Abstract
Rift Valley fever virus (RVFV) encodes one major virulence factor, the NSs protein. NSs suppresses host general transcription, including interferon (IFN)-β mRNA synthesis, and promotes degradation of the dsRNA-dependent protein kinase (PKR). We generated a novel RVFV mutant (rMP12-NSsR173A) specifically lacking the function to promote PKR degradation. rMP12-NSsR173A infection induces early phosphorylation of eIF2α through PKR activation, while retaining the function to inhibit host general transcription including IFN-β gene inhibition. MP-12 NSs but not R173A NSs binds to wt PKR. R173A NSs formed filamentous structure in nucleus in a mosaic pattern, which was distinct from MP-12 NSs filament pattern. Due to early phosphorylation of eIF2α, rMP12-NSsR173A could not efficiently accumulate viral proteins. Our results suggest that NSs-mediated host general transcription suppression occurs independently of PKR degradation, while the PKR degradation is important to inhibit the phosphorylation of eIF2α in infected cells undergoing host general transcription suppression.
Keywords: Rift Valley fever virus, bunyavirus, NSs protein, PKR, interferon, MP-12, reverse genetics
Introduction
Rift Valley fever virus (RVFV) belonging to the family Bunyaviridae, genus Phlebovirus is a mosquito-borne zoonotic pathogen endemic to sub-Saharan Africa, and has spread into Egypt, Madagascar, Saudi Arabia and Yemen (Pepin et al., 2010; Swanepoel and Coetzer, 2004). Humans infected with RVFV suffer from febrile illness with occasional complications such as hemorrhagic fever, encephalitis or blindness (Ikegami and Makino, 2011). RVFV infection of pregnant ruminants causes high rates of abortion and fetal malformation (Swanepoel and Coetzer, 2004). Because of its potential impact on public health and agriculture, RVFV is classified as Category A Priority pathogen by NIH/NIAID, and overlap select agent by CDC/USDA in the U.S. (Bird et al., 2009; Mandell and Flick, 2010). Currently, there are no commercially available vaccines outside endemic countries, and there are no effective therapeutics to treat RVF patients. MP-12 is the only strain excluded from select agent rule, and can be handled in BSL-2 laboratories.
RVFV genome is comprised of a tripartite negative-stranded RNA genome named S-, M-, and L-segments (Schmaljohn and Nichol, 2007). In addition to viral structural proteins; i.e., N, L, Gn and Gc proteins, RVFV encodes two nonstructural proteins (NSs and NSm) and the less characterized 78 kD protein. NSs protein, encoded in the S-segment, is a major virulence factor of RVFV, and has three functions; 1) suppression of the general host transcription by sequestering TFIIH p44 subunit proteins (Le May et al., 2004) and by promoting the degradation of TFIIH p62 subunit proteins (Kalveram, Lihoradova, and Ikegami, 2011), 2) degradation of dsRNA-dependent protein kinase (PKR) (Habjan et al., 2009; Ikegami et al., 2009), and 3) inhibition of the IFN-β promoter activation through sin3A-associated protein (SAP30) (Le May et al., 2008). MP-12 strain encodes functional NSs gene, which inhibits host general transcription and promotes degradation of PKR (Billecocq et al., 2008; Ikegami et al., 2009; Ikegami et al., 2006; Kalveram, Lihoradova, and Ikegami, 2011). In addition, NSs is responsible for cell cycle arrest at either G0/G1 or S phase, as well as DNA damage response via ataxia-telangiectasia mutated (ATM) (Austin et al., 2012; Baer et al., 2012). NSs interacts with pericentromeric γ-satellite sequence and induces defect of chromosome cohesion and segregation (Mansuroglu et al., 2009).
Little is known about the role of PKR degradation in RVFV life cycle. Habjan et al. showed that the RVFV clone 13 strain, lacking a functional NSs, replicates efficiently in PKR-knockout mice (Habjan et al., 2009). Our study demonstrated that cells infected with rMP12-rLuc (lacking NSs) resulted in increased levels of eIF2α phosphorylation, as compared to those infected with parental MP-12 (encoding NSs), in the presence of actinomycin D (ActD; transcription inhibitor) (Ikegami et al., 2009). On the other hand, phosphorylation of eIF2α was significantly suppressed in the presence of MP-12 NSs or PKRΔE7 (a dominant-negative form of human PKR). These results suggest that the PKR degradation by MP-12 NSs alleviates the negative effect of host transcription suppression to maintain viral translation. Although RVFV NSs encodes both host transcription suppression function and PKR degradation function, it is not clear whether host transcription suppression occurs independently of PKR degradation, or vice versa, and NSs-mediated transcription suppression creates cellular environment which requires PKR degradation for an efficient viral translation.
We hypothesize that host transcription suppression and IFN-β gene suppression occur independently of PKR degradation, while PKR degradation is important to inhibit eIF2α phosphorylation under NSs-mediated host transcription suppression. In this study, we generated and characterized a novel NSs mutant that does not promote PKR degradation while inhibiting host general transcription. Our results suggest that NSs-mediated host transcription suppression occurs independently of PKR degradation, and cells undergoing host general transcription by NSs induce early eIF2α phosphorylation by viral replication when PKR degradation does not occur. Thus, PKR degradation plays an important role for an efficient viral protein synthesis in RVFV-infected cells.
Results
Generation of MP-12 NSs mutants by alanine substitution
To study the significance of PKR degradation in cells infected with RVFV, we tried to generate an MP-12 encoding a mutant NSs that does not promote PKR degradation yet inhibits host general transcription. Since no structural analyses are available for the NSs protein, we performed random screening of NSs mutants. Initially, 11 serial in-frame truncations of 25 amino acids were introduced into MP-12 NSs, and the resulting NSs mutants were functionally analyzed. Those NSs truncated mutants lacked PKR degradation function, as well as IFN-β gene suppression (Head, Kalveram, and Ikegami, 2012). In an attempt to generate mutants lacking only one of known functions of NSs, we next randomly substituted one or two charged residues of NSs with alanine to create single or double point mutants. Among 10 recombinant MP-12 viruses characterized (R15A/R16A, R47A, R103A, D124A, E156A/D157A, R164A, E169A, R173A, D184A, E188A), R15A/R16A, R47A, R103A, D124A, R164A and R173A mutants inhibited IFN-β gene in MRC-5 cells. Among the mutants inhibiting IFN-β gene, only R173A did not promote degradation of PKR in VeroE6 cells, while R15A/R16A, R47A, R103A, D124A and R164A still promoted PKR degradation in VeroE6 cells (data not shown). We designated the R173A mutant as rMP12-NSsR173A, and further characterized the phenotypes to know the NSs functions in detail. We first characterized the plaque phenotype because MP-12 forms varied sizes of plaques with clear cytopathic effect (CPE), while MP-12 lacking NSs forms turbid plaques with minimum CPE (Ikegami et al., 2006). R15A/R16A, R47A, D124A, E156A/D157A, R164A, E188A formed MP-12-like plaques, while E169A and D184A formed uniformly turbid plaques, and R103A formed a mixture of MP-12-like plaques and turbid plaques (data not shown). The rMP12-NSsR173A formed uniformly smaller plaques than those of MP-12 with clear cytopathic effect (CPE) (Fig.1).
