Abstract
Glycoprotein B (gB) facilitates HCMV entry into cells by binding receptors and mediating membrane fusion. The crystal structures of gB ectodomains from HSV-1 and EBV are available, but little is known about the HCMV gB structure. Using multiangle light scattering and electron microscopy, we show here that HCMV gB ectodomain is a trimer with the overall shape similar to HSV-1 and EBV gB ectodomains. HCMV gB ectodomain forms rosettes similar to rosettes formed by EBV gB and the postfusion forms of other viral fusogens. Substitution of several bulky hydrophobic residues within the putative fusion loops with more hydrophilic residues reduced rosette formation and abolished cell fusion. We propose that like gB proteins from HSV-1 and EBV, HCMV gB has two internal hydrophobic fusion loops that likely interact with target membranes. Our work establishes structural and functional similarities between gB proteins from three subfamilies of herpesviruses.
Keywords: herpesvirus, cytomegalovirus, entry, membrane fusion, glycoprotein B, glycosylation, rosette, fusion loop
Introduction
Herpesviruses are a large and diverse family of dsDNA, enveloped viruses that cause lifelong, latent infections. These viruses are classified into three subfamilies, alpha-, beta-, and gamma-, based on their replication cycle, tropism, and other characteristics. Eight herpesviruses from all three subfamilies are human pathogens. Human cytomegalovirus (HCMV), a betaherpesvirus, is notable for its extremely high prevalence in the general population as well as the ability to cause serious infections in neonates and the immunocompromised (Colugnati et al., 2007).
HCMV can infect a wide range of mammalian cells (Compton and Feire, 2007), which correlates with its ability to infect most organs and tissues. This broad cellular tropism suggests that HCMV may bind a number of receptors or a common surface molecule (Compton and Feire, 2007). HCMV enters cells by fusing its envelope with either the plasma membrane (fibroblasts) (Compton et al., 1992) or the endosomal membrane (epithelial and endothelial cells) (Ryckman et al., 2006). HCMV initiates cell entry by attaching to the cell surface heparan sulfate proteoglycans using envelope glycoprotein M (gM) or gB (Boyle and Compton, 1998; Boyle et al., 1999). This step is followed by interaction with cell surface receptors that trigger entry or initiate intracellular signaling (Compton and Feire, 2007). HCMV has been reported to engage PDGFR (Soroceanu et al., 2008) and cellular integrins (Feire et al., 2004b), probably through their interaction with gB (Feire et al., 2004a; Soroceanu et al., 2008). In support of this, a disintegrin-like consensus sequence, an integrin-binding motif, was recently identified within residues 92–111 of HCMV gB (Feire et al., 2004a) and shown to interact with a β1 integrin domain (Feire et al., 2010). EGFR, initially reported to be necessary for HCMV cell entry (Wang et al., 2003), was subsequently shown not to be important for either viral entry or HCMV-induced signaling (Isaacson et al., 2007).
Interaction of gB with PDGFR and integrins probably engages cellular signaling pathways instead of directly participating in viral entry because gB expression does not interfere with entry into either epithelial cells or fibroblasts (Ryckman et al., 2008). The entry receptor function might instead be provided by gH/gL glycoprotein complexes. Different gH/gL complexes are known to facilitate entry into epithelial cells, endothelial cells, or fibroblasts. While entry into fibroblasts requires gH/gL heterodimer (Wille et al., 2010), entry into epithelial and endothelial cells requires the pentameric complex gH/gL/UL128/UL130/UL131 (or gH/gL/UL128-131) (Ryckman et al., 2006), in addition to gH/gL (Ryckman et al., 2010; Wille et al., 2010). Expression of gH/gL/UL128-131 complex, but not gH/gL, in epithelial cells causes cells to become resistant to HCMV infection (Ryckman et al., 2008), which suggests that gH/gL/UL128–131 complex binds HCMV receptor on epithelial cells. By contrast, HCMV infection of fibroblasts can be blocked by co-expression of gH/gL and gO (Vanarsdall et al., 2011) but not gH/gL/UL128-131. Thus, different gH/gL complexes engage distinct entry receptors on epithelial/endothelial cells and fibroblasts, but they have not yet been identified.
Receptor engagement is followed by membrane fusion, a process mediated by gB and gH/gL. Early antibody studies have supported critical roles for both gB and gH/gL in HCMV entry (Britt, 1984; Cranage et al., 1986; Simpson et al., 1993; Urban et al., 1996). gB is essential for entry and cell spread, as shown by studies on an HCMV gB-null mutant in fibroblasts (Isaacson and Compton, 2009). gB and gH/gL are necessary and sufficient for cell fusion (Vanarsdall et al., 2008) and thus constitute the “core fusion machinery” of HCMV, which is conserved among other herpesviruses (Eisenberg et al., 2012). HCMV gB and gH/gL can mediate fusion either in cis (from the same membrane) or in trans (from opposing membranes) (Vanarsdall et al., 2008), which has also been observed for HSV-1 (Atanasiu et al., 2010). The precise fusion trigger in HCMV has not yet been identified. Receptor binding by gH/gL/UL128-131 probably does not trigger fusion because the expression of gH/gL/UL128–131 does not block cell fusion (Vanarsdall et al., 2008). Alternatively, receptor binding by gH/gL/UL128-131 functions as a fusion trigger only during endosomal entry but not entry at the plasma membrane or cell fusion.
Relatively little is known about the specific role of gB in HCMV entry and cell fusion apart from its requirement for both processes and its engagement of cellular signaling receptors. By contrast, more mechanistic details are available for homologous gB proteins from HSV-1, an alphaherpesvirus, and EBV, a gammaherpesvirus. Crystal structures of gB from HSV-1 (Heldwein et al., 2006) and EBV (Backovic et al., 2009) revealed strong protein fold similarities between the two ectodomains (Connolly et al., 2011) despite low sequence identity (25.9%) and provided important functional insights. gB proteins are structurally similar to the viral fusion proteins from unrelated viruses vesicular stomatitis virus G (VSV G) and baculovirus gp64, both of which are necessary and sufficient for viral entry and cell fusion. This structural similarity, in the absence of any sequence similarity, implicated gB as the fusion protein of herpesviruses capable of undergoing large conformational changes to effect fusion (Heldwein et al., 2006). Together, HSV-1 gB, EBV gB, VSV G, and gp64 formed a new class of viral fusion proteins, class III, that is distinct from class I, exemplified by influenza hemagglutinin, and class II, represented by flavivirus envelope protein E (Harrison, 2008; Steven and Spear, 2006).
Structural similarity with VSV G helped pinpoint the location of the two putative fusion loops in HSV-1 gB (Heldwein et al., 2006). Mutations of specific residues in these loops abolished fusion (Hannah et al., 2009; Hannah et al., 2007) as well as interaction with model membranes (liposomes) (Hannah et al., 2009), providing biochemical and functional evidence for the possible involvement of these residues in fusion, most likely in membrane insertion. Hydrophobic residues in fusion loops in EBV gB were shown to be necessary for fusion (Backovic et al., 2007a) and also responsible for the formation of rosettes by the recombinant EBV gB ectodomain (Backovic et al., 2007b). Such rosette formation has been observed with the ectodomains of several viral fusion proteins, e.g., influenza hemagglutinin (Ruigrok et al., 1988) and SFV E1 (Gibbons et al., 2004) whereby the exposed hydrophobic fusion peptides or loops interacted, causing aggregation. Finally, unlike gH/gL that can participate in fusion as a soluble protein, gB has to be membrane anchored (Atanasiu et al., 2010), which is consistent with it being a fusion protein. Taken together, these data support the hypothesis that HSV-1 and EBV gB are viral fusion proteins.
By comparison, little structural and mechanistic information is available on HCMV gB. Its ectodomain shares 24.2% sequence identity with HSV-1 gB and 30.3% with EBV gB. Previously, the ectodomain of HCMV gB was expressed in insect cells and shown to be proteolytically processed and to react with conformational antibodies (Carlson et al., 1997), justifying its use in functional experiments. However, no additional structural characterization has been carried out, and it remains unclear to what extent HCMV gB resembles its alpha- and gammaherpesvirus counterparts.
Here we show that the HCMV gB ectodomain is a trimer with an overall architecture resembling the ectodomains of HSV-1 and EBV gB, which supports the idea that gB proteins from all three herpesvirus subfamilies share similar structures. The ectodomain of HCMV gB forms rosette-like aggregates, similar to rosettes formed by the EBV gB ectodomain. By generating and analyzing a homology model of HCMV gB ectodomain based on HSV-1 gB structure (Heldwein et al., 2006), we identified four exposed hydrophobic residues within the putative fusion loops, Y155, I156, Y157, and W240. Substitution of these residues with their more hydrophilic HSV-1 counterparts reduced rosette formation and abolished cell fusion. We propose that HCMV gB has two internal hydrophobic fusion loops, functionally similar to those of HSV-1 and EBV gB, that are required for membrane fusion. Together with the structural information, these results support the hypothesis that gB is the fusion protein of HCMV. Our findings establish structural and functional similarities between gB proteins from three different herpesvirus subfamilies.
