal region, including this nucleosome, remains largely unaddressed. We have established a fully defined in vitro assay system to delineate mechanisms for Pol II transit across the +1 nucleosome. Our results reveal cooperative functions of multiple cofactors, particularly of Mediator and elongation factor SII, in transcribing into this nucleosome. This is achieved, in part, through an unusual activity of SII that alters the intrinsic catalytic properties of promoter-proximal Pol II and, in concert with the Mediator, leads to enhancement in transcription of nucleosomal DNA. Our data provide additional mechanistic bases for Mediator function after recruitment of Pol II and, potentially, for regulation of genes controlled via nucleosome-mediated promoter-proximal pausing.
INTRODUCTION
The Mediator has emerged as the key universal regulator of the eukaryotic transcriptional preinitiation complex (PIC), which is composed of RNA polymerase II (Pol II) and its associated general transcription factors (GTFs) (Kornberg, 2005; Malik and Roeder, 2010; Roeder, 1998). Its ability to physically interact with both Pol II and activators initially suggested that Mediator fulfills its coactivator role by acting as an adaptor. However, it is now apparent that it can play a more active role by delivering calibrated output levels of gene activity (Malik and Roeder, 2010). Thus, Mediator function may be manifested not just at the level of PIC formation, but also at post-recruitment steps, which include maturation of the PIC into a full-fledged transcription elongation complex following escape of Pol II from the promoter. Functional interactions between Mediator and a series of transcriptional elongation factors have pointed to a direct role of the Mediator in regulating the elongation step of transcription. We showed that Mediator counteracts the negative effects on Pol II of DSIF (Spt4/Spt5) (Malik et al., 2007). Studies in yeast systems revealed a functional link between Mediator and the elongation factor SII (TFIIS) (Guglielmi et al., 2007; Kim et al., 2007). More recently, direct physical interactions between a Mediator subunit (MED26) and P-TEFb and other elongation factors that form transient complexes have also been reported (Takahashi et al., 2011).
The physiological significance of regulating transcription at a post-PIC recruitment level is best attested by recent global profiling studies in metazoan cells that have suggested that regulation of many genes may be achieved subsequent to recruitment of Pol II (Gilmour, 2009; Margaritis and Holstege, 2008). As for the prototypical Drosophila heat shock gene (Saunders et al., 2006), Pol II molecules at these promoter complexes might exist in a poised state awaiting appropriate signals (transduced via activators) that would allow synthesis of the full-length transcript. Potentially reflecting superimposed regulatory mechanisms, it has also become evident that at many loci the PIC assembles within a nucleosome free region (NFR) near the well-positioned +1 nucleosome downstream of the transcription start site (TSS). Studies in yeast have revealed that the location of the +1 nucleosome dictates the distinct activation strategies of various genes (Rhee and Pugh, 2012). In some cases, the PIC and the +1 nucleosome might even constitute a single extended nucleoprotein complex. However, the precise mechanisms by which Pol II is regulated in these contexts remain unclear, especially in metazoans and, importantly, any potential role of Mediator remains unaddressed.
Here we have established an in vitro system, reconstituted from homogeneously pure components, to assess how Mediator regulates Pol II transit across the +1 nucleosome. Our basic assay system relies on generating templates through fusion of pre-assembled mononucleosomes to defined positions and recapitulates dependencies on previously characterized chromatin cofactors. We focus on an interplay between Mediator and SII, which cooperate to overcome the negative effects of the nucleosome. Our analysis reveals an SII-induced catalytic mode of promoter-bound Pol II, which in conjunction with Mediator effects facilitates Pol II transcription into the nucleosome.
RESULTS
An assay system to assess effect of nucleosomes positioned at specified distances downstream of the TSS
Most previous in vitro studies on activation of chromatin templates, including our own (Barrero and Malik, 2006), rely on chromatinized plasmids. Whereas such templates display physiological spacing of assembled nucleosomes, they are comprised of heterogeneous populations, especially with respect to nucleosome locations relative to the TSS. Studies that have yielded more detailed mechanistic insights have utilized well-positioned mononucleosomal templates (Bondarenko et al., 2006; Carey et al., 2006; Kireeva et al., 2005). While these latter systems efficiently allow monitoring of Pol II passage across the nucleosome, the Pol II typically has not been “loaded” on the template through bona fide promoter-dependent pathways. The potential involvement of the +1 nucleosome both in specifying how a PIC assembles and in regulating post-initiation fates of Pol II (e.g., promoter-proximal pausing) have emphasized the need for developing in vitro assay systems that allow dissection of Pol II mechanisms in the context of an NFR specified by a precisely positioned +1 nucleosome.
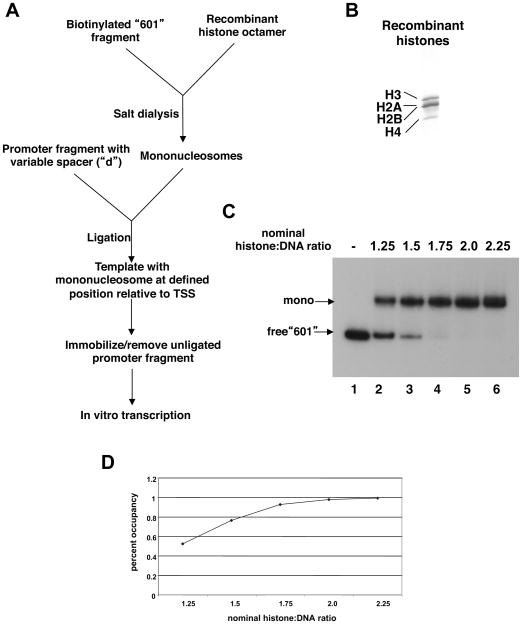
We therefore first developed an experimental system that allows site-specific installation of a mononucleosome downstream of a TSS on a template that can be transcribed in vitro in a promoter- and activator-dependent fashion. The overall scheme (Fig. 1A) entails salt dialysis-mediated assembly of a nucleosome (consisting of recombinant histones [Fig. 1B]) on a biotinylated derivative of 601, the strong nucleosome positioning sequence. We used EMSA (Fig. 1C) to ensure that the 601 fragment was efficiently chromatinized; saturation was observed at a nominal histone:DNA ratio of 2.0 as evidenced by shifting of close to 100% of the 601 band (Fig. 1D). The mononucleosome is then ligated into specified restriction enzyme sites downstream of the core promoter (Ad ML) and activator binding sequences (four repeats of HNF4–cognate site A; Fig. 2A). The ligated templates (Fig. S1), bearing a variable but well-defined NFR (“d”) that is followed by a precisely positioned nucleosome (the +1 nucleosome), are then immobilized via the biotin tag and subjected to in vitro transcription.
Fig. 1. Experimental system for site-specific installation of a mononucleosome.
A. The overall scheme for in vitro generation of templates in which mononucleosomes are assembled on biotinylated 601 sequence and ligated into promoter fragments with variable distances, “d”, between the TSS and the ligatable end (see also Fig. 2A).
B. Purified recombinant core histones (H2A, H2B, H3, H4) were analyzed by SDS-PAGE and Coomassie staining.
C. EMSA analysis of in vitro assembled mononucleosomes. Mononucleosome assembly reactions (salt dialysis) were performed with a fixed amount of end-labeled 601 DNA and increasing amounts of the histone octamer. Nominal histone:DNA ratio refers to the molar ratio deduced from Bradford concentrations of the octamer.
D. EMSA bands for each reaction from (C) were quantified by phosphorimaging and percent occupancy (nucleosome-bound 601/free 601) was plotted as a function of the histone:DNA ratio to assess the ratio that gives complete saturation of the 601 DNA.