Fig. 1. rMP12-NSsR173A virus does not promote PKR degradation.
Plaque phenotype of MP-12 (left) and rMP12-NSsR173A virus (right) on VeroE6 cells at 3 dpi.
The rMP12-NSsR173A induces PKR-mediated eIF2α phosphorylation
We examined the viral protein synthesis and eIF2α phosphorylation level in cells infected with rMP12-NSsR173A. Type-I IFN-competent mouse embryonic fibroblast cells (MEF/wt cells) were mock-infected or infected with MP-12, rMP12-C13type, rMP12-NSsR173A, or rMP12-mPKRN167 virus at an moi of 3. Cell lysates were collected at 6 and 12 hpi, and analyzed by Western blot (Fig. 2A and 2B). We included rMP12-C13type or rMP12-mPKRN167(Lihoradova et al., 2012) which lack all NSs functions or specifically inhibit mouse PKR while still inducing IFN-β, respectively, as controls. We used mouse MEF cells to be able to further characterize the effect in cells lacking PKR (MEF/PKR0/0 cells) in later experiments. As expected, cells infected with MP-12 caused a degradation of PKR at 6 hpi, and phosphorylation status of eIF2α was not significantly changed at 6 or 12 hpi. Cells infected with rMP12-C13type increased the abundance of phosphorylated eIF2α (1.63 times) at 12 hpi, and the accumulation of Gn and Gc was 25% and 31% less than those of MP-12-infected cells, respectively, while accumulation of N protein was still similar to that in cells that were infected with MP-12. On the other hand, cells that were infected with rMP12-NSsR173A showed a visible band of PKR, while phosphorylated eIF2α was significantly increased by 1.87 times (p=0.0185, vs. MP-12) at 6 hpi, and the accumulation of Gn, Gc, and N proteins was 53%, 41%, and 38% less than those of MP-12-infected cells at 6 hpi, respectively. At 12 hpi, cells infected with rMP12-NSsR173A still had abundant phosphorylated eIF2α and less abundant Gn, Gc, and N. Levels of phosphorylated eIF2α in cells infected with rMP12-mPKRN167 virus were not significantly increased, and the abundance of Gn, Gc, and N proteins were similar to those of MP-12-infected cells. These results suggested that rMP12-NSsR173A does not induce PKR degradation, and increases the abundance of phosphorylated eIF2α faster than rMP12-C13type.
Fig. 2. Status of eIF2α and viral translation in MEF cells.
MEF/wt cells were mock-infected or infected with indicated viruses at an moi of 3, and the cell lysates were harvested at indicated time points, and Western blotting was performed to detect indicated protein accumulations. (A) Abundance of PKR, eIF2α, phosphorylated eIF2α (Ser51), viral N and NSs proteins, Flag-mPKRN167, and β-actin in cells mock-infected or infected with MP-12, rMP12-C13type (C13type), rMP12-NSsR173A (R173A), or rMP12-mPKRN167 (mPKRN167) at 6 and 12 hpi. Signal intensity of viral proteins (N, Gn and Gc) in cells infected with MP-12 was used as the scale to measure the others and considered 100%. Asterisk (*) represents a nonspecific band. (B) The relative abundance of phosphorylated eIF2α at 6 and 12 hpi. The density of phosphorylated eIF2α normalized to total eIF2α in mock-infected cells is represented as 100%. Asterisk represents statistical significance (unpaired t-test, *p<0.05, **p<0.01). (C) Abundance of PKR, eIF2α, phosphorylated eIF2α (Ser51), viral N and NSs proteins and β-actin in cells mock-infected or infected with MP-12, rMP12-NSsR173A (R173A) at 3, 4 and 5 hpi. (D) The relative abundance of PKR, phosphorylated eIF2α, N and NSs in cells infected with MP-12 or rMP12-NSsR173A at 3, 4 and 5 hpi. The density of PKR, phosphorylated eIF2α (normalized to total eIF2α) in mock-infected cells, or that of N or NSs in MP-12-infected cells at 3 hpi were represented as 100% for PKR, eIF2αP, N or NSs, respectively. Asterisk represents statistical significance (unpaired t-test, *p<0.05, ns; not significant). In B and D, means +/− standard deviation of three independent experiments are shown as a graph, while representative data of three independent experiments are shown in A and C.
Since the abundance of R173A NSs proteins was lower than that of MP-12 NSs at 6 and 12 hpi, it was possible that low level of R173A NSs accumulation could result in a lack of PKR degradation. To address this, we analyzed early time points (i.e., 3, 4 and 5 hpi in MEF/wt cells mock-infected or infected with MP-12 or rMP12-NSsR173A). At 3 hpi, PKR abundance was lower in cells infected with MP-12 than those infected with rMP12-NSsR173A at 3 hpi (unpaired t-test, p=0.029) due to an increase of PKR abundance in cells infected with rMP12-NSsR173A (Fig.2C and D). However, the abundance of PKR was clearly decreased at 4 hpi in cells infected with MP-12 compared to mock-infected cells (unpaired t-test, p=0.033 vs. mock-infected cells at 4 hpi). Thus, MP-12 NSs could start degrading PKR as early as 4 hpi in MEF/wt cells. We compared the PKR level of MP-12-infected cells at 4 hpi and that of rMP12-NSsR173A-infected cells at 5 hpi. The abundance of R173A N and NSs at 5 hpi was approximately 82.6% and 55.9% of that of MP-12 N and NSs at 4 hpi, respectively. The abundance of PKR in cells infected with rMP12-NSsR173A at 5 hpi was significantly higher than that in cells infected with MP-12 at 4 hpi (unpaired t-test, p=0.033). Compared to mock-infected cells at 4 hpi or 5 hpi, the PKR abundance was decreased by 69% in MP-12-infected cells at 4 hpi, and it was increased by 9% in rMP12-NSsR173A-infected cells at 5 hpi, respectively. In addition, cells infected with MP-12 at 3 hpi showed 84% of NSs abundance compared to R173A NSs at 5 hpi, yet slightly decreased PKR abundance by 12% although the difference was not statistically significant. Thus, a lack of PKR degradation was most probably not due to abundance of R173A NSs. We also confirmed that rMP12-NSsR173A could induce accumulation of phosphorylated eIF2α as early as 5 hpi (Fig.2D). The results suggest that rMP12-NSsR173A does not promote PKR degradation and induces the phosphorylation of eIF2α by 5 hpi, which inhibits the accumulation of viral proteins.