Results
Expression and purification of recombinant HCMV gB ectodomain
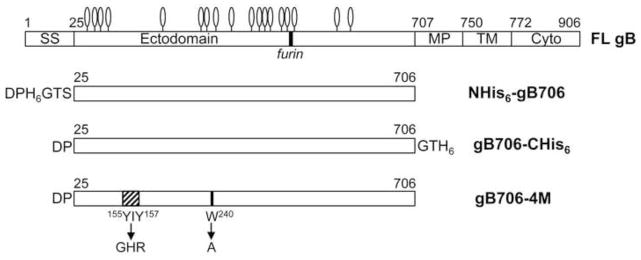
All gB ectodomain constructs were expressed in insect cells infected by recombinant baculoviruses. Initially, three gB constructs were created, all encoding the mature ectodomain of HCMV (strain AD169) gB, residues 25–706, with a honeybee melittin signal sequence and either an N-terminal His6 tag, a C-terminal His6 tag, or no tag (Fig. 1). All constructs lacked the hydrophobic membrane-proximal region, the transmembrane region, and the cytoplasmic domain. HCMV gB residues 25–706 correspond to the HSV-1 gB residues 31–730. Previously expressed version of the HCMV gB ectodomain, residues 1–692, was shorter and contained a C-terminal His6 tag (Carlson et al., 1997) yet could not be purified using metal affinity chromatography (Carlson et al., 1997). HCMV gB residue 692 corresponds to HSV-1 gB residue 715, which is buried in the crystal structure (Heldwein et al., 2006; Stampfer et al., 2010), so a His6 tag immediately following this residue would not be fully exposed. To avoid this problem, our constructs were extended to residue 706, which corresponds to HSV-1 gB residue 730.
Fig. 1.
Schematic view of the full-length HCMV gB and constructs used in this work. SS – signal sequence, MP – membrane-proximal region, TM – transmembrane region, Cyto – cytoplasmic domain. 18 predicted N-linked glycosylation sites are shown as lollipops.
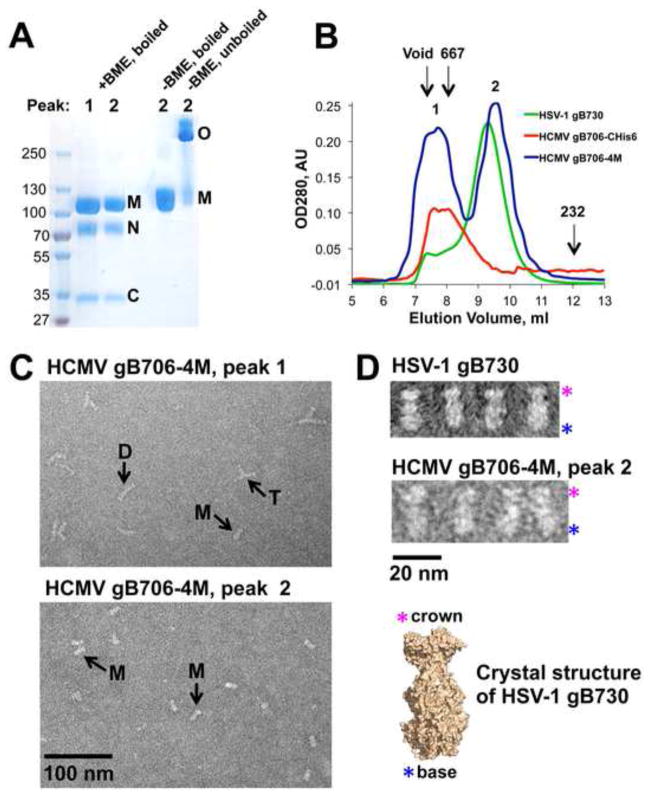
Initially, His6-tagged gB706 proteins were purified using metal affinity chromatography. Due to poor protein purity and yield, we switched to immunoaffinity purification with the mAb 27–39, which recognizes a conformational epitope within the oligomeric form of the HCMV gB ectodomain (Britt and Vugler, 1992). Subsequent size-exclusion chromatography yielded pure HCMV gB ectodomain (Fig. 2A). Purity and yield, ~10–20 μg from 1 liter of insect cell supernatant, were highest with gB706-CHis6, and it was used in subsequent work.
Fig. 2.
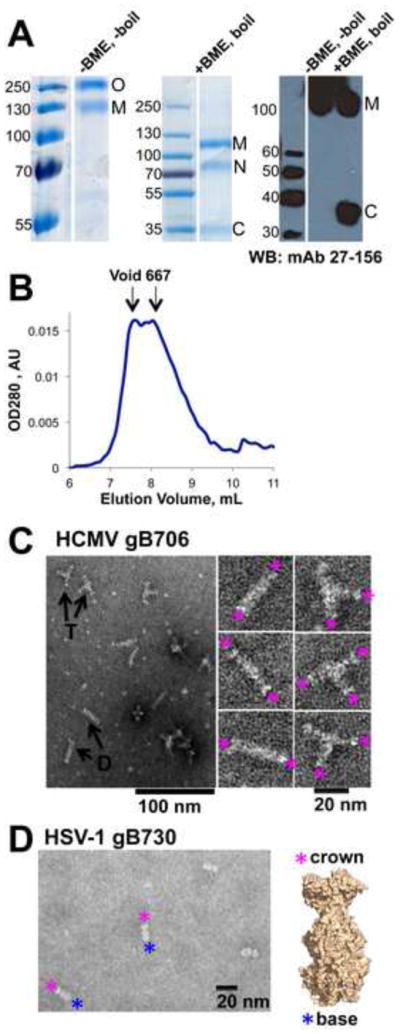
Properties of the HCMV gB706-CHis6. (A). Coomassie-stained gels and a Western blot of purified HCMV gB706-CHis6. Under non-reducing mildly denaturing conditions (-BME, unboiled), the protein migrates as a mixture of monomer and trimer. Under reducing denaturing conditions (+BME, boiled), some protein migrates as an intact monomer while a fraction separates into the N-terminal and C-terminal fragments. O – oligomer, M – monomer, N – N-terminal fragment, C – C-terminal fragment. Molecular size standards are labeled. (B). Size-exclusion chromatogram of purified gB706-CHis6. gB elutes as a broad peak near the void volume. The elution volumes of the void and the 667 kDa standard are shown (arrows). (C). Electron micrograph of purified gB706-CHis6. Arrows indicate long rods (D, dimer of trimers) and three-armed particles (T, trimer of trimers). Close-up view of selected rods and three-armed particles is shown below. Pink asterisks mark crown ends. (D). Electron micrograph of HSV-1 gB730 along with its crystal structure, in surface representation. Pink asterisks mark crown ends, and blue asterisks mark fusion-loop-containing base ends.
HCMV gB is proteolytically processed into disulfide-linked N and C termini
HCMV gB contains a furin cleavage site, RTRR, residues 456–459 (Figs. 1 and 3). In cells infected with HCMV or transfected with a plasmid encoding HCMV gB, the 160-kDa precursor is cleaved into an N-terminal product of ~116 kDa and a C-terminal product of ~58 kDa, albeit incompletely (Britt and Vugler, 1989; Vey et al., 1995). gB incorporated into HCMV virions is likewise incompletely processed (Britt, 1984). Since insect cells express furin-like proteases (Cieplik et al., 1998), we tested if HCMV gB ectodomain expressed in insect cells underwent proteolytic processing by analyzing purified gB706-CHis6 protein by SDS-PAGE in the presence or absence of the reducing agent, beta-mercaptoethanol (BME).
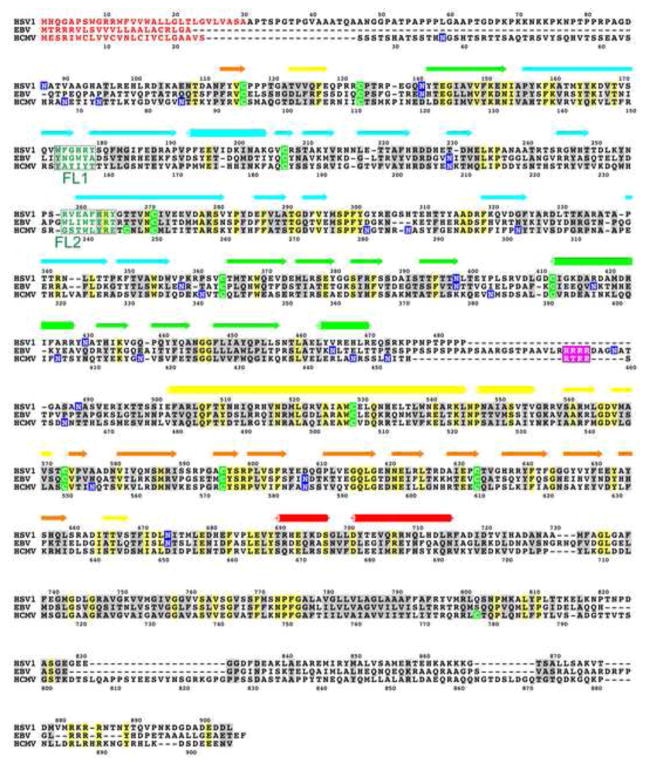
Fig. 3.
