SUMMARY
The mammalian target of rapamycin (mTOR) integrates signals from nutrients and insulin via two distinct complexes, mTORC1 and mTORC2. Disruption of mTORC2 impairs the insulin-induced activation of Akt, an mTORC2 substrate. Here we found that mTORC2 can also regulate insulin signaling at the level of insulin receptor substrate-1 (IRS-1). Despite phosphorylation at the mTORC1-mediated serine sites, which supposedly triggers IRS-1 downregulation, inactive IRS-1 accumulated in mTORC2-disrupted cells. Defective IRS-1 degradation was due to attenuated expression and phosphorylation of the ubiquitin ligase substrate-targeting subunit, Fbw8. mTORC2 stabilizes Fbw8 by phosphorylation at Ser86, allowing the insulin-induced translocation of Fbw8 to the cytosol where it mediates IRS-1 degradation. Thus, mTORC2 negatively feeds back to IRS-1 via control of Fbw8 stability and localization. Our findings reveal that in addition to persistent mTORC1 signaling, heightened mTORC2 signals can promote insulin resistance due to mTORC2-mediated degradation of IRS-1.
INTRODUCTION
mTOR responds to the presence of nutrients, energy and growth factors to link cellular metabolism with growth and proliferation. The rapamycin-sensitive mTOR protein complex 1 (mTORC1) activates the translational regulator S6K leading to increased protein synthesis in response to amino acids (Zoncu et al., 2011). On the other hand, mTORC2 composed of mTOR, rictor, SIN1 and mLST8, which is neither directly or acutely sensitive to rapamycin, responds to growth factors such as insulin (Oh and Jacinto, 2011). Insulin stimulation induces PI3K activation and subsequent increase of phosphatidylinositol-3,4,5-trisphosphate (PIP3). This phospholipid second messenger recruits Akt to the plasma membrane where Akt becomes phosphorylated and activated by PDK1 and mTORC2. Activated Akt mediates the metabolic actions of insulin such as augmenting glucose transport and promoting mTORC1 signaling to drive protein synthesis and cell growth (Zoncu et al., 2011).
Proximal to the insulin receptor (IR), the insulin receptor substrate (IRS) mediates signals triggered by insulin. Inhibition of IRS function contributes to desensitization of cells to insulin that can occur in type 2 diabetes and other metabolic disorders (White, 2002). IRS (family of proteins IRS-1–6; referred herein as the well-studied IRS-1) is recruited to the IR and phosphorylated at Tyr residues upon IR stimulation. IRS-1 contains up to 20 potential Tyr phosphosites. Tyr-phosphorylated IRS-1 is associated with positive regulation of insulin signaling as it recruits a number of effector molecules that bind to the pTyr via their SH2 domains. However, IRS-1 is also regulated via Ser phosphorylation at numerous sites (Gual et al., 2005). Although Ser phosphorylation at specific sites can enhance insulin signaling (Giraud et al., 2004; Luo et al., 2007; Paz et al., 1999), most studies support that hyper-Ser/Thr phosphorylation serves as a negative feedback to downregulate IRS-1 function and can eventually promote IRS-1 degradation and insulin resistance (Hotamisligil et al., 1996; Liu et al., 2004; Shah and Hunter, 2006; Tremblay et al., 2007). Ser phosphorylation of IRS-1 at critical sites can block Tyr phosphorylation and prevent IRS-1 binding to the IR (Hartman et al., 2001; Liu et al., 2004). Thus, a balance of negative and positive signals via counteracting Ser and Tyr phosphorylation serves to regulate IRS-1.
Numerous kinases including mTOR have been linked to IRS-1 downregulation (Gual et al., 2005). In line with the role of mTORC1 in IRS-1 downregulation, rapamycin inhibits IRS-1 mobility upshift in SDS-PAGE, which corresponds to increased Ser phosphorylation but whether it blocks IRS-1 degradation is controversial (Haruta et al., 2000; Pederson et al., 2001; Sun et al., 1992; Zhande et al., 2002). Moreover, mTORC1 is linked to phosphorylation of IRS-1 at Ser636/639 (murine Ser632/635) and raptor knockdown can stabilize IRS-1 (Tzatsos, 2009). IRS-1 is also phosphorylated at Ser302 by S6K. Increased phosphorylation at this site along with Ser632 and Ser635 has been observed in mouse models of obesity and insulin resistance (Harrington et al., 2004; Um et al., 2004; Werner et al., 2004). Importantly, elevated activation of the mTORC1/S6K pathway that can occur in the absence of TSC1/TSC2, elicits a negative feedback signal that hyperphosphorylates IRS-1 at Ser residues thereby diminishing insulin signals (Harrington et al., 2004; Shah et al., 2004; Tremblay and Marette, 2001; Um et al., 2004). Thus, abundant nutrients that augment mTORC1 signaling could generate insulin resistance via the mTORC1/S6K-mediated phosphorylation and downregulation of IRS-1.
IRS-1 degradation following chronic insulin exposure occurs in a PI3K-dependent manner and involves the ubiquitin (Ub)-proteasome pathway (Egawa et al., 2000; Haruta et al., 2000; Pederson et al., 2001; Smith et al., 1995; Sun et al., 1999). PI3K inhibition blocks the 26S proteasome-mediated degradation of IRS-1 (Lee et al., 2000). 26S proteasome inhibitors also prevent IRS-1 degradation (Lee et al., 2000; Sun et al., 1999). The proteasome degradation cascade is initiated by activation of E1 ubiquitin activating enzyme, followed by activation of E2 Ub-conjugating enzymes that transfer ubiquitin from E1 to E3 that then catalyzes substrate ubiquitylation. Ub-tagged proteins are recognized by the 26S proteasome and undergo degradation. Recently, the Cullin 7 (CUL7) E3 Ub-ligase complex containing the substrate recognition subunit F-box protein Fbw8 (also called Fbxw8 and Fbx29) and ROC1 RING finger protein was identified to target IRS-1 in an mTOR/S6K dependent manner (Xu et al., 2008). The precise mechanism as to how this Ub-ligase complex can target IRS-1 for degradation remains to be further elucidated.
Whereas the role of mTORC1 in downregulating insulin signals is relatively well characterized, it is unclear if mTORC2 can also signal to IRS-1 via feedback regulation. mTORC2 promotes the allosteric activation and stability of AGC kinases and some of these kinases can regulate IRS-1 as part of a feedback loop (Gual et al., 2005; Oh and Jacinto, 2011). Whether enhanced mTORC2 signals could promote insulin resistance via mechanisms other than AGC kinase regulation has not been addressed. Here we found that mTORC2 can negatively feed back to IRS-1 to diminish insulin signaling. We uncover that mTORC2 functions in the regulation of Fbw8 that allows Fbw8 to mediate IRS-1 turnover following insulin stimulation.
RESULTS
IRS-1 protein levels are increased upon mTORC2 disruption
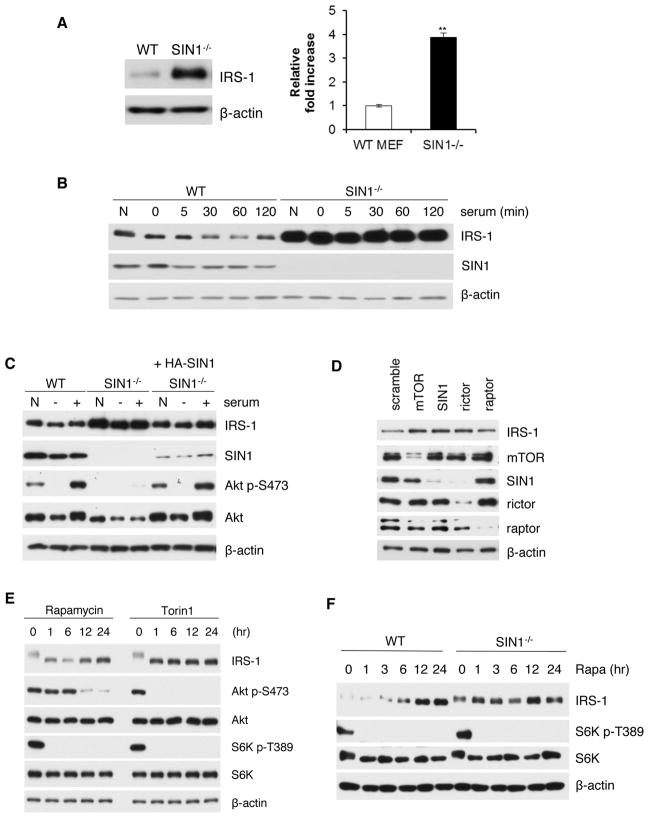
Akt and PKC phosphorylation are defective in mTORC2-disrupted cells whereas S6K activation is normal (Oh and Jacinto, 2011). Since Akt and PKC have also been reported to control IRS-1, we investigated how insulin/IRS-1 signaling is affected in mTORC2-disrupted cells. Unexpectedly, IRS-1 levels were highly elevated in the absence of SIN1 (Figure 1A). Following serum restimulation, IRS-1 levels were decreased by 30 minutes in wild type (WT) MEFs, whereas they remained elevated in SIN1−/− cells (Figure 1B). The increase in IRS-1 levels can be attenuated upon re-expression of SIN1, confirming that the defect is specific to SIN1 deletion (Figure 1C). We further verified using siRNA how mTORC2 components can modulate IRS-1 levels. The expression of IRS-1 in MEFs was enhanced when each mTORC2 component was silenced (Figure 1D). Knockdown of raptor can also slightly increase IRS-1 levels as reported previously (Tzatsos, 2009). To examine the contribution of mTORC1 versus mTORC2 in controlling IRS-1 levels, we treated MEFs with rapamycin. Acute treatment (1–6 hr), which abolished mTORC1-mediated phosphorylation of S6K, did not significantly enhance IRS-1 expression levels (Figure 1E). Prolonged (12–24 hr) rapamycin treatment that can disrupt mTORC2, supported by diminished Akt phosphorylation (Figure 1E), led to elevation of IRS-1 levels (Figure 1E and 1F). The mTOR active site inhibitor, Torin1, which inhibits mTORC1 and mTORC2, increased IRS-1 levels both during acute and prolonged treatment (Figure 1E). Furthermore, the elevated IRS-1 levels in SIN1−/− were only slightly enhanced by rapamycin (Figure 1F). Collectively, these results support that mTORC2 plays a role in controlling IRS-1 expression levels.