MP-12 replication is negatively affected by a lack of PKR degradation
We next tested viral replication levels of MP-12, rMP12-C13type, rMP12-NSsR173A, or rMP12-mPKRN167 virus in MEF/wt cells and MEF/PKR0/0 cells (MEF cells lacking PKR) (Fig.3). The replication of rMP12-NSsR173A or rMP12-C13type in MEF/wt cells was not significantly decreased compared to that of MP-12 at an moi of 1.0, while it was significantly decreased at an moi of 0.01 (Fig.3A). On the other hand, rMP12-mPKRN167 could replicate as efficiently as MP-12 in MEF/wt cells at both high and low moi (Fig.3A). The replication of rMP12-NSsR173A or rMP12-C13type was not decreased in MEF/PKR0/0 cells both at an moi of 0.01 or 1.0 (Fig.3B). We also tested the replication of rMP12-NSsR173A in human MRC-5 cells. In MRC-5 cells, replication of rMP12-NSsR173A or rMP12-C13type was decreased both at high and low moi compared to that of MP-12 (Fig.3C). Thus, the results suggest that the replication levels of rMP12-NSsR173A and rMP12-C13type are decreased due to a lack of PKR degradation in infected cells. We also noted that the replication of rMP12-NSsR173A was less efficient in human MRC-5 cells than in mouse MEF/wt cells.
Fig. 3. Replication of rMP12-NSsR173A.
MEF/wt cells (A) or MEF/PKR0/0 cells (B) were mock-infected or infected with MP-12, rMP12-C13type, rMP12-NSsR173A, or rMP12-mPKRN167 at an moi of 1.0 or 0.01, and culture supernatants were collected at 16 hpi or 72 hpi, respectively, to determine the viral titer by plaque assay of VeroE6 cells. (C) MRC-5 cells were infected with MP-12, rMP12-C13type or rMP12-NSsR173A at moi of 3.0 or 0.01, and virus titer at 16 hpi or 72 hpi were measured, respectively. Mean +/− standard deviations of three independent experiments are shown in the graph. Unpaired Student’s t-test was performed to compare to the value of MP-12: **p<0.01, *p<0.05.
The rMP12-NSsR173A NSs does not interact with PKR
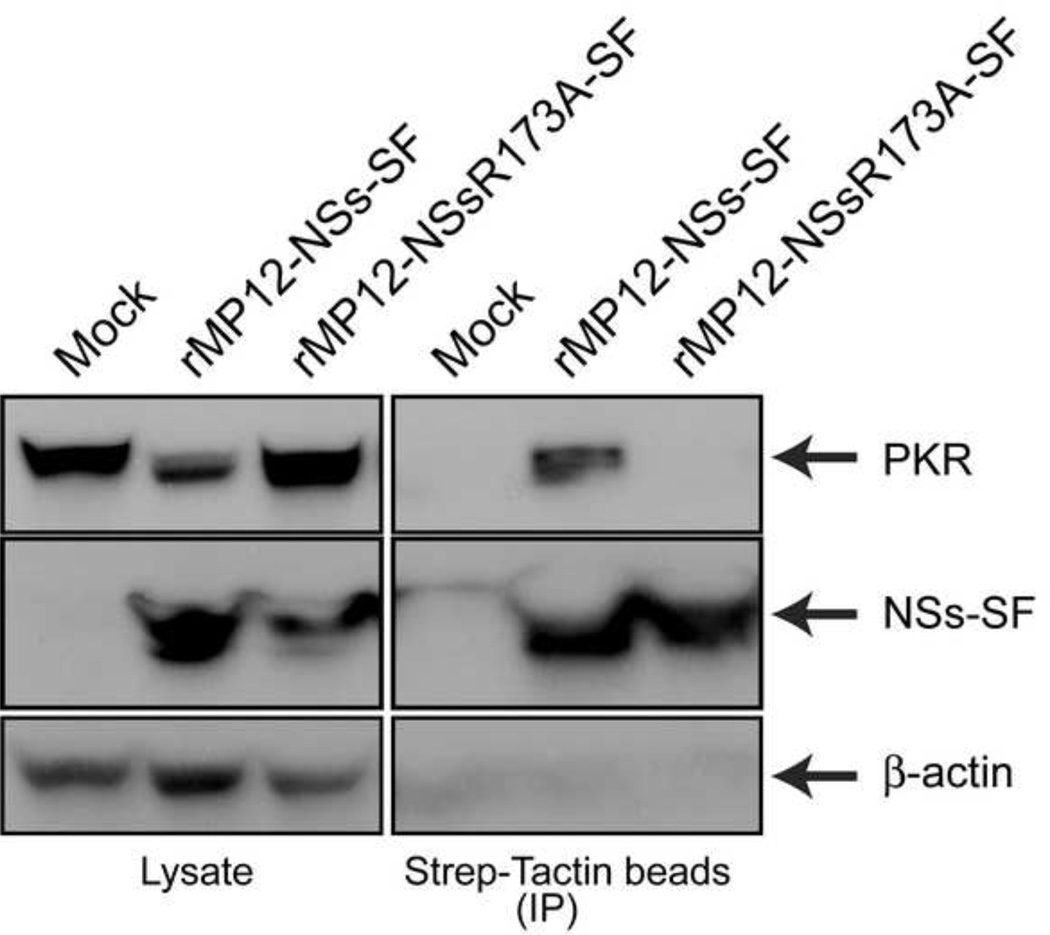
To further characterize the lack of PKR degradation by rMP12-NSsR173A, we analyzed the molecular interaction of NSs with PKR (Fig.4). 293 cells were mock-infected or infected with rMP12-NSs-SF (a tandem repeat of Strep-tag and Flag-tag at the C-terminus of NSs) or rMP12-NSsR173A-SF at an moi of 3. Cell lysates were collected at 5 hpi, and NSs-SF was precipitated by using Strep-Tactin magnetic beads. We collected cells at 5 hpi because cells infected with rMP12-NSs-SF do not completely degrade PKR at this time point. We confirmed abundant precipitation of NSs-SF and R173 NSs-SF, while the amount of PKR input was much smaller in rMP12-NSs-SF samples than rMP12-NSsR173A-SF samples. PKR was co-precipitated with MP-12 NSs-SF, suggesting the interaction of NSs with PKR (Fig.4). On the other hand, R173A NSs-SF did not co-precipitate PKR, in spite of an abundant precipitation of R173A NSs and higher levels of PKR than cells infected with rMP12-NSs-SF (Fig.4). The result suggests that R173A NSs lacks the ability to interact with PKR.
Fig. 4. Co-precipitation of PKR with rMP12-NSs-SF and rMP12-NSsR173A-SF.
293 cells were mock-infected or infected with rMP12-NSs-SF (a tandem repeat of Strep-tag and Flag-tag at the C-terminus of NSs) or rMP12-NSsR173A-SF at an moi of 3. Cell lysates were collected at 5 hpi, and NSs-SF was precipitated by using Strep-Tactin magnetic beads. Western blotting was performed to detect, NSs-SF (anti-Flag antibody) or β-actin.