Sequence alignment of gB proteins from HSV-1 (strain KOS), EBV (strain B958), and HCMV (strain AD169), generated using ClustalW2 (Larkin et al., 2007). Residue numbers for HSV-1 gB and HCMV gB are shown above and below their respective sequences. Secondary structure of HSV-1 gB is shown above the sequence alignment, and its coloring matches that in reference (Heldwein et al., 2006). Signal sequences, absent from mature glycoproteins, are shown in red. Identical residues are highlighted in yellow; similar residues, in grey; cysteines, in green; and putatively glycosylated asparagines, in blue. Furin cleavage sites in HCMV and EBV are highlighted in purple. Residues in two fusion loop regions are shown in teal. Figure was generated in ALSCRIPT (Barton, 1993).
In the absence of BME, gB706-CHis6 migrated as a ~120–130 kDa band, consistent with a monomer, as well as a higher MW species (discussed below) (Fig. 2A). In contrast, in the presence of BME, we observed two additional bands migrating at ~70 kDa and ~35 kDa (Fig. 2A). The ~35-kDa band reacted with a mAb 27–156 (Fig. 2A), which recognizes antigenic domain 1 (AD-1) (Spaete et al., 1988), the dominant antibody-binding site within residues 552–635 of HCMV gB (Britt et al., 2005; Wagner et al., 1992). Thus, the ~70 kDa and the 35-kDa bands corresponded to the N- and C-terminal fragments, respectively. The smaller size of the C-terminal fragment in the gB706-CHis6 relative to the virion gB is due to the absence of residues 707–906 while the smaller size of the N-terminal fragment can be explained by the presence of less complex carbohydrate chains that insect cells use to modify glycoproteins (Jarvis, 2003). Similar results were previously obtained with HCMV gB692-CHis6 expressed in TN5 insect cells (Carlson et al., 1997). The recombinant HCMV gB ectodomain expressed in insect cells is processed similarly to the gB incorporated into HCMV virions, although with a lower efficiency. Lower efficiency of furin cleavage, probably due to low levels of endogenous furin, has been observed with other glycoproteins expressed in insect cells (Cieplik et al., 1998).
HCMV gB ectodomain forms rosette-like aggregates
Recombinant gB ectodomains from HSV-1 and EBV are trimeric (Backovic et al., 2009; Heldwein et al., 2006). HCMV gB692-CHis6 was reported to migrate as a dimer on a mildly denaturing SDS-PAGE (Carlson et al., 1997). Likewise, we found that under non-reducing, mildly denaturing conditions, approximately half of gB706-CHis6 migrated as a high-molecular-weight band (Fig. 2A). Under similar conditions, the trimeric HSV-1 gB ectodomain also migrates as an apparent dimer (Silverman et al., 2010; Stampfer et al., 2010). This seeming anomaly is probably due to incomplete denaturation, which precludes accurate estimation of the molecular size based on electrophoretic mobility. Unfortunately, we could not assess the oligomeric state of the HCMV ectodomain in solution using size-exclusion chromatography because the protein aggregated and eluted as a broad double peak near the void volume (Fig. 2B).
To gain insight into the nature of oligomerization and aggregation of the HCMV gB ectodomain, we examined the purified gB706-CHis6 samples using negative-stain electron microscopy (EM). We observed primarily long rods and three-armed particles (Fig. 2C), plus occasional rosettes with 4 or 5 arms. The length of each arm in the three-armed particles, 16.9 ± 1.3 nm (n=6), was consistent with the length of HSV-1 gB ectodomain calculated from its atomic coordinates (16.7 nm) (Heldwein et al., 2006) or measured from EM images (16.1 nm) (Stampfer et al., 2010). Thus, each arm was likely composed of HCMV gB706-CHis6 ectodomain, such that the three-armed particle contained three ectodomains. The long rods were 34.2 ± 1.5 nm (n=5) in length, approximately twice as long as the HSV-1 gB ectodomain, and were likely formed by two HCMV gB ectodomains joined end-to-end. The characteristic “crown” shape at the tip of each arm in the three-armed particles and at both ends of the long rods (Fig. 2C) resembled the “crown” end of the HSV-1 gB730 molecule (Fig. 2D) (Heldwein et al., 2006; Silverman et al., 2010; Stampfer et al., 2010). Both three-armed particles and long rods appear to associate by the “base” ends, which in HSV-1 and EBV gB contain the fusion loops.
Fusion loops (and fusion peptides) are important functional regions in viral fusion proteins. Typically rich in aromatic and hydrophobic residues, fusion loops are thought to insert into target cellular membranes during fusion. Exposed hydrophobic fusion loops in the postfusion forms of several viral fusion proteins can lead to the formation of rosette-like aggregates (Gibbons et al., 2004; Ruigrok et al., 1988). In these rosettes, ectodomains associated by their fusion loop regions (Gibbons et al., 2004; Ruigrok et al., 1988). EBV gB ectodomain also formed rosettes that depend on interactions between the hydrophobic fusion loops (Backovic et al., 2007b). Specifically, replacement of hydrophobic residues WY112–113 and WLIW193–196 in fusion loops 1 and 2 of EBV gB with their less hydrophobic HSV-1 gB counterparts HR and RVEA, respectively, abolished rosette formation (Backovic et al., 2007b) and allowed crystallization (Backovic et al., 2009). In contrast to EBV gB, HSV gB ectodomain did not aggregate (Heldwein et al., 2006) (Fig. 2D); nevertheless, the fusion loops of HSV-1 gB enabled liposome association (Hannah et al., 2009). Since HCMV gB rosette-like particles were reminiscent of the rosettes formed by EBV gB ectodomain and by the ectodomains of other viral fusion proteins, we hypothesized that the putative fusion loops of HCMV gB were involved in rosette formation.
Formation of HCMV gB rosette-like aggregates is mediated by hydrophobic residues within the fusion loops
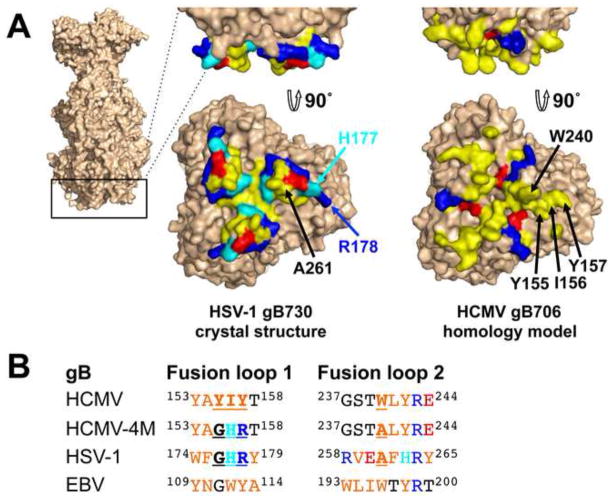
The putative fusion loops of HCMV gB, YAYIYT153–158 and GSTWLYRE237–244, contain several hydrophobic residues (Figs. 3 and 4). To identify surface-exposed hydrophobic residues within the fusion loops, we generated a homology model of HCMV gB ectodomain (Fig. 4) using the structure of HSV-1 gB730 (Heldwein et al., 2006). Residues YIY155–157 and W240 were prominently exposed on the surface of the HCMV gB706 homology model (Fig. 4A). To determine their role in rosette formation, we substituted residues YIY155–157 and W240 with their less hydrophobic HSV-1 gB counterparts GHR and A, respectively (Figs. 1 and 4B), generating gB706-4M mutant (Y155G/I156H/Y157R/W240A).
Fig. 4.
