Abstract
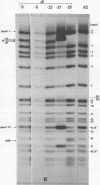
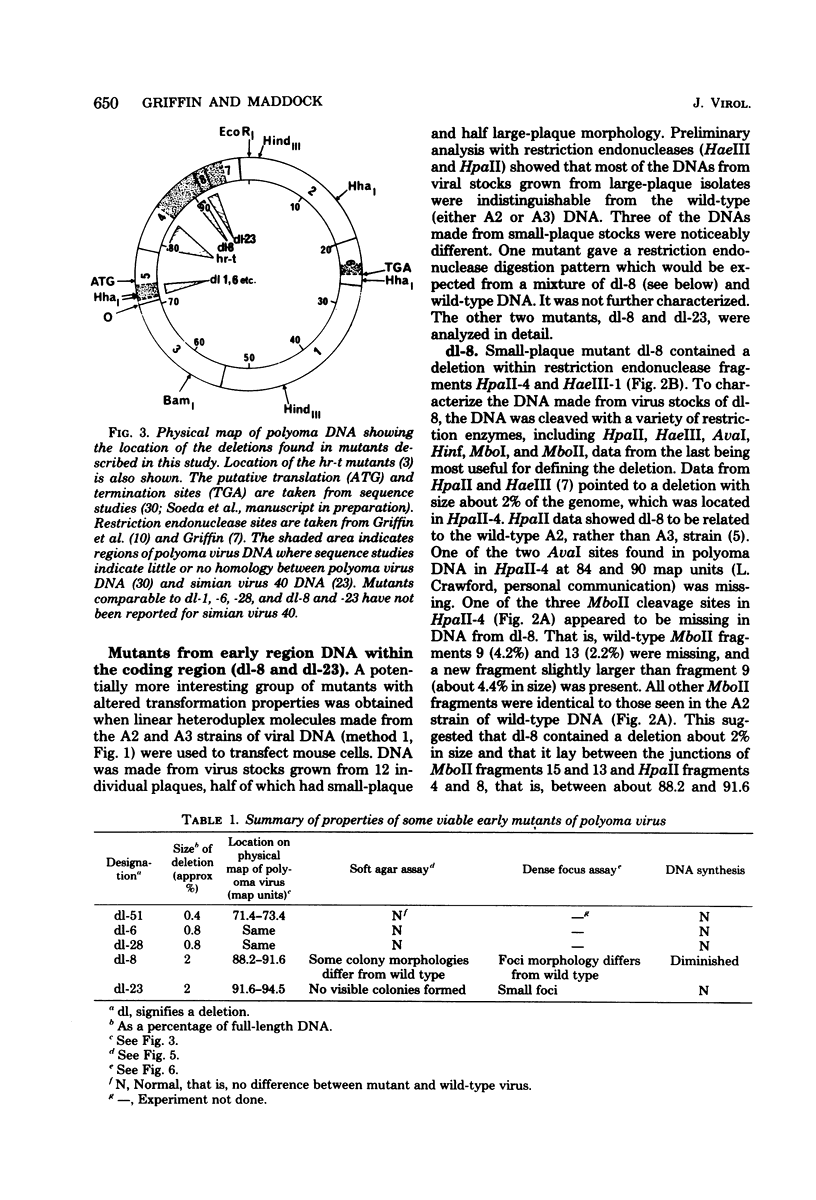
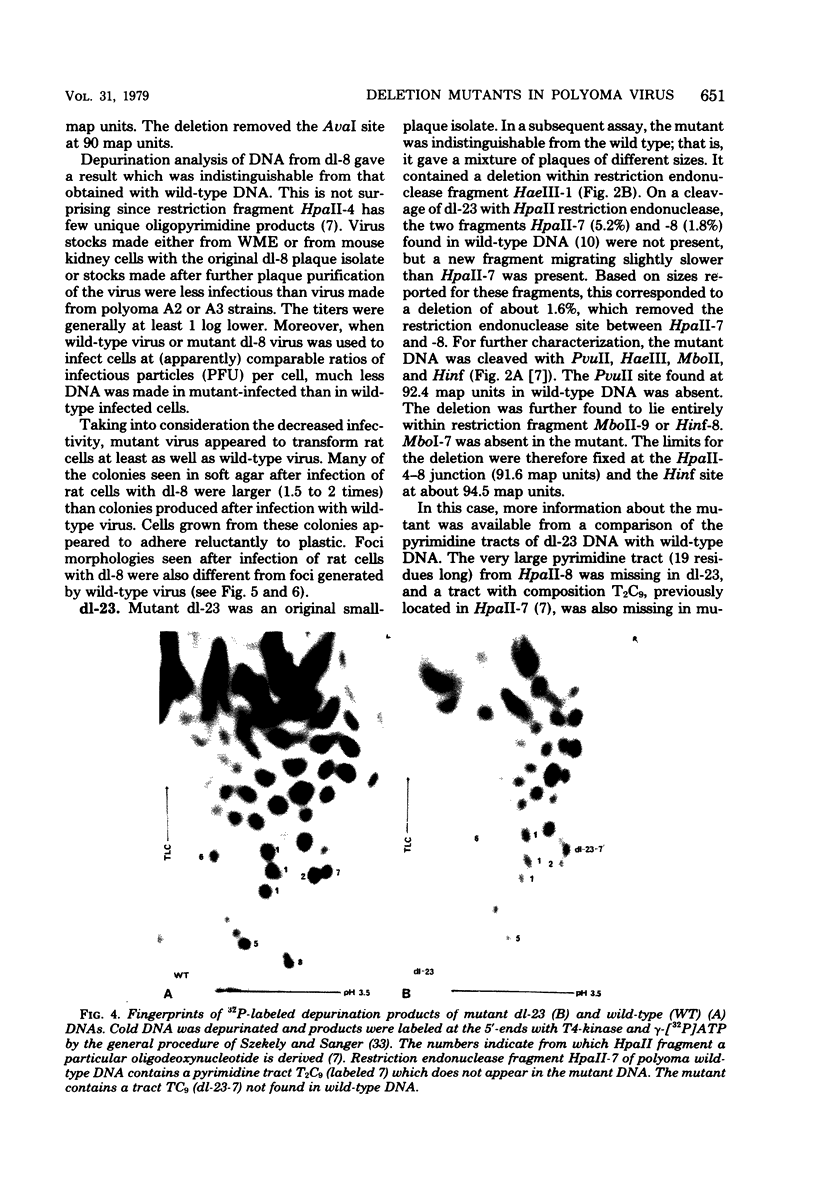
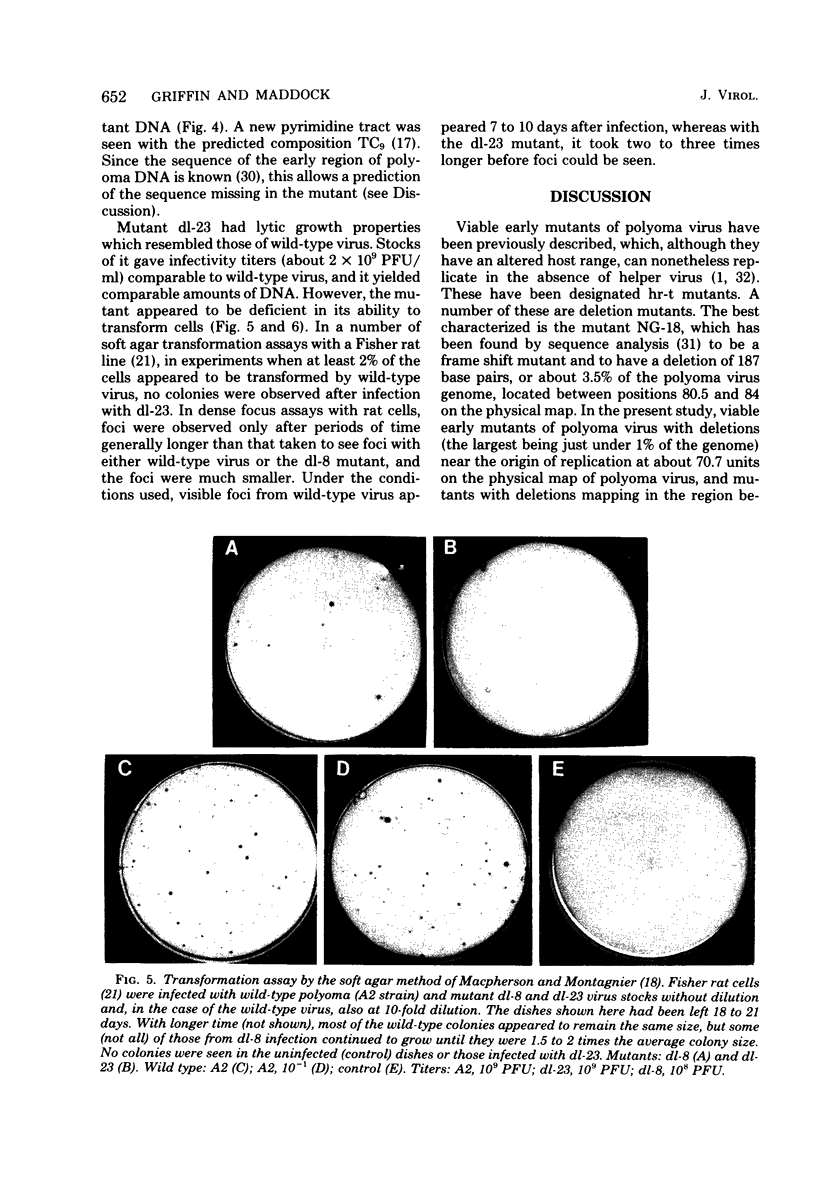
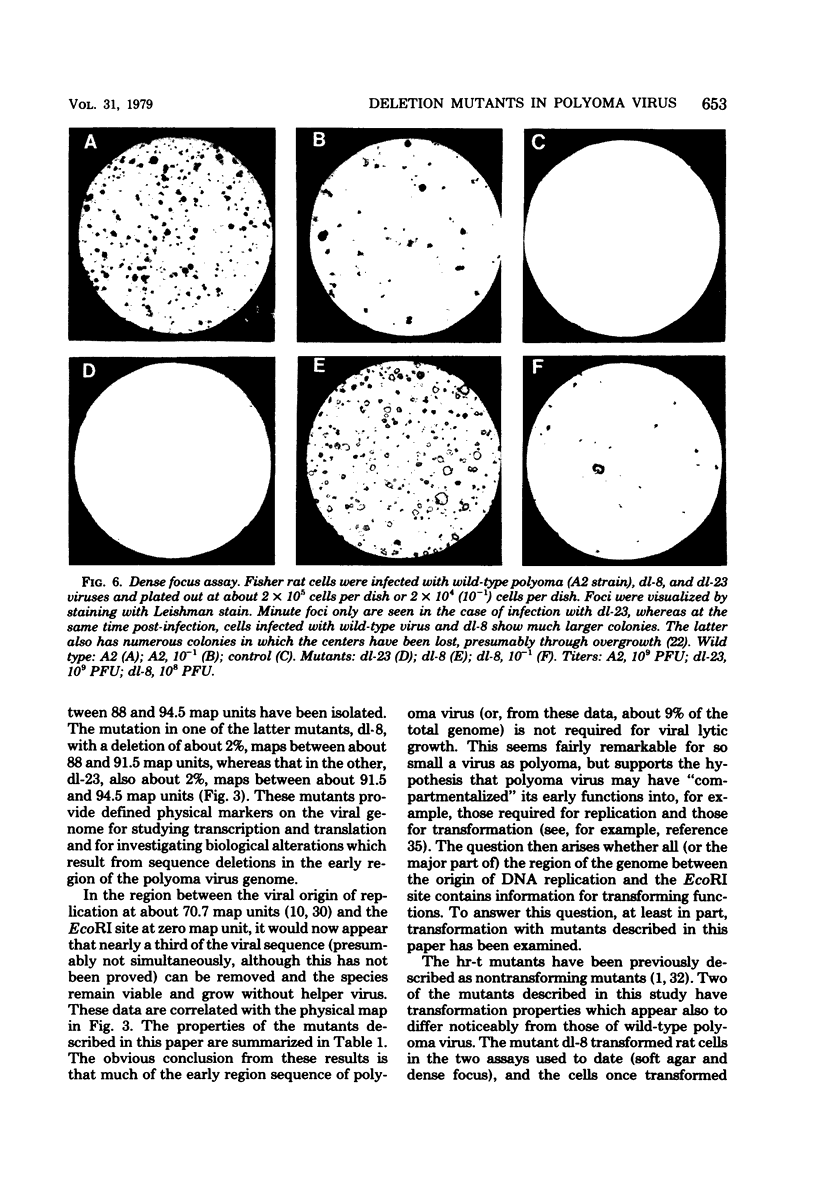
Viable mutants of polyoma virus have been isolated which have deletions in defined parts of the early region of the genome. One class of mutants has deletions (less than 1% of viral genome length) located between 71.5 and 73.5 on the physical map of polyoma virus DNA, near the origin of replication. These mutants appear to grow and to transform cells in a manner indistinguishable from wild-type virus. A second type of mutant with deletions (about 2% of viral genome length) located between about 88 and 94.5 units on the physical map of polyoma virus DNA have altered transformation properties. One of the latter (which maps between 88 and 91.5 units) also has altered growth characteristics, whereas another (which maps between 91.5 and 94.5 units) resembles wild-type virus in its growth properties. The regions with deleted sequences have been defined by cleaving mutant DNAs with restriction endonucleases and analyzing pyrimidine tracts.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benjamin T. L. Host range mutants of polyoma virus. Proc Natl Acad Sci U S A. 1970 Sep;67(1):394–399. doi: 10.1073/pnas.67.1.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhart W. Complementation between temperature-sensitive (ts) and host range nontransforming (hr-t) mutants of polyoma virus. Virology. 1977 Apr;77(2):589–597. doi: 10.1016/0042-6822(77)90484-6. [DOI] [PubMed] [Google Scholar]
