Abstract
Over the course of our lives, humans are colonized by a tremendous diversity of commensal microbes, which comprise the human microbiome. The collective genetic potential (metagenome) of the human microbiome is orders of magnitude more than the human genome, and it profoundly affects human health and disease in ways we are only beginning to understand. Advances in computing and high-throughput sequencing have enabled population-level surveys such as MetaHIT and the recently-released Human Microbiome Project, detailed investigations of the microbiome in human disease, and mechanistic studies employing gnotobiotic model organisms. The resulting knowledge of human microbiome composition, function, and range of variation across multiple body sites has begun to assemble a rich picture of commensal host-microbe and microbe- microbe interactions as well as their roles in human health and disease and their potential as diagnostic and therapeutic tools.
Keywords: Human Microbiome Project, microbiota, metagenomics
Understanding our microbial majority
Humans consist only of our own somatic cells until birth, but over the first several years of life, our bodies, including the skin surface, oral cavity, and gut, are colonized by an enormous variety of bacteria, archaea, fungi, and viruses, which form a community collectively known as the human microbiome or microbiota [1–3]. Our microbiome contains ten times as many cells as the rest of our bodies, and orders of magnitude more genes than the human genome [1, 4]. Normally these microbes are commensal or mutualists, helping to digest our food and maintain our immune systems. Although the human microbiome has long been understood to influence human health and disease, low-cost high-throughput sequencing has only recently given us the experimental tools to investigate the breadth of its involvement. This has resulted in an explosion of recent work illuminating the role of the microbiome in conditions ranging from obesity [5, 6] to inflammatory bowel disease (IBD) [7, 8] to kidney stones [9]. Large-scale surveys such as the Metagenomics of the Human Intestinal Tract (MetaHIT) consortium [10] and the recent Human Microbiome Project (HMP) [1, 11] have shown that these are only the beginning of a richer functional understanding of the human microbiome.
From Van Leeuwenhoek to high-throughput sequencing: enabling meta'omic studies of the human microbiome
Although human-associated microbes were among the first investigated by microscopy, most are difficult or impossible to culture under convenient laboratory conditions. The vast majority of microbial taxa were uncultured and thus minimally studied until the development of DNA-based techniques. The earliest high-throughput technique for microbial ecology (and still the most common) is to sequence the highly-conserved 16S rRNA gene. Also known as 16S gene (or amplicon) profiling, this provides a rapid and very cost-effective survey of the bacteria present within a community [12], but it is rarely informative with regard to non-bacterial microbes and offers little functional information. Historically, this gene was amplified with universal bacterial primers, and the individual sequences were cloned and determined using Sanger sequencing. More recent 16S sequencing techniques are very high-throughput, generating up to millions of reads per sample, but with typically very short read lengths [13].
Metagenomic studies, in which the DNA of an entire community is sequenced, provide a much broader and more complex picture of microbial communities by simultaneously cataloging community organisms (including the non-bacterial component), genes, and genomes (Figure 1). The earliest large-scale metagenomic studies began soon after the turn of the century [14, 15], but the expense of clone library construction and Sanger sequencing made metagenomic human microbiome studies unfeasible for most groups. The advent of high-throughput sequencing made much larger-scale metagenomic efforts, including MetaHIT [10] and the HMP [1, 11], economically feasible. Here we review what these and recent disease- and model-organism studies have shown us about the composition and genetic capabilities of the human microbiome and discuss the future of the field of human microbiome research.
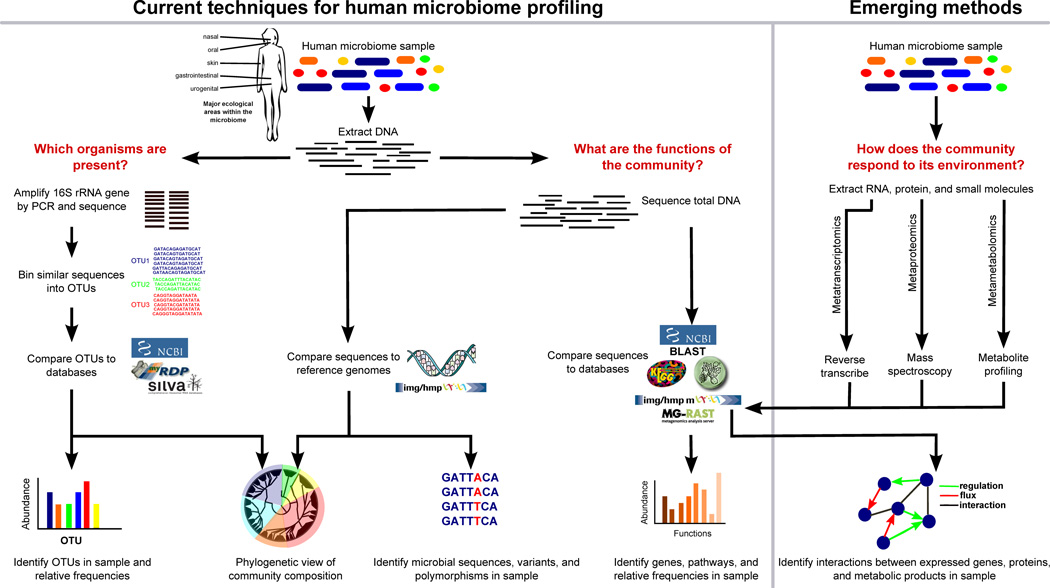
Figure 1. Overview of bioinformatic methods for functional metagenomics.