The rMP12-NSsR173A NSs inhibits IFN-β gene expression
To understand the functional difference of cells infected with MP-12 lacking NSs from those infected with rMP12-NSsR173A, we next characterized the residual transcription suppression function of rMP12-NSsR173A. At first, we tested whether rMP12-NSsR173A virus inhibits the activation of host IFN-β promoter. MEF/wt cells or MEF/PKR0/0 cells were infected at an moi of 3 with MP-12, rMP12-C13type, rMP12-NSsR173A or rMP12-mPKRN167, and total RNA was extracted at 7 hpi. We included MEF/PKR0/0 cells and rMP12-mPKRN167 virus to determine if PKR is required for IFN-β gene up-regulation. Northern blot was performed to detect the accumulation of IFN-β or ISG56 mRNA (Fig.5). Both rMP12-C13type and rMP12-mPKRN167, which lack functional NSs expression, induced accumulation of IFN-β and ISG56 mRNA in MEF/wt cells and MEF/PKR0/0 cells. On the other hand, MEF/wt cells or MEF/PKR0/0 cells infected with MP-12 or rMP12-NSsR173A virus did not induce IFN-β or ISG56 mRNA. The results suggested that R173A NSs inhibits IFN-β mRNA synthesis in the absence or presence of PKR, while PKR is dispensable for IFN-β gene activation by MP-12 lacking NSs.
Fig. 5. rMP12-NSsR173A virus inhibits IFN-β mRNA synthesis.
(A) MEF/wt cells or MEF/PKR0/0 cells were mock-infected or infected with MP-12, rMP12-C13type (C13type), rMP12-NSsR173A (R173A), or rMP12-mPKRN167 (N167), or (C) MRC-5 cells were mock-infected or infected with rMP12-rLuc, MP-12 or rMP12-NSsR173A virus at an moi of 3. Total RNA was harvested at 7 hpi. Northern blotting was performed with the strand-specific RNA probes to detect IFN-β or ISG56 mRNA, or RVFV anti-sense S-segment / N mRNA.
We also tested IFN-β mRNA induction in human MRC-5 cells. MRC-5 cells were mock-infected or infected with MP-12, rMP12-rLuc (a control lacking NSs), which encodes rLuc in place of NSs, or rMP12-NSsR173A at an moi of 3. At 7 hpi., total RNA was extracted. IFN-β mRNA and ISG56 mRNA were detected in cells infected with rMP12-rLuc, while they were not induced in cells infected with MP-12 or rMP12-NSsR173A (Fig.5B).
We noted slowly migrating RNA bands derived from MEF/wt cells and MEF/PKR0/0 cells infected with MP-12, rMP12-NSsR173A and rMP12-mPKRN167 as well as MRC-5 cells infected with rMP12-NSsR173A. On the other hand, ISG56 mRNA, which is inducible by IFN-β, was not increased in MEF/wt, MEF/PKR0/0 or MRC-5 cells infected with MP-12 or rMP12-NSsR173A. We did not further characterize the slowly migrating bands reactive to IFN-β probe in this study because it is beyond the scope of study.
The rMP12-NSsR173A inhibits host general transcription
We subsequently analyzed the effect of rMP12-NSsR173A on host general transcription. MEF/wt cells were mock-infected or infected with MP-12, rMP12-C13type, rMP12-NSsR173A or rMP12-mPKRN167 viruses at an moi of 3 (Fig. 6A). Cells were treated with the uridine analog 5-ethynyluridine (EU) at 16 hpi, and fixed at 17 hpi. The incorporated EU was covalently linked to Alexa Fluor 594 coupled-azide (red signal) by click reaction (Jao and Salic, 2008), and cells were further reacted with anti-RVFV antibodies (green signal). Mock-infected cells actively incorporated the EU into nascent RNA in the nucleus or nucleoli, as shown by strong red signals. Such incorporation of EU was not detectable in the presence of ActD, suggesting that the incorporation of EU is specific to RNA transcription. Cells infected with rMP12-C13type or rMP12-mPKRN167 actively incorporated EU, while those infected with MP-12 or rMP12-NSsR173A virus did not incorporate EU as efficiently as cells infected with rMP12-C13 type or rMP12-mPKRN167 virus. These results suggested that both MP-12 and rMP12-NSsR173A suppress host general transcription, while rMP12-C13type and rMP12-mPKRN167 lack the ability to suppress host transcription in MEF/wt cells.
Fig. 6. Host general transcriptional suppression by RVFV MP-12 mutants.
(A) Incorporation of uridine analog 5-ethynyluridine (EU) into nascent RNA is tested in MEF/wt cells. MEF/wt cells were mock-infected or infected with MP-12, rMP12-C13type, rMP12-NSsR173A, or rMP12-mPKRN167 at an moi of 3, and 1 mM EU was added into the culture at 16 hpi for 1 hour. Then, cells were fixed and stained with Alexa Fluor 594-coupled azide (red). The cells were further stained with anti-RVFV antibodies and Alexa Fluor 488-conjugated anti-mouse IgG (green). As a control, mock-infected cells were co-treated with actinomycin D (ActD) at 5 µg/ml with 1 mM EU for 1 hour at 16 hpi. (B) Fluorescence-activated cell sorting (FACS) analysis was performed in 293 cells. 293 cells were mock-infected or infected with MP-12, rMP12-C13type, or rMP12-NSsR173A at an moi of 3 and treated with 0.5 mM EU at 8 hpi for 1 hour. The control cells were co-treated with ActD (5 µg/ml) at 8 hpi for 1 hour. Incorporated EU was stained with Alexa Fluor 647-azide, and viral antigens were stained with anti-RVFV antibodies. Cells were then analyzed by flow cytometry on the LSRII Fortessa.
We also analyzed the level of EU incorporation quantitatively by using 293 cells as described previously (Kalveram, Lihoradova, and Ikegami, 2011). Mock-infected 293 cells or those infected with MP-12, rMP12-C13type, or rMP12-NSsR173A were treated with EU at 8 hpi, fixed at 9 hpi, and stained with Alexa Fluor 647 coupled-azide and anti-RVFV antibody before analysis by fluorescence-activated cell sorting (FACS) (Fig. 6B). We also used rMP12-PKRΔE7, which encodes dominant-negative human PKR in place of NSs (Ikegami et al., 2009). The rationale to use rMP12-PKRΔE7 instead of rMP12-mPKRN167 for the control is that dominant-negative suppression of PKR is host species-specific (Li and Koromilas, 2001). Most cells infected with rMP12-NSsR173A showed reduced EU incorporation, while 19.3% of total cells (22% of RVFV-infected cell population) incorporated EU at a similar level to that of rMP12-C13type -infected cells. Thus, we concluded that rMP12-NSsR173A retains host general transcription suppression function, as opposed to rMP12-C13type which lacks all NSs functions.