Figure 1.
IRS-1 levels are increased upon mTORC2 disruption. A. Protein expression in wild type (WT) or SIN1−/− MEFs was assessed by immunoblotting. Right panel, quantification of immunoblots; results are expressed as mean total IRS-1± S.E.M. relative to total IRS-1 of WT. ** statistically significant B. WT and SIN1−/− were grown under normal growing (N) conditions, starved (0) or restimulated with serum at the indicated times. C. SIN1−/− MEFs were transfected with 1 μg of plasmid expressing HA- SIN1β. WT, SIN1−/− or HA-SIN1-reconstituted SIN1−/− MEFs were normally grown (N), starved (−) or starved then restimulated with serum (+) for 30 min. D. WT MEFs were transfected with 100 nM siRNA against mTOR, SIN1, rictor, raptor or control scramble. E. WT MEFs were starved and restimulated with insulin in the presence of rapamycin (100 nM) or Torin1 (250 nM). F. Growing MEFs were incubated with rapamycin (100 nM) at the indicated times.
IRS-1 synthesis is similar in WT and SIN1−/− MEFs
We next examined why IRS-1 levels are increased in the absence of SIN1. First, we determined whether the elevated expression could be due to enhanced transcription. However, total IRS-1 mRNA levels were comparable between WT and SIN1−/− (Figure S1A). In addition, quantitative PCR analysis revealed no significant difference in IRS-1 mRNA levels (Figure S1B). Next, we performed metabolic labeling to determine the amount of newly translated IRS-1. Despite a slight decrease in the amount of total labeled proteins as previously reported in SIN1−/− (Oh et al., 2010), there was no significant difference in the amount of newly synthesized IRS-1 between WT and SIN1−/− (Figure S1C). Thus, increased IRS-1 levels upon mTORC2 disruption are not due to enhanced transcription or translation.
IRS-1 has a slower turnover rate in the absence of SIN1
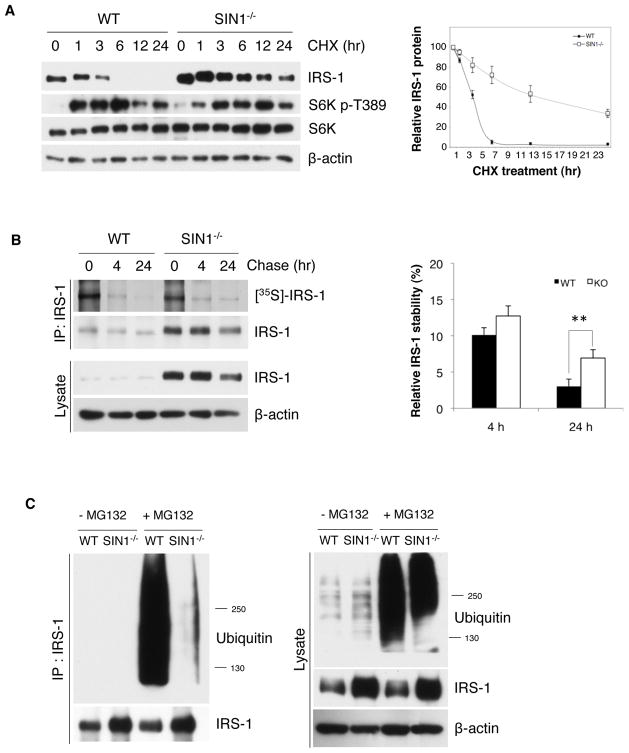
We then compared the turnover rate of total IRS-1. We incubated cells with CHX to block de novo IRS-1 synthesis (Figure 2A). Whereas IRS-1 had a half-life of roughly 4 hr in WT, IRS-1 in SIN1−/− remained substantial even at 24 hr (Figure 2A) and the half-life was approximately 15 hr (Figure 2A, right panel). CHX-induced S6K activation remained robust in SIN1−/−, further supporting that the decreased IRS-1 turnover is not due to defective mTORC1 signaling. To analyze the specific turnover of newly synthesized IRS-1, we performed pulse-chase analysis. Consistent with the decreased turnover of total IRS-1, metabolically labeled IRS-1 remained detectable at 24 hr of chase in SIN1−/− (Figure 2B). We then examined if the decreased turnover of IRS-1 is due to abnormal proteasomal degradation. We used MG132 to inhibit proteasome degradation and assessed the accumulation of ubiquitylated IRS-1. In WT, abundant ubiquitylated IRS-1 accumulated upon MG132 treatment. In contrast, minimal ubiquitylated IRS-1 in SIN1−/− was observed despite robust accumulation of ubiquitylated total proteins in these cells (Figure 2C). Taken together, our results indicate that there is decreased turnover of IRS-1 when mTORC2 is disrupted.
Figure 2.
IRS-1 turnover is defective in mTORC2-disrupted cells. A. WT and SIN1−/− were treated with 10 μM cycloheximide (CHX) for the indicated times. Decreasing amounts of pre-existing IRS-1 protein levels were analyzed by immunoblotting, quantitated and plotted relative to non-CHX treated (right panel). Results are expressed as mean ± S.E.M. B. WT and SIN1−/− MEFs were starved of serum in Met/Cys-free media then pulse-labeled with 35S-Met/Cys and chased for the indicated times. Cell lysates were immunoprecipitated with anti-IRS-1 and the labeled newly synthesized IRS-1 were analyzed by SDS-PAGE and visualized by autoradiography. Right panel, quantification of immunoblots; results are expressed as mean levels of labeled IRS-1 ± S.E.M. relative to 0 hr of chase. ** p<0.01 C. WT and SIN1−/− MEFs were treated with or without 20 μM MG132 for 4 hours prior to harvest. Total cell lysates (right panel) or immunoprecipitated IRS-1 (left) from lysates were analyzed by immunoblotting. See also Figure S1.