Fusion loop regions in HCMV gB and HSV-1 gB ectodomains. (A). Crystal structure of HSV-1 gB ectodomain, gB730, is shown side-by-side with a homology model of HCMV gB ectodomain, gB706, in surface representations. Fusion loop regions are shown in side and face-on views. Hydrophobic residues (yellow), positively charged residues (blue), negatively charged residues (red) and histidines (cyan) are shown. Side chains of mutated residues in HCMV gB706-4M mutant and their HSV-1 counterparts are labeled (in one protomer only, for clarity). Figures were generated using PyMol (www.pymol.org). (B). Sequence alignment of the fusion loop regions in HCMV gB, HCMV gB706-4M mutant, HSV-1 gB, and EBV gB. Hydrophobic residues (orange), positively charged residues (blue), negatively charged residues (red), and histidines (cyan) are colored. Mutated residues in HCMV gB and their HSV-1 gB counterparts are underlined.
The gB706-4M mutant ectodomain was expressed and purified in the same way as the wt gB706-CHis6, but its yield was significantly higher, ~500 μg from 1 liter of insect cells. Like the wt ectodomain, the mutant migrated as an oligomer on a mildly denaturing SDS-PAGE and was partially processed into disulfide-linked N- and C-terminal fragments (Fig. 5A). N-terminal sequencing of the C-terminal fragment revealed the sequence STSDNN, which matched the sequence following the furin cleavage site. This result unequivocally confirmed that the HCMV gB ectodomain was cleaved at the furin site during expression in insect cells.
Fig. 5.
Properties of the quadruple mutant gB706-4M. (A). Coomassie-stained gel of purified gB706-4M. Under non-reducing mildly denaturing conditions (-BME, unboiled), the protein migrates as a mixture of monomer and trimer. Under non-reducing denaturing conditions (-BME, boiled), the protein migrates as a monomer. Under reducing denaturing conditions (+BME, boiled), some protein migrates as an intact monomer while a fraction separates into the N-terminal and C-terminal fragments. O – oligomer, M – monomer, N – N-terminal fragment, C – C-terminal fragment. Molecular size standards are labeled. (B). Overlay of size-exclusion chromatograms of wt HCMV gB706-CHis6, HCMV gB706-4M mutant, and HSV-1 gB730. The elution volumes of the void and the 667 kDa and 232 kDa standards are marked with arrows. (C). Electron micrographs of peaks 1 and 2 with different particles marked with arrows: long rods (D, dimer of trimers), three-armed particles (T, trimer of trimers), and a single short rod (M, monodisperse trimer). (D) Close-up views of individual particles in HCMV gB706-4M, peak 2 and HSV-1 gB730. The crystal structure of HSV-1 gB730 in surface representation is shown for comparison. Pink asterisks mark crown ends, and blue asterisks mark fusion-loop containing base ends.
On a size-exclusion column, gB706-4M eluted as two peaks (Fig. 5B). Peak 1 eluted in the void volume of the column and contained aggregated protein while peak 2 eluted in the same volume as the monodisperse HSV-1 gB730 and likely contained the monodisperse gB706-4M protein (Fig. 5B). In support of this, negative-stain EM revealed that peak 1 contained rosette-like aggregates, mostly long rods but also three-armed particles, plus an occasional short rod (Fig. 5C). By contrast, peak 2 contained only short rods, 17.8 ± 0.8 nm (n=4) in length, representing monodisperse gB706-4M ectodomains (Fig. 5C).
Thus, replacement of several hydrophobic residues in the putative fusion loop regions with neutral or charged residues prevented aggregation of the HCMV gB ectodomain. This finding confirmed that the rosette-like aggregates of HCMV gB were held together by interactions between hydrophobic residues in the fusion loop regions.
Hydrophobic residues in fusion loops are necessary for cell fusion
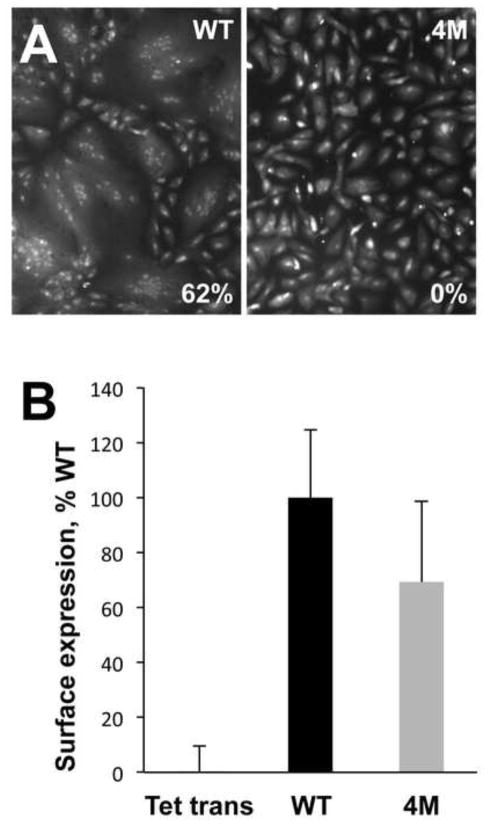
The importance of residues Y155, I156, Y157, and W240 for rosette formation suggested that they might be necessary for fusion. To evaluate their contribution to fusion directly, we tested the gB-4M mutant in a cell fusion using an established adenovirus transduction assay (Vanarsdall et al., 2008). The mutations were introduced into HCMV TR gB, generating mutant gB-4M (Y156G/I157H/H158R/W241A, TR numbering). Unlike the WT HCMV TR gB, the gB-4M mutant did not mediate any cell fusion of ARPE-19 cells in the presence of gH/gL (Fig. 6A). Lack of fusion was not due to insufficient surface expression because surface expression level of the gB-4M mutant was comparable to the expression level of the WT HCMV TR gB, as detected by cell-based ELISA (Fig. 6B). The gB-4M mutant also became largely endoglycosidase H resistant in a similar fashion to the WT gB (data not shown), confirming that this mutant gB passed ER quality control. Thus, substitution of four residues in the putative fusion loops abrogated fusion without affecting the expression or the trafficking of the mutant to the cell surface. On the basis of these observations, we propose that residues 153–158 and 237–244 constitute functional fusion loops in HCMV gB, with residues Y155, I156, Y157, and W240 (AD169 numbering) likely playing important roles in fusion.
Fig. 6.
gB-4M mutant does not mediate cell fusion despite adequate surface expression. (A) SYTO 13 green fluorescent nucleic acid staining was used to quantify fusion of ARPE-19 cells mediated by either WT gB or gB-4M mutant, in the presence of gH/gL. The level of fusion was quantified by counting the total number of cell nuclei in syncytia divided by the total number of nuclei in the same field and expressed as the percentage of cells fused. A syncytium was defined as such when membrane enclosed 10 or more nuclei. Reported values represent an average of three experiments. (B) Surface expression in HDFn cells of WT gB and gB-4M mutant was assessed by cell-based ELISA with the mAb 27–156. Non-specific binding of the antibody to cells was measured with cells infected with recombinant adenovirus encoding Tet transactivator instead of gB. In each experiment, the Tet transactivator control was subtracted from the sample. Six replicate samples were averaged, and the signal was expressed as a percentage of WT (100%). Error bars represent standard deviations. Relatively large standard deviations are likely due to the fact that at any time, only a small fraction of total HCMV gB is present on the cell surface, especially when compared to HSV gB or HCMV gB lacking the endocytic motif within the cytoplasmic domain (data not shown).
HCMV gB ectodomain is a trimer
Monodisperse HCMV gB706-4M particles were very similar to those of trimeric HSV-1 gB730 in both their length and shape (Fig. 5D). Moreover, the mutant eluted in the same volume as the HSV-1 gB730 on a size-exclusion column (Fig. 5B), suggesting that like HSV-1 gB ectodomain, the HCMV gB ectodomain is trimeric.