PKR plays an important role for eIF2α phosphorylation induced by rMP12-NSsR173A
Although we observed that rMP12-NSsR173A induced an early eIF2α phosphorylation (Fig.2), it might be due to cellular stress responses induced by an accumulation of misfolded NSs proteins. To know the involvement of PKR in eIF2α phosphorylation, we analyzed the effect of R173A NSs expression on eIF2α phosphorylation. Human Hec1B cells were mock-infected or infected with rMP12-rLuc or rMP12-PKRΔE7 at an moi of 3. Then, cells were mock-transfected or immediately transfected with in vitro synthesized RNA encoding R173A NSs. We used in vitro synthesized RNA rather than plasmid DNA because R173A NSs can inhibit the transcription of own gene. Cells were collected at 16 hpi. We used type-I IFN unresponsive Hec1B cells to avoid PKR activation by RNA transfection (Constantinescu et al., 1995). The expression of R173A NSs without RVFV replication did not induce PKR phosphorylation, while it induced slight increase in eIF2α phosphorylation compared to mock-treated cells (Fig.7A). The result suggests that PKR activation is negligible by the transfection of in vitro synthesized RNA in Hec1B cells. On the other hand, a combination of R173A NSs expression and the replication of rMP12-rLuc induced clearly detectable PKR and eIF2α phosphorylation, and slightly decreased the synthesis of R173A NSs from transfected R173A NSs RNA (Fig.7A). The phosphorylation of PKR was inhibited in a combination of R173A NSs expression and the replication of rMP12-PKRΔE7, and the level of eIF2α phosphorylation was significantly decreased (Fig.7A and B). These results suggest that PKR is responsible for inducing eIF2α phosphorylation in cells infected with rMP12-NSsR173A.
Fig. 7. R173A NSs-mediated eIF2α phosphorylation is decreased in the presence of dominant-negative PKR.
(A) Human Hec1B cells were mock-infected or infected with rMP12-rLuc or rMP12-PKRΔE7 at an moi of 3. Then, cells were mock-transfected or immediately transfected with in vitro synthesized RNA encoding R173A NSs. Cells were collected at 16 hpi, and the abundance of PKR, phosphorylated eIF2α (Ser51), total eIF2α, R173A NSs, N and β-actin was analyzed by Western blot. (B) The relative abundance of phosphorylated eIF2α and NSs were analyzed. The density of phosphorylated eIF2α (normalized to total eIF2α) or NSs in mock-infected cells which are transfected with R173A NSs RNA is represented as 100%. The mean +/− standard deviation of three independent experiments is shown as a graph. Asterisk represents statistical significance (unpaired t-test, *p<0.05, **p<0.01).
The R173A NSs forms filamentous structure in nucleus
Since the filamentous structure of NSs may play an important role in host general transcription suppression and IFN-β gene inhibition (Le May et al., 2004; Le May et al., 2008), we next characterized cellular localization of R173A NSs. However, the synthesis of R173A NSs could be inhibited at transcriptional level (Fig.6) and translational level via PKR-mediated eIF2α phosphorylation (Fig.2). To maximize the expression of R173A NSs in cells, MEF/PKR0/0 cells were mock-transfected or transfected with in vitro synthesized RNA encoding MP-12 NSs or R173A NSs. At 14 hours post transfection, cells were fixed with methanol, and subjected into indirect immunofluorescent assay using anti-RVFV antibody. As shown in Fig.8, anti-RVFV antibody did not react to mock-transfected cells (Fig.8A). Cells transfected with MP-12 NSs RNA was specifically stained with anti-RVFV antibody (Fig.8B and C). Interestingly, the shade of NSs filament could be detected in DAPI staining only (Fig.8B, arrows), suggesting MP-12 NSs sequesters chromatin structure. MP-12 NSs also accumulated in cytoplasm (Fig.8C and D), which is consistent with previous study (Yadani et al., 1999). Although the accumulation level was low, R173A NSs could be detected in both nucleus and cytoplasm, and formed filamentous structure inside nucleus, which was different in shape from that of MP-12 NSs, and showed irregular mosaic-like pattern (Fig.8E and F).
Fig. 8. Cellular localization of MP-12 NSs and R173A NSs.
MEF/PKR0/0 cells were mock-transfected or transfected with in vitro synthesized RNA encoding MP-12 NSs or R173A NSs. At 14 hours post transfection, cells were fixed with methanol, and indirect immunofluorescent assay using anti-RVFV antibody was performed. Mock-transfected cells (A) or cells transfected with MP-12 NSs RNA (B–D) or R173A NSs RNA (E and F) were incubated with anti-RVFV antibody (A and C–F) or normal mouse serum (B). Nuclei were visualized by DAPI. Scale (50 µm) is shown as while bars, and enlarged image is shown as an inset in E. White arrow(s) indicate the area unstained with DAPI with filamentous shadow (B), or cells corresponding to inset image (E).
Discussion
In this study, we hypothesized that host transcription suppression and IFN-β gene suppression occur independently of PKR degradation, while PKR degradation is important to inhibit eIF2α phosphorylation under NSs-mediated host transcription suppression. We screened out a novel recombinant MP-12 NSs point mutants named rMP12-NSsR173A. The rMP12-NSsR173A forms small plaques in VeroE6 cells (Fig.1), does not promote PKR degradation (Fig.2), yet induces host general transcription suppression (Fig.6) including IFN-β gene (Fig.5). We found that MP-12 NSs but not R173A NSs interacts with wt PKR (Fig.4), while R173A NSs forms filamentous structure with a mosaic pattern which is distinct from MP-12 NSs filamentous structure (Fig.8). Cells supporting the replication of rMP12-NSsR173A but not MP-12 induced early phosphorylation of eIF2α through PKR activation (Fig.2 and 7). On the other hand, the replication of rMP12-NSsR173A was inefficient in MEF/wt cells, but not in MEF/PKR0/0 cells (Fig.3). Thus, our results suggest that NSs-mediated PKR degradation is important to inhibit early phosphorylation of eIF2α and contributes to an efficient viral translation in infected cells that undergo host general transcription suppression.
The small plaque of rMP12-NSsR173A is distinct from the large turbid plaque of rMP12-C13type (Ikegami et al., 2006). The CPE (typically change in cell shape and cell detachment from plate) of rMP12-NSsR173A is similar to that of MP-12, while rMP12-C13type only causes minimum cell detachment. Since R173A NSs but not C13type NSs retains host transcription suppression activity, it is likely that MP-12 cytopathic effect might be caused by host general transcription suppression. Alternatively, uncharacterized NSs functions may contribute to such CPE in infected cells. Among all the mutants tested, only rMP12-NSsR173A formed uniform small plaques in VeroE6 cells. This suggests that a combination of host general transcription suppression and active PKR in infected cells affects the spread of RVFV.
We observed that rMP12-C13type and rMP12-NSsR173A replicate inefficiently in MEF/wt cells and MRC-5 cells (Fig.3); (1) rMP12-C13type induces type-I IFNs and other antiviral genes including PKR in response to IFN stimulation (Habjan et al., 2009), while also inducing eIF2α phosphorylation at relatively late time in infection (Fig.2B) (2) rMP12-NSsR173A does not induce type-I IFNs and does not respond to type-I IFN signaling due to host general transcription suppression, while inhibiting viral protein synthesis by inducing eIF2α phosphorylation at early time in infection. Although it is difficult to analyze all the factors in two different cellular environment which affect infectious virus production, our data showed that rMP12-NSsR173A replication was recovered due to a lack of PKR activation (Fig.3A). Thus, it indicates that PKR degradation plays a role in promoting efficient viral replication in cells undergoing general transcription suppression, and the result is consistent with previous studies using RVFV and that encoding nonfunctional NSs (Habjan et al., 2009; Ikegami et al., 2009).