Proximal IRS-1 signaling is defective in mTORC2-disrupted cells
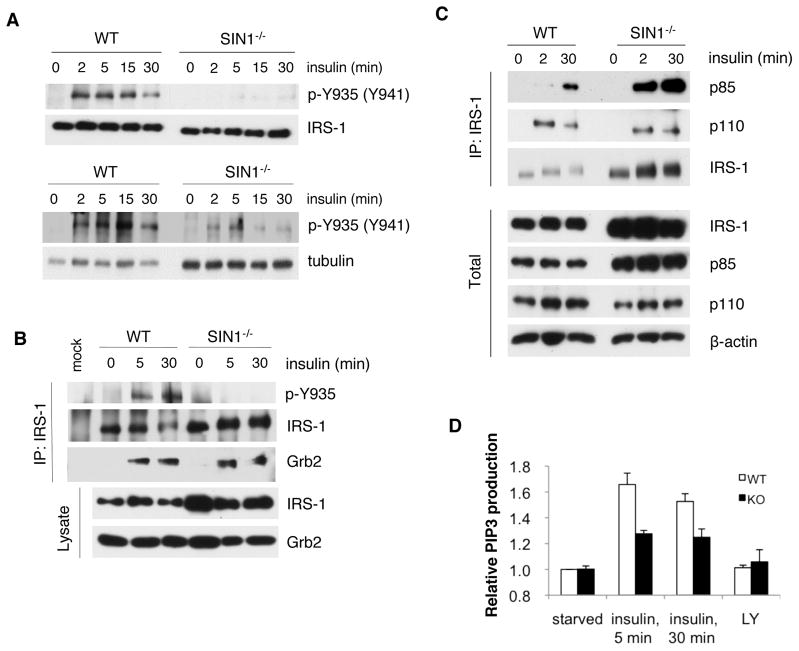
Previous studies have shown that blocking IRS-1 degradation can sustain insulin signaling (Haruta et al., 2000). Although downstream insulin signaling, such as Akt activation, is defective in mTORC2-disrupted cells, it is possible that signals upstream of mTORC2 or proximal to the IR and IRS-1 could be enhanced in SIN1−/−. Therefore, we examined if the elevated IRS-1 levels are associated with enhanced IRS-1 signaling. We evaluated whether IRS-1 undergoes phosphorylation at Tyr935 (human Tyr941), one of several sites where PI3K can bind upon insulin stimulation (Taniguchi et al., 2006). Whereas Tyr935 phosphorylation was highly induced after insulin restimulation in WT, there was hardly any discernible phosphorylation in SIN1−/− when IRS-1 levels were normalized to WT levels (Figure 3A, upper panel) or even upon loading increased amounts of protein from SIN1−/− extracts (Figure 3A, lower panel). IGF-1 stimulation led to more robust Tyr935 phosphorylation in SIN1−/− albeit at lower levels than WT (Figure S2). Because of the attenuated IRS-1 Tyr phosphorylation, we then examined the association of Grb2 and PI3K, two SH2 domain-containing proteins that bind to IRS-1 (Taniguchi et al., 2006). Immunoprecipitation of IRS-1 further confirmed low Tyr935 phosphorylation in SIN1−/− (Figure 3B). Interaction of Grb2 with IRS-1 was induced upon insulin restimulation but appeared to be similar in both WT and SIN1−/− despite increased amounts of immunoprecipitated IRS-1 from SIN1−/− (Figure 3B). In contrast to Grb2, p85 binding to IRS-1 was increased in SIN1−/−, corresponding to enhanced total p85 expression. Interestingly, despite elevated p85 expression and association with IRS-1, the binding of p110 was not increased (Figure 3C). Since inactive monomeric p85 can compete with the catalytically active p85–p110 heterodimer for IRS-1 binding (Taniguchi et al., 2006), our results suggest that there could be attenuated PI3K activity and thereby diminished amount of PIP3 production in SIN1−/−. Indeed, PIP3 production was blunted upon insulin restimulation of starved cells (Figure 3D). Together, these results reveal that although IRS-1 levels are elevated, IRS-1 signaling is dampened and suggest diminished signaling capacity of IRS-1 in the absence of mTORC2.
Figure 3.
IRS-1 phosphotyrosine and PI3K signaling is defective in SIN1−/− MEFs. A. WT and SIN1−/− were starved then restimulated with insulin for the indicated times. Upper panel, total extracts were normalized for total IRS-1; lower panel, total extracts were normalized for tubulin. B–C. WT and SIN1−/− were starved (0) then restimulated with insulin. IRS-1 was immunoprecipitated and the co-immunoprecipitated proteins were detected by immunoblotting. D. Starved MEFs were untreated or pretreated for 1 hr with 50 μM of the PI3K inhibitor LY294002 (LY). Cells were then left unstimulated or restimulated with insulin for 5 and 30 min. PIP3 levels relative to starved condition were plotted + S.E.M. See also Figure S2.
IRS-1 undergoes phosphorylation at the mTORC1/S6K-mediated serine sites in mTORC2-disrupted cells
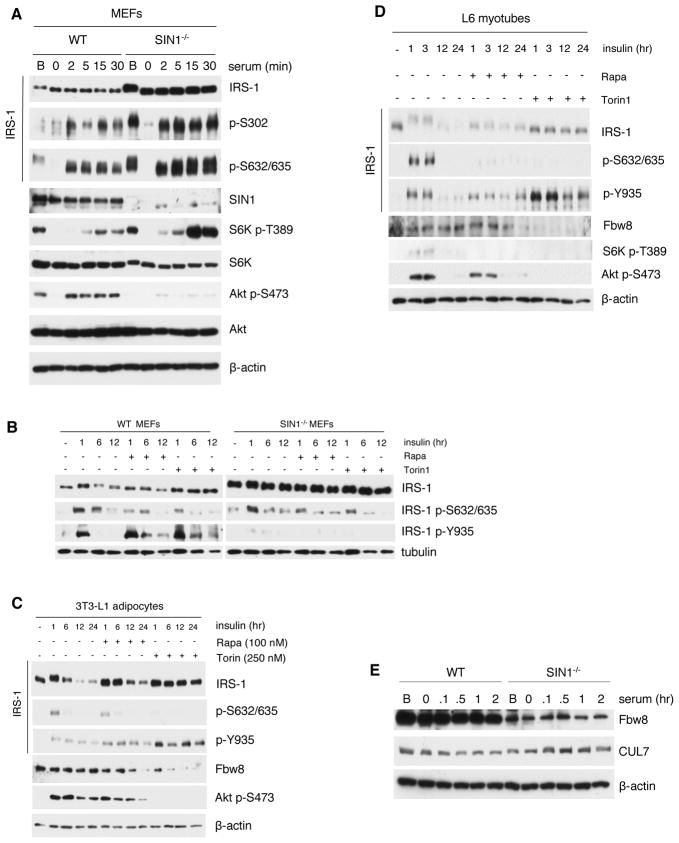
We then examined Ser phosphorylation, which is linked to IRS-1 degradation (Liu et al., 2004; Shah and Hunter, 2006; Tremblay et al., 2007). The phosphorylation levels of Ser302 and Ser632/635 were proportional to the enhanced IRS-1 expression levels in SIN1−/− (Figure 4A). Phosphorylation of these sites was abolished during starvation and induced upon serum restimulation. In agreement with these sites being phosphorylated via mTORC1/S6K, S6K activation was triggered and considerably enhanced in SIN1−/−. Prolonged insulin stimulation reduced IRS-1 levels in WT but not in SIN1−/− (Figure 4B). Rapamycin attenuated phosphorylation at Ser632/635 in both WT and SIN1−/−. Furthermore, inhibition of both mTOR complexes by Torin1 also reduced phosphorylation at these sites. Surprisingly, despite the decreased Ser phosphorylation, there was no increase in Tyr phosphorylation in SIN1−/−, further supporting that the accumulated IRS-1 in SIN1−/− has decreased signaling capacity (Figure 3). Together, these results suggest that Ser phosphorylation at sites mediated by mTORC1/S6K is linked to decreased IRS-1 signaling but can be uncoupled from its degradation.
Figure 4.
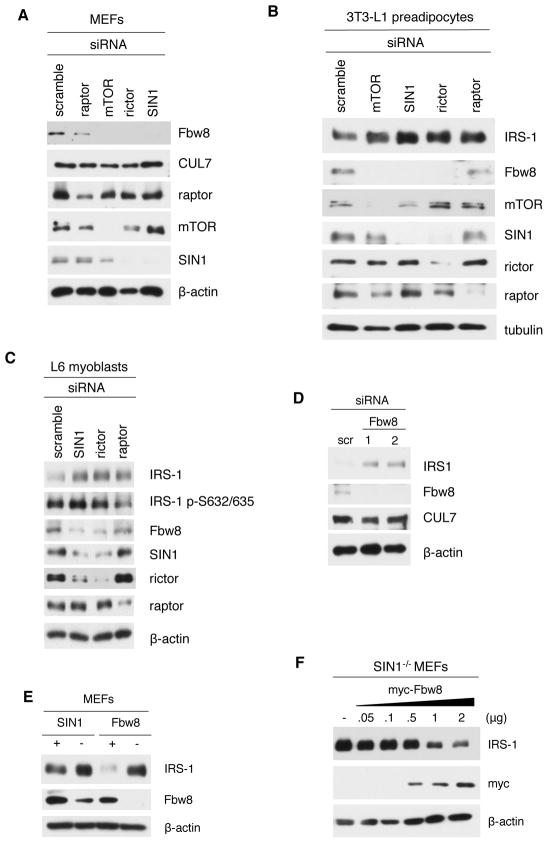
mTORC1-mediated serine phosphorylation is increased while Fbw8 levels are attenuated upon mTORC2 disruption. A–B. WT and SIN1−/− MEFs grown under basal (B) conditions were starved then restimulated with serum or insulin at the indicated times in the absence or presence of rapamycin (100 nM) or Torin1 (250 nM). Total extracts were subjected to SDS-PAGE and immunoblotting. C–D. Differentiated 3T3-L1 adipocytes (C) or L6 myotubes (D) were treated as in B. E. WT and SIN1−/− growing under basal (B) conditions were starved then unstimulated or restimulated with serum. See also Figure S3.