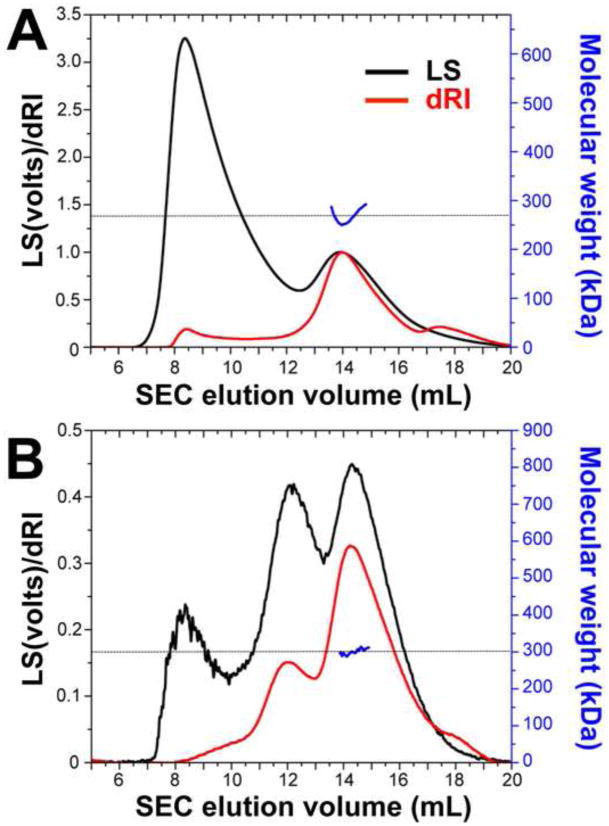
To determine the oligomeric state of HCMV gB706-4M directly, we used multi-angle light scattering (MALS). This technique is independent of molecular shape and can be used to determine molecular weight of non-globular proteins. HSV-1 gB730, a known trimer, was used as a control. The precise molecular weights of the glycosylated HSV-1 gB730 and HCMV gB706-4M polypeptides were determined by MALDI-TOF mass spectrometry. The experimentally determined molecular weight of HSV-1 gB730 was 85.6 kDa, larger than that calculated from its amino acid sequence 78.8 kDa due to glycosylation of 6 predicted N-linked glycosylation sites (Fig. 3). The molecular weight of the HSV-1 gB730 ectodomain in solution, as determined by MALS, was 266.1 kDa (Fig. 7A), which is consistent with a trimer. The experimentally determined molecular weight of HCMV gB706-4M polypeptide was 101.3 kDa, larger than that calculated molecular weight of 78.4 kDa. The difference of 22.9 kDa suggested that all 18 predicted N-linked glycosylation sites were glycosylated (Fig. 3). The molecular weight of HCMV gB706-4M ectodomain in solution, as determined by MALS, was 300.3 kDa (Fig. 7B), which corresponds to a trimer. Thus, the HCMV gB ectodomain is a trimer like HSV-1 and EBV homologs.
Fig. 7.
HCMVgB706-4M is a trimer in solution. SEC-coupled multi-angle light scattering (MALS) analysis of HSV-1 gB730 (A) and HCMV gB706-4M (B) is shown. The signal from the 90°-scattering detector is shown in black and the signal from the refractive index detector is shown in red; both are plotted against the left Y-axis. Average molecular weights are plotted in blue against the right Y-axis. Molecular weight corresponding to that of a trimer is shown as a horizontal dashed black line.
Discussion
HCMV gB shares structural similarities with gB proteins from HSV-1 and EBV
gB is a key entry protein of HCMV, yet very little is known about its structure and its specific role in HCMV entry. Here, we carried out a detailed biochemical characterization of the HCMV gB ectodomain. The recombinant HCMV gB ectodomain expressed in insect cells was proteolytically cleaved and glycosylated similarly to virion HCMV gB and was used to investigate the structural and biochemical properties of HCMV gB. We obtained direct evidence that HCMV gB ectodomain is a trimer and that its overall structure resembles the ectodomains of HSV-1 and EBV gB. This novel finding demonstrates that despite relatively low sequence identity, the three proteins share fundamentally similar structures. More detailed structural comparisons await the crystal structure of HCMV gB.
HCMV gB has functional fusion loops
We found that HCMV gB ectodomain formed rosette-like aggregates associated by its presumed fusion loop regions. By generating and analyzing a homology model of HCMV gB706, based on the crystal structure of HSV-1 gB730 (Heldwein et al., 2006), we identified several exposed hydrophobic residues in the putative fusion loops. Replacement of the most prominently exposed residues Y155, I156, Y157, and W240 with their hydrophilic HSV-1 counterparts significantly decreased aggregation, suggesting that these residues confer the ability to form rosettes on HCMV gB ectodomain. The propensity of HCMV gB ectodomain to aggregate via its fusion loop regions resembles EBV gB and is consistent with both proteins containing multiple hydrophobic residues within these loops. Due to aggregation of the WT HCMV gB ectodomain, we were unable to test its ability to associate with membranes or to evaluate the effect of fusion loop mutations on this ability. Instead, we tested the importance of these four residues for cell fusion. The quadruple mutant gB-4M was unable to mediate cell fusion despite adequate surface expression, confirming the importance of mutated residues for fusion. Analogous substitutions within the fusion loops of EBV gB eliminated rosette formation (Backovic et al., 2007b) and abrogated its fusion activity (Backovic et al., 2007a). We propose that HCMV gB has two functional fusion loops and that residues Y155, I156, Y157, and W240 are necessary for fusion. Future work will assess individual contributions of these residues to fusion.
Possible role of hydrophobic residues in HCMV gB fusion loops in fusion
Fusion loops, found in class II and class III viral fusion proteins, are typically rich in aromatic and hydrophobic residues. Fusion loops insert into and perturb the outer leaflet of the target cellular membranes thereby facilitating membrane fusion. Aromatic residues are thought to play an essential role in the destabilization of the outer leaflet because they position themselves at an interface between the hydrocarbon side chains and the polar head groups. In VSV G, three aromatic residues in fusion loops are required for fusion (Sun et al., 2008). The fusion loops of EBV gB contain four aromatic residues (Backovic et al., 2007a). Residues W174 and Y179 within relatively hydrophilic fusion loops of HSV-1 gB are critical for fusion (Hannah et al., 2007). Therefore, we anticipate that all aromatic residues within HCMV gB fusion loops will be essential for fusion, likely for insertion into the target membrane.
The recombinant HCMV gB ectodomain likely adopts the postfusion conformation
The conformation adopted by the recombinant gB ectodomains from HSV-1, EBV, and HCMV probably represents the postfusion form. Indeed, the structures of HSV-1 and EBV gB ectodomains more closely resemble the extended postfusion rather than the globular prefusion structure of VSV G. Also, in both structures, the fusion loops are located in proximity to the C termini leading into the transmembrane region, which is a typical feature of the postfusion forms of all other fusion proteins. Therefore, we conclude that like HSV-1 and EBV gB, the HCMV gB ectodomain expressed in insect cells adopts the trimeric postfusion conformation. Of course, the structure of the prefusion form of gB is necessary to definitively confirm this. By analogy with other viral fusion proteins, gB is hypothesized to undergo large-scale refolding during fusion, although evidence for refolding has not yet been obtained. The recombinantly expressed gB ectodomain adopts the postfusion conformation whereas on the viral envelope or on cell surface gB is presumably in its prefusion conformation. Thus, any future work with the HCMV gB ectodomain should take into account the fact that the conformation of the recombinantly expressed gB ectodomain may differ from that of the full-length, membrane-bound gB.
gB is the likely fusion protein of HCMV
The most conserved among herpesvirus entry glycoproteins, gB is thought to be the fusion protein of HSV and EBV. Here we showed that HCMV gB ectodomain shares overall structure with the HSV-1 gB ectodomain and that several residues in its fusion loops mediate rosette formation and are necessary for fusion. These results support the hypothesis that gB is the fusion protein of HCMV. Taken together, our findings establish structural and functional similarities between gB proteins from three different herpesvirus subfamilies.
Materials and Methods
Cloning and mutagenesis
To generate a HCMV gB ectodomain expression construct, DNA encoding the mature ectodomain of HCMV (strain AD169) gB, residues 25–706, was amplified using forward primer 5′-CGCGGATCCGTCTTCTAGTACTTCCCATGC-3′, BamHI site underlined, and reverse primer 5′-CGCGGTACCTCAGGGCGGTAGCGGGTCGACTA-3′, KpnI site underlined. The resulting PCR product was cloned into the BamHI and KpnI-digested pFastBac1 vector (Invitrogen) modified to contain melittin signal sequence to generate plasmid pKH74, which encodes HCMV gB706. To generate gB706 with a C-terminal His6 tag, PCR product was generated using forward primer 5′-CGCGCGCATATGAAAAATACGGAAACGTGTC-3′, NdeI site underlined, reverse primer 5′-CGCAAGCTTTCAATGGTGATGGTGATGGTGGGTACCGGGCGGTAGCGGGTCGACTA-3′, HindIII site underlined, and pKH74 plasmid as a template. This product was subcloned into the NdeI and HindIII-digested pKH74 plasmid yielding plasmid pKH75 encoding gB706-CHis6. To generate gB706 with an N-terminal His6 tag, PCR product was generated using forward primer 5′-CGCGGATCCGCACCATCACCATCACCATGGTACTAGTTCTTCTAGTACTTCCCATGC-3′, BamHI site underlined, reverse primer 5′-CGCGCGCATATGTTTGATTGTATGAAGTATTG-3′, NdeI site underlined, and pKH74 plasmid as a template. This product was subcloned into the BamHI and NdeI-digested pKH74 plasmid yielding plasmid pKH76 encoding gB706-NHis6.