- Feunteun J., Sompayrac L., Fluck M., Benjamin T. Localization of gene functions in polyoma virus DNA. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4169–4173. doi: 10.1073/pnas.73.11.4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluck M. M., Staneloni R. J., Benjamin T. L. Hr-t and ts-a: two early gene functions of polyoma virus. Virology. 1977 Apr;77(2):610–624. doi: 10.1016/0042-6822(77)90486-x. [DOI] [PubMed] [Google Scholar]
- Fried M., Griffin B. E., Lund E., Robberson D. L. Polyoma virus--a study of wild-type, mutant and defective DNAs. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):45–52. doi: 10.1101/sqb.1974.039.01.009. [DOI] [PubMed] [Google Scholar]
- Fried M., Griffin B. E. Organization of the genomes of polyoma virus and SV40. Adv Cancer Res. 1977;24:67–113. doi: 10.1016/s0065-230x(08)61013-1. [DOI] [PubMed] [Google Scholar]
- Griffin B. E. Fine structure of polyoma virus DNA. J Mol Biol. 1977 Dec 5;117(2):447–471. doi: 10.1016/0022-2836(77)90137-1. [DOI] [PubMed] [Google Scholar]
- Griffin B. E., Fried M. Amplification of a specific region of the polyoma virus genome. Nature. 1975 Jul 17;256(5514):175–179. doi: 10.1038/256175a0. [DOI] [PubMed] [Google Scholar]
- Griffin B. E., Fried M., Cowie A. Polyoma DNA: a physical map. Proc Natl Acad Sci U S A. 1974 May;71(5):2077–2081. doi: 10.1073/pnas.71.5.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Hunter T., Hutchinson M. A., Eckhart W. Translation of polyoma virus T antigens in vitro. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5917–5921. doi: 10.1073/pnas.75.12.5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson M. A., Hunter T., Eckhart W. Characterization of T antigens in polyoma-infected and transformed cells. Cell. 1978 Sep;15(1):65–77. doi: 10.1016/0092-8674(78)90083-1. [DOI] [PubMed] [Google Scholar]