See also Fig. S1
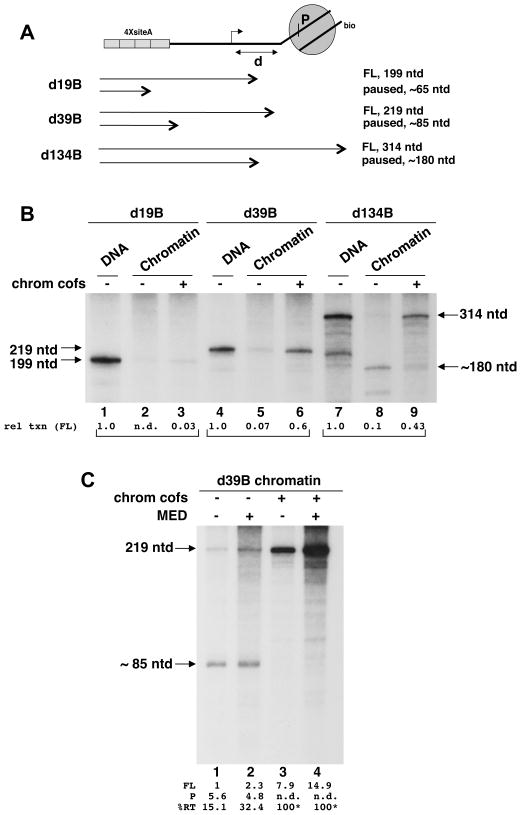
Fig. 2. In vitro transcription assays of defined mononucleosomal templates.
A. Expected products for the d19B, d39B, and d134B templates in which the mononucleosome has been placed at defined positions (d:19 bp, 39 bp, 134 bp) relative to the TSS. Two products are shown for each: one resulting from Pol II pausing at a previously mapped site (P) within the nucleosome and the other from transcription through the entire nucleosome (FL, full-length).
B. Indicated templates (chromatinized or in naked DNA form) were transcribed in vitro in the presence of purified Pol II, the GTFs (TFIIA, B, D, E, F, H), and the cofactor PC4. All reactions contained HNF4α and Mediator. Complete complement (PBAF, p300, FACT, SII) of chromatin cofactors (chrom cofs), as well as acetyl CoA (10 μM), was added to reactions containing chromatin templates (lanes 3, 6, 9). Product sizes for the d19B and d39B templates are indicated to the left; product sizes for the d134B template are indicated to the right. Following PIC assembly in the presence of the factors, transcription was allowed to proceed by adding labeled NTPs for 60 min. Uniformly radiolabeled RNA products were analyzed by electrophoresis. Numbers underneath show relative transcription levels (rel txn) of the full-length (FL) RNA. Transcriptional levels were separately normalized for each template. See also Fig. S2 and S3.
C. In vitro transcription analysis of the chromatinized d39B template. Reactions containing the template were reconstituted and analyzed as in (B) except that all reactions contained acetyl CoA and Mediator and chromatin cofactors (PBAF, p300, FACT, SII) were added as indicated. The full-length (FL, 219 ntd) and paused (P, ~85 ntd) transcripts generated by this template are identified. Percent read-through (RT) is also shown. Asterisks in lanes 3 and 4 signify that the 85-mer could not be accurately quantified (“n.d.”) and that RT levels are visual estimates.
Although high-resolution data for metazoan genes comparable to that for yeast (Rhee and Pugh, 2012) was not available at the time we began this study, there were indications that in CD4+ T cells, the upstream boundary of the +1 nucleosome on active genes is ca. 40 bp from the TSS (Schones et al., 2008). We thus generated a collection of templates that differed in the downstream spans of their NFRs (Fig. 2A). In d19B, the upstream edge of the nucleosome (assuming a 147 bp footprint centered at the dyad axis) is 19 bp from the TSS, whereas in d39B and d134B, the NFR extends to +39 and +134, respectively. Thus, in this series, d39B closely simulated a documented NFR for an active gene whereas in d19B the upstream edge of the nucleosome coincided with the downstream edge of Pol II in the PIC; d134B was intended as a control template in which the nucleosome is too distal to impinge on the PIC.
We subjected these chromatinized templates and corresponding control DNA templates, which were generated following ligation with mock assembled 601, to in vitro transcription in an assay system reconstituted from pure preparations of Pol II, the GTFs (TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH), Mediator, the cofactor PC4 and the orphan nuclear receptor HNF4α (Malik and Roeder, 2003) (Fig. S2). As expected, each of the DNA templates was efficiently transcribed by this combination of factors yielding the appropriate-sized RNA transcript (Fig. 2B, lanes 1, 4, and 7). However, when chromatinized versions of the same templates were assayed, transcription of full-length products was greatly reduced; instead, smaller transcripts were observed in the case of the d39B (Fig. 2B, lane 5; but see also Fig. 2C, lanes 1,2) and d134B templates (Fig. 2B, lane 8) but not the d19B template (Fig. 2B, lane 2). The smaller transcripts corresponded exactly to the truncated products expected when transcribing Pol II stalls at the major pause site (P; Fig. 2A; see also Fig. 2B, lane 8 vs. lane 7; Fig. S3) located 45–50 bp into the 601 mononucleosome (Bondarenko et al., 2006; Carey et al., 2006). These results indicate that Pol II PICs can assemble on the NFRs of d39B and d134B mononucleosomal templates and that after initiation and promoter escape, Pol II can read into the downstream nucleosome at experimentally detectable efficiencies. By contrast, the d19B NFR does not support transcription under these conditions (see below).
When a cocktail of chromatin cofactors (discussed in detail below) was added to the reactions, substantial full-length transcription was recovered from the d39B (Fig. 2B, lane 6; see also Fig. 2C, lanes 3 and 4) and d134B (Fig. 2B, lane 9) mononucleosomal templates, but not from d19B (Fig. 2B, lane 3). Thus, not only is our system suitable for scoring authentic promoter-initiated and nucleosome-impacted transcription but it also can be used to assess effects of chromatin cofactors. In this regard, the behavior of the d19B nucleosomal template was unanticipated since we had expected that in concert with various chromatin cofactors, a functional PIC would assemble on the d19B, albeit in very close juxtaposition with the nucleosome. Even “spiking” the reactions with total unfractionated nuclear extract did not activate this template (not shown). However, replacing TFIID, whose footprint extends well past +19 (Nakajima et al., 1988), with TBP, whose footprint is limited to the TATA box, allowed efficient transcription of this template in the presence of the chromatin coactivators (Fig. S3). In view of emerging ideas on the intimate association between the +1 nucleosome and TAFs in TFIID, this result has important implications for our understanding of how PICs might be established in contexts where the promoter is occluded by a nucleosome. However, in this study we have focused on the d39B template, which as demonstrated above nicely approximates a natural NFR-containing template in which a functional PIC can form and project transcription into a downstream +1 nucleosome that abuts it.
Cofactor requirements for a template containing a +1 nucleosome
Our overall strategy was to first assess how our d39B system responds to previously described chromatin factors so as to also allow us to define a minimal set of factors needed for efficient function of a PIC that is confronting a nucleosome. A second aim was to investigate how Mediator might affect PIC formation and function on the d39B template when the nucleosome was in effect “clamped” in place through omission of chromatin remodeling factors.
The chromatin cofactor cocktail that was finally used in the assays shown in Fig. 2 emerged from a systematic screen of various classes of chromatin cofactors. In this section, we demonstrate that a combination of p300, PBAF, FACT, and SII (Fig. S4) suffice to fully overcome the major blocks imposed by the +1 nucleosome on the d39B template. We had previously identified p300 as a critical HNF4α coactivator (Barrero and Malik, 2006). PBAF also had been reported as a nuclear receptor coactivator (Lemon et al., 2001); we found additionally that it physically interacts with HNF4α (Fig. S5). FACT had previously been identified as a general facilitator of transcription through nucleosomes (Sims et al., 2004). SII was of particular interest not only because it facilitates transcription through nucleosomes (Guermah et al., 2006; Kireeva et al., 2005) but also because of indications that it functions closely with Mediator (above).