To further examine how IRS-1 levels and phosphorylation are affected upon mTOR inhibition, we utilized adipocytes and muscle cells, which are highly insulin-responsive cells. Upon insulin restimulation of differentiated 3T3-L1 adipocytes, IRS-1 had decreased mobility accompanied by increased Ser632/635 phosphorylation (Figure 4C and Figure S3A). After 6 hours of insulin restimulation, IRS-1 acquired faster mobility and had attenuated levels. Rapamycin blocked Ser phosphorylation of IRS-1 and upon prolonged treatment (6–24 hr), it delayed IRS-1 degradation in adipocytes (Figure 4C) and myotubes (Figure 4D). Torin1 treatment prevented mobility upshift and Ser phosphorylation (Figures 4C and 4D; S3A). Furthermore, IRS-1 expression levels remained high even at 24 hours of insulin stimulation. Unlike in SIN1−/− (Figure 4B), the attenuated Ser phosphorylation upon Torin1 treatment corresponded to prolonged phosphorylation of Tyr935 (Figures 4C, 4D and S3A). Thus, inhibition of both mTOR complexes can prolong IRS-1 Tyr phosphorylation and prevent its degradation in adipocytes and myotubes.
Fbw8 protein expression is decreased upon mTORC2 disruption
We next investigated whether the Fbw8/CUL7 ubiquitin ligase complex that targets IRS-1 (Xu et al., 2008) could be involved in the aberrant degradation of IRS-1 in SIN1−/−. When we examined the protein expression of CUL7 and Fbw8, there was a marked attenuation of Fbw8 levels in SIN1−/− whereas CUL7 expression was comparable (Figure 4E). Torin1 and prolonged rapamycin treatment also diminished Fbw8 levels in adipocytes and myotubes (Figure 4C–4D). Thus, reduced levels of Fbw8 could contribute to defective IRS-1 degradation and suggest that Fbw8 is subject to regulation by mTORC2.
We then further examined how silencing expression of mTORC2 components can affect Fbw8 and IRS-1 levels. Knockdown of mTORC2 components in MEFs significantly reduced Fbw8 levels without decreasing CUL7 expression (Figure 5A). Knockdown of both mTORC1 and mTORC2 components augmented IRS-1 levels in adipocytes and myoblasts (Figure 5B and 5C). Strikingly, knockdown of mTORC2 components decreased Fbw8 expression (Figures 5A–5C). When Fbw8 expression was silenced, IRS-1 levels became dramatically enhanced (Figure 5D). Using Fbw8-deficient MEFs (Tsutsumi et al., 2008), we also observed enhancement of IRS-1 levels similar to SIN1−/− MEFs (Figure 5E). Next, we overexpressed Fbw8 in SIN1−/− to determine if this would diminish IRS-1 expression. Increasing amounts of Fbw8 led to a decline in IRS-1 levels in SIN1−/− (Figure 5F). Together, these results reveal that mTORC2 disruption can prevent IRS-1 turnover due to Fbw8 deregulation.
Figure 5.
Knockdown of mTORC2 components increases IRS-1 while decreasing Fbw8 expression levels. A–C. WT MEFs (A), 3T3-L1 pre-adipocytes (B), L6 myoblasts (C) were transfected with indicated siRNAs. D. WT MEFs were transfected with 100 (1) or 200 (2) nM siRNA against Fbw8 or 100 nM of a control scramble siRNA. E. SIN1- or Fbw8-deficient MEFs, along with corresponding WT MEFs were grown normally and analyzed for expression of the indicated proteins. F. SIN1−/− MEFs were transfected with either empty vector (−) or increasing amounts of myc-Fbw8.
mTORC2 stabilizes Fbw8 by phosphorylation at Ser86
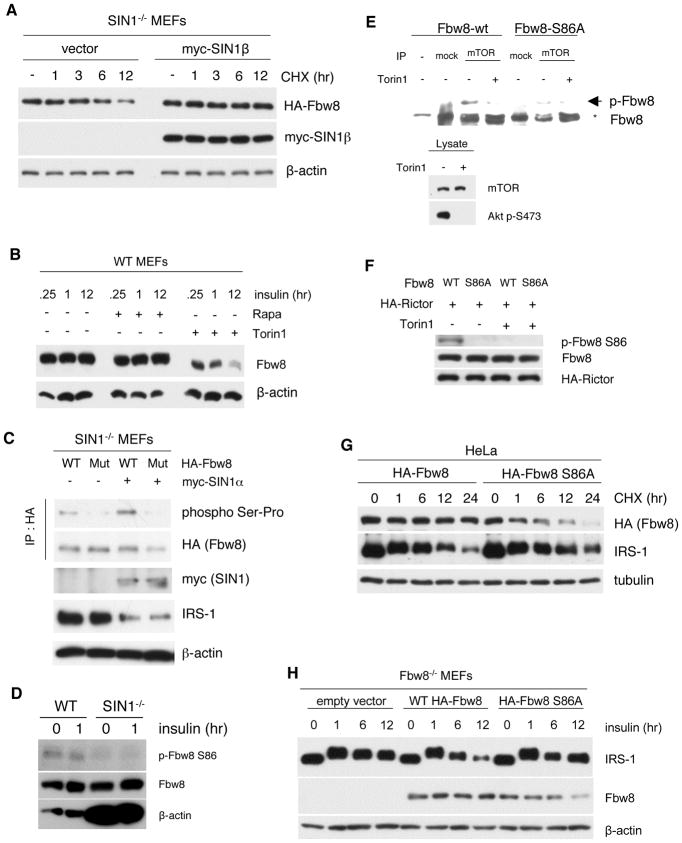
Next, we investigated how mTORC2 can regulate Fbw8. Upon CHX treatment, exogenous HA-Fbw8 declined dramatically from 6–12 hr whereas it remained stable in the presence of SIN1 (Figure 6A). These observations suggest that mTORC2 could promote Fbw8 protein stability. We then examined how mTOR inhibition can affect Fbw8 levels and phosphorylation. Whereas rapamycin treatment from 15 min to 12 hr did not affect Fbw8 levels, Torin1 treatment progressively decreased Fbw8 expression levels over time (Figure 6B). Furthermore, Fbw8 mobility in Phos-tag gel was faster upon Torin1 treatment (Figure 6B), suggesting Fbw8 dephosphorylation upon mTORC2 inhibition. Since Fbw8 was pulled out as a putative mTOR target in a recent phosphoproteomic analysis (Hsu et al., 2011; Yu et al., 2011), we examined how phosphorylation could stabilize Fbw8. First, we mutated the possible mTORC target Ser phosphorylation site (Ser86) to Ala. In the absence of SIN1, HA-Fbw8 had decreased phosphorylation as detected using phospho-Ser-Pro antibody (Figure 6C). This phospho-antibody did not detect the mutant Fbw8 both in the absence or presence of SIN1, strongly suggesting that phosphorylation of Ser86 is dependent on mTORC2. We generated a phospho-antibody against this site and confirmed that phosphorylation of Ser86 in SIN1−/− is abolished (Figure 6D). Second, we performed an in vitro kinase assay to determine if mTOR can directly phosphorylate Fbw8. Robust phosphorylation of Fbw8, but not the mutant Ser86Ala, was observed when immunoprecipitated mTOR (Figure 6E) or HA-rictor (Figure 6F) from HeLa cells was used as the kinase source. Little to no phosphorylation occurred when mTOR or rictor was immunoprecipitated from Torin1-treated cells. These results indicate that mTORC2 can phosphorylate Fbw8 at Ser86.
Figure 6.
mTORC2 mediates IRS-1 degradation by phosphorylation of Fbw8 at Ser86 to promote its stability. A. SIN1−/− MEFs were transfected with empty vector or myc-SIN1β. 24 hr post-transfection, cells were untreated or treated with 10 μM CHX for the indicated hr. See quantitation in Figure S4A B. Insulin-restimulated WT MEFs were treated with either rapamycin or Torin1 for the indicated times. Total cellular extracts were fractionated using Phos-tag SDS-PAGE. C. SIN1−/− MEFs were co-transfected with 2 μg of either wild-type or S86A mutant HA-Fbw8 (Mut) and myc-SIN1α (+) or empty vector (−). Immunoprecipitation with anti-HA was followed by immunoblot analysis using a phospho Ser-Pro-specific antibody. D. Insulin-restimulated MEFs were immunoblotted with phospho-S86 antibody. Total Fbw8 levels were normalized between WT and SIN1−/− MEFs by using more extracts from the latter as reflected by β-actin blotting. E. In vitro translated HA-Fbw8 wild type or S86A mutant was incubated with mock, mTOR immunoprecipitates from HeLa cells that were untreated or treated with Torin1 (lower panels). Proteins were separated using Phos-tag gel. Phosphorylated Fbw8 (arrowhead), which migrates slower than non-phosphorylated Fbw8 (asterisk) was detected using anti Fbw8 antibody. F. Kinase assay as in E was performed using HA-rictor expressed from HeLa cells. Phosphorylation of Fbw8 was detected using phospho-S86 Fbw8 antibody. G. HeLa cells were transfected with either HA-tagged wild type or S86A mutant of Fbw8. 48-hours post-transfection, cells were incubated with 10 μM CHX for the indicated hr. See quantitation in Figure S4B–C H. Fbw8-deficient MEFs were transfected with either empty vector, HA-Fbw8 or HA-Fbw8-S86A. Transfected cells were starved and restimulated with insulin for the indicated times. See quantitation in Figure S4D–E.