- Ito Y., Brocklehurst J. R., Dulbecco R. Virus-specific proteins in the plasma membrane of cells lytically infected or transformed by pol-oma virus. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4666–4670. doi: 10.1073/pnas.74.10.4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y., Spurr N., Dulbecco R. Characterization of polyoma virus T antigen. Proc Natl Acad Sci U S A. 1977 Mar;74(3):1259–1263. doi: 10.1073/pnas.74.3.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling V. Fractionation and sequences of the large pyrimidine oligonucleotides from bacteriophage fd DNA. J Mol Biol. 1972 Feb 28;64(1):87–102. doi: 10.1016/0022-2836(72)90322-1. [DOI] [PubMed] [Google Scholar]
- MACPHERSON I., MONTAGNIER L. AGAR SUSPENSION CULTURE FOR THE SELECTIVE ASSAY OF CELLS TRANSFORMED BY POLYOMA VIRUS. Virology. 1964 Jun;23:291–294. doi: 10.1016/0042-6822(64)90301-0. [DOI] [PubMed] [Google Scholar]
- Miller L. K., Cooke B. E., Fried M. Fate of mismatched base-pair regions in polyoma heteroduplex DNA during infection of mouse cells. Proc Natl Acad Sci U S A. 1976 Sep;73(9):3073–3077. doi: 10.1073/pnas.73.9.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller L. K., Fried M. Construction of the genetic map of the polyoma genome. J Virol. 1976 Jun;18(3):824–832. doi: 10.1128/jvi.18.3.824-832.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad I., Zouzias D., Basilico C. State of the viral DNA in rat cells transformed by polyoma virus. I. Virus rescue and the presence of nonintergrated viral DNA molecules. J Virol. 