When the d39B mononucleosomal template is subjected to transcription in a standard reaction (no additional cofactors), the predominant (>80%) product is an 85-nucleotide long RNA species (Fig. 2C, lane 1). This reflects Pol II that stalls upon encountering the site of the strongest interactions between the 601 DNA and the nucleosome (Bondarenko et al., 2006; Carey et al., 2006). A small fraction of the Pol II does yield the full-length (219 nucleotide) product, which reflects the stochastic tendency of transcription to proceed through this barrier (Bondarenko et al., 2006) and not transcription from an un-chromatinized subpopulation since saturating levels of histones were used in the assembly reactions (Fig. 1D). In the presence of the complete set of chromatin factors, including p300, PBAF, FACT, and SII, efficient transcription of the full-length product (100% read-through, RT) was elicited indicating that together these factors suffice to quantitatively eliminate nucleosome-specific pausing of Pol II (Fig. 2C, lane 3 vs. lane 1). Under these conditions, Mediator had a modest (less than 2-fold), but reproducible, effect on the total level of transcription either in the absence (Fig. 2C, lane 2 vs. lane 1) or presence (Fig. 2C, lane 4 vs. lane 3) of the chromatin cofactors. Similarly, Mediator also affected the RT efficiency (2-fold) through the P site in the absence of these cofactors (Fig. 2C, lane 2 vs. lane 1).
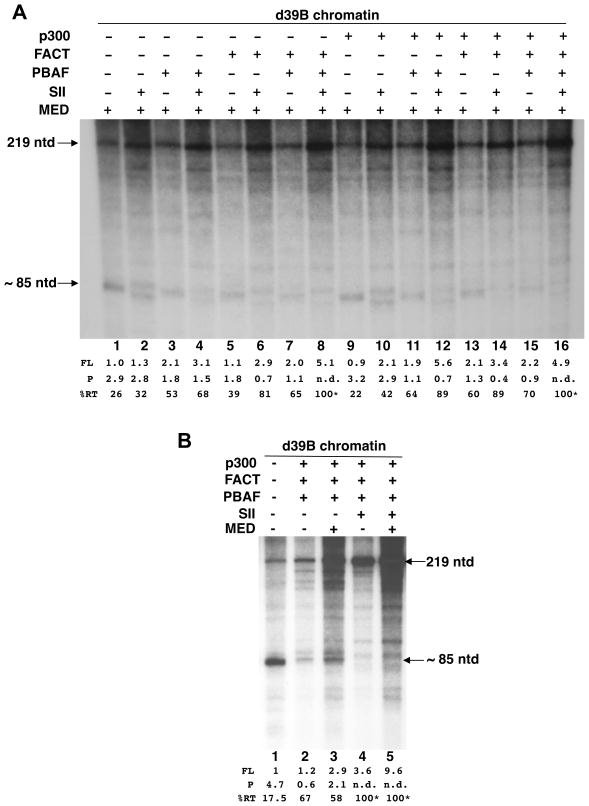
To assess the individual contributions of the cofactors in eliminating nucleosome-induced stalling of Pol II, we performed the analyses with all possible combinations of the chromatin factors (Fig. 3A). To simplify the analyses, all reactions in the first experiment contained Mediator and acetyl CoA, the p300 substrate. When only PBAF was added to the reactions, stalling at the P site was markedly reduced as the relative distribution (RT) of the 219-mer and 85-mer increased to 53% from 26% in its absence (Fig. 3A, lane 3 vs. lane 1). Consistent with previous data for Pol II elongation through the 601 nucleosome (Bondarenko et al., 2006), addition of FACT alone resulted in reduced enhancement of RT (39%; Fig. 3A, lane 5 vs. lane 3). When PBAF and FACT were added together, the RT efficiency was 65% (Fig. 3A, lane 7). However, when SII was added to reactions containing either PBAF alone (Fig. 3A, lane 4) or FACT alone (Fig. 3A, lane 6), significant additional enhancement in the RT efficiency was seen. Importantly, when all three factors were added to the system, RT efficiency approached 100% and overall transcription was stimulated by about 70% (Fig. 3A, lane 8). These data suggest that SII eliminates residual stalling at the P site, presumably by chasing Pol II molecules that had paused here despite the stimulatory effects of PBAF and FACT operating through disparate mechanisms. These results are consistent with and extend earlier data that showed that SII alone does not suffice for efficient read-through from this site (Bondarenko et al., 2006; Luse et al., 2011).
Fig. 3. Chromatin cofactor dependencies for activation of the chromatinized d39B template.
In vitro transcription reactions in each of the panels A and B were done as in Fig. 2C with individual cofactor additions as indicated. Note that in (A), all reactions contained Mediator; hence the baseline for the %RT is relatively elevated.
See also Fig. S4–S6.
When p300 was also included in the reactions (Fig. 3A, lanes 9–16), only modest additional enhancement in RT efficiency was seen. In one scenario, when p300 was added to reactions containing PBAF and SII, potentially more significant effects (89% RT) may be noted (Fig. 3A, lane 12 vs. lane 4), perhaps reflecting previously observed stimulation of remodeling activity in response to acetylation of histones (Carey et al., 2006). Otherwise, p300 (and acetyl CoA; Fig. S6) effects are not very pronounced and may even be redundant to the contributions of the other factors. Overall, this conclusion is consistent with the design of our d39B template in which the PIC is allowed to form on an NFR.
Next we evaluated the contribution of the Mediator to chromatin cofactor-stimulated transcription of the chromatinized d39B template (Fig. 3B). When PBAF, FACT, and p300 were added to the reactions in the absence of Mediator, the total amount of transcription was significantly reduced (3-fold) relative to reactions that lacked these factors (Fig. 3B, lane 2 vs. lane 1). Addition of Mediator (which has modest effects in the absence of these cofactors [Fig. 2C]), restored transcription to the expected levels (Fig. 3B, lane 3) although without significant effects on the relative synthesis of the 219-mer and 85-mer transcripts (Fig. 3B, lane 3 vs. lane 2). This result suggests that the Mediator stabilizes the PIC in the face of nucleosome mobilization by the chromatin cofactors (presumably PBAF). As in Fig. 3A, addition of SII, in the absence and presence (Fig. 3B, lanes 4 and 5) of Mediator mainly had the effect of increasing RT efficiency to nearly 100% and overall transcription by about two-fold.
The above data define a minimal set of factors that allow transcription to efficiently proceed through the +1 nucleosome following initiation from a PIC that forms in an NFR and are in general agreement with previously deduced factor requirements for chromatin transcription in vitro. The results thus validate our system as being suitable for use in dissecting Mediator mechanisms vis a vis the +1 nucleosome.