Microbial community samples typically contain many species of bacteria and other microorganisms, here indicated by different colors. After total DNA has been extracted, the composition of the community is determined by amplifying and sequencing the 16S rRNA gene. Highly similar sequences are grouped into Operational Taxonomic Units (OTUs), and these OTUs are labeled by comparison with databases of recognized organisms. OTUs can then be analyzed in terms of presence/absence, abundance, or phylogenetic diversity. In order to determine biomolecular and metabolic functions present in the community, the total metagenomic DNA may be sequenced and compared with function-oriented databases. Alternatively, sequenced community DNA can be compared to reference genomes. This allows identification of microbial sequence variants and polymorphisms, as well as provides an alternative means of detecting the presence and abundance of specific organisms. In the future, the emerging fields of metatranscriptomics, metaproteomics, and metametabolomics will help build an integrated picture of gene expression and interactions in microbial communities.
Early human microbiome studies
The largest early efforts at measuring the composition of the human microbiome were 16S cloning-based studies of the gut that included fecal and mucosal samples from up to a dozen individuals [16, 17]. These resulted in 10–20,000 sequences each, corresponding
to between 400 and 4,000 taxa (this range of species counts reflects largely bioinformatic rather than biological differences between studies). These investigations showed that gut alpha diversity was much higher than most previous estimates and that beta diversity was high both between subjects as well as between mucosal and stool samples from the same subject [18]. This, combined with preliminary associations between altered microbiota and disease, highlighted the need for much larger-scale studies to determine the range of microbial variation and function in the human gut across entire populations.
Large-scale human microbiome studies
The first population-scale metagenomic study of the human microbiome was conducted by the MetaHIT consortium [10]. Stool sequenced from 124 Spanish and Danish subjects were estimated to contain 1,150 common bacterial species in the human gut with a collective 3.3 million genes. However, only 75 of these organisms and 294,000 genes were shared by more than half of subjects, even within a population with limited genetic and environmental diversity. In addition to essential housekeeping genes, common gut bacteria were enriched for pathways that metabolized complex sugars from the host diet and intestinal lining, as well as for adhesion and vitamin and xenobiotic processing. The abundance of archaeal and fungal components was very low (<1%), although as in all such studies, this is influenced by sample collection and DNA extraction [10].
The HMP was by far the largest human microbiome study to date, a five-year effort that determined the range of microbial diversity in disease-free individuals by sequencing over 5,000 samples from approximately 250 healthy volunteers in two U.S. cities. Each subject was sampled at 15 or 18 body sites (nine oral, four skin, one nasal, one stool, three vaginal), and roughly half of subjects were sampled at up to two additional time points. Most samples were profiled using 16S amplicon sequencing to determine community composition; approximately 15% were subjected to metagenomic sequencing to determine community function [1, 11]. In addition to expanding the MetaHIT gut gene catalogue by nearly 1.8 million genes and identifying a microbiome-wide count of over 10 million total unique microbial genes, the HMP allowed detection of between-body-site microbial associations.
Diversity in the human microbiome
Estimating a community's species richness is bioinformatically challenging. Thus, the total human microbiome assessed by the HMP contained between 3,500 and 35,000 species-level Operational Taxonomic Units (OTUs) depending on parameter choices [1]. These, however, confidently spanned roughly 600 genera and covered 90% of the phylogenetic range of microbes expected in this population [1, 19]. Several signature bacterial genera were observed across nearly all individuals and represented the plurality (and often majority) of particular body site communities (Figure 2), but specific taxa were highly variable and almost never universal [1]. The gastrointestinal tract, including both the oral cavity and stool, was the most diverse microbiome among the sampled body habitats, estimated to contain several thousand OTUs corresponding to approximately 150 genera [20]. The well-studied gut microbiome exemplified inter-individual variability; the Bacteroidetes and Firmicutes phyla dominated as expected, but abundance ranged from >90% Bacteroidetes to >90% Firmicutes in different subjects. Only some half dozen gut taxa were near-universal (present in >=95% of subjects) [21].
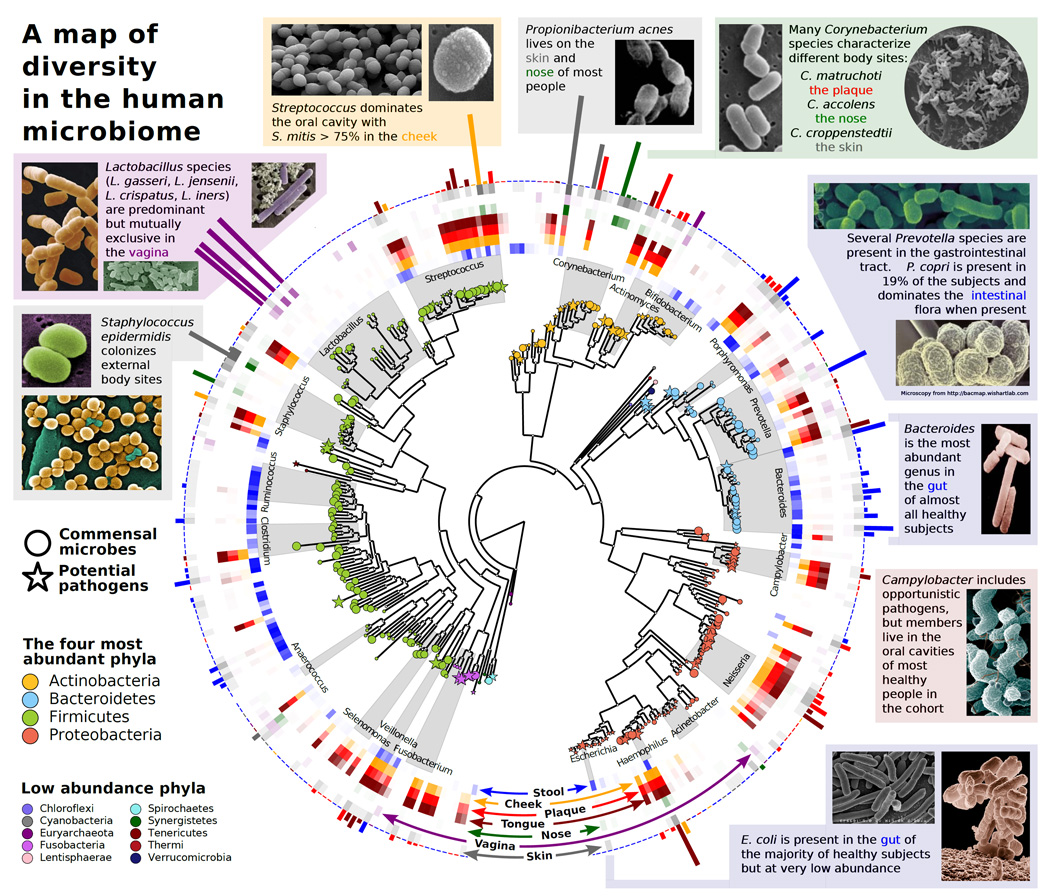
Figure 2. A map of diversity in the human microbiome.
