Abstract
Targeted delivery of imaging agents to cells can be optimized with the understanding of uptake and efflux rates. Cellular uptake of macromolecules is studied frequently with fluorescent probes. We hypothesized that the internalization and efflux of fluorescently labeled macromolecules into and out of mammalian cells could be quantified by confocal microscopy to determine the rate of uptake and efflux, from which the mass transfer coefficient is calculated. The cellular influx and efflux of a third generation poly(amido amine) (PAMAM) dendrimer labeled with an Alexa Fluor 555 dye was measured in Capan-1 pancreatic cancer cells using confocal fluorescence microscopy. The Capan-1 cells were also labeled with 5-chloromethylfluorescein diacetate (CMFDA) green cell tracker dye to delineate cellular boundaries. A dilution curve of the fluorescently labeled PAMAM dendrimer enabled quantification of the concentration of dendrimer in the cell. A simple mass transfer model described the uptake and efflux behavior of the PAMAM dendrimer. The effective mass transfer coefficient was found to be 0.054 ± 0.043 μm/min, which corresponds to a rate constant of 0.035 ± 0.023 min−1 for uptake of the PAMAM dendrimer into the Capan-1 cells. The effective mass transfer coefficient was shown to predict the efflux behavior of the PAMAM dendrimer from the cell if the fraction of labeled dendrimer undergoing non-specific binding is accounted for. This work introduces a novel method to quantify the mass transfer behavior of fluorescently labeled macromolecules into mammalian cells.
Keywords: Confocal microscopy, drug delivery, internalization, efflux, PAMAM dendrimer, mass transfer coefficient
1. Introduction
The rate of transport of biological macromolecules into mammalian cells is of significant interest for drug delivery. Intensive investigations have been directed toward developing novel methods to affect macromolecular transport into mammalian cells [1–6], but comparatively little work has been done to quantitatively understand the time course of uptake into or efflux from mammalian cells. Dendrimers are a class of macromolecules applied widely as delivery vehicles for cellular internalization of biomolecules into cells [7]. For example, dendrimers have been used as an antibacterial agent [8], as a transfection agent for oligonucleotides [9–13], and a delivery vehicle for biologically active molecules such as anti-arthritis drugs [14], corticosteroids [15], and ibuprofen [16].
The uptake of dendrimers by mammalian cells is mostly limited to a qualitative understanding, where the dendrimer’s charge governs its behavior [17, 18]. The mechanism of internalization has been studied through scanning electron microscopy [19] and fluorescence [20–23], showing that internalization occurs through endocytosis [19, 22, 23], in particular adsorptive endocytosis [22]. The exact mechanism appears to depend on the surface charge of the PAMAM dendrimer itself [24], where PAMAM dendrimers with an anionic surface are internalized via caveolae-mediated endocytosis, while neutral and cationic PAMAM dendrimers are taken up via non-clathrin, non-caveolae-mediated endocytosis [23]. Other work appears to indicate uptake through a cholesterol-dependent pinocytotic pathway in the case of cationic dendrimers [20, 21]. It has also been shown that PAMAM dendrimers exhibit dose-dependent cytotoxicity [25, 26], most likely due to their ability to perforate lipid bilayers [27, 28]. The exocytosis of dendrimers has only been studied over short durations of about an hour [20]. The surface charge of the dendrimer has been shown to affect the rate of uptake, where uptake rate increases from the neutral to anionic case, while the cationic dendrimers are taken up fastest [23]. Semi-quantitative analysis of the uptake of an intrinsically fluorescently dendrimer by Caco-2 human colon adenocarcinoma cells showed that within 5 minutes the dendrimer was present in the cytoplasm, and after 35 minutes it had reached the nucleus [29]. Only one study has quantitatively investigated the binding of PAMAM dendrimers to the surface of NA1B fibroblast cells from the mouse, and determined a dissociation constant of the dendrimer from the cell surface [30].