Effect of the +1 nucleosome on early transcription steps
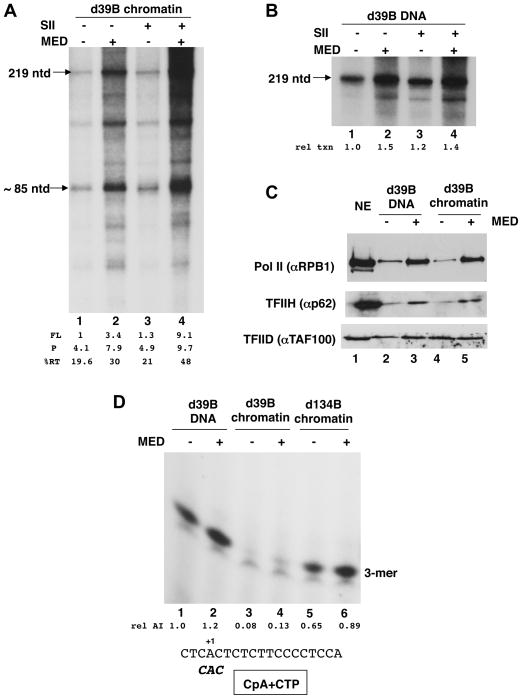
We performed the next series of experiments in the absence of chromatin factors (p300, PBAF, and FACT) to ensure that the translational position of an intact +1 nucleosome remains fixed. When transcription from chromatinized d39B templates was allowed to take place for relatively short (12 min) periods following an extended (45 min) PIC formation step in the absence of NTPs, total transcription (219-mer plus 85-mer) was found to be stimulated 2.2-fold when Mediator was added to the reactions (Fig. 4A, lane 2 vs. lane 1). Since formation of even the 85-mer, which arises from the first strong intra-nucleosomal pause encountered by the escaped Pol II, was also enhanced (1.9-fold) in the presence of Mediator, its effect is manifested at an earlier step. We further found that SII and Mediator act cooperatively to enhance total transcription under these conditions (Fig. 4A, lane 4 vs. lanes 3 and 2). The synergistic function of these factors is particularly evident for the full-length product for a net stimulation of more than 9-fold (Fig. 4A, lane 4 vs. lane 1). Again, since formation of even the 85-mer was also enhanced more than 2-fold in the presence of SII and Mediator, their cooperativity is likely first manifested at the earliest steps. By contrast, production of full-length transcripts on unchromatinized d39B template under the same conditions was relatively immune to the combined effects of these factors (Fig. 4B). We conclude that the +1 nucleosome imposes the conditions for eliciting the synergy between SII and Mediator.
Fig. 4. Involvement of Mediator and SII at early steps in transcription of the d39B template.
A. In vitro transcription reactions using the chromatinized d39B template were done as in Fig. 2C, except that following PIC formation, transcription was allowed to proceed for only 12 min in the presence of radiolabeled NTPs. Mediator and SII were included as indicated. Other chromatin cofactors (PBAF, p300, FACT) were not added.
B. In vitro transcription reactions as in (A) except that the d39B was not chromatinized.
C. Immobilized template recruitment assay for selected PIC components. Scaled up transcription reactions were assembled on d39B templates (DNA, lanes 2,3; chromatin, lanes 4,5) as in (A). After PIC formation in the absence or presence of Mediator, templates were washed, and bound material was immunoblotted with the indicated antibodies. Chromatin cofactors (PBAF, p300, FACT, SII) were not included. Note that although pure factors were used in the binding assay, HeLa nuclear extract (NE, lane 1) served as immunoblot reference.
D. For abortive initiation reactions on the indicated templates, PICs were assembled (+/− Mediator) as for standard reactions (Fig. 2C) but reactions contained only the CpA dinucleotide primer, α32P-CTP and dATP. The trimeric product was visualized by autoradiography (25% PAG) and is marked (3-mer). The sequence around the Ad ML start site and the expected abortive product is shown.
See also Fig. S7.
We also examined PIC formation on the d39B template by incubating Pol II and GTFs with either naked DNA or chromatinized derivatives of the templates that had been bead immobilized (Fig. 4C). As expected from the presence of the NFR on the chromatinized d39B, PIC formation (as evidenced by recruitment of Pol II and TFIIH) on both the DNA and chromatin versions of the template were very similar (compare Fig. 4C, lanes 2 and 4). Importantly, even though Mediator enhanced the amount of Pol II and TFIIH retained on the beads, the final extents of PIC formation were nearly equivalent for the two templates (Fig. 4C, lane 5 vs. lane 3).
We next investigated the competence of the PICs formed on the d39B template for transcription initiation per se. For this purpose, we formed the PICs as before and monitored a trimeric RNA product that is abortively synthesized on the AdML core promoter by Pol II when it is presented with just the dinucleotide CpA and α32P–CTP. Under the conditions of the assay, PICs formed on naked d39B efficiently generate the abortive CpApC trimer (Fig. 4D) (Holstege et al., 1997); Mediator marginally enhanced formation of this trimer by 20% (Fig. 4D, lane 2 vs. lane 1). Similarly, the chromatinized d134B template supported efficient CpApC synthesis (Fig. 4D, lanes 5,6 vs. lanes 1,2). By contrast, chromatinized d39B was severely compromised in its ability to synthesize the trimer, with or without the Mediator (Fig. 4D, lanes 3 and 4). Furthermore, its abortive initiation capacity was not restored by addition of the various chromatin cofactors that we used above (Fig. S7). These results strongly indicate that despite the fact that PICs efficiently form on the d39B NFR, the nucleosome nonetheless restricts actual function of the PIC, including at the level of initiation. While this is rather surprising in view of the results above (Fig. 2A; Fig. 4A) that under appropriate conditions transcription levels through the d39B nucleosome can be relatively high, it points to mechanisms that underlie maturation into a functionally active complex of a nucleosome-impacted PIC (Discussion). Because total productive transcription levels through the nucleosome are significantly higher in the presence of Mediator and SII, the results suggest that while Mediator stabilizes PICs on the d39B template to some extent, additional effects in transcribing through the +1 nucleosome, are likely to be exerted at a subsequent step in concert with SII.
SII-induced reiterative synthesis of a small RNA product at a promoter-proximal location
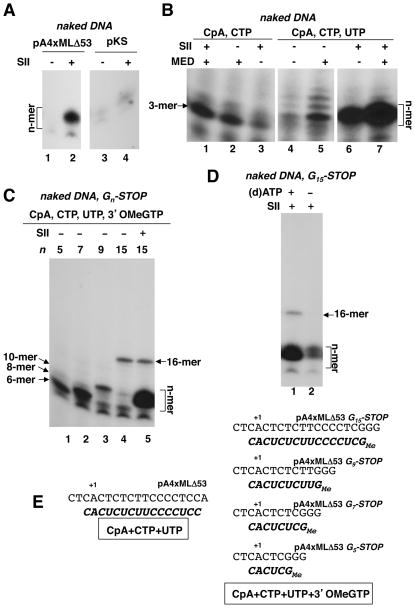
Since SII is recruited to promoters (Kim et al., 2007; Kim et al., 2010), it might functionally interact with Mediator at an early step following PIC recruitment. We investigated if SII has any effects on the earliest steps in transcription and found that inclusion of SII in the standard steady state transcription reactions with DNA templates results in the build-up of high levels of small (8–10 nucleotides-long “n-mer”) RNA species (Fig. 5A, lane 2 vs. lane 1). To investigate the origin of the n-mer RNA, we first ascertained in control experiments that its production is dependent on a promoter-bound PIC. Thus, a plasmid lacking a eukaryotic promoter did not serve as a template for its synthesis (Fig. 5A, compare lanes 3, 4 with lanes 1, 2). Furthermore, n-mer synthesis was found to be dependent upon Pol II and the various GTFs ( not shown).
Fig. 5. An SII-induced oligomer resulting from reiterative RNA synthesis at a promoter-proximal location.
A. Standard in vitro transcription reactions with a complete set of nucleotides using naked pA4XMLΔ53 (lanes 1,2) or pKS (lanes 3,4) templates. Reactions were reconstituted from basic components plus HNF4α. SII was added as indicated. Analysis on 25% PAG.
B. In vitro transcription reactions using naked pA4XMLΔ53 template contained only subsets of nucleotides. All reactions were primed with CpA; those in lanes 1–3 additionally contained α32P-CTP whereas those in lanes 4–7 contained α32P-CTP and UTP. dATP was used as energy source. Reactions were reconstituted from basic components plus HNF4α and SII and Mediator were added as indicated. Analysis on 25% PAG.