The human microbiome is dominated by four phyla: Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria. In the center is a phylogenetic tree of organisms abundant in the human microbiome. Commensal microbes are indicated by circles, and potential pathogens are indicated by stars. The middle ring corresponds to body sites at which the various taxa are abundant and is color-coded by site (e.g., Ruminococcus (blue) is found mostly in the gut, whereas Lactobacillus (purple) is found mostly in the vagina). The bar heights on the outside of the circle are proportional to taxa abundance at the body site of greatest prevalence (e.g., Streptococcus mitis (yellow) dominates the inside of the cheek, whereas the gut is abundant in a variety of Bacteroides). The intensity of external colors corresponds to species prevalence in each body site. Data from the Human Microbiome Project [11] and MetaPhlan [79], and microscopy images from BacMap [80].
All body areas examined by the HMP dramatically differed not only in composition, but in ecological organization as well (see Text Box 1). The oral microbiome was essentially as diverse as the gut but, by contrast, more of this diversity was shared between individuals. Signature clades also varied strikingly even between very similar oral surfaces (e.g., mucosa, saliva, and plaque); abundant genera included Streptococcus, Haemophilus Veillonella, Actinomyces, and Fusobacterium. Skin diversity (sampled at the inner elbows and behind the ear) was low both within and between subjects, perhaps due to exposure and frequent disruption [1]. The human body community of lowest alpha diversity was that of the vagina; most women were dominated by one of four single species of Lactobacillus [21]. The vaginal microbiome becomes even less diverse during pregnancy [22], and it is emerging as a useful human-associated model community due to its uniqueness relative to other primates [23] and highly structured temporal fluctuations [24, 25].
The HMP discovered and validated several novel taxa, mostly at the genus level, but hinted at a possible novel family within the order Clostridiales [26]. Novel OTUs were generally of low abundance (<2%), a level comparable to that of E. coli in healthy subjects [1], but were carried across multiple subjects and multiple visits. The novel taxa were mainly from the largely-uncharacterized Barnesiella genus, but also included novel Dorea Oscillibacterand Desulfovibriogenera which are associated broadly with colon cancer, dietary shifts, and opportunistic infections, respectively [26]. Low-abundance community members can affect health even at low numbers [27] and in the worst case, under certain circumstances, can overgrow to the detriment of host health (e.g., Clostridium difficile). Further study may reveal important roles for these novel taxa. However, as the microbial variation between individuals was greater than that of samples from the same subject at different points in time [1], these low-abundance microbes at the very least contribute to a uniquely personalized human microbiome when considered in terms of microbial presence and abundance.
Beyond the baseline: microbial variation in children, the elderly, and global populations
Human microbiome composition appears to be particularly dependent on age and geography, although both are confounded by a variety of dietary, developmental, environmental, and genetic factors. The HMP included only American adults between the ages of 18 to 40, and MetaHIT extended this somewhat with Spanish and Danish individuals up to age 70. The infant microbiome is of particular interest with respect to acquisition, immune training, and extreme dietary changes over the first few years of life [28, 29]. Some of the earliest studies of microbiome structure and function indicated that immediate acquisition occurs from the environment and is dependent on mode of delivery [29], with stabilization to a more adult-like community occurring over two to three years [28]. Functionally, the shift from the newborn to adult-like gut microbiome includes enrichment for anaerobic fermentation, complex carbohydrate degradation (e.g., glycosyl hydrolases), and a depletion of simple sugar (lactose/galactose/sucrose) transport and metabolism proteins [28]. Recent studies of the microbiota of the elderly found that frailty is correlated with an overall decrease in microbial diversity, with overall increases in Bacteroides and decreases in Firmicutes that corresponded to a decrease in glutarate and the anti-inflammatory short-chain fatty acid butyrate [30].
Another recent comprehensive study addressed several of these questions simultaneously by surveying the stool microbiomes of a highly-diverse population that included 314 Americans, 114 Malawians, and 100 Amerindians. Roughly two-thirds of the subjects were under the age of 17, while the remaining third were between the ages of 18 and 70 [31]. In all three populations of this study, inter-subject variability was higher in children than in adults, infant microbiomes were dominated by Bifidobacteriaand microbiomes reached an adult-like configuration by age three. Microbial diversity increased with age in all populations, but both Malawian and Amerindian adults achieved higher diversity than American adults. The Malawian and Amerindian microbiomes were also more similar to one another than to the U.S. microbiome, in both adults and children [31].