Overall, many of the examples cite a more rapid uptake of a dendrimer-complexed or conjugated active molecular agent over the active molecule alone. Therefore, we quantitated the uptake rate constant and mass transfer coefficient of a third generation PAMAM dendrimer fluorescently labeled with an Alexa Fluor 555 dye as a model system. Hence, this study generates an additional baseline study of PAMAM dendrimers as a cellular transfection agent. More importantly, we provide proof of concept for the novel use of confocal fluorescence microscopy for the quantitative measurement of cellular uptake and efflux rates of fluorescently labeled molecules. Further, we develop a simple, lumped mass transfer model that is used to extract the effective mass transfer coefficients from the quantitative uptake experiments and determination of the cell volume and surface area. As the mass transfer coefficient gives the fundamental rate of transport of a species independent of the area available for transport, these mass transfer coefficients can be used in physiologically-based pharmacokinetic models, e. g. [31]. Determining a mass transfer coefficient for a compound allows for a fundamental understanding of how the compound is internalized. The method introduced here will also be used to study the uptake of genetic imaging nanoparticles used to visualize and quantitate oncogene mRNA expression in pancreatic cancer cells. [32–35]
2. Materials and Methods
2.1 Preparation of Alexa Fluor 555-PAMAM G3
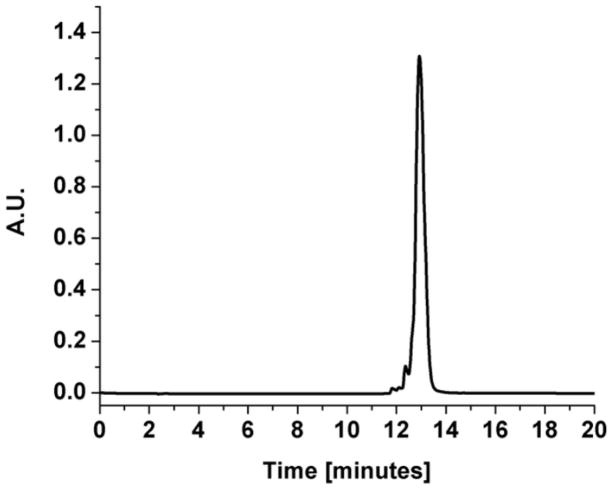
The Alexa Fluor 555-PAMAM G3 dendrimers were synthesized following a protocol developed by Dr. Nariman Amirkhanov, adapted from the “Alexa Fluor 555 protein labeling kit” instructions (Molecular Probes) [36]. The third generation PAMAM (Sigma), bearing 32 primary amines per dendrimer, was used as received at 20% w/v in methanol. For the labeling reaction, 20 μL of the stock PAMAM G3 solution was combined with 0.6 mg of Alexa Fluor 555 succinimide ester dissolved in 60 μL of acetonitrile to achieve a molar ratio of one to one. Additionally, 30 μL of 0.1 M sodium bicarbonate and 190 μL of deionized water were added to the reaction mixture, yielding a final pH of ~9. The reaction was allowed to proceed for 3 hours. Stoichiometric amounts of PAMAM G3 dendrimer and Alexa Fluor 555 succinimide ester were used, in order to label an average of one primary amine on the surface of each PAMAM dendrimer, out of 32 available amines. The product was purified using a Centricon Centrifugal Filter Unit (Millipore) with a 3000 Dalton cutoff for 20 hours at 2000×g. The absorption spectra of the upper and lower fractions collected after filtration were measured, indicating a molar ratio of Alexa Fluor 555-PAMAM dendrimer in the upper fraction vs. free Alexa Fluor 555 in the lower fraction of 96:4. This result was interpreted as a 96% yield of PAMAM dendrimer labeling. The upper fraction was analyzed using reverse-phase HPLC with a linear gradient from 0 to 80% acetonitrile in aqueous 0.1% trifluoroacetic acid, at 50°C, monitored at 555 nm, the absorption peak of the Alexa Fluor 555 dye. The resulting chromatograph showed a single peak (Figure 1). Due to the hydrophobic nature of the Alexa Fluor 555 dye, the labeled Alexa Fluor 555-PAMAM peak can be expected to elute at higher acetonitrile concentration than any unlabeled PAMAM dendrimer. Unconjugated Alexa Fluor 555 eluted during the washing step at 100% acetonitrile with 0.1% trifluoroacetic acid.