C. In vitro transcription reactions using naked “Gn-STOP” templates to measure n-mer length. Reactions were primed with CpA and additionally contained α32P-CTP, UTP, 3′O-methyl-GTP and dATP. Reactions were reconstituted from basic components plus HNF4α and SII was added to reactions with the G15-STOP template (lane 5). Indicated products (6-mer, 8-mer, 10-mer, 16-mer and the n-mer) were analyzed on a 25% PAG.
D. In vitro transcription reactions using naked “G15-STOP” template, as described in (C). Both reactions contained SII but dATP was omitted in the reaction in lane 2. The data in lane 1 was quantified to gauge the relative stoichiometries of the n- and 16-mers (see main text).
E. Nucleotide sequence around the transcription start site of the pA4XMLΔ53 template. The expected RNA products when transcription is carried out with the indicated subsets of nucleotides are also shown.
Next we tried to fully delineate nucleotide requirements for n-mer synthesis. We found that SII did not appreciably affect abortive synthesis of the CpApC in the presence of CpA and α32P–CTP (Fig. 5B, lane 1 vs. lane 2) suggesting that events leading up to this process are not affected by SII. Therefore, we carried out transcription reactions by priming with CpA but in the presence of defined subsets of nucleotides. Additional inclusion of UTP in the reactions, which should allow the formation of a product no longer than a 16-mer (Fig. 5E), led to synthesis of a nested set of products (Fig. 5B, lane 5). Of these, the major species was a longer oligomer that reflects the predominant abortive initiation product at this location (Fig. 5B, lanes 4,5). In the presence of SII, this complement of nucleotides (CpA, CTP, UTP) predominantly yielded a single product that migrated like the major abortive product in reactions that did not contain SII (Fig. 5B, lane 6 vs. lane 4, lane 7 vs. lane 5) but it was dramatically enriched; Mediator marginally stimulated its synthesis (Fig. 5B, lane 7 vs. lane 6).
To eliminate the possibility that there might be some “leaky” transcription past register 15, we also utilized a G15-STOP derivative of the template in which tandem G-residues were inserted at this position (Fig. 5E). This template was subjected to in vitro transcription in the presence of the nucleotides CpA, UTP, CTP and the chain blocker 3′O-methyl GTP. Under these conditions, the expected 16-mer is readily formed (Fig. 5C, lane 4) and, in agreement with a promoter-restricted location for its generation, the n-mer was also efficiently generated in response to SII (Fig. 5C, lane 5 vs. lane 4). Formation of the 16-mer, which reflects an escaped product, remained relatively unaffected in the presence of SII, at least on this DNA template. We used additional Gn-STOP templates (G5, G7, and G9) to generate short RNAs of defined lengths (6-mer, 8-mer, and 10-mer, respectively) to more precisely measure the length of the n-mer (Fig. 5C, lanes 1–4). These templates were transcribed in the presence of the same combination of nucleotides as the G15 template and the resulting products were run alongside the n-mer. Based on this analysis, the SII-induced n-mer cluster was found to be composed of a 9-mer and a 10-mer species.
Since production of an abortive trimer requires neither TFIIH nor energy under these conditions (Goodrich and Tjian, 1994), we investigated the energy requirements for n-mer formation. When dATP (which is used in lieu of ATP in these reactions with subsets of nucleotides; see Supplemental Experimental Procedures) was omitted from the SII-containing reaction, formation of both the 16-mer, and also of the n-mer was abolished (Fig. 5D, lane 2 vs. lane 1). Thus, n-mer formation requires energy either for the extended melting necessary for its synthesis or because it may likely be linked to promoter escape, also an energy-dependent step (Lin et al., 2005) (Discussion). These data further confirm that n-mer formation is a bona fide promoter-centered event and rule out alternative explanations such as generation of the n-mer as a result of SII-dependent cleavage of longer RNAs that are formed subsequent to promoter clearance by Pol II.
Because of the close similarity in the deduced size of the n-mer (9 ntds) and the 16-mer, which reflects the productively escaped transcript that gives rise to a full-length RNA, we were also able to use the data in Fig. 5D to estimate that the n-mer is produced at a 49-fold molar excess relative to the full-length RNA. This indicates that n-mer formation is a reiterative process even if one assumes that not all templates support productive PIC function. [Under our conditions, upto 25% of these DNA templates may be active (Holstege et al., 1997)].
Overall, these results are consistent with the idea that Pol II molecules tend to become arrested when prevented from elongating (e.g., by withholding a nucleotide or through perturbations from a nearby nucleosome) and that SII resets the active site to allow Pol II to restart transcribing (Saunders et al., 2006). But since the situation here pertains to Pol II that is still in a promoter-proximal location, our results further imply that the Pol II active site keeps resuming transcription from the TSS and leads to superstoichiometric production of the n-mer.
Reiterative n-mer synthesis at the promoter correlates with facilitated transcription into the +1 nucleosome
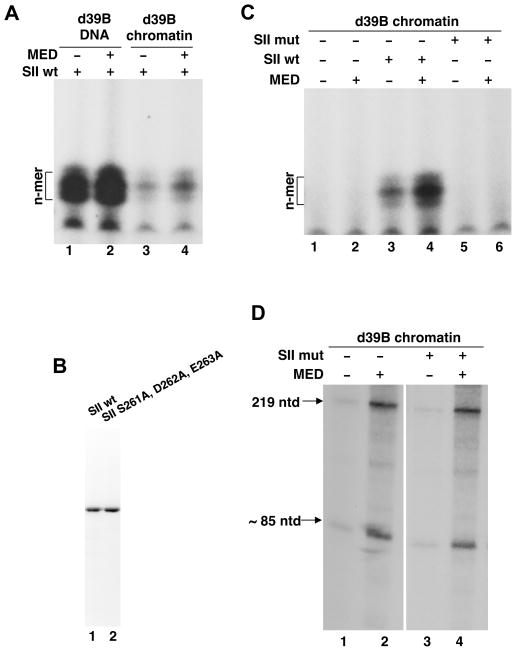
We next asked if the n-mer formation that we characterized on DNA templates plays any role in the context of the nucleosome. Chromatinized and control DNA versions of the d39B template were therefore subjected to a standard transcription reaction in the presence of SII and the resulting products were analyzed for small RNAs (Fig. 6A). As in the preceding analyses, the n-mer formed efficiently from DNA templates both in the absence or presence (Fig. 6A, lanes 1 and 2) of Mediator. It also formed, albeit at a reduced efficiency, on chromatinized d39B (Fig. 6A, lane 3) and was only moderately stimulated by Mediator (Fig. 6A, lane 4 vs. lane 3).
Fig. 6. SII-induced abortive initiation by Pol II is dependent on the catalytic activity of SII and is required for cooperativity with Mediator.
A. In vitro transcription of d39B templates as in Fig. 2C except that reaction products were analyzed on a 25% PAG. The basic reconstituted system (plus HNF4α) was supplemented with SII; Mediator was added as indicated. Other chromatin cofactors (PBAF, p300, FACT) were not added.
B. Coomassie-stained purified bacterially expressed SII and a mutant derivative (SII S261A,D262A,E263A) used in this study.
C. Chromatinized d39B template was subjected to in vitro transcription exactly as in (A). Mediator and either wild-type or mutant SII were added as indicated.
D. Chromatinized d39B template was subjected to in vitro transcription exactly as described for Fig. 4A except that mutant SII was used as indicated.