We then analyzed the role of Ser86 phosphorylation in Fbw8 turnover. Fbw8-S86A mutant had increased turnover compared to Fbw8-wt (Figure 6G). Decreased levels of Fbw8-S86A corresponded to enhancement of IRS-1 levels at prolonged time points. When Fbw8 was reconstituted in Fbw8-deficient MEFs, overexpression of wild type Fbw8 led to downregulation of IRS-1 after prolonged insulin stimulation (Figure 6H). In contrast, IRS-1 levels remained elevated after overexpression of the mutant, corresponding to diminishing Fbw8-S86A expression at 12 hr. These results support that sufficient Fbw8 levels are required to prevent IRS-1 accumulation and that mTOR could directly phosphorylate Fbw8 to promote its stability, which is important for IRS-1 degradation.
Lack of Fbw8 phosphorylation by mTORC2 excludes Fbw8 from the cytosol and prevents degradation of cytosolic IRS-1
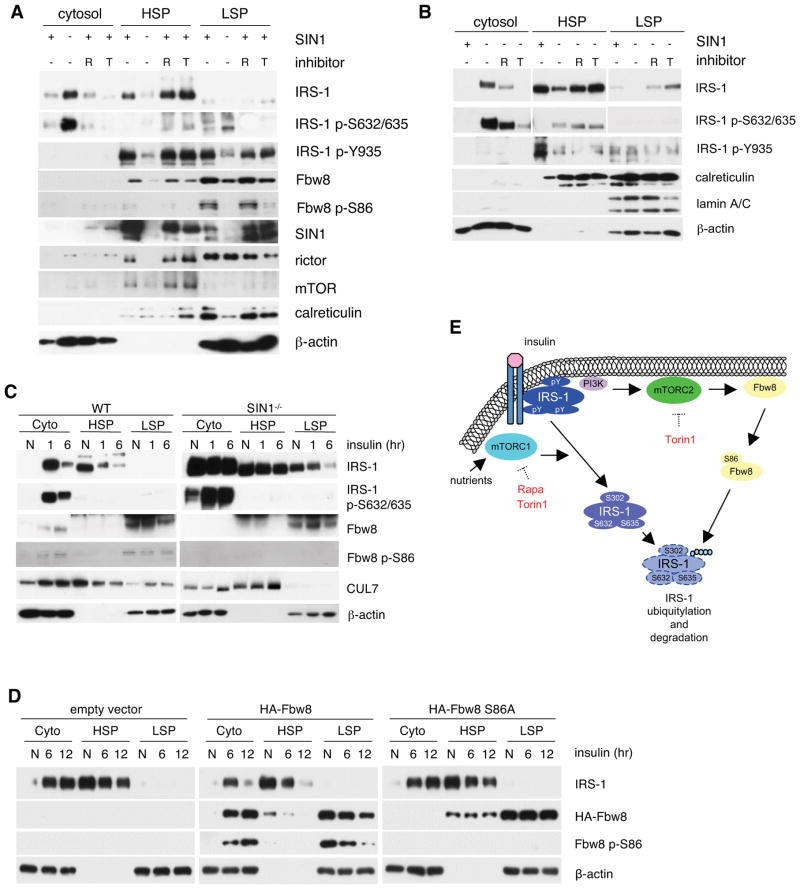
Cytosolic IRS-1 is distinct from insulin-responsive IRS-1 and becomes degraded (Clark et al., 1998; Luo et al., 2005). We postulated that decreased levels and defective phosphorylation of Fbw8 in SIN1−/− cells could prevent Fbw8 translocation to the cytosol where it targets IRS-1. In growing WT MEFs, the majority of total IRS-1 associated with the membrane-containing fractions (HSP and LSP) whereas most IRS-1 from SIN1−/− partitioned with the cytosolic fraction (Figures 7A–C). Treatment with rapamycin or Torin1 of both WT (Figure 7A) and SIN1−/− (Figure 7B) retained IRS-1 in the HSP. While IRS-1 in the membrane fractions is phosphorylated at Tyr935, IRS-1 from the cytosolic fractions is phosphorylated at Ser632/635 (Figure 7A and 7B). However, despite the presence of IRS-1 in the membrane in SIN1−/−, phosphorylation at Tyr935 remained low (Figure 7B). This is consistent with results from Figure 4B demonstrating that inhibiting Ser phosphorylation in SIN1−/− does not prolong Tyr935 phosphorylation. In WT, the majority of mTORC2 components localized to the membrane, whereas in SIN1−/−, rictor and mTOR were more discernible in the cytosol (Figure 7A). On the other hand, Fbw8 was absent in the cytosol but found exclusively in the membrane fractions where it is phosphorylated at Ser86 (Figure 7A). These observations suggest that active IRS-1 colocalizes with Fbw8 and mTORC2 at the membrane.
Figure 7.
Disruption of mTORC2 promotes accumulation of inactive IRS-1 in the cytosol and exclusion of Fbw8 from this compartment. A–B. Growing WT or SIN1−/− MEFs were treated with either 100 nM rapamycin (R) or 250 nM Torin1 (T). Cells were homogenized and extracts were fractionated into cytosol, high-speed (HSP) and low-speed pellet (LSP). Fractions were analyzed by Western blotting with 20 μg of protein loaded in each lane. C. WT or SIN1−/− were grown in normal media (N), starved and restimulated with insulin for either 1 or 6 hrs. Cells were processed as in A. D. Fbw8−/− MEFs were transfected with either empty vector or Fbw8 constructs as indicated. Cells were processed as in C above. E. Model for IRS-1 downregulation by mTORC1 vs mTORC2. Tyrosine-phosphorylated-IRS-1 on the membrane becomes Ser phosphorylated at several residues including the mTORC1/S6K-mediated sites after prolonged insulin stimulation. mTORC2 and Fbw8 colocalize at the membrane under basal conditions where mTORC2 phosphorylates Ser86 to stabilize Fbw8 and promotes its cytosolic localization upon insulin stimulation. During prolonged insulin stimulation, inactive serine-phosphorylated IRS-1 and Fbw8 colocalize to the cytosol where the former becomes ubiquitylated via Fbw8 and undergoes proteasomal degradation.
Although IRS-1 Ser phosphorylation is implicated in its downregulation, it is unclear whether IRS-1 degradation occurs in the cytosol or membrane compartment. Since Fbw8 is only present in membrane-containing fractions under basal conditions (Figure 7A), we asked if it could colocalize with IRS-1 in the cytosol during prolonged insulin stimulation. Upon one-hour insulin stimulation of WT, IRS-1 was depleted from the HSP and accumulated in the cytosol (Figure 7C). After 6 hours, IRS-1 in the cytosol was dramatically diminished as compared to 1 hr treatment. Interestingly, CUL7 and phosphorylated Fbw8 translocated to this compartment upon insulin stimulation. In contrast, Fbw8 was hardly detectable in the cytosol of SIN1−/− even after stimulation. However, since Fbw8 expression in SIN1−/− is lower, we then further verified if the mTORC2-mediated phosphorylation at Ser86 plays a role in Fbw8 cytosolic localization upon insulin stimulation. To this end, we expressed wt versus Ser86Ala mutant Fbw8 in Fbw8−/− MEFs. Lack of Ser86 phosphorylation prevented cytosolic translocation of Fbw8 upon insulin stimulation with concomitant accumulation of IRS-1 in the cytosol (Figure 7D). Together, these results reveal that in the absence of the mTORC2-mediated phosphorylation, Fbw8 is incapable of cytosolic translocation thereby preventing degradation of cytosolic IRS-1.