The introduction of alanine might cause the misfolding of NSs, and induce ER stress responses in infected cells (Zhao and Ackerman, 2006), which might induce eIF2α phosphorylation earlier than rMP12-C13type independent of PKR action (Fig.2). We found that there was no dramatic increase in PKR and eIF2α phosphorylation caused by the expression of R173A NSs itself (Fig.7). On the other hand, cells infected with rMP12-rLuc in the presence of R173A NSs induced eIF2α phosphorylation, while it did not occur by infection with rMP12-PKRΔE7, which expresses a dominant-negative form of PKR (Fig.7). These results suggest that eIF2α phosphorylation induced by replication of rMP12-NSsR173A occurs by PKR-dependent manner.
Since rMP12-NSsR173A replicated less efficiently than MP-12, it was difficult to know whether the absence of PKR degradation is due to an insufficient accumulation of NSs required for PKR degradation. However, we did not observe any significant decrease of PKR in cells infected with rMP12-NSsR173A in spite of the accumulation of R173A NSs to comparable level to that of MP-12 NSs (Fig.2C and D). We also tested the overexpression of R173A NSs, yet the over-expression of R173A NSs did not promote PKR degradation (Fig.7). Furthermore, we confirmed that R173A NSs does not interact with wt PKR (Fig.4). Thus, our results indicate that rMP12-NSsR173A indeed lacks the PKR degradation function, and that the decreased expression of R173A NSs is the effect, rather than the cause, of the increased phosphorylation. On the other hand, it is not clear why R173A mutation resulted in a loss of PKR degradation function. It is possible that NSs exerts PKR degradation through domain containing aa.173. Since arginine is a substrate of methylation or acetylation, the involvement such post-translational modification with R173A mutation might be also important for PKR degradation.
In this study, we developed and characterized a novel recombinant MP-12 that does not promote degradation of PKR, while inhibiting host general transcription including IFN-β gene suppression. Host general transcription suppression including IFN-β gene occurs independently of PKR degradation, and PKR degradation plays an important role in inhibiting early eIF2α phosphorylation in infected cells undergoing host general transcription suppression by NSs.
Materials and Methods
Media, Cells, and Viruses
Mouse embryonic fibroblast (MEF) cells (kindly provided by Dr. Bryan R.G. Williams at Monash University), MRC-5 (human diploid hung embryonic) cells, VeroE6 (African green monkey kidney) cells, HEK 293 (human embryonic kidney) cells and Hec1B (human endometrial carcinoma) cells were maintained in Dulbecco’s modified minimum essential medium (DMEM) containing 10% fetal calf serum (FCS). BHK/T7-9 cells that express T7 RNA polymerase (Ito et al., 2003) were grown in MEM-alpha containing 10% FCS. Penicillin (100 U/ml) and streptomycin (100 µg/ml) were added to the media. BHK/T7-9 cells were selected in medium containing 600 µg/ml hygromycin. MP-12 viral stock was passaged twice in VeroE6 cells. The rMP12-C13type, virus encoding the NSs gene with a 69% truncation (Ikegami et al., 2006), has been described previously. The rMP12-NSsR173A encodes NSs with an alanine substitution at aa.173. The rMP12-mPKRN167 encodes the N-terminus of mouse PKR from aa.1 to 167 with an N-terminal Flag tag in place of MP-12 NSs (Lihoradova et al., 2012). These recombinant MP-12s were recovered from BHK/T7-9 cells as described previously (Ikegami et al., 2006) and passaged once in VeroE6 cells. Virus titers were determined by plaque assay in VeroE6 cells. Lyophilized MP-12 vaccine was reconstituted and passaged once in MRC-5 cells.
Plasmid
To make a PCR fragment encoding the NSs ORF of rMP12-NSsR173A, PCR fragments was performed with the following primer sets: 1) HpavNSF: AGT TGT TAA CAT GGA TTA CTT TCC TGT GAT ATC TGT TGA TTT GCA G and 173AR: GGT CAA TCC CTG CGA GGA TGG CCT CAG, 2) 173AF: CTG AGG CCA TCC TCG CAG GGA TTG ACC and SpevNSR: CGC TAC TAG TCT AAT CAA CCT CAA CAA ATC CAT C (restriction enzyme sites are underlined). The second PCR was performed with the two PCR fragments obtained above with the primer set of HpavNSF and SpevNSR. The PCR fragment encoding NSs of R173A was digested with HpaI and SpeI and inserted into the NSs ORF of pProT7-vS(+), cut with HpaI and SpeI as described previously (Ikegami et al., 2006). The resulting plasmid was designated pProT7-S(+)-NS/R173A. To generate the RNA probes to detect mouse IFN-β mRNA or mouse ISG56 mRNA, we cloned a part of mouse IFN-β or mouse ISG56 between KpnI and HindIII of the PSPT18 plasmid after amplification of PCR fragments from first-strand cDNA from wild-type MEF cells that were infected with rMP12-C13type (7 hpi). The primers that were used for PCR amplification of mouse IFN-β or mouse ISG56 are as follows; KpnmIFNb-F: GAA GGG TAC CGC TCG GAC CAC CAT CCA GGC GTA G, HindmIFNb-R: GGC TAA GCT TAT GAA CAA CAG GTG GAT CCT CCA CGC, KpnmISG56F: GGG TGG TAC CGC TCC ACT TTC AGA GCC TTC GCA AAG CAG, HindmISG56R: TAC AAA GCT TAT GGG AGA GAA TGC TGA TGG TGA CCA GG (restriction enzyme sites are underlined). Human ISG56 probe was synthesized by using pCRII-ISG56 plasmid (kindly provided by Dr.Kui Li, University of Tennessee Health Science Center).
Recovery of recombinant MP-12
The rMP12-NSsR173A viruses was recovered by using a plasmid combination of pProT7-S(+)-NS/R173A, pProT7-M(+), pProT7-L(+), pT7-IRES-vN, and pT7-IRES-vL in BHK/T7-9 cells, as described previously (Ikegami et al., 2006).
Northern blot analysis
Total RNA was extracted from infected or mock-infected cells using TRIzol reagent. Denatured RNA was separated on 1% denaturing agarose-formaldehyde gels and transferred onto a nylon membrane (Roche Applied Science). Northern blot analysis was performed as described previously with strand-specific RNA probes to detect RVFV anti-sense S-segment / N mRNA, IFN-β mRNA (human or mouse), or ISG56 mRNA (human or mouse) (Ikegami et al., 2009; Ikegami et al., 2005).