We also confirmed that synthesis of the n-mer was dependent on the catalytic activity of SII by introducing mutations (S261A,D262A,E263A) in the conserved “RSADE” domain (Guglielmi et al., 2007; Kim et al., 2007). The purified mutant protein (Fig. 6B, lane 2) was tested both for its ability to induce n-mer synthesis (Fig. 6C) and to facilitate transcription from the chromatinized d39B template (Fig. 6D). The results showed a dramatic elimination of the ability to induce n-mer synthesis (Fig. 6C, lanes 5, 6 vs. lanes 3, 4). Importantly, this loss correlated well with the failure of the mutant SII to further enhance Mediator-stimulated transcription across the nucleosomal template (Fig. 6D) and rules out alternative explanations that n-mer synthesis reflects a futile side reaction.
From the above series of experiments in which the +1 nucleosome was maintained in a fixed position, we infer that (i) the +1 nucleosome adversely impacts the ability of a PIC formed in an NFR to initiate transcription; (ii) SII induces Pol II to enter a mode in which it reiteratively synthesizes the n-mer; (iii) Mediator further facilitates the SII-reactivated Pol II to efficiently escape and form longer transcripts; and (iv) that Pol II molecules that initiate in the presence of Mediator display a higher propensity to read through a downstream nucleosome-induced pause.
DISCUSSION
In this paper we describe a transcription system in which Mediator stimulates transcription from a template bearing a precisely positioned +1 nucleosome. Our data implicate Mediator in linking the formation of a PIC that forms in an NFR to its maturation into an elongating complex that efficiently traverses the juxtaposed nucleosome. We also uncover an activity of SII by which it induces reiterative oligomeric RNA synthesis at the promoter and show that Mediator contributes to translating this activity into efficient transcription into the +1 nucleosome.
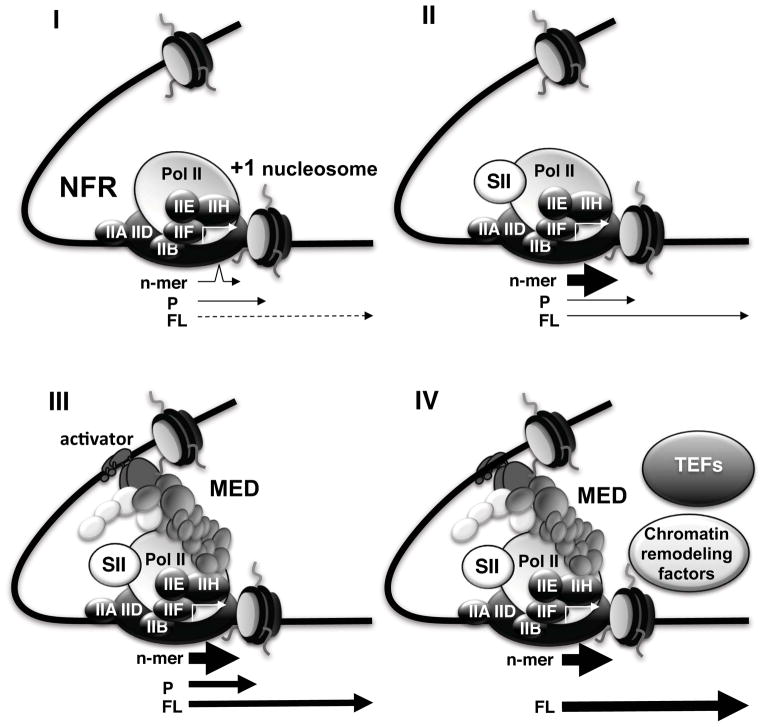
Cumulatively, our data suggest the following series of events at an NFR-embedded promoter (Fig. 7). First, a PIC is formed on the NFR in the template, analogously to one on a naked DNA template. However, PIC function is impacted by the adjacent +1 nucleosome. While low levels of transcription might occur in the absence of any cofactors, most of the Pol II molecules stall at the main pause site within the nucleosome. Whereas the chromatin factors (p300, PBAF, FACT) enhance overall transcription levels through direct effects on the nucleosome, the elongation factor SII is needed to restart the paused subpopulation of Pol II. Overlaid on this chromatin cofactor interplay, especially at earlier steps, is the contribution of the Mediator and SII, which together act to drive higher levels of transcription into the nucleosome, as discussed below. Although the fate of the +1 nucleosome was not investigated, it is reasonable to expect that the combined effects of the chromatin cofactors lead to mobilization of the nucleosome. One outcome of this mobilization appears to be a dynamic occlusion of the promoter and a consequent additional requirement of Mediator (Fig. 3B). This suggests that the PIC recruitment functions of the Mediator manifest not just at the level of loading Pol II but also at the level of preventing destabilization in contexts where nucleosomes are being actively mobilized.
Fig. 7. Pathways for transcriptional activation in the context of a +1 nucleosome.
In I, PICs that form on the NFR are functionally compromised because of the abutting +1 nucleosome (kinked arrow). Some low level of productive transcription occurs but most Pol II molecules pause at the site of the tightest interaction between histones and DNA. In II, SII-induced reiterative initiation by Pol II “jump-starts” transcription by stabilizing the early elongation complex. In III, where Mediator has additionally been recruited (by the activator), this leads to more efficient transcription into the nucleosome, likely as a result of the Mediator’s role in escape; a substantial fraction of Pol II still pauses within the nucleosome. In IV, where additional factors are present, transcription is most efficient both at the earliest steps and through the nucleosome.
A more intriguing role of the Mediator uncovered here pertains to its role in “jump-starting” PICs that fail to efficiently fire in the face of negative interference from the +1 nucleosome. This nucleosome - as in the case of our d39B template – is closely juxtaposed against the PIC and, in view of potential TFIID-nucleosome interactions, is likely a part of the same extended nucleoprotein complex. Unexpectedly, nucleosomal interference manifests already at the level of abortive synthesis of a trimer, which is among the earliest functional hallmarks of an active PIC. Although dinucleotide-primed abortive synthesis may not entail stable promoter melting (Holstege et al., 1997), it would appear that the nucleosome can severely limit even the transient template breathing events that are necessary for trimer formation.
Our finding that SII induces Pol II to reiteratively synthesize a promoter-proximal 9- or 10-mer RNA species following arrest at a promoter-proximal location suggests that this is part of a bypass mechanism that is used by the initiating Pol II to establish a better foothold in this destabilizing environment prior to transitioning productively into the elongation phase. This property seems to be a promoter-proximal manifestation of the known ability of SII to re-start Pol II molecules arrested along the body of the gene (Saunders et al., 2006; Sims et al., 2004). Although this property is highly reminiscent of the phenomenon of abortive initiation it is not an autonomous Pol II activity. Rather, this activity arises from the ability of SII to stimulate Pol II’s intrinsic RNA cleavage activity and may be particularly relevant for short nascent transcripts that are prone to mis-synthesis (Liu et al., 2011; Pal and Luse, 2002). Presumably, cleavage of short nascent RNAs results in release of the n-mers and frees up the Pol II active site to resume RNA synthesis from the +1 register of the TSS. This function could reflect the proposed “tunability” of the Pol II active site by Pol II-interacting factors (Kettenberger et al., 2003). In the present situation, the active site is, in essence, being retuned to alter the length of the favored abortive initiation product (from less than 6 nucleotide long to a 9- or 10-mer). This raises the issue of how induction of this Pol II mode translates into more efficient transcription of the gene body, and more generally, of the role of abortive initiation in the process. It has further been suggested that abortive initiation is a proofreading device that ensures formation of a stable ternary complex that signifies a bona fide PIC (Liu et al., 2011). Analogously, the SII-induced retuning of the active site to increase initial product length might represent an additional mechanism to ensure appropriate formation of a PIC and its eventual maturation into an elongating complex in the destabilizing nucleosomal environment.