To generate the quadruple mutant gB ectodomain, Y155G/I156H/Y157R/W240A (gB706-4M), mutation W240A was first introduced into pKH74 plasmid by site-directed mutagenesis using forward primer 5′-GGCAGCACCGCGCTCTATCGTGAGACCTGT-3′ and reverse primer 5′-ACAGGTCTCACGATAGAGCGCGGTGCTGCC-3′, generating plasmid pSS1. Three mutations Y155G/I156H/Y157R were then introduced simultaneously by splicing-by-overlap-extension PCR (SOE-PCR) method. Forward primer 5′-CGGTCTAGAACCATGAAATTCTT-3′, XbaI site underlined, and reverse primer 5′-GCAGATAAGTGGTGCGGTGGCCAGCGTAGCTACG-3′ were used to generate the 5′ SOE PCR fragment. Forward primer 5′-CGTAGCTACGCTGGCCACCGCACCACTTATCTGC-3′ and reverse primer 5′-CGCGCATATGTTTGATTGTAT-3′, NdeI site underlined, were used to generate the 3′ SOE PCR fragment. The resulting PCR product was subcloned into XbaI and NdeI-digested plasmid pSS1 to yield plasmid pSS2 encoding HCMV gB706-4M mutant.
All clones were sequenced and verified to contain the correct reading frame and appropriate sequences. Due to the cloning strategy, all mature proteins contain two extra residues (DP) at the N terminus. The His6 tag was left out from the gB706-4M construct because it was not necessary for immunoaffinity purification.
The full-length HCMV gB-4M mutant was generated by the SOE-PCR method using the entire HCMV gB (strain TR) gene as a template. Three PCR fragments were made separately using three primer pairs: 5′-AAAAAATCTAGAATGGAATCCAGGATCTGGTGCC-3′ (XbaI site underlined) and 5′-GCAGATAAGTGGTGCGGTGGCCAGCGTAGCTACG-3′; 5′-CGTAGCTACGCTGGCCACCGCACCACTTATCTGC-3′ and 5′-ACAGGTCTCACGATAGAGCGCGGTGCTGCC-3′; and 5′-GGCAGCACCGCGCTCTATCGTGAGACCTGT-3′ and 5′-AAAAAACTCGAGTCAGACGTTCTCTTCTTCGTCAGAG-3′ (XhoI site underlined). The three PCR fragments were spliced together, and the resulting PCR product was subcloned into XbaI and XhoI-digested pCAGGS vector to yield plasmid pSS3 encoding HCMV TR gB-4M mutant, Y156G/I157H/H158R/W241A. The plasmid also has an additional, unintended, mutation E610G. gB-4M sequence was then excised from the pSS3 plasmid and inserted into plasmid pCMV-SPORT6 (Invitrogen).
Antibodies
Hybridoma cell lines expressing anti-HCMV gB monoclonal antibodies 27–39 or 27–156 were a gift from William J. Britt (University of Alabama). The monoclonal antibodies were purified at the GRASP facility at Tufts Medical Center. 27–39 is a conformational mAb that recognizes the oligomeric form of HCMV gB ectodomain (Britt and Vugler, 1992). 27–156 recognizes antigenic domain 1 (AD-1) (Spaete et al., 1988), the dominant antibody-binding site within HCMV gB that maps to residues 552–635 (Wagner et al., 1992).
Viruses and cells
Spodoptera frugiperda (Sf9) cells were grown in SF-900 II SFM (Invitrogen) in suspension at 27ºC. Trichoplasia Ni (HiFive) cells were grown in Express Five SFM (Invitrogen). Recombinant baculoviruses of all HCMV gB ectodomain constructs were generated using Bac-to-Bac technology (Invitrogen). After two rounds of amplification, passage 3 (P3) stocks of baculoviruses were harvested and stored at 4ºC in the dark and in the presence of 2% Fetal Bovine Serum (FBS, Invitrogen). Human retinal pigmented epithelial (ARPE-19) cells were obtained from ATCC and grown in DMEM/F-12 medium (Hyclone) in the presence of 10% FBS (Hyclone). Human dermal fibroblasts (HDFn, Invitrogen) were grown in DMEM (HyClone) containing 10% FBS, 5% Bovine Growth Serum Supplemented Calf (HyClone), 3mM glutamine, and Pen/Strep. 911 retinoblastoma cells were maintained in DMEM containing 10% FBS and Pen/Strep.
Protein expression and purification
For protein expression, P3 stocks of baculoviruses were used to infect Sf9 or Hi5 cells. The amount of viral inoculum was optimized to yield the highest protein expression. Both Sf9 and Hi5 cells were tested, but Sf9 cells yielded a higher expression of all constructs and were used for all subsequent expression. Typically, 10 mL of viral stock was added to 1 L of SF9 cells at 2×106 cells/mL, and cell supernatants were harvested 72 hours post-infection. Cells were pelleted using centrifugation at 3750 rpm at 4ºC for 20 minutes. The supernatant was collected, filtered through a 0.45 um filter, and concentrated using a Tangential Flow Filtration system (Millipore) using a 30-kDa PLTK cartridge (Millipore). Buffer exchange was done three times into phosphate-buffered saline, pH 7.4, to remove media components. 1 mM benzamidine and 0.1 mM PMSF were added to the extract to inhibit proteolytic activity.
The gB ectodomain constructs, gB706-CHis6, gB706-NHis6, and the gB706-4M mutant were purified using immunoaffinity chromatography using monoclonal antibody 27–39 coupled to a CNBr-activated Sepharose 4B (GE Healthcare). After extensive washing with 10 mM Tris (pH 8.0) and 500 mM NaCl, the protein was eluted with 3M KSCN and concentrated on an Amicon Ultra-15 concentrator (Millipore) with a 30-kDa MW cutoff.
The protein was further purified by size-exclusion chromatography using Superdex S200 column (GE Healthcare) equilibrated with 20 mM Tris (pH 8.0), 150 mM NaCl, and 1 mM EDTA. The column was calibrated using blue dextran (~2000 kDa), thyroglobulin (667 kDa), ferritin (440 kDa), catalase (232 kDa), and aldolase (158 kDa). The final yield of gB706-CHis6 and gB706-4M was ~10–20 μg and ~500 μg, respectively, from 1 L of insect cell supernatant.
HSV-1 gB730 was purified as described previously (Heldwein et al., 2006; Stampfer et al., 2010).
SDS-PAGE and Western blot
Three conditions were used in preparation of samples for SDS-PAGE: mildly denaturing (0.1% SDS, no reducing agent, no boiling), non-reducing denaturing (1% SDS, no reducing agent, boiling), and reducing denaturing (1% SDS, reducing agent, boiling).
For Western blot, proteins were resolved by SDS-PAGE (4–15% acrylamide gel, Bio-Rad) and transferred onto nitrocellulose membrane. The membrane was blocked with 5% skimmed milk in Tris-buffered saline containing 0.05% Tween-20 and probed with mAb 27–156 at 1:1000 dilution. Following incubation with secondary antibody, anti-mouse IgG conjugated to horseradish peroxidase, at a 1:10,000 dilution, and blots were developed using HRP substrate chemiluminescence kit (Bio-Rad).
Electron microscopy
Purified protein samples of gB706-CHis6 and gB706-4M constructs (5 μg/mL) were incubated on glow-discharged formvar/carbon-coated 200 mesh copper grids (SPI) at room temperature and stained with 0.75% solution of uranyl formate. EM images were obtained using a Tecnai G2 Spirit BioTWIN transmission electron microscope at the Harvard Medical School EM core facility.
N-terminal sequencing
For N-terminal sequencing, protein samples were resolved by a 4–15% SDS-PAGE and transferred to a PVDF membrane. The membrane was stained with Coomassie R-250. The protein bands of interest were cut out and submitted for sequencing by Edman degradation at the Tufts University Core Facility.
Mass Spectrometry
For mass spectrometry analysis, the HSV-1 gB730 and HCMV gB706-4M proteins were analyzed using sinapinic acid (Agilent Technologies, US) as matrix. MS measurements were performed on a Voyager DE-Pro MALDI-TOF Mass Spectrometer (Applied Biosystems).
Size-exclusion chromatography (SEC)-coupled multiangle light scattering (MALS)
For SEC-coupled MALS, the HSV-1 gB730 and HCMV gB706-4M proteins (~2.6 mg/mL and ~1.5 g/mL injected concentration) were subjected to SEC using Superose 6 column (GE Healthcare) equilibrated overnight in 20 mM Tris (pH 8.0), 300 mM NaCl, and 1 mM EDTA. The chromatography system connected in line to a DAWN Heleos II light-scattering instrument equipped with a 658 nm laser, and Optilab T-rEX interferometric refractometer (Wyatt Technology). Data were analyzed with ASTRA 6 software (Wyatt Technology), yielding the molar mass and mass distribution (polydispersity) of the sample. Due to glycosylation, the data were analyzed using the protein conjugate analysis option within ASTRA 6 software. To account for glycosylation, the dn/dc value of 0.13 mL/g was used instead of 0.185 mL/g, which is used for unmodified proteins. For normalization of the light scattering detectors and data quality control, monomeric BSA was used.