When the authors examined functional gene families and enzyme classes within these global gut metagenomes, no enzyme classes were wholly unique to adults or babies, and the total number of enzyme classes was similar in both groups. However, the adults had many more uncharacterized enzymes than babies, likely reflecting the age-related increase in diversity. The infant microbiome had higher levels of folate-producing enzymes than that of adults, likely reflecting adults’ ability to obtain folate from diet. Conversely, biosynthetic capacity for the vitamins cobalamin, thiamine, and biotin increased with age. Urease genes were more frequently observed in Malawian and Amerindian infant microbiomes, possibly corresponding to greater probability of dietary nitrogen deficiency. Malawian and Amerindian adult microbiomes were enriched in glutamate synthase relative to the American microbiomes, the latter instead carrying an increased capacity for degradation of glutamine and other amino acids. Interestingly, this functional difference has also been observed when comparing the gut microbiota of herbivores and carnivores [32
Habitat-specific functionality as a principle of human microbiome organization
Several human microbiome studies have reported the metagenomic distribution of pathways within each body habitat to be much more consistent among individuals than are microbial abundances [1, 6, 10, 33]. In each case, microbial abundances within the same habitat (gut, oral cavity, etc.) varied tremendously among subjects, but two types of "core" pathways were stable in abundance. The first, corresponding to a more-or-less universal set of housekeeping genes like those reported by MetaHIT [10], were present in every habitat and represent processes necessary for human-associated microbial life, such as the ribosomal machinery, ATP synthesis, and glycolysis.
More surprisingly, certain sets of site-specific gene function were maintained within each habitat regardless of the taxa present. The stool microbiome was particularly abundant in genes related to complex carbohydrate degradation despite highly variable Bacteroidetes:Firmicutes ratios; hydrogen sulfide production and methionine degradation were also site-specific to the gut at low abundance [1]. Individual profiles of carbohydrate-active enzymes within each body habitat were also very similar by site. The oral cavity microbiome, for example, was optimized for simple sugar metabolism and particularly for dextran, whereas the vaginal microbiome was optimized for glycogen and peptidoglycan degradation [34]. Much like individual bacterial genomes, each habitat thus seems to have a core metagenome present in most hosts, in addition to a pan- metagenome of more flexible auxiliary genes carried by each habitat’s community.
Gnotobiotics and model systems for the human microbiome
Much of our knowledge of microbiome influence in human health comes from gnotobiotic systems, which predate the field of metagenomics [35]. Gnotobiotics are animals (typically mice or zebrafish) in which normal host microbiota has been replaced by a defined set of microbes, allowing studies of how microbial colonization influences host health and development or other processes of interest. Gnotobiotic animals are born under germ-free conditions and subsequently colonized (typically by gavage) with a single species [36], a defined set of species such as the eight-species Schaedler flora, or entire gut communities from mice or humans with phenotypes of interest [35, 37]. Gnotobiotic studies have shown that gut microbiota affects the host in many ways, including fertility, the development of the heart, lungs, and liver [35], and even brain gene expression and behavior. For example, a recent study found that germ-free mice were less anxious and more active than mice with normal gut microbiota, but these effects were largely reversed by exposure to gut microbiota at an early age [38]. Three areas of particularly intense focus are the heavy influence of the microbiome on nutrition [35] and obesity [5], immune system development [36, 39–41], and inflammatory bowel disease [7, 8, 42].
The gut microbiome: diet and obesity
The earliest attempts at raising germ-free animals demonstrated the vital contribution of the gut microbiota to nutrition, as the animals rapidly succumbed to vitamin deficiencies and required extensive supplementation [35]. When germ-free mice were colonized with gut bacteria from conventionally-raised adult mice, their food intake decreased, but their body fat increased due to increased monosaccharide availability from food signaling the liver to increase fat storage [5]. Studies of mice genetically predisposed to obesity found that they had more Firmicutes and fewer Bacteroidetes than their genetically-lean siblings, and when a group of germ-free mice were colonized with microbiota from obese mice, they gained more weight than those colonized with microbiota from lean mice [5]. Furthermore, mice colonized with human gut microbiota showed large and rapid shifts in gut microbiome composition in response to diet; these were accompanied by stable changes in microbial taxa abundance and gene expression [43].
The results of human obesity studies have proven more variable than the mouse studies; Bacteroides have been reported by various groups to increase, decrease, and not change during weight loss [5]. Other controlled feeding studies suggest that short-term dietary changes can modify taxon abundances [44] or transcriptional levels [45] but seldom change the presence or absence of specific taxa [44]. Thus, it has initially been difficult to interpret the extreme changes often seen in gnotobiotic animals relative to the more subtle behavior in the human gut.
A study comparing the gut microbiota of normal weight, obese, and post-gastric-bypass subjects noted that obese individuals were more likely to harbor both Prevotella and methanogenic archaea [46]. The authors proposed that methanogens removed hydrogen produced by Prevotella, allowing Prevotella to more efficiently produce short-chain fatty acids, which were absorbed by the host, contributing to host obesity [46]. This proposal is supported by evidence from gnotobiotic mouse studies; mice co-colonized with Bacteroides thetaiotamicron (which, like Prevotellaferments carbohydrates into short- chain fatty acids and produces hydrogen) and the methanogen Methanobrevibacter smithii had higher numbers of total gut bacteria, higher acetate levels in the intestinal lumen and blood, and more body fat than monocolonized mice or mice co-cultured with B. thetaiotamicron and the sulfate-reducing bacteria Desulfovibrio piger [47].
Host immune interactions in the gut and body-wide
In addition to nutrient absorption, the gut microbiome also greatly affects immune development. The immune systems of germ-free mice are strikingly abnormal, with smaller lymph nodes, lower serum immunoglobin levels, and lower levels of leukocytes than conventially-raised mice [35]. Germ-free mice are particularly deficient in CD4+ T- cells [48, 49], but colonization with Bacteroides fragilis or even exposure to its polysaccharide restores CD4+ cells to normal levels [48]. Monocolonization of germ- free mice with segmented filamentous bacteria (SFB), a mouse-commensal Clostridiasimultaneously stimulated many kinds of T-cells, including Th1, Th2, Th17, and Treg [36]. This immune activation is not universally beneficial; SFB also triggers autoimmune arthritis in genetically-predisposed mice in a Th17-dependent manner, but they do not became arthritic if raised in germ-free conditions [50].