DISCUSSION
IRS-1 serves to integrate multiple intracellular inputs that can either upregulate or downregulate insulin signaling (Gual et al., 2005). Hence, the spatial and temporal regulation of its expression is tightly controlled. The hyperphosphorylation of IRS-1 at several serine residues, including the mTORC1/S6K-mediated phosphorylation, is a key event that has been linked to IRS-1 downregulation and degradation (Gual et al., 2005). In our studies, we have found that mTORC2 is additionally required for proper turnover of IRS-1 and to prevent the accumulation of inactive IRS-1 in the cytosol. We provide evidence that mTORC2 can stabilize Fbw8, the substrate-targeting subunit of the CUL7 E3 ligase complex that has been shown to mediate degradation of IRS-1 (Xu et al., 2008). We present a model whereby mTORC2 phosphorylates Fbw8 to modulate its stability and enable its translocation to the cytosol upon insulin stimulation (Figure 7E). In the cytosol, Fbw8 mediates the ubiquitylation and degradation of IRS-1.
Our findings strongly support a role for mTORC2 in controlling IRS-1 cellular levels. Previous studies have demonstrated that mTORC1 inhibition by rapamycin prevents IRS-1 Ser phosphorylation (Haruta et al., 2000; Pederson et al., 2001; Takano et al., 2001), although whether it blocks degradation has been controversial (Zhande et al., 2002). Since chronic rapamycin treatment can also disrupt mTORC2 in some cell lines (Zoncu et al., 2011), it is possible that mTORC2 inhibition could explain the effect on IRS-1 expression levels in some reports (Pederson et al., 2001; Shah and Hunter, 2006; Yu et al., 2011). We have shown here that whereas acute rapamycin treatment has slight effects on IRS-1 expression levels, prolonged exposure can dramatically enhance IRS-1 and diminish Fbw8 levels similar to Torin1 treatment (Figure 1D–E, 4C–D). These results are further substantiated in SIN1-deficient cells and by silencing expression of mTORC2 components. We note however that a previous report failed to observe changes in IRS-1 levels upon rictor or SIN1 knockdown (Tzatsos, 2009). Elevated IRS-1 expression levels were also observed in different cancer cell lines even after acute rapamycin treatment (O’Reilly et al., 2006). The reason for the discrepancy with our findings is unclear but it would be interesting to determine Fbw8 regulation under the conditions used in those studies. Enhanced mTORC2 signals likely serve as a negative feedback to diminish insulin signaling and prevent chronic activation of this pathway that can lead to uncontrolled cell growth.
IRS-1 downregulation during persistent insulin stimulation is coupled to a PI3K-dependent pathway (Hartley and Cooper, 2002; Smith et al., 1995). IRS-1 Ser phosphorylation can be blocked using PI3K inhibitors (Haruta et al., 2000; Lee et al., 2000; Pederson et al., 2001; Sun et al., 1999; Zhande et al., 2002). Furthermore, expression of a constitutively active form of PI3K induces IRS-1 Ser phosphorylation and degradation (Egawa et al., 2000; Haruta et al., 2000; Takano et al., 2001). Given that mTORC2 can be activated by conditions that enhance PIP3 signals (Zinzalla et al., 2011), our findings place mTORC2 as the missing link connecting PI3K activation to IRS-1 degradation. Inhibiting mTOR simultaneously with PI3K resulted in a similar increase in IRS-1 levels compared to mTOR inhibition alone (Figure S3B–D, Figure 4), supporting that mTOR and PI3K act on the same pathway. How PI3K could promote mTORC2 activation remains elusive but speculatively could allow mTORC2 to phosphorylate Fbw8 at the membrane where they colocalize (Figure 7A). Since we have not observed enhanced Fbw8 phosphorylation during insulin stimulation (Figure 6D), it would be interesting to determine if this mTORC2-mediated phosphorylation is also constitutive and occurs cotranslationally (Oh et al., 2010). Whereas mTOR or PI3K inhibition up to 24 hr can prolong IRS-1 Tyr phosphorylation (Figure 4B–D, 7A–B) (Haruta et al., 2000), we did not observe such rescue in SIN1−/− MEFs (Figure 4B and 7B). Consistent with this, we found diminished expression of IR in SIN1−/− (Figure S6A). Extended (48 hr) Torin1 treatment of WT could mimic SIN1 deficiency, eventually reducing both IRS-1 Tyr phosphorylation and IR expression (Figure S6B). In fact, other signals triggered by insulin were altered as well upon mTORC2 disruption such as reduced PIP3 production and enhanced p85 expression (Figure 3). The former would be consistent with reduced IR signals and the latter could inactivate IRS-1 in sequestered compartments (Luo et al., 2005; Taniguchi et al., 2006). Thus, mTORC2 not only couples PI3K to IRS-1 downregulation but has other yet-to-be characterized roles in insulin signaling.
What is the role of mTORC2 in IRS-1 downregulation? Our results reveal that mTORC2 is required to prevent the accumulation of inactive IRS-1 in the cytosol. The compartment where inactive IRS-1 becomes degraded has been uncertain. Proteasome inhibition led to cytosolic accumulation of IRS-1 in adipocytes (Takano et al., 2001) whereas it was retained in the membrane in HEK293T cells (Shah and Hunter, 2006), suggesting the compartment where it is degraded could be cell type-specific. Here we demonstrate that mTORC2-regulated Fbw8 is crucial for degradation of cytosolic IRS-1 in MEFs. Previous studies by Xu et al already demonstrated that IRS-1 is a proteolytic target of the CUL7 E3 ligase and that Fbw8 interacts with IRS-1 (Xu et al., 2008). How Fbw8 is regulated and the compartment where it degrades IRS-1 has remained unclear but they have shown that the Fbw8 and IRS-1 interaction requires phosphorylation of IRS-1 by mTOR/S6K. However, inhibition of the Fbw8-mediated degradation of IRS-1 occurred upon 24 hr treatment with rapamycin (Xu et al., 2008), raising a possible role for mTORC2 instead of mTORC1. Our findings now further elucidate how Fbw8 is regulated by mTORC2 via phosphorylation. Ser86 conforms to a Ser-Pro-Pro motif that is reminiscent of the Thr-Pro-Pro motif in the conserved turn motif of AGC kinases, which we have shown to be phosphorylated by mTORC2 for stability (Facchinetti et al., 2008; Oh et al., 2010). Ser86 phosphorylation may not directly affect binding of Fbw8 with CUL7 (Figure S5) but is critical for promoting the stability and insulin-induced cytosolic localization of Fbw8 (Figure 7C and 7D). Indeed, F-box proteins can undergo auto-ubiquitylation and the binding of cognate substrates can prevent their self-ubiquitylation (Pashkova et al., 2010). Because Fbw8 in mTORC2-disrupted cells is excluded from the cytosol, the decreased interaction of Fbw8 with IRS-1 could subsequently predispose Fbw8 for auto-ubiquitylation. Fbw8 phosphorylation by mTORC2 would thus prevent its premature self-degradation and favor interaction with IRS-1 in the cytosol in the presence of insulin. Thus, although mTORC2 is predominantly in the membrane, it prevents accumulation of inactive IRS-1 in the cytosol by ensuring Fbw8 accessibility to this pool of IRS-1. How Fbw8 becomes sequestered in the cytosol in the presence of insulin remains to be elucidated.
Our studies highlight the importance of mTORC2 in controlling cellular insulin sensitivity. mTORC2 disruption in mice led to insulin resistance (Cybulski et al., 2009; Kumar et al., 2010) and yet increased lifespan was also reported under mTORC2-disrupted conditions (Lamming et al., 2012). It would be interesting to determine how defective IRS-1 turnover could contribute to the metabolic defects in these mice. Dampening the negative feedback regulatory signals from mTORC2 could enhance insulin sensitivity and have beneficial effects on lifespan extension.
EXPERIMENTAL PROCEDURES
Plasmids and Reagents
Myc-Fbw8 plasmid (Xu et al., 2008) was a gift from Dr. Z-Q. Pan (Mount Sinai School of Medicine). The full clone of mouse Fbw8 (mFbw8) gene, isolated from MEFs using PCR based methods and mutant Ser86Ala, generated by a PCR-based site-directed mutagenesis method, were subcloned into pCI-HA vector. Fbw8 phospho-S86 antibody (SRSRpSPPDRDA) was custom generated by arrangement with Bio-Synthesis, Inc. (Lewisville, TX). All chemicals were purchased from Sigma (St Louis, MO) unless otherwise noted. MG132 and Torin1 were purchased from Tocris (Ellisville, MO). SIN1 plasmid and sources for most antibodies were described previously (Oh et al., 2010). Antibodies to Fbw8 were from Dr. J. DeCaprio (Dana Farber Cancer Inst); p-IRS-1 (S636/639, S302), p-Ser-Pro, p85, and p110 were from Cell Signaling (Danvers, MA); IRS-1, p-IRS-1 (Tyr 941), and SIN1 were from Millipore (Billerica, MA).