Homology model generation
Homology model of HCMV gB ectodomain, gB706, was generated using standard protocols in the Modeller software (Eswar et al., 2006) and the crystal structure of HSV-1 gB ectodomain, gB730 (PDB ID 2GUM) as the template.
Preparation of recombinant replication-defective adenovirus
HCMV TR gB-4M in pCMV-SPORT6 was used for BP-mediated recombination (Invitrogen) with the Adeno-X-ccdB bacterial artificial chromosome (BAC) according to manufacturer’s instructions. After in-vitro recombination, BAC DNA was electroporated into DH10B bacterial cells, and bacteria selected using chloramphenicol and ampicillin. Individual bacterial colonies were screened for the presence of recombinant sequences by restriction analysis of extracted BAC DNA with BsrGI. Purified BAC DNA was linearized with PacI (New England Biolabs), and the resulting adenovirus vector encoding HCMV TR gB-4M was transfected into 911 retinoblastoma cells using calcium phosphate method. Transfected cells were monitored for 4–6 days for the presence of adenovirus-induced cytopathic effect, at which point cells were harvested and sonicated to obtain total cell lysates containing infectious adenovirus. The recombinant adenovirus encoding HCMV TR gB-4M was titered on 911 retinoblastoma cells as described previously (Vanarsdall et al., 2008).
Cell-cell fusion assay
ARPE-19 cells were infected with recombinant adenoviruses encoding HCMV TR gH, gL, and either WT gB or gB-4M mutant at 30 PFU/cell in growth media containing 2% FBS. Cells were concomitantly infected with a recombinant adenovirus supplying the Tet transactivator, required to activate protein expression, at one-fifth of the total PFU. At 4 h post infection, virus was removed and cells washed twice with growth media, then incubated in growth media at 37°C for additional 24–60 h. To assess the level of fusion, cells were fixed in PBS containing 2% formaldehyde, permeabilised with PBS containing 0.2% Triton X-100, and stained with a 5 μm stock of SYTO 13 green fluorescent nucleic acid stain (Invitrogen) diluted 1:20,000 in PBS. Images were captured on Nikon Eclipse TS100 microscope fitted with a Qiacam digital CCD camera. The level of fusion was quantified by counting the total number of cell nuclei in syncytia divided by the total number of nuclei in the same field and expressed as the percentage of cells fused. A syncytium was defined as such when one fluorescent membrane enclosed 10 or more nuclei.
Cell-based ELISA assay
HDFn cells were seeded at 2×104 cells per well of a 96 well plate. The following day, cells were infected with adenoviral vector encoding WT gB or gB-4M mutant at 100 pfu/cell in media containing 2% FBS. 6 h post infection, the cells were washed twice and low serum media was replaced. 48 h post infection, mAb 27–156 was added to the cells in media containing 2% donkey serum at 4°C. Cells were incubated with the antibody for 2 h at 4°C, washed extensively with cold PBS +/+, then fixed with 4% PFA in PBS+/+ for 1 hr at 4°C. After rehydration in PBS+/+, the samples were incubated with donkey anti-mouse HRP (Jackson ImmunoResearch) for 30 min at RT, washed, and TMB substrate (Sigma) was added. Reaction was stopped with the addition of 2M H2SO4 and plates read in a SpectraMAX190 (Molecular Devices) at 450 nm.
Acknowledgments
We thank William Britt (University of Alabama) for providing monoclonal antibodies and hybridomas used in this work, Maria Eriksson at the HMS EM Core Facility for help with sample preparation and data acquisition, Michael Berne at the Tufts University Core Facility for N-terminal protein sequencing and MALDI-TOFF mass spectrometry, Ayman Ismail for help with MALS experiments, Janna Bigalke for MALS data analysis and Fig. 7, and Motti Hakim at the Weizmann Institute for homology modeling. We also thank Gary Cohen, Roselyn Eisenberg, and Abraham Sonenshein for critical reading of the manuscript. This work was funded by the NIH grant 1DP20D001996, by the Pew Scholar Program in Biomedical Sciences, by the CFAR Developmental Research Fund grant (E.E.H.), and by the NIH grant R01AI081517 (D.C.J.).
Footnotes
Author Contributions
E.E.H. designed the experiments, S.S. and T.W.W. carried out the experiments, S.S., T.W.W., and E.E.H. contributed new reagents, all authors analyzed the data, and E.E.H. wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Atanasiu D, Saw WT, Cohen GH, Eisenberg RJ. Cascade of events governing cell-cell fusion induced by herpes simplex virus glycoproteins gD, gH/gL, and gB. Journal of virology. 2010;84:12292–12299. doi: 10.1128/JVI.01700-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backovic M, Jardetzky TS, Longnecker R. Hydrophobic residues that form putative fusion loops of Epstein-Barr virus glycoprotein B are critical for fusion activity. J Virol. 2007a;81:9596–9600. doi: 10.1128/JVI.00758-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backovic M, Leser GP, Lamb RA, Longnecker R, Jardetzky TS. Characterization of EBV gB indicates properties of both class I and class II viral fusion proteins. Virology. 2007b;368:102–113. doi: 10.1016/j.virol.2007.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backovic M, Longnecker R, Jardetzky TS. Structure of a trimeric variant of the Epstein-Barr virus glycoprotein B. Proc Natl Acad Sci U S A. 2009;106:2880–2885. doi: 10.1073/pnas.0810530106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton GJ. Alscript: a tool to format multiple sequence alignments. Protein Eng. 1993;6:37–40. doi: 10.1093/protein/6.1.37. [DOI] [PubMed] [Google Scholar]
- Boyle KA, Compton T. Receptor-binding properties of a soluble form of human cytomegalovirus glycoprotein B. Journal of virology. 1998;72:1826–1833. doi: 10.1128/jvi.72.3.1826-1833.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle KA, Pietropaolo RL, Compton T. Engagement of the cellular receptor for glycoprotein B of human cytomegalovirus activates the interferon-responsive pathway. Mol Cell Biol. 1999;19:3607–3613. doi: 10.1128/mcb.19.5.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britt WJ. Neutralizing antibodies detect a disulfide-linked glycoprotein complex within the envelope of human cytomegalovirus. Virology. 1984;135:369–378. doi: 10.1016/0042-6822(84)90193-4. [DOI] [PubMed] [Google Scholar]
- Britt WJ, Jarvis MA, Drummond DD, Mach M. Antigenic domain 1 is required for oligomerization of human cytomegalovirus glycoprotein B. J Virol. 2005;79:4066–4079. doi: 10.1128/JVI.79.7.4066-4079.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britt WJ, Vugler LG. Processing of the gp55-116 envelope glycoprotein complex (gB) of human cytomegalovirus. J Virol. 1989;63:403–410. doi: 10.1128/jvi.63.1.403-410.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britt WJ, Vugler LG. Oligomerization of the human cytomegalovirus major envelope glycoprotein complex gB (gp55-116) J Virol. 1992;66:6747–6754. doi: 10.1128/jvi.66.11.6747-6754.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson C, Britt WJ, Compton T. Expression, purification, and characterization of a soluble form of human cytomegalovirus glycoprotein B. Virology. 1997;239:198–205. doi: 10.1006/viro.1997.8892. [DOI] [PubMed] [Google Scholar]
- Cieplik M, Klenk HD, Garten W. Identification and characterization of spodoptera frugiperda furin: a thermostable subtilisin-like endopeptidase. Biol Chem. 1998;379:1433–1440. doi: 10.1515/bchm.1998.379.12.1433. [DOI] [PubMed] [Google Scholar]
- Colugnati FA, Staras SA, Dollard SC, Cannon MJ. Incidence of cytomegalovirus infection among the general population and pregnant women in the United States. BMC Infect Dis. 2007;7:71. doi: 10.1186/1471-2334-7-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton T, Feire A. Early events in human cytomegalovirus infection. In: Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K, editors. Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis. Cambridge; 2007. [Google Scholar]
- Compton T, Nepomuceno RR, Nowlin DM. Human cytomegalovirus penetrates host cells by pH-independent fusion at the cell surface. Virology. 1992;191:387–395. doi: 10.1016/0042-6822(92)90200-9. [DOI] [PubMed] [Google Scholar]