Gut/immune system interactions are not limited to CD4+ cells; natural killer (NK) cells accumulate in large numbers in the lungs and colons of germ-free mice, making them much more susceptible to experimental asthma and colitis. This NK accumulation cannot be reversed by exposing mice to germs later in development but is prevented by very early exposure to normal mouse gut flora [39]. The continuum between early immune development, ongoing training, and acute or localized inflammation is still being explored. Some specific microbes, primarily Faecalibacterium prausnitziihave been suggested to act as suppressors of inflammation by short-chain fatty acid production and stimulation of the anti-inflammatory cytokine IL-10 [7].
The microbiome in disease: studies from IBD
In addition to its roles in obesity and immune response, it is increasingly evident that the microbiome influences the development of many other diseases (Table 1). IBD is only one example of a human disease that apparently does not follow Koch’s postulates, yet is intimately connected to host microbiome. It has long been known that IBD is both etiologically and microbially distinct, and that antibiotics, particularly rifampicin, are sometimes beneficial in its treatment [51]. Rodent studies showed that rats and mice genetically-predisposed to IBD would not develop the disease if they were kept germ-free [51] or preemptively treated with antibiotics.[42]. Furthermore, fostering or co-caging healthy mice with IBD-predisposed mice was sufficient to cause IBD in the healthy mice; this correlated with the transfer of the Enterobacteriaceae species Klebsiella pneumonia and Proteus mirabilis from the IBD mice to the healthy mice [42]. Intriguingly, this colitis was attenuated by the administration of probiotic fermented milk containing Bifidobacterium spp. [52].
Table 1.
Conditions recently associated with changes in the human microbiome:
| Disease | Microbiome–Related Findings | Reference |
|---|---|---|
| Asthma | Large increase in airway microbial diversity; Comamonadaceae, Sphingomonadaceae, Oxalobacteraceae increases correlate with bronchial hyperresponsiveness. | [73] |
| Atopic dermatitis | Decreased microbial diversity, increased Staphylococcus aureus; therapy restored commensals. | [74] |
| Colorectal cancer | Tumors were enriched in typical gut microbes relative to surrounding healthy tissue; healthy tissue enriched in Citrobacter, Cronobacter, Salmonella, and Shigella | [75] |
| Kidney stones | Lack of Oxalobacter formigenes increases urinary oxalate levels and risk of kidney stones. Supplementation may be beneficial; results mixed. | [9, 76] |
| Periodontitis | Shift from Actinomyces and Streptococcus to Haemophilus and Selenomonas | [77] |
| Psoriasis | Decrease in commensal Staphylococcus and Propionibacterium and increase in Proteobacteria. | [78] |
The relationships between human gut microbiota and IBD are less well understood, but it is widely accepted that human IBD is not caused by a specific pathogen, but rather an immune system imbalance with respect to normal gut bacteria. Metagenomic studies of IBD in the human gut have consistently shown that IBD is associated with gut dysbiosis [8] and reduced microbial diversity [10]. A lack of Faecalibacterium prausnitzii was associated with high postoperative recurrence of Crohn’s disease, and F. prausnitzii supplementation attenuated experimental colitis in mice [7], but no human IBD F. prausnitzii supplementation studies have been reported to date. Many groups have observed decreases in members of anti-inflammatory Clostridia from clades IV and XIVa and increases in Escherichia coli [53], but it has not yet been determined whether these microbiome changes are a cause of IBD in humans, or are merely caused by IBD. Two probiotics, E. coli strain Nissle (isolated from an exceptionally- gastroenteritis-resistant World War 1 soldier) and VSL#3 (eight strains of Bifidobacterium Lactobacillusand Streptococcus) have been shown to be effective treatments for maintaining and inducing remission in ulcerative colitis [54], but the underlying molecular mechanisms are not yet well understood. Further probiotic research may shed light on how short- or long-term “ecosystem therapy” may restore balance to dysbiosis.
Future directions in human microbiome research
In addition to understanding the basic ecological principles of human microbiome organization and its life-long epidemiology, two major areas of microbiota function have recently begun to be explored. The first is diagnostic: does microbiota composition or genetic function predict human disease onset, progression, or outcome? The past decade has seen great strides in assessing disease history and predicting risk using high- dimensional gene expression and genetic biomarkers; the next decade is likely to see the same with respect to the microbiome [55, 56]. The microbiome's second area of tremendous potential is in therapeutic intervention; human genetic defects are now detectable but difficult to modify, whereas the microbiome is both measurable and plastic. Targeted narrow-spectrum antibiotics [57], drug-microbe interactions [58], probiotics [45, 52, 54], and dietary interventions [59] all show promise for beneficial microbiome modulation. Remarkably, the roles and mechanisms by which host genetics, early life events, geographical location, or transmission of the microbiota might operate all represent largely open questions.
The technologies available to realize these research goals continue to develop at a remarkable pace, and are rapidly approaching diagnostic utility. Sanger sequencing has been replaced by Roche 454, and this is in turn giving way to ultra-high-throughput Illumina sequencing [13, 60], with longer reads for microbial isolate genomes and community metagenomes on the horizon [61, 62]. Metagenomes measure only what a community is capable of doing, but developing functional assays will better assess how it dynamically responds to its environment. Metatranscriptomics presents additional technical challenges such as depletion of ribosomal RNA, but has already been used to describe marine [63] and fecal [64] communities. Metaproteomics, or quantification of community peptides, has likewise been steadily developing to describe, for example, the gut microbiota [65, 66]. Metametabolomics, or quantification of community small- molecule metabolites, has been used to compare differences in host blood plasma between germ-free and conventionally-raised mice [67] and to compare fecal samples from healthy individuals and those with Crohn’s disease [68]. The immense computational challenge of assembling these vast and diverse meta’omic data to describe entire ecosystems, identifying how the genes interact with one another within and between bacteria to produce metabolites, and quantifying and understanding how these processes change over time and in response to the environment is the goal of the emerging field of molecular ecosystems biology [69]. Integrating these new technologies with model organisms and culture-based approaches will help us to unravel our relationship with our microbial majority.