Western blot analysis
Western blot analysis was performed as described previously (Ikegami et al., 2009). The membranes were incubated with anti-human PKR monoclonal antibody (BD biosciences), anti-mouse PKR antibody (B-10, Santa Cruz Biotech.), anti-PKR Phospho (pT446) antibody (Epitomics Inc.), anti-eIF2-alpha antibody (Cell Signaling Tech.), anti-phospho-eIF2-alpha (ser51) antibody (Cell Signaling Tech.), anti-RVFV mouse polyclonal antibody (a kind gift from Dr. R.B.Tesh, UTMB), anti-DYKDDDDK tag antibody (Cell Signaling Tech.), anti-Flag M2 monoclonal antibody (Sigma), or anti-actin goat polyclonal antibody (I-19; Santa Cruz Biotech.) overnight at 4°C and with secondary antibodies for 1 hr at room temperature.
Analysis of viral growth
MEF cells were infected with viruses at a multiplicity of infection (moi) of 3, 1 or 0.01 at 37°C for 1 hr. Culture supernatants were harvested at 16 hpi (moi of 3 or 1) or 72 hpi (moi of 0.01), and the viral titer was measured by plaque assay. The growth titers are shown as mean +/− standard deviation from three independent experiments.
Analysis of general transcription
MEF/wt cells were mock-infected or infected with MP-12, C13type, R173A, or N167 at an moi of 3. At 16 hpi, 1 mM 5-ethynyluridine (EU) was added directly into the cell culture, and cells were incubated for 1 hour. As a control, cells were treated with 5 µg/ml of ActD to inhibit cellular RNA synthesis for 1 hour with 1 mM EU (Ikegami et al., 2009). Cells were washed with PBS and fixed with 125 mM Pipes pH 6.8, 10 mM EGTA, 1 mM magnesium chloride, 0.2% Triton X-100, and 3.7% formaldehyde for 30 min at room temperature. After rinsing with TBS once, cells were stained at room temperature for 30 min with 100 mM Tris pH 8.5, 1 mM CuSO4, 10 µM Alexa Fluor 594-coupled azide (Invitrogen), and 100 mM ascorbic acid. The addition of ascorbic acid will reduce copper (II) and produce copper (I) in situ, where copper catalyzes a covalent reaction between a fluorescent azide and an alkyne of EU (click reaction) (Jao and Salic, 2008). After staining, cells were washed 4 times with TBS containing 0.5% Triton X-100 and then reacted with anti-RVFV antibody that was kindly provided by Dr.R.B.Tesh (1:500), followed by the staining with Alexa Fluor 488-conjugated anti-mouse IgG and DAPI. Cells were imaged with fluorescent microscopy to detect cellular RNA that incorporated EU in 1 hour. Fluorescence-activated cell sorting (FACS) analysis was performed in 293 cells. 293 cells were mock-infected or infected with MP-12, rMP12-C13type, or rMP12-NSsR173A at an moi of 3 and treated with 0.5 mM EU at 8 hpi for 1 hour. The control cells were co-treated with ActD (5 µg/ml) at 8 hpi for 1 hour. Incorporated EU was stained with Alexa Fluor 647-azide, and viral antigens were stained with anti-RVFV antibodies. Cells were then analyzed by flow cytometry on the LSRII Fortessa (BD Biosciences).
Co-affinity precipitation
SF (Strep-Flag)-tagged protein was precipitated with Strep-Tactin magnetic beads (Qiagen). 293 cells were mock-infected or infected with either rMP12-NSs-SF or rMP12-NSsR173A-SF at an moi of 3 and harvested at 5 hpi. Lysis was performed in 20 mM Tris pH 7.6, 0.1% Triton X-100, 150 mM NaCl, 1 mM EDTA, 1× complete protease inhibitors (Roche) and 100 U/ml Benzonase using 3 cycles of freeze-thaw. Cleared lysates (15 min, 13000 rpm, 4 °C) were incubated with 50 µl Strep-Tactin magnetic beads (Qiagen) for 2h at 4°C in an end-over-end rotor. Precipitates were washed 3 times in lysis buffer, boiled in SDS-PAGE sample buffer and analyzed by Western blot.
In vitro RNA synthesis
The pcDNA3.1mycHisA plasmid encoding MP-12 NSs or R173A NSs was linearized, and in vitro transcribed by using mMESSAGE mMACHINE T7 Ultra kit (Ambion) according to manufacturer’s instruction.
Transfection
Transfection of in vitro synthesized RNA was performed by using TransIT-mRNA Transfection Kit (Mirus) according to manufacturer’s instruction.
Indirect immunofluorescent assay (IFA)
MEF/PKR0/0 cells were mock-transfected or transfected with in vitro synthesized RNA encoding MP-12 NSs or R173A NSs. At 14 hours post transfection, cells were fixed with methanol at room temperature for 20 min. Then, cells were dried for 5 min, and blocked with PBS containing normal goat IgG (Santa Cruz Biotech, sc-2028, 1:1000) at 37°C for 30 min. Cells were incubated with anti-RVFV mouse antibody (kind gift from Dr.R.B.Tesh, University of Texas Medical Branch) (1:500 in PBS with normal goat IgG 1:1000) at 37°C for 1 hour, and subsequently incubated with Alexa Fluor 594 goat anti-mouse IgG (H+L) (Life Technologies) (1:1000 in PBS) at 37°C for 1 hour. After washing with PBS, DNA was stained with 4’,6’-diamidino-2-phenylindole (DAPI) (SlowFade Gold Antifade Reagent with DAPI, Life Technologies) to visualize nuclei. Cells were observed under Olympus IX71 microscope with a DP71 camera, and the image was detected in DP manager software.
Statistical analysis
The statistical analyses were performed by using the Graphpad Prism 5.03 program (Graphpad Software Inc.). Unpaired Student’s t-test was used for the comparison of two subjects.
Highlights.
RVFV NSs inhibits host transcription independently of PKR degradation.
R173A mutation in NSs gene specifically abolishes PKR degradation function.
Early eIF2α phosphorylation is induced by MP-12 encoding NSs R173A mutation.
MP-12 NSs but not R173A NSs interacts with wt PKR.