Cell Culture, gene silencing, transfection and metabolic labeling
Wild-type and SIN1−/− MEFs or Fbw8−/− MEFs were cultured as previously described (Oh et al., 2010; Tsutsumi et al., 2008). siRNA against mTOR and SIN1 were purchased from Dharmacon (Lafayette, CO); rictor and raptor were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Cells were transfected with indicated amounts of plasmid DNA or siRNA. Cells were harvested 48 hours post-transfection. For metabolic labeling, cells were first starved for 90 min in Met/Cys-free media, labeled with 100 μCi of a [35S] Met/Cys mixture (Perkin-Elmer, Waltham, MA) for 4 hr and chased with 10% FBS-containing media for 90 min. 3T3-L1 fibroblasts obtained from ATCC (Manassas, VA) were cultured at 5% CO2 in DMEM containing 100 U/ml penicillin, 100 mg/ml streptomycin and 10% FCS. 3T3-L1 fibroblasts were differentiated to adipocytes by addition of differentiation medium (DMEM containing 10% FBS, 500 μM IBMX, 250 nM dexamethasone and 870 nM insulin). 3T3-L1 adipocytes were used between 7–9 days after initiation of differentiation, when more than 90% of the cells exhibited an adipocyte-like morphology. The differentiation to mature adipocytes was confirmed by Oil Red O staining of lipid vesicles and by immunoblotting using the specific adipocyte marker PPARγ. L6 myoblasts were cultured at 5% CO2 in DMEM containing 100 U/ml penicillin, 100 mg/ml streptomycin and 10% FBS. At 100% confluency, media was replaced with 2.5% horse serum-containing DMEM and changed at 2-day intervals. By day 7, cells express the myotube phenotype. Cells were used for experiments between days 7 and 10.
Immunoprecipitation and Immunoblotting
Cells were rinsed with ice-cold phosphate buffered saline (PBS) and harvested using lysis buffer (40 mM HEPES, pH 7.9, 120 mM NaCl, 1 mM EDTA) containing 0.3% CHAPS or RIPA buffer for analysis of ubiquitylation. Lysates (300–500 μg) were incubated for 4 hours with indicated antibodies, except for IRS-1 which was immunoprecipitated overnight at 4°C, followed by incubation with protein A/G-agarose or Sepharose beads for 2 hr. After incubation, beads were washed three times with lysis buffer. To analyze total extracts, cells were lysed in RIPA buffer (150 mM NaCl, 25 mM Tris pH 7.4, 1% NP-40, 0.25% sodium deoxycholate, 1 mM EDTA, 1 mM Na3V04, 1 mM NaF, and protease inhibitor cocktail). The lysates were centrifuged at 16,000g for 30 minutes at 4°C. Total extracts or immunoprecipitated proteins were fractionated by SDS-PAGE followed by immunoblotting. Phos-tag acrylamide gels (Wako, Richmond, VA) containing 200 μM MnCl2 were used for analysis of phosphorylated proteins. Blots were visualized using enhanced chemiluminescence. Quantification of immunoblots was performed using Image J analysis.
PIP3 Measurement
Wild-type and SIN1−/− MEFs were starved overnight, pretreated with LY294002 for 1 hr as indicated, then restimulated with serum. Cells were washed twice with ice-cold PBS and PIP3 was extracted and measured using PIP3 Mass ELISA kit (Echelon Biosciences, Inc., Salt Lake City, UT).
Subcellular Fractionation
Cells were washed twice using ice-cold PBS and homogenized in buffer (10 mM Tris, pH 7.4, 0.5 mM EDTA, 25 mM sucrose, supplemented with a mixture of protease and phosphatase inhibitors) by passing through a 27G1/2 needle (Becton Dickinson & Co, Franklin Lakes, NJ) 15 times. The homogenate was centrifuged at 1,000g for 10 min at 4°C to separate LSP and the supernatant was recentrifuged at 200,000g for 1 h at 4°C to obtain cytosol and HSP fractions. The LSP and HSP were resuspended with RIPA and IP buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100), respectively.
Kinase assay
mTOR or HA-rictor was immunoprecipitated from HeLa cells that were untreated or treated with Torin1. In vitro-translated and purified HA-Fbw8 or HA-Fbw8-S86A was used as substrates and added to immunoprecipitates along with kinase buffer (50 mM HEPES pH7.4, 200 mM KOAc, 2mM MgCl2) and 500 μM cold ATP, incubated at 37°C at 500 rpm for 30 min. Reaction was stopped by addition of SDS-PAGE loading dye.
Supplementary Material
Highlights.
mTORC2 is required to prevent accumulation of inactive IRS-1.
Inactive IRS-1 accumulates due to decreased expression of Fbw8.
mTORC2 phosphorylates Fbw8 to promote its stability and cytosolic translocation.
Fbw8 translocates to the cytosol where it mediates IRS-1 degradation.
Acknowledgments
We thank Dr. James DeCaprio for generously providing Fbw8−/− MEFs and Fbw8 antibody; Dr. Zhen-Qiang Pan for sharing plasmids, Po-Chien Chou for helpful discussions. We acknowledge support from the NIH (GM079176), ACS (RSG0721601TBE), and SU2C-AACR-IRG0311 (E.J.).
Footnotes
Supplemental Information includes six figures and one method.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Clark SF, Martin S, Carozzi AJ, Hill MM, James DE. Intracellular localization of phosphatidylinositide 3-kinase and insulin receptor substrate-1 in adipocytes: potential involvement of a membrane skeleton. J Cell Biol. 1998;140:1211–1225. doi: 10.1083/jcb.140.5.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cybulski N, Polak P, Auwerx J, Ruegg MA, Hall MN. mTOR complex 2 in adipose tissue negatively controls whole-body growth. Proc Natl Acad Sci U S A. 2009;106:9902–9907. doi: 10.1073/pnas.0811321106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egawa K, Nakashima N, Sharma PM, Maegawa H, Nagai Y, Kashiwagi A, Kikkawa R, Olefsky JM. Persistent activation of phosphatidylinositol 3-kinase causes insulin resistance due to accelerated insulin-induced insulin receptor substrate-1 degradation in 3T3-L1 adipocytes. Endocrinology. 2000;141:1930–1935. doi: 10.1210/endo.141.6.7516. [DOI] [PubMed] [Google Scholar]
- Facchinetti V, Ouyang W, Wei H, Soto N, Lazorchak A, Gould C, Lowry C, Newton AC, Mao Y, Miao RQ, et al. The mammalian target of rapamycin complex 2 controls folding and stability of Akt and protein kinase C. EMBO J. 2008;27:1932–1943. doi: 10.1038/emboj.2008.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud J, Leshan R, Lee YH, White MF. Nutrient-dependent and insulin-stimulated phosphorylation of insulin receptor substrate-1 on serine 302 correlates with increased insulin signaling. J Biol Chem. 2004;279:3447–3454. doi: 10.1074/jbc.M308631200. [DOI] [PubMed] [Google Scholar]
- Gual P, Le Marchand-Brustel Y, Tanti JF. Positive and negative regulation of insulin signaling through IRS-1 phosphorylation. Biochimie. 2005;87:99–109. doi: 10.1016/j.biochi.2004.10.019. [DOI] [PubMed] [Google Scholar]
- Harrington LS, Findlay GM, Gray A, Tolkacheva T, Wigfield S, Rebholz H, Barnett J, Leslie NR, Cheng S, Shepherd PR, et al. The TSC1-2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. J Cell Biol. 2004;166:213–223. doi: 10.1083/jcb.200403069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley D, Cooper GM. Role of mTOR in the degradation of IRS-1: Regulation of PP2A activity. J Cell Biochem. 2002;85:304–314. doi: 10.1002/jcb.10135. [DOI] [PubMed] [Google Scholar]
- Hartman ME, Villela-Bach M, Chen J, Freund GG. Frap-dependent serine phosphorylation of IRS-1 inhibits IRS-1 tyrosine phosphorylation. Biochem Biophys Res Commun. 2001;280:776–781. doi: 10.1006/bbrc.2000.4214. [DOI] [PubMed] [Google Scholar]
- Haruta T, Uno T, Kawahara J, Takano A, Egawa K, Sharma PM, Olefsky JM, Kobayashi M. A rapamycin-sensitive pathway down-regulates insulin signaling via phosphorylation and proteasomal degradation of insulin receptor substrate-1. Mol Endocrinol. 2000;14:783–794. doi: 10.1210/mend.14.6.0446. [DOI] [PubMed] [Google Scholar]
- Hotamisligil GS, Peraldi P, Budavari A, Ellis R, White MF, Spiegelman BM. IRS-1-mediated inhibition of insulin receptor tyrosine kinase activity in TNF-alpha- and obesity-induced insulin resistance. Science. 1996;271:665–668. doi: 10.1126/science.271.5249.665. [DOI] [PubMed] [Google Scholar]
- Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MB, et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011;332:1317–1322. doi: 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Lawrence JC, Jr, Jung DY, Ko HJ, Keller SR, Kim JK, Magnuson MA, Harris TE. Fat cell-specific ablation of rictor in mice impairs insulin-regulated fat cell and whole-body glucose and lipid metabolism. Diabetes. 2010;59:1397–1406. doi: 10.2337/db09-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamming DW, Ye L, Katajisto P, Goncalves MD, Saitoh M, Stevens DM, Davis JG, Salmon AB, Richardson A, Ahima RS, et al. Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science. 2012;335:1638–1643. doi: 10.1126/science.1215135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AV, Gooch JL, Oesterreich S, Guler RL, Yee D. Insulin-like growth factor I-induced degradation of insulin receptor substrate 1 is mediated by the 26S proteasome and blocked by phosphatidylinositol 3′-kinase inhibition. Mol Cell Biol. 2000;20:1489–1496. doi: 10.1128/mcb.20.5.1489-1496.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YF, Herschkovitz A, Boura-Halfon S, Ronen D, Paz K, Leroith D, Zick Y. Serine phosphorylation proximal to its phosphotyrosine binding domain inhibits insulin receptor substrate 1 function and promotes insulin resistance. Mol Cell Biol. 2004;24:9668–9681. doi: 10.1128/MCB.24.21.9668-9681.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Field SJ, Lee JY, Engelman JA, Cantley LC. The p85 regulatory subunit of phosphoinositide 3-kinase down-regulates IRS-1 signaling via the formation of a sequestration complex. J Cell Biol. 2005;170:455–464. doi: 10.1083/jcb.200503088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Langlais P, Yi Z, Lefort N, De Filippis EA, Hwang H, Christ-Roberts CY, Mandarino LJ. Phosphorylation of human insulin receptor substrate-1 at Serine 629 plays a positive role in insulin signaling. Endocrinology. 2007;148:4895–4905. doi: 10.1210/en.2007-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly KE, Rojo F, She QB, Solit D, Mills GB, Smith D, Lane H, Hofmann F, Hicklin DJ, Ludwig DL, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–1508. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh WJ, Jacinto E. mTOR complex 2 signaling and functions. Cell Cycle. 2011;10:2305–2316. doi: 10.4161/cc.10.14.16586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh WJ, Wu CC, Kim SJ, Facchinetti V, Julien LA, Finlan M, Roux PP, Su B, Jacinto E. mTORC2 can associate with ribosomes to promote cotranslational phosphorylation and stability of nascent Akt polypeptide. Embo J. 2010;29:3939–3951. doi: 10.1038/emboj.2010.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashkova N, Gakhar L, Winistorfer SC, Yu L, Ramaswamy S, Piper RC. WD40 repeat propellers define a ubiquitin-binding domain that regulates turnover of F box proteins. Mol Cell. 2010;40:433–443. doi: 10.1016/j.molcel.2010.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz K, Liu YF, Shorer H, Hemi R, LeRoith D, Quan M, Kanety H, Seger R, Zick Y. Phosphorylation of insulin receptor substrate-1 (IRS-1) by protein kinase B positively regulates IRS-1 function. J Biol Chem. 1999;274:28816–28822. doi: 10.1074/jbc.274.40.28816. [DOI] [PubMed] [Google Scholar]
- Pederson TM, Kramer DL, Rondinone CM. Serine/threonine phosphorylation of IRS-1 triggers its degradation: possible regulation by tyrosine phosphorylation. Diabetes. 2001;50:24–31. doi: 10.2337/diabetes.50.1.24. [DOI] [PubMed] [Google Scholar]
- Shah OJ, Hunter T. Turnover of the active fraction of IRS1 involves raptor-mTOR- and S6K1-dependent serine phosphorylation in cell culture models of tuberous sclerosis. Mol Cell Biol. 2006;26:6425–6434. doi: 10.1128/MCB.01254-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah OJ, Wang Z, Hunter T. Inappropriate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. Curr Biol. 2004;14:1650–1656. doi: 10.1016/j.cub.2004.08.026. [DOI] [PubMed] [Google Scholar]
- Smith LK, Vlahos CJ, Reddy KK, Falck JR, Garner CW. Wortmannin and LY294002 inhibit the insulin-induced down-regulation of IRS-1 in 3T3-L1 adipocytes. Mol Cell Endocrinol. 1995;113:73–81. doi: 10.1016/0303-7207(95)03622-e. [DOI] [PubMed] [Google Scholar]
- Sun XJ, Goldberg JL, Qiao LY, Mitchell JJ. Insulin-induced insulin receptor substrate-1 degradation is mediated by the proteasome degradation pathway. Diabetes. 1999;48:1359–1364. doi: 10.2337/diabetes.48.7.1359. [DOI] [PubMed] [Google Scholar]
- Sun XJ, Miralpeix M, Myers MG, Jr, Glasheen EM, Backer JM, Kahn CR, White MF. Expression and function of IRS-1 in insulin signal transmission. J Biol Chem. 1992;267:22662–22672. [PubMed] [Google Scholar]
- Takano A, Usui I, Haruta T, Kawahara J, Uno T, Iwata M, Kobayashi M. Mammalian target of rapamycin pathway regulates insulin signaling via subcellular redistribution of insulin receptor substrate 1 and integrates nutritional signals and metabolic signals of insulin. Mol Cell Biol. 2001;21:5050–5062. doi: 10.1128/MCB.21.15.5050-5062.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi CM, Emanuelli B, Kahn CR. Critical nodes in signalling pathways: insights into insulin action. Nat Rev Mol Cell Biol. 2006;7:85–96. doi: 10.1038/nrm1837. [DOI] [PubMed] [Google Scholar]
- Tremblay F, Brule S, Hee Um S, Li Y, Masuda K, Roden M, Sun XJ, Krebs M, Polakiewicz RD, Thomas G, Marette A. Identification of IRS-1 Ser-1101 as a target of S6K1 in nutrient- and obesity-induced insulin resistance. Proc Natl Acad Sci U S A. 2007;104:14056–14061. doi: 10.1073/pnas.0706517104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay F, Marette A. Amino acid and insulin signaling via the mTOR/p70 S6 kinase pathway. A negative feedback mechanism leading to insulin resistance in skeletal muscle cells. J Biol Chem. 2001;276:38052–38060. doi: 10.1074/jbc.M106703200. [DOI] [PubMed] [Google Scholar]
- Tsutsumi T, Kuwabara H, Arai T, Xiao Y, Decaprio JA. Disruption of the Fbxw8 gene results in pre- and postnatal growth retardation in mice. Mol Cell Biol. 2008;28:743–751. doi: 10.1128/MCB.01665-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzatsos A. Raptor binds the SAIN (Shc and IRS-1 NPXY binding) domain of insulin receptor substrate-1 (IRS-1) and regulates the phosphorylation of IRS-1 at Ser-636/639 by mTOR. J Biol Chem. 2009;284:22525–22534. doi: 10.1074/jbc.M109.027748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um SH, Frigerio F, Watanabe M, Picard F, Joaquin M, Sticker M, Fumagalli S, Allegrini PR, Kozma SC, Auwerx J, Thomas G. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature. 2004;431:200–205. doi: 10.1038/nature02866. [DOI] [PubMed] [Google Scholar]
- Werner ED, Lee J, Hansen L, Yuan M, Shoelson SE. Insulin resistance due to phosphorylation of insulin receptor substrate-1 at serine 302. J Biol Chem. 2004;279:35298–35305. doi: 10.1074/jbc.M405203200. [DOI] [PubMed] [Google Scholar]
- White MF. IRS proteins and the common path to diabetes. Am J Physiol Endocrinol Metab. 2002;283:E413–422. doi: 10.1152/ajpendo.00514.2001. [DOI] [PubMed] [Google Scholar]
- Xu X, Sarikas A, Dias-Santagata DC, Dolios G, Lafontant PJ, Tsai SC, Zhu W, Nakajima H, Nakajima HO, Field LJ, et al. The CUL7 E3 ubiquitin ligase targets insulin receptor substrate 1 for ubiquitin-dependent degradation. Mol Cell. 2008;30:403–414. doi: 10.1016/j.molcel.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Yoon SO, Poulogiannis G, Yang Q, Ma XM, Villen J, Kubica N, Hoffman GR, Cantley LC, Gygi SP, Blenis J. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science. 2011;332:1322–1326. doi: 10.1126/science.1199484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhande R, Mitchell JJ, Wu J, Sun XJ. Molecular mechanism of insulin-induced degradation of insulin receptor substrate 1. Mol Cell Biol. 2002;22:1016–1026. doi: 10.1128/MCB.22.4.1016-1026.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzalla V, Stracka D, Oppliger W, Hall MN. Activation of mTORC2 by Association with the Ribosome. Cell. 2011;144:757–768. doi: 10.1016/j.cell.2011.02.014. [DOI] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.