- Connolly SA, Jackson JO, Jardetzky TS, Longnecker R. Fusing structure and function: a structural view of the herpesvirus entry machinery. Nature reviews Microbiology. 2011;9:369–381. doi: 10.1038/nrmicro2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cranage MP, Kouzarides T, Bankier AT, Satchwell S, Weston K, Tomlinson P, Barrell B, Hart H, Bell SE, Minson AC, Smith GL. Identification of the human cytomegalovirus glycoprotein B gene and induction of neutralizing antibodies via its expression in recombinant vaccinia virus. EMBO J. 1986;5:3057–3063. doi: 10.1002/j.1460-2075.1986.tb04606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg RJ, Atanasiu D, Cairns TM, Gallagher JR, Krummenacher C, Cohen GH. Herpes virus fusion and entry: a story with many characters. Viruses. 2012;4:800–832. doi: 10.3390/v4050800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics. 2006;Chapter 5(Unit 5):6. doi: 10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feire AL, Koss H, Compton T. Cellular integrins function as entry receptors for human cytomegalovirus via a highly conserved disintegrin-like domain. Proc Natl Acad Sci U S A. 2004a;101:15470–15475. doi: 10.1073/pnas.0406821101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feire AL, Koss H, Compton T. Cellular integrins function as entry receptors for human cytomegalovirus via a highly conserved disintegrin-like domain. Proceedings of the National Academy of Sciences of the United States of America. 2004b;101:15470–15475. doi: 10.1073/pnas.0406821101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feire AL, Roy RM, Manley K, Compton T. The glycoprotein B disintegrin-like domain binds beta 1 integrin to mediate cytomegalovirus entry. Journal of virology. 2010;84:10026–10037. doi: 10.1128/JVI.00710-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons DL, Vaney MC, Roussel A, Vigouroux A, Reilly B, Lepault J, Kielian M, Rey FA. Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus. Nature. 2004;427:320–325. doi: 10.1038/nature02239. [DOI] [PubMed] [Google Scholar]
- Hannah BP, Cairns TM, Bender FC, Whitbeck JC, Lou H, Eisenberg RJ, Cohen GH. Herpes simplex virus glycoprotein B associates with target membranes via its fusion loops. J Virol. 2009;83:6825–6836. doi: 10.1128/JVI.00301-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannah BP, Heldwein EE, Bender FC, Cohen GH, Eisenberg RJ. Mutational evidence of internal fusion loops in herpes simplex virus glycoprotein B. J Virol. 2007;81:4858–4865. doi: 10.1128/JVI.02755-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldwein EE, Lou H, Bender FC, Cohen GH, Eisenberg RJ, Harrison SC. Crystal structure of glycoprotein B from herpes simplex virus 1. Science. 2006;313:217–220. doi: 10.1126/science.1126548. [DOI] [PubMed] [Google Scholar]
- Isaacson MK, Compton T. Human cytomegalovirus glycoprotein B is required for virus entry and cell-to-cell spread but not for virion attachment, assembly, or egress. Journal of virology. 2009;83:3891–3903. doi: 10.1128/JVI.01251-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacson MK, Feire AL, Compton T. Epidermal growth factor receptor is not required for human cytomegalovirus entry or signaling. Journal of virology. 2007;81:6241–6247. doi: 10.1128/JVI.00169-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis DL. Developing baculovirus-insect cell expression systems for humanized recombinant glycoprotein production. Virology. 2003;310:1–7. doi: 10.1016/s0042-6822(03)00120-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Ruigrok RW, Aitken A, Calder LJ, Martin SR, Skehel JJ, Wharton SA, Weis W, Wiley DC. Studies on the structure of the influenza virus haemagglutinin at the pH of membrane fusion. J Gen Virol. 1988;69 (Pt 11):2785–2795. doi: 10.1099/0022-1317-69-11-2785. [DOI] [PubMed] [Google Scholar]
- Ryckman BJ, Chase MC, Johnson DC. HCMV gH/gL/UL128-131 interferes with virus entry into epithelial cells: evidence for cell type-specific receptors. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:14118–14123. doi: 10.1073/pnas.0804365105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryckman BJ, Chase MC, Johnson DC. Human cytomegalovirus TR strain glycoprotein O acts as a chaperone promoting gH/gL incorporation into virions but is not present in virions. Journal of virology. 2010;84:2597–2609. doi: 10.1128/JVI.02256-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryckman BJ, Jarvis MA, Drummond DD, Nelson JA, Johnson DC. Human cytomegalovirus entry into epithelial and endothelial cells depends on genes UL128 to UL150 and occurs by endocytosis and low-pH fusion. J Virol. 2006;80:710–722. doi: 10.1128/JVI.80.2.710-722.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JL, Sharma S, Cairns TM, Heldwein EE. Fusion-deficient insertion mutants of herpes simplex virus type 1 glycoprotein B adopt the trimeric postfusion conformation. J Virol. 2010;84:2001–2012. doi: 10.1128/JVI.01791-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JA, Chow JC, Baker J, Avdalovic N, Yuan S, Au D, Co MS, Vasquez M, Britt WJ, Coelingh KL. Neutralizing monoclonal antibodies that distinguish three antigenic sites on human cytomegalovirus glycoprotein H have conformationally distinct binding sites. Journal of virology. 1993;67:489–496. doi: 10.1128/jvi.67.1.489-496.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soroceanu L, Akhavan A, Cobbs CS. Platelet-derived growth factor-alpha receptor activation is required for human cytomegalovirus infection. Nature. 2008;455:391–395. doi: 10.1038/nature07209. [DOI] [PubMed] [Google Scholar]
- Spaete RR, Thayer RM, Probert WS, Masiarz FR, Chamberlain SH, Rasmussen L, Merigan TC, Pachl C. Human cytomegalovirus strain Towne glycoprotein B is processed by proteolytic cleavage. Virology. 1988;167:207–225. doi: 10.1016/0042-6822(88)90071-2. [DOI] [PubMed] [Google Scholar]
- Stampfer SD, Lou H, Cohen GH, Eisenberg RJ, Heldwein EE. Structural basis of local, pH-dependent conformational changes in glycoprotein B from herpes simplex virus type 1. Journal of virology. 2010;84:12924–12933. doi: 10.1128/JVI.01750-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steven AC, Spear PG. Viral glycoproteins and an evolutionary conundrum. Science. 2006;313:177–178. doi: 10.1126/science.1129761. [DOI] [PubMed] [Google Scholar]
- Sun X, Belouzard S, Whittaker GR. Molecular architecture of the bipartite fusion loops of vesicular stomatitis virus glycoprotein G, a class III viral fusion protein. The Journal of biological chemistry. 2008;283:6418–6427. doi: 10.1074/jbc.M708955200. [DOI] [PubMed] [Google Scholar]
- Urban M, Klein M, Britt WJ, Hassfurther E, Mach M. Glycoprotein H of human cytomegalovirus is a major antigen for the neutralizing humoral immune response. The Journal of general virology. 1996;77 (Pt 7):1537–1547. doi: 10.1099/0022-1317-77-7-1537. [DOI] [PubMed] [Google Scholar]
- Vanarsdall AL, Chase MC, Johnson DC. Human cytomegalovirus glycoprotein gO complexes with gH/gL, promoting interference with viral entry into human fibroblasts but not entry into epithelial cells. Journal of virology. 2011;85:11638–11645. doi: 10.1128/JVI.05659-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanarsdall AL, Ryckman BJ, Chase MC, Johnson DC. Human cytomegalovirus glycoproteins gB and gH/gL mediate epithelial cell-cell fusion when expressed either in cis or in trans. Journal of virology. 2008;82:11837–11850. doi: 10.1128/JVI.01623-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vey M, Schafer W, Reis B, Ohuchi R, Britt W, Garten W, Klenk HD, Radsak K. Proteolytic processing of human cytomegalovirus glycoprotein B (gpUL55) is mediated by the human endoprotease furin. Virology. 1995;206:746–749. doi: 10.1016/s0042-6822(95)80002-6. [DOI] [PubMed] [Google Scholar]
- Wagner B, Kropff B, Kalbacher H, Britt W, Sundqvist VA, Ostberg L, Mach M. A continuous sequence of more than 70 amino acids is essential for antibody binding to the dominant antigenic site of glycoprotein gp58 of human cytomegalovirus. J Virol. 1992;66:5290–5297. doi: 10.1128/jvi.66.9.5290-5297.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Huong SM, Chiu ML, Raab-Traub N, Huang ES. Epidermal growth factor receptor is a cellular receptor for human cytomegalovirus. Nature. 2003;424:456–461. doi: 10.1038/nature01818. [DOI] [PubMed] [Google Scholar]
- Wille PT, Knoche AJ, Nelson JA, Jarvis MA, Johnson DC. A human cytomegalovirus gO-null mutant fails to incorporate gH/gL into the virion envelope and is unable to enter fibroblasts and epithelial and endothelial cells. Journal of virology. 2010;84:2585–2596. doi: 10.1128/JVI.02249-09. [DOI] [PMC free article] [PubMed] [Google Scholar]