1976 May;18(2):436–444. doi: 10.1128/jvi.18.2.436-444.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy V. B., Thimmappaya B., Dhar R., Subramanian K. N., Zain B. S., Pan J., Ghosh P. K., Celma M. L., Weissman S. M. The genome of simian virus 40. Science. 1978 May 5;200(4341):494–502. doi: 10.1126/science.205947. [DOI] [PubMed] [Google Scholar]
- Schaffhausen B. S., Silver J. E., Benjamin T. L. Tumor antigen(s) in cell productively infected by wild-type polyoma virus and mutant NG-18. Proc Natl Acad Sci U S A. 1978 Jan;75(1):79–83. doi: 10.1073/pnas.75.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seif R., Cuzin F. Temperature-sensitive growth regulation in one type of transformed rat cells induced by the tsa mutant of polyoma virus. J Virol. 1977 Dec;24(3):721–728. doi: 10.1128/jvi.24.3.721-728.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenk T. E., Carbon J., Berg P. Construction and analysis of viable deletion mutants of simian virus 40. J Virol. 1976 May;18(2):664–671. doi: 10.1128/jvi.18.2.664-671.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenk T. E., Rhodes C., Rigby P. W., Berg P. Biochemical method for mapping mutational alterations in DNA with S1 nuclease: the location of deletions and temperature-sensitive mutations in simian virus 40. Proc Natl Acad Sci U S A. 1975 Mar;72(3):989–993. doi: 10.1073/pnas.72.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenk T. A biochemical method for increasing the size of deletion mutations in simian virus 40 DNA. J Mol Biol. 1977 Jul 5;113(3):503–515. doi: 10.1016/0022-2836(77)90235-2. [DOI] [PubMed] [Google Scholar]