How does Mediator aid in translating this reiterative n-mer synthesis mode into productive transcription into the nucleosome? Although the catalytic activity of the yeast ortholog of SII is not required for its promoter-restricted functions, genetic analyses in yeast suggested that SII and the MED31 Mediator subunit function at the same early step in transcription (Guglielmi et al., 2007). Additional insights into how Mediator might act come from our observation that when transcription reactions are performed for extended periods of time, Mediator effects are partially amortized. Furthermore, it has been shown previously that PICs at a given promoter fire at different rates, presumably because some complexes are diverted off-line (Luse and Jacob, 1987); nonetheless, such complexes eventually fire and in extended incubations catch up with the faster-initiating complexes. We therefore propose that PICs that form at the d39B NFR are functionally diverse, with a significant fraction incapable of overcoming nucleosomal interference. A post-recruitment role of Mediator is either to speed up the transition of these complexes to active forms or to prevent descent into non-functional states in the first place. This parallels the above role of SII in reviving failing Pol II molecules at the promoter. Indeed, without the Mediator, the SII-induced reiterative RNA synthesis is not effectively channeled into productive RNA synthesis. It may even be that through Mediator’s known role in PIC stability, PICs being acted upon by SII are held together long enough to ensure a properly initiated transcript.
The net outcome is that Mediator and SII facilitate the initiation to elongation transition. We propose that in the course of promoting PIC formation, Mediator also gives rise to Pol II molecules that are particularly amenable to promoter escape, especially in the context of natural (chromatin) templates. It is potentially relevant that the size and ATP-dependence of the reiteratively synthesized RNA described here is reminiscent of the requirements for promoter escape (Pal et al., 2005). The physical basis of this effect might entail: (i) Mediator-induced conformational changes (e.g., clamp movement) in Pol II (Cai et al., 2012); (ii) Mediator stabilization in the PIC of TFIIB (Baek et al., 2006), the GTF that oversees the transition from the abortive phase to escape and productive elongation (Pal et al., 2005); or (iii) modulation through Mediator interactions with TFIIH, the GTF implicated in both promoter melting and escape (Esnault et al., 2008; Lin et al., 2005; Roeder, 1998). Although any combination of these mechanisms might prevail, the first possibility is particularly attractive in light of the observation that Mediator effects persist at distal locations for some Pol II molecules that initiated in its presence and which continue to display enhanced processivity with respect to the P site (Fig. 4A).
The mechanisms discussed here are relevant for gene regulation at the level of the promoter-proximally paused Pol II. Although interplay between negatively and positively acting elongation factors is a major determinant in regulating the pause (Gilmour, 2009), a critical role of the +1 nucleosome is anticipated (Brown et al., 1996). Paused Pol II molecules generally map to a region 50 to 100 bp downstream of the promoter in genome-wide studies, but cases of Pol II localizing much closer to the promoter are also known (Nechaev et al., 2010). Our data would predict that a “poised” Pol II might exist in a state in which it would continuously generate small RNAs in conjunction with SII (Fig. 7). Upon transduction of appropriate signals via activator and Mediator recruitment, Pol II would be redirected into a productive pathway to synthesize the full-length transcript. Because it very efficiently elicits bona fide Pol II pausing 85 bp downstream of the TSS, our system is also a good model for cases in which pausing occurs in this region. Importantly, our system is responsive to a combination of factors that counteract the nucleosome-induced pause even as it is clear that additional factors are involved in vivo. We note that although MED26 has been implicated in interactions with P-TEFb and associated factors, which potentially might be involved in regulating the paused Pol II (Takahashi et al., 2011), the Mediator effects that we report are mainly exerted at an earlier step and are independent of these elongation factors.
EXPERIMENTAL PROCEDURES
Protein purification
Recombinant core histones, Pol II, GTFs, Mediator and p300 were purified essentially as described (Barrero and Malik, 2006; Malik and Roeder, 2003). Purification of these and other factors are also presented in the Supplement.
Template constructions
The d19B, d39B and d134B templates were assembled from modular components consisting of an arm that contained the HNF4 binding sites and the core promoter and a corresponding “601” sequence (Lowary and Widom, 1998). For the d39B and d134B templates, promoter arms were derived from the HNF4 cognate template pA4XMLΔ53 (Malik et al., 2007) by introducing an Ava I site (CTCGGG) either 15 bp or 110 bp downstream from the TSS. The products were subcloned into pGEM-T vectors (Promega). The arms were released by digestion with Sca I and Ava I and gel-purified. PCR primers that introduced the Ava I site at the 5′ end and a biotinylated residue at the 3′ end were used to generate 601 fragments for ligation to the arms. The Ava I site was 97 bp upstream from the dyad center (Hha I site) of the 601 sequence. Since 73 bp on either side of the dyad are protected by the nucleosome, fusion with an arm with Ava I at +15 places the upstream nucleosome edge at +39 for the d39B template and fusion with an arm with Ava I at +110 places it at +134 in the case of the d134B template. The “B” orientation as used here is defined by the 5′-StyI-HhaI-NotI-3′ arrangement of restriction sites (Bondarenko et al., 2006; Carey et al., 2006). Following PCR, the products were digested with Ava I and gel-purified. For the d19B template, the Ava I site was introduced at −4 in the core promoter and the arms were obtained as described above. The corresponding 601 fragment was obtained by using as PCR primer a sequence that introduced an Ava I site as well as the core promoter sequences from the pA4XMLΔ53 template that spanned from −4 to +19; ligation with the corresponding arm places the upstream nucleosome edge at +19. Additional details are presented in Supplemental Procedures.
The “Gn-STOP” templates were also derived from pA4XMLΔ53 by using a PCR primer that introduced an Ava I site (which introduces 3 tandem G residues) at the appropriate location. The PCR products were cloned into pGEM-T.
Chromatin assembly
Mononucleosome assembly was achieved by salt-dialysis. Typically, the appropriate Ava I-digested 601 fragment (600 ng) was mixed with 1.4 μg recombinant histones in 50 μl buffer containing 2 M NaCl; for naked DNA templates, histones were omitted. Samples were placed in Millipore Slide-a-Lyzer tubes and dialyzed to 30 mM NaCl. Dialysis was carried out over 20 hr using a peristaltic pump to slowly deliver the diluent. See also Supplemental Procedures.
In vitro transcription and PIC recruitment assays
For in vitro transcription, ligated templates were diluted 4-fold and immobilized to M280 Dynabeads. After washing, in vitro transcription reactions with these templates were done using pure components essentially as described (Malik and Roeder, 2003). Basic reactions were reconstituted with all the GTFs, Pol II and PC4; Mediator, SII and other chromatin cofactors were added during PIC formation as specified in figure legends. For standard reactions, PIC formation was first allowed to take place (45 min, 30°C) and transcription was initiated by NTP addition (details in Supplemental Procedures). Reactions were stopped by proteinase K treatment of bead-bound material. RNA was analyzed on 8% PAG/urea. To visualize oligomeric RNAs, supernatants from the transcription reactions were cipped and analyzed directly by electrophoresis on 25% PAG/urea. Gels were autoradiographed; selected experiments were quantified using Molecular Dynamics phosphorimaging except data in Fig. 4D, Fig. 5D and Fig. S7, which were scanned as TIFF files and quantified by ImageQuant software. After background adjustment, the data were normalized to the full-length product made in the absence of any cofactors. Because products were uniformly labeled, the 85-mer band was additionally corrected for its C-content by multiplying by 2.46.
Recruitment assays were done using our standard methods, which are described in Supplemental Procedures.
Supplementary Material
HIGHLIGHTS.