Concluding remarks
It is an exciting time for work on human-associated microbial communities, both for basic biological discovery and for translational research. Experimental and computational tools are at the cusp of widespread affordability and availability, driven both by large-scale projects such as the HMP and by the democratization of high- throughput genomics and bioinformatics. Large-scale metagenomic studies have shown that the human microbiome is highly variable between individuals, and its genetic capacity is far greater than the human genome. Bacterial community function is highly conserved between corresponding body sites in human hosts, while community gene transcription is much more variable; it all affects health in many ways we are just beginning to understand. The most productive future research will integrate the metagenome, metatranscriptome, metaproteome, and metametabolome to model entire ecosystems.
Text Box 1: Class discovery in the microbiome.
The human body comprises a set of unique ecosystems with a broad range of configurations. At one extreme, the low-diversity vaginal microbiome typically occurs in one of five discrete types: four are each dominated by a different signature species of Lactobacillus [70], and the fifth is characterized by the absence of Lactobacillus. At the other extreme, the highly-diverse human mouth microbiome has a complex core of prevalent but variably-abundant taxa [21]. In communities such as the vagina, it is easy to perform class discovery by community clustering or typing; organizing the complexity of other human body habitats in this manner has been the subject of much recent work.
In the gut in particular, it has been hypothesized that the microbiome forms "enterotypes," or discrete, stable clusters of similar microbiome configurations [71]. Like the mouth, the gut microbiome is highly diverse; a combination of environmental effects, early exposures, and long-term diet determine the overall community profile. Short-term dietary changes induce more modest and transient effects [72] realized primarily at the transcriptional level [45]. The existence of human gut enterotypes and their associations with factors such as genetics or diet are much less straightforward than vaginal ecotypes and are an area of active research.
A recent study of individuals from six nations initially proposed three gut microbiome types, dominated by Prevotella Ruminococcus, and Bacteroides [71]. Subsequent studies [1, 72] also observed a distinct Prevotella type, but suggested that the Bacteroides and Ruminococcus types instead represented two ends of a single continuum between abundant Bacteroidetes and abundant Firmicutes. An associated controlled feeding study associated the Prevotella enterotype with a high-carbohydrate diet, and Bacteroides with higher animal fat and protein consumption. Short-term enforced dietary shifts from high- fat and high-protein to high-carbohydrate (and vice versa) somewhat modified organismal abundances, but rarely changed the presence and absence of specific subjects' gut microbes [44].
The largest currently published study (528 Americans, Malawians, and Amerindians) found that diverse Prevotella OTUs, distinct from the proposed Prevotella enterotype, were highly predictive in distinguishing between the populations, as the genus was more prevalent in Amerindians and Malawians (who ate maize-based diets) than in Americans, who typically had higher levels of Bacteroides. Although Bacteroides and Prevotella were almost mutually exclusive in most subjects, a considerable minority of subjects harbored substantial populations of both genera, raising the intriguing possibility that even the Prevotella and Bacteroides enterotypes may not represent wholly discrete "energy minima" in gut community configurations [31].
Glossary
- Taxon
the microbes belonging to a taxonomic category, typically of low level such as genus or species.
- Clade
a group of organisms descended from a common ancestor, corresponding to a single branch on the tree of life; often used to refer to higher-level taxa or phylogenetically related groups of microbes.
- Operational Taxonomic Unit (OTU)
a cluster of organisms similar at a defined sequence identity threshold (e.g. 95%) within some region (e.g. the whole 16S gene), used in the absence of a named species, genus, etc.
- Alpha diversity
the organismal diversity within a sample. Quantification metrics may emphasize richness (total number of organisms in sample) and/or evenness (whether they are evenly distributed, in contrast to some more abundant than others).
- Beta diversity
the organismal diversity shared between two or more communities, e.g. two or more different people sampled at the same body site. Can be measured either taxonomically (counting shared microbes) or phylogenetically (quantifying shared phylogenetic branches).
- 16S rRNA gene
the small subunit of the bacterial and archaeal ribosome; the DNA sequence of this gene is the most commonly-used taxonomic marker for microbial communities. Other markers include Hsp65, a highly conserved heat shock protein, and RpoB, which encodes the â subunit of RNA polymerase.
- Gnotobiotic
a host animal containing a defined set of microorganisms, either synthetically implanted or transferred from another host; often used to refer to model organisms with microbiota humanized by fecal transfer or gavage.
- Metagenome/metatranscriptome
the total genomic DNA or RNA transcripts of all organisms within a community.
- Microbiota
the total collection of microbial organisms within a community, here generally referring to the resident microbes of the human body.
- Microbiome
the total microbial community, including biomolecules, within a defined environment such as the human body or individual body site habitats.
- Commensal bacteria/microbes
members of a host-associated microbial community that are present in a "typical" state independent of disease, considered to be beneficial, harmless, or non-pathogenic under ordinary circumstances.