- Smart J. E., Ito Y. Three species of polyoma virus tumor antigens share common peptides probably near the amino termini of the proteins. Cell. 1978 Dec;15(4):1427–1437. doi: 10.1016/0092-8674(78)90066-1. [DOI] [PubMed] [Google Scholar]
- Soeda E., Arrand J. R., Smolar N., Griffin B. E. Sequence from early region of polyoma virus DNA containing viral replication origin and encoding small, middle and (part of) large T antigens. Cell. 1979 Jun;17(2):357–370. doi: 10.1016/0092-8674(79)90162-4. [DOI] [PubMed] [Google Scholar]
- Soeda E., Griffin B. E. Sequences from the genome of a non-transforming mutant of polyoma virus. Nature. 1978 Nov 16;276(5685):294–298. doi: 10.1038/276294a0. [DOI] [PubMed] [Google Scholar]
- Staneloni R. J., Fluck M. M., Benjamin T. L. Host range selection of transformation-defective hr-t mutants of polyoma virus. Virology. 1977 Apr;77(2):598–609. doi: 10.1016/0042-6822(77)90485-8. [DOI] [PubMed] [Google Scholar]
- Székely M., Sanger F. Use of polynucleotide kinase in fingerprinting non-radioactive nucleic acids. J Mol Biol. 1969 Aug 14;43(3):607–617. doi: 10.1016/0022-2836(69)90362-3. [DOI] [PubMed] [Google Scholar]
- Vogt V. M. Purification and further properties of single-strand-specific nuclease from Aspergillus oryzae. Eur J Biochem. 1973 Feb 15;33(1):192–200. doi: 10.1111/j.1432-1033.1973.tb02669.x. [DOI] [PubMed] [Google Scholar]
- Weinberg R. A. How does T antigen transform cells? Cell. 1977 Jun;11(2):243–246. doi: 10.1016/0092-8674(77)90041-1. [DOI] [PubMed] [Google Scholar]