Development of an in vitro system to analyze transcription through a +1 nucleosome
There is functional interplay between Mediator and chromatin cofactors
SII induces reiterative oligomeric RNA synthesis by promoter-bound Pol II
Mediator translates this activity of SII into processive transcription
Acknowledgments
We are indebted to Dr. R.G. Roeder for constant support. This work was supported, in part, by NIH grant RO1 DK060764 to SM. JMA was supported by a Ruth Kirschstein postdoctoral fellowship (F32 DK082140-01A1) and MJB by a Rockefeller University Women & Science fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baek HJ, Kang YK, Roeder RG. Human mediator enhances basal transcription by facilitating recruitment of transcription factor IIB during preinitiation complex assembly. J Biol Chem. 2006;281:15172–15181. doi: 10.1074/jbc.M601983200. [DOI] [PubMed] [Google Scholar]
- Barrero MJ, Malik S. Two functional modes of a nuclear receptor-recruited arginine methyltransferase in transcriptional activation. Mol Cell. 2006;24:233–243. doi: 10.1016/j.molcel.2006.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondarenko VA, Steele LM, Ujvari A, Gaykalova DA, Kulaeva OI, Polikanov YS, Luse DS, Studitsky VM. Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Mol Cell. 2006;24:469–479. doi: 10.1016/j.molcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Brown SA, Imbalzano AN, Kingston RE. Activator-dependent regulation of transcriptional pausing on nucleosomal templates. Genes & Dev. 1996;10:1479–1490. doi: 10.1101/gad.10.12.1479. [DOI] [PubMed] [Google Scholar]
- Cai G, Chaban YL, Imasaki T, Kovacs JA, Calero G, Penczek PA, Takagi Y, Asturias FJ. Interaction of the mediator head module with RNA polymerase II. Structure. 2012;20:899–910. doi: 10.1016/j.str.2012.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey M, Li B, Workman JL. RSC exploits histone acetylation to abrogate the nucleosomal block to RNA polymerase II elongation. Mol Cell. 2006;24:481–487. doi: 10.1016/j.molcel.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C, Ghavi-Helm Y, Brun S, Soutourina J, Van Berkum N, Boschiero C, Holstege F, Werner M. Mediator-dependent recruitment of TFIIH modules in preinitiation complex. Mol Cell. 2008;31:337–346. doi: 10.1016/j.molcel.2008.06.021. [DOI] [PubMed] [Google Scholar]
- Gilmour DS. Promoter proximal pausing on genes in metazoans. Chromosoma. 2009;118:1–10. doi: 10.1007/s00412-008-0182-4. [DOI] [PubMed] [Google Scholar]
- Goodrich JA, Tjian R. Transcription factors IIE and IIH and ATP hydrolysis direct promoter clearance by RNA polymerase II. Cell. 1994;77:145–156. doi: 10.1016/0092-8674(94)90242-9. [DOI] [PubMed] [Google Scholar]
- Guermah M, Palhan VB, Tackett AJ, Chait BT, Roeder RG. Synergistic functions of SII and p300 in productive activator-dependent transcription of chromatin templates. Cell. 2006;125:275–286. doi: 10.1016/j.cell.2006.01.055. [DOI] [PubMed] [Google Scholar]
- Guglielmi B, Soutourina J, Esnault C, Werner M. TFIIS elongation factor and Mediator act in conjunction during transcription initiation in vivo. Proc Natl Acad Sci USA. 2007;104:16062–16067. doi: 10.1073/pnas.0704534104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege FC, Fiedler U, Timmers HT. Three transitions in the RNA polymerase II transcription complex during initiation. EMBO J. 1997;16:7468–7480. doi: 10.1093/emboj/16.24.7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettenberger H, Armache KJ, Cramer P. Architecture of the RNA polymerase II-TFIIS complex and implications for mRNA cleavage. Cell. 2003;114:347–357. doi: 10.1016/s0092-8674(03)00598-1. [DOI] [PubMed] [Google Scholar]
- Kim B, Nesvizhskii AI, Rani PG, Hahn S, Aebersold R, Ranish JA. The transcription elongation factor TFIIS is a component of RNA polymerase II preinitiation complexes. Proc Natl Acad Sci USA. 2007;104:16068–16073. doi: 10.1073/pnas.0704573104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Guermah M, Roeder RG. The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell. 2010;140:491–503. doi: 10.1016/j.cell.2009.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kireeva ML, Hancock B, Cremona GH, Walter W, Studitsky VM, Kashlev M. Nature of the nucleosomal barrier to RNA polymerase II. Mol Cell. 2005;18:97–108. doi: 10.1016/j.molcel.2005.02.027. [DOI] [PubMed] [Google Scholar]
- Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci. 2005;30:235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Lemon B, Inouye C, King DS, Tjian R. Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature. 2001;414:924–928. doi: 10.1038/414924a. [DOI] [PubMed] [Google Scholar]
- Lin YC, Choi WS, Gralla JD. TFIIH XPB mutants suggest a unified bacterial-like mechanism for promoter opening but not escape. Nat Struct Mol Biol. 2005;12:603–607. doi: 10.1038/nsmb949. [DOI] [PubMed] [Google Scholar]
- Liu X, Bushnell DA, Silva DA, Huang X, Kornberg RD. Initiation complex structure and promoter proofreading. Science. 2011;333:633–637. doi: 10.1126/science.1206629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- Luse DS, Jacob GA. Abortive initiation by RNA polymerase II in vitro at the adenovirus 2 major late promoter. J Biol Chem. 1987;262:14990–14997. [PubMed] [Google Scholar]
- Luse DS, Spangler LC, Ujvari A. Efficient and rapid nucleosome traversal by RNA polymerase II depends on a combination of transcript elongation factors. J Biol Chem. 2011;286:6040–6048. doi: 10.1074/jbc.M110.174722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Barrero MJ, Jones T. Identification of a regulator of transcription elongation as an accessory factor for the human Mediator coactivator. Proc Natl Acad Sci USA. 2007;104:6182–6187. doi: 10.1073/pnas.0608717104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Roeder RG. Isolation and functional characterization of the TRAP/Mediator complex. Methods Enzymol. 2003;364:257–284. doi: 10.1016/s0076-6879(03)64015-2. [DOI] [PubMed] [Google Scholar]
- Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nature Rev Gen. 2010;11:761–772. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margaritis T, Holstege FC. Poised RNA polymerase II gives pause for thought. Cell. 2008;133:581–584. doi: 10.1016/j.cell.2008.04.027. [DOI] [PubMed] [Google Scholar]
- Nakajima N, Horikoshi M, Roeder RG. Factors involved in specific transcription by mammalian RNA polymerase II: purification, genetic specificity, and TATA box-promoter interactions of TFIID. Mol Cell Biol. 1988;8:4028–4040. doi: 10.1128/mcb.8.10.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S, Fargo DC, dos Santos G, Liu L, Gao Y, Adelman K. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science. 2010;327:335–338. doi: 10.1126/science.1181421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal M, Luse DS. Strong natural pausing by RNA polymerase II within 10 bases of transcription start may result in repeated slippage and reextension of the nascent RNA. Mol Cell Biol. 2002;22:30–40. doi: 10.1128/MCB.22.1.30-40.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal M, Ponticelli AS, Luse DS. The role of the transcription bubble and TFIIB in promoter clearance by RNA polymerase II. Mol Cell. 2005;19:101–110. doi: 10.1016/j.molcel.2005.05.024. [DOI] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature. 2012;483:295–301. doi: 10.1038/nature10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder RG. Role of general and gene-specific cofactors in the regulation of eukaryotic transcription. Cold Spring Harb Symp Quant Biol. 1998;63:201–218. doi: 10.1101/sqb.1998.63.201. [DOI] [PubMed] [Google Scholar]
- Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes & Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Parmely TJ, Sato S, Tomomori-Sato C, Banks CA, Kong SE, Szutorisz H, Swanson SK, Martin-Brown S, Washburn MP, et al. Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell. 2011;146:92–104. doi: 10.1016/j.cell.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.