- Koch’s postulates
a set of criteria used to determine if a microorganism is the causative agent of a particular disease, including presence in disease, absence in health, isolation, induction of disease upon introduction, and subsequent reisolation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peterson J, et al. The NIH Human Microbiome Project. Genome Res. 2009;19(12):2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reyes A, et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466(7304):334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ley RE. Obesity and the human microbiome. Curr Opin Gastroenterol. 2010;26(1):5–11. doi: 10.1097/MOG.0b013e328333d751. [DOI] [PubMed] [Google Scholar]
- 6.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sokol H, et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflammatory bowel diseases. 2009;15(8):1183–1189. doi: 10.1002/ibd.20903. [DOI] [PubMed] [Google Scholar]
- 8.Joossens M, et al. Dysbiosis of the faecal microbiota in patients with Crohn's disease and their unaffected relatives. Gut. 2011;60(5):631–637. doi: 10.1136/gut.2010.223263. [DOI] [PubMed] [Google Scholar]
- 9.Kaufman DW, et al. Oxalobacter formigenes may reduce the risk of calcium oxalate kidney stones. Journal of the American Society of Nephrology : JASN. 2008;19(6):1197–1203. doi: 10.1681/ASN.2007101058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.A framework for human microbiome research. Nature. 2012;486(7402):215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tringe SG, Hugenholtz P. A renaissance for the pioneering 16S rRNA gene. Curr Opin Microbiol. 2008;11(5):442–446. doi: 10.1016/j.mib.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 13.Lazarevic V, et al. Metagenomic study of the oral microbiota by Illumina high-throughput sequencing. J Microbiol Methods. 2009;79(3):266–271. doi: 10.1016/j.mimet.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tyson GW, et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428(6978):37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- 15.Venter JC, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304(5667):66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 16.Gill SR, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312(5778):1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ley RE, et al. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444(7122):1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 18.Eckburg PB, et al. Diversity of the Human Intestinal Microbial Flora. Science. 2005;308(5728):1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fodor A, Earl A. "Most Wanted" Taxa from the Human Microbiome. Plos one. 2012 doi: 10.1371/journal.pone.0041294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li K, et al. Analyses of the Microbial Diversity across the Human Microbiome. PloS one. 2012;7(6):e32118. doi: 10.1371/journal.pone.0032118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huse SM, et al. A core human microbiome as viewed through 16S rRNA sequence clusters. PloS one. 2012;7(6):e34242. doi: 10.1371/journal.pone.0034242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ratner AJ, et al. A Metagenomic Approach to Characterization of the Vaginal Microbiome Signature in Pregnancy. PloS one. 2012;7(6):e36466. doi: 10.1371/journal.pone.0036466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rivera AJ, et al. Differences between the normal vaginal bacterial community of baboons and that of humans. American Journal of Primatology. 2011;73(2):119–126. doi: 10.1002/ajp.20851. [DOI] [PubMed] [Google Scholar]
- 24.Gajer P, et al. Temporal dynamics of the human vaginal microbiota. Sci Transl Med. 2012;4(132):132ra52. doi: 10.1126/scitranslmed.3003605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ravel J, et al. Vaginal microbiome of reproductive-age women. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(Suppl 1):4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wylie KM, et al. Novel Bacterial Taxa in the Human Microbiome. Plos one. 2012;7(6):e35294. doi: 10.1371/journal.pone.0035294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hajishengallis G, et al. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell host & microbe. 2011;10(5):497–506. doi: 10.1016/j.chom.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koenig JE, et al. Succession of microbial consortia in the developing infant gut microbiome. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(Suppl 1):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dominguez-Bello MG, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107(26):11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Claesson MJ, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488(7410):178–184. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- 31.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ley RE, et al. Evolution of mammals and their gut microbes. Science. 2008;320(5883):1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burke C, et al. Bacterial community assembly based on functional genes rather than species. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(34):14288–14293. doi: 10.1073/pnas.1101591108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cantarel BL, Lombard V, Henrissat B. Complex Carbohydrate Utilization by the Healthy Human Microbiome. PloS one. 2012;7(6):e28742. doi: 10.1371/journal.pone.0028742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.J Rahija R. Chapter 7 - Gnotobiotics. In: James GF, et al., editors. The Mouse in Biomedical Research. Second Edition. Burlington: Academic Press; 2007. pp. 217–233. [Google Scholar]
- 36.Gaboriau-Routhiau Vr, et al. The Key Role of Segmented Filamentous Bacteria in the Coordinated Maturation of Gut Helper T Cell Responses. Immunity. 2009;31(4):677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 37.Faith JJ, et al. Creating and characterizing communities of human gut microbes in gnotobiotic mice. The ISME journal. 2010;4(9):1094–1098. doi: 10.1038/ismej.2010.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heijtz RD, et al. Normal gut microbiota modulates brain development and behavior. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(7):3047–3052. doi: 10.1073/pnas.1010529108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olszak T, et al. Microbial Exposure During Early Life Has Persistent Effects on Natural Killer T Cell Function. Science. 2012;336(6080):489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ivanov II, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell host & microbe. 2008;4(4):337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Atarashi K, et al. ATP drives lamina propria T(H)17 cell differentiation. Nature. 2008;455(7214):808–812. doi: 10.1038/nature07240. [DOI] [PubMed] [Google Scholar]