Acknowledgments
We thank Dr. R.B. Tesh at UTMB for providing anti-RVFV mouse polyclonal antibody, Dr.Kui Li at The University of Tennessee Health Science Center for providing pCRII-ISG56 plasmid, and Dr. Bryan R.G. Williams at Monash University, Australia, for providing MEF/PKR0/0 cells. The study was supported by useful suggestions at monthly meetings organized by the Western Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research. This work was supported by U54 AI057156 (NIAID, the Western Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research), R01 AI08764301-A1 (NIAID), and funding from the Sealy Center for Vaccine Development at UTMB. BK was supported by the James W. McLaughlin Fellowship Fund at UTMB. OL was supported by Maurice R.Hilleman Early-Stage Career Investigator Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Austin D, Baer A, Lundberg L, Shafagati N, Schoonmaker A, Narayanan A, Popova T, Panthier JJ, Kashanchi F, Bailey C, Kehn-Hall K. p53 activation following Rift Valley fever virus infection contributes to cell death and viral production. PLoS ONE. 2012;7(5):e36327. doi: 10.1371/journal.pone.0036327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer A, Austin D, Narayanan A, Popova T, Kainulainen M, Bailey C, Kashanchi F, Weber F, Kehn-Hall K. Induction of DNA Damage Signaling upon Rift Valley Fever Virus Infection Results in Cell Cycle Arrest and Increased Viral Replication. J Biol Chem. 2012;287(10):7399–7410. doi: 10.1074/jbc.M111.296608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billecocq A, Gauliard N, Le May N, Elliott RM, Flick R, Bouloy M. RNA polymerase I-mediated expression of viral RNA for the rescue of infectious virulent and avirulent Rift Valley fever viruses. Virology. 2008;378(2):377–384. doi: 10.1016/j.virol.2008.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird BH, Ksiazek TG, Nichol ST, Maclachlan NJ. Rift Valley fever virus. J Am Vet Med Assoc. 2009;234(7):883–893. doi: 10.2460/javma.234.7.883. [DOI] [PubMed] [Google Scholar]
- Constantinescu SN, Croze E, Murti A, Wang C, Basu L, Hollander D, Russell-Harde D, Betts M, Garcia-Martinez V, Mullersman JE, Pfeffer LM. Expression and signaling specificity of the IFNAR chain of the type I interferon receptor complex. Proc Natl Acad Sci U S A. 1995;92(23):10487–10491. doi: 10.1073/pnas.92.23.10487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habjan M, Pichlmair A, Elliott RM, Overby AK, Glatter T, Gstaiger M, Superti-Furga G, Unger H, Weber F. NSs protein of rift valley fever virus induces the specific degradation of the double-stranded RNA-dependent protein kinase. J Virol. 2009;83(9):4365–4375. doi: 10.1128/JVI.02148-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head JA, Kalveram B, Ikegami T. Functional analysis of Rift Valley fever virus NSs encoding a partial truncation. PLoS ONE. 2012;7:e45730. doi: 10.1371/journal.pone.0045730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami T, Makino S. The Pathogenesis of Rift Valley Fever. Viruses. 2011;3(5):493–519. doi: 10.3390/v3050493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami T, Narayanan K, Won S, Kamitani W, Peters CJ, Makino S. Rift Valley fever virus NSs protein promotes post-transcriptional downregulation of protein kinase PKR and inhibits eIF2alpha phosphorylation. PLoS Pathog. 2009;5(2) doi: 10.1371/journal.ppat.1000287. e1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami T, Won S, Peters CJ, Makino S. Rift Valley fever virus NSs mRNA is transcribed from an incoming anti-viral-sense S RNA segment. J Virol. 2005;79(18):12106–12111. doi: 10.1128/JVI.79.18.12106-12111.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami T, Won S, Peters CJ, Makino S. Rescue of infectious rift valley fever virus entirely from cDNA, analysis of virus lacking the NSs gene, and expression of a foreign gene. J Virol. 2006;80(6):2933–2940. doi: 10.1128/JVI.80.6.2933-2940.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito N, Takayama-Ito M, Yamada K, Hosokawa J, Sugiyama M, Minamoto N. Improved recovery of rabies virus from cloned cDNA using a vaccinia virus-free reverse genetics system. Microbiol Immunol. 2003;47(8):613–617. doi: 10.1111/j.1348-0421.2003.tb03424.x. [DOI] [PubMed] [Google Scholar]
- Jao CY, Salic A. Exploring RNA transcription and turnover in vivo by using click chemistry. Proc Natl Acad Sci U S A. 2008;105(41):15779–15784. doi: 10.1073/pnas.0808480105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalveram B, Lihoradova O, Ikegami T. NSs Protein of Rift Valley Fever Virus Promotes Post-Translational Downregulation of the TFIIH Subunit p62. J Virol. 2011;85:6234–6243. doi: 10.1128/JVI.02255-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le May N, Dubaele S, Proietti De Santis L, Billecocq A, Bouloy M, Egly JM. TFIIH transcription factor, a target for the Rift Valley hemorrhagic fever virus. Cell. 2004;116(4):541–550. doi: 10.1016/s0092-8674(04)00132-1. [DOI] [PubMed] [Google Scholar]
- Le May N, Mansuroglu Z, Leger P, Josse T, Blot G, Billecocq A, Flick R, Jacob Y, Bonnefoy E, Bouloy M. A SAP30 complex inhibits IFN-beta expression in Rift Valley fever virus infected cells. PLoS Pathog. 2008;4(1):e13. doi: 10.1371/journal.ppat.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Koromilas AE. Dominant negative function by an alternatively spliced form of the interferon-inducible protein kinase PKR. J Biol Chem. 2001;276(17):13881–13890. doi: 10.1074/jbc.M008140200. [DOI] [PubMed] [Google Scholar]
- Lihoradova O, Kalveram B, Indran SV, Lokugamage N, Juelich TL, Hill TE, Tseng CT, Gong B, Fukushi S, Morikawa S, Freiberg AN, Ikegami T. The Dominant-Negative Inhibition of Double-Stranded RNA-Dependent Protein Kinase PKR Increases the Efficacy of Rift Valley Fever Virus MP-12 Vaccine. J Virol. 2012;86(14):7650–7661. doi: 10.1128/JVI.00778-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell RB, Flick R. Rift Valley fever virus: An unrecognized emerging threat? Hum Vaccin. 2010;6(7) doi: 10.4161/hv.6.7.11761. [DOI] [PubMed] [Google Scholar]
- Mansuroglu Z, Josse T, Gilleron J, Billecocq A, Leger P, Bouloy M, Bonnefoy E. Non structural NSs protein of Rift Valley Fever Virus interacts with pericentromeric DNA sequences of the host cell inducing chromosome cohesion and segregation defects. J Virol. 2009;84:928–939. doi: 10.1128/JVI.01165-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepin M, Bouloy M, Bird BH, Kemp A, Paweska J. Rift Valley fever virus (Bunyaviridae: Phlebovirus): an update on pathogenesis, molecular epidemiology, vectors, diagnostics and prevention. Vet Res. 2010;41(6):61. doi: 10.1051/vetres/2010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmaljohn C, Nichol ST. Bunyaviridae. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. In Fields Virology. 5th ed. Philadelphia, PA: Lippincott, Williams & Wilkins; 2007. pp. 1741–1789. [Google Scholar]
- Swanepoel R, Coetzer JAW. Rift Valley fever. In: Coetzer JAW, Thompson GR, Tustin RD, et al., editors. Infectious diseases of livestock with special reference to southern Africa. 2nd ed. Cape Town, South Africa: Oxford University Press; 2004. pp. 1037–1070. 2004. [Google Scholar]
- Yadani FZ, Kohl A, Prehaud C, Billecocq A, Bouloy M. The carboxy-terminal acidic domain of Rift Valley Fever virus NSs protein is essential for the formation of filamentous structures but not for the nuclear localization of the protein. J Virol. 1999;73(6):5018–5025. doi: 10.1128/jvi.73.6.5018-5025.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Ackerman SL. Endoplasmic reticulum stress in health and disease. Curr Opin Cell Biol. 2006;18(4):444–452. doi: 10.1016/j.ceb.2006.06.005. [DOI] [PubMed] [Google Scholar]