- 42.Garrett WS, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8(3):292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turnbaugh PJ, et al. The Effect of Diet on the Human Gut Microbiome: A Metagenomic Analysis in Humanized Gnotobiotic Mice. Science Translational Medicine. 2009;1(6):6ra14–6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu GD, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334(6052):105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McNulty NP, et al. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Science translational medicine. 2011;3(106):106ra106. doi: 10.1126/scitranslmed.3002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang H, et al. Human gut microbiota in obesity and after gastric bypass. Proceedings of the National Academy of Sciences. 2009;106(7):2365–2370. doi: 10.1073/pnas.0812600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Samuel BS, Gordon JI. A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(26):10011–10016. doi: 10.1073/pnas.0602187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mazmanian SK, et al. An Immunomodulatory Molecule of Symbiotic Bacteria Directs Maturation of the Host Immune System. Cell. 2005;122(1):107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 49.Macpherson AJ, et al. IgA responses in the intestinal mucosa against pathogenic and non-pathogenic microorganisms. Microbes and infection / Institut Pasteur. 2001;3(12):1021–1035. doi: 10.1016/s1286-4579(01)01460-5. [DOI] [PubMed] [Google Scholar]
- 50.Wu H-J, et al. Gut-Residing Segmented Filamentous Bacteria Drive Autoimmune Arthritis via T Helper 17 Cells. Immunity. 2010;32(6):815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khan KJ, et al. Antibiotic therapy in inflammatory bowel disease: a systematic review and meta-analysis. Am J Gastroenterol. 2011;106(4):661–673. doi: 10.1038/ajg.2011.72. [DOI] [PubMed] [Google Scholar]
- 52.Veiga P, et al. Bifidobacterium animalis subsp. lactis fermented milk product reduces inflammation by altering a niche for colitogenic microbes. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1011737107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Joossens M, et al. High-throughput method for comparative analysis of denaturing gradient gel electrophoresis profiles from human fecal samples reveals significant increases in two bifidobacterial species after inulin-type prebiotic intake. FEMS microbiology ecology. 2011;75(2):343–349. doi: 10.1111/j.1574-6941.2010.01008.x. [DOI] [PubMed] [Google Scholar]
- 54.Jonkers D, et al. Probiotics in the management of inflammatory bowel disease: a systematic review of intervention studies in adult patients. Drugs. 2012;72(6):803–823. doi: 10.2165/11632710-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 55.Oh PL, et al. Characterization of the ileal microbiota in rejecting and nonrejecting recipients of small bowel transplants. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2012;12(3):753–762. doi: 10.1111/j.1600-6143.2011.03860.x. [DOI] [PubMed] [Google Scholar]
- 56.Farrell JJ, et al. Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut. 2012;61(4):582–588. doi: 10.1136/gutjnl-2011-300784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Louie TJ, et al. Fidaxomicin versus vancomycin for Clostridium difficile infection. N Engl J Med. 2011;364(5):422–431. doi: 10.1056/NEJMoa0910812. [DOI] [PubMed] [Google Scholar]
- 58.Okuda H, et al. A possible mechanism of eighteen patient deaths caused by interactions of sorivudine, a new antiviral drug, with oral 5-fluorouracil prodrugs. The Journal of pharmacology and experimental therapeutics. 1998;287(2):791–799. [PubMed] [Google Scholar]
- 59.Spencer MD, et al. Association between composition of the human gastrointestinal microbiome and development of fatty liver with choline deficiency. Gastroenterology. 2011;140(3):976–986. doi: 10.1053/j.gastro.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456(7218):53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eid J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323(5910):133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 62.Rothberg JM, et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature. 2011;475(7356):348–352. doi: 10.1038/nature10242. [DOI] [PubMed] [Google Scholar]
- 63.Gilbert JA, et al. Detection of Large Numbers of Novel Sequences in the Metatranscriptomes of Complex Marine Microbial Communities. PloS one. 2008;3(8):e3042. doi: 10.1371/journal.pone.0003042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Booijink CC, et al. Metatranscriptome analysis of the human fecal microbiota reveals subject-specific expression profiles, with genes encoding proteins involved in carbohydrate metabolism being dominantly expressed. Applied and environmental microbiology. 2010;76(16):5533–5540. doi: 10.1128/AEM.00502-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li X, et al. A metaproteomic approach to study human-microbial ecosystems at the mucosal luminal interface. Plos one. 2011;6(11):e26542. doi: 10.1371/journal.pone.0026542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Verberkmoes NC, et al. Shotgun metaproteomics of the human distal gut microbiota. The ISME journal. 2009;3(2):179–189. doi: 10.1038/ismej.2008.108. [DOI] [PubMed] [Google Scholar]
- 67.Wikoff WR, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(10):3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jansson J, et al. Metabolomics reveals metabolic biomarkers of Crohn's disease. PLoS One. 2009;4(7):e6386. doi: 10.1371/journal.pone.0006386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Raes J, Bork P. Molecular eco-systems biology: towards an understanding of community function. Nat Rev Microbiol. 2008;6(9):693–699. doi: 10.1038/nrmicro1935. [DOI] [PubMed] [Google Scholar]
- 70.Ravel J, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Arumugam M, et al. Enterotypes of the human gut microbiome. Nature. 2011 doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu GD, et al. Linking Long-Term Dietary Patterns with Gut Microbial Enterotypes. Science. 2011;334(6052):105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huang YJ, et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. The Journal of allergy and clinical immunology. 2011;127(2):372–381. e1–e3. doi: 10.1016/j.jaci.2010.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kong HH, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Research. 2012;22(5):850–859. doi: 10.1101/gr.131029.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Marchesi JR, et al. Towards the human colorectal cancer microbiome. PloS one. 2011;6(5):e20447. doi: 10.1371/journal.pone.0020447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hoppe B, et al. Efficacy and safety of Oxalobacter formigenes to reduce urinary oxalate in primary hyperoxaluria. Nephrol Dial Transplant. 2011;26(11):3609–3615. doi: 10.1093/ndt/gfr107. [DOI] [PubMed] [Google Scholar]
- 77.Liu B, et al. Deep sequencing of the oral microbiome reveals signatures of periodontal disease. PloS one. 2012;7(6):e37919. doi: 10.1371/journal.pone.0037919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fahlen A, et al. Comparison of bacterial microbiota in skin biopsies from normal and psoriatic skin. Archives of dermatological research. 2012;304(1):15–22. doi: 10.1007/s00403-011-1189-x. [DOI] [PubMed] [Google Scholar]
- 79.Segata N, et al. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods. 2012 doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stothard P, et al. BacMap: an interactive picture atlas of annotated bacterial genomes. Nucleic acids research. 2005;33(Database issue):D317–D320. doi: 10.1093/nar/gki075. [DOI] [PMC free article] [PubMed] [Google Scholar]