Figure 1.
Reversed-phase HPLC chromatograph of Alexa Fluor 555-PAMAM G3, eluted with a 20 minute linear gradient from 0 to 80% acetonitrile in aqueous 0.1% trifluoroacetic acid, at 50°C, monitored at 555 nm.
2.2 Cell culture
Human Capan-1 pancreatic cancer cells (ATCC) were maintained in complete medium (RPMI 1640 with 20% fetal bovine serum and 1% penicillin/streptomycin, Sigma Aldrich) at 37°C under 5% CO2. The cell line was chosen because the method described here will be used to quantitate the uptake behavior of genetic imaging nanoparticles into pancreatic cancer cells [32–35]. The cells were grown to ~80% confluency in 75 cm2 flasks (BD Biosciences) before plating them for an experiment. The experiments were performed in Nunc Lab-Tek II No. 1.5 chambered coverslips (Thermo Fisher Scientific) with eight wells. The Nunc chambers were incubated with 0.01% 150–300 kDa Poly-L-Lysine (Thermo Fisher Scientific) in sterile water for 12 hours to increase binding of the cells to the glass surface and to prevent aggregation of the PAMAM on the glass surface [37]. The chambers were then washed with phosphate buffered saline (PBS) and deionized water, and were allowed to dry for at least 24 hr. before use. Approximately 3000 cells were plated in each 0.7 cm2 well, approximately 24 hr. before an experiment. This number of cells was chosen to ensure that the cells remained separate and did not form a confluent monolayer or clusters. The cells were allowed to attach to the slide for about 14 hours prior to CMFDA labeling below.
2.3 Green tracker dye labeling of cells
The cell surface and hence, internal volume, was defined by labeling the cells in each well with 200 μL of 10 μM CellTracker™ Green 5-chloromethylfluorescein diacetate (CMFDA) (Invitrogen) in serum-free medium containing 0.1% dimethylsulfoxide for 40 minutes, as instructed by the manufacturer [38]. Following the incubation with the dye were another 30 minutes of incubation in complete medium. Following the second incubation, the cells were washed once with PBS. After staining, the chambered coverslips were wrapped in aluminum foil to prevent photobleaching and incubated at 37°C until the experiment was started. The cytoplasmic fluorescence within the cells was used for analysis to determine the boundaries as well as the surface area and volume of the cell, as demonstrated previously [30]. CMFDA labeling of the cells was completed 5–6 hours prior to the start of each PAMAM uptake experiment.
2.4 Fluorescent PAMAM uptake measurements
Alexa Fluor 555-PAMAM G3 uptake experiments were performed with a Zeiss LSM 510 NLO on an Axiovert 200M with a motorized stage. Samples were maintained at 37°C and 5% CO2 in a humid environment using a PECON Environmental Incubation System along with a PECON 22.5 mm Objective Heater. To visualize the samples a 20× Plan-Apochromat (NA 0.75) lens was used. An Argon laser (25 mW) at 488 nm and a HeNe laser (1 mW) at 543 nm were used to excite the fluorophores introduced into the samples, and a HeNe laser (5 mW) at 633 nm was used to auto-focus the coverslip as a reference in reflection mode. The Argon and HeNe lasers were attenuated via software control of the acousto-optic tunable filter to avoid light induced cell damage and eliminate measurable photobleaching. The pinhole diameter was set to 114 μm for the green channel and 98 μm for the red channel. These settings were chosen to ensure that the slice thickness (2.5 μm) was the same for both channels, and images were taken at 2.5 μm z-intervals, and typically 12–15 slices per time point were taken to cover the height from the coverslip to above the cells to reconstruct the cell volume. Signal collection from each fluorophore was achieved with a 500–530 nm band pass filter for the green channel and a 560 nm long pass filter for the red channel. To reduce crosstalk between the channels, samples were imaged in multi-track fast-line switch mode, which resulted in 1.7 second scan time per slice of 1024 x 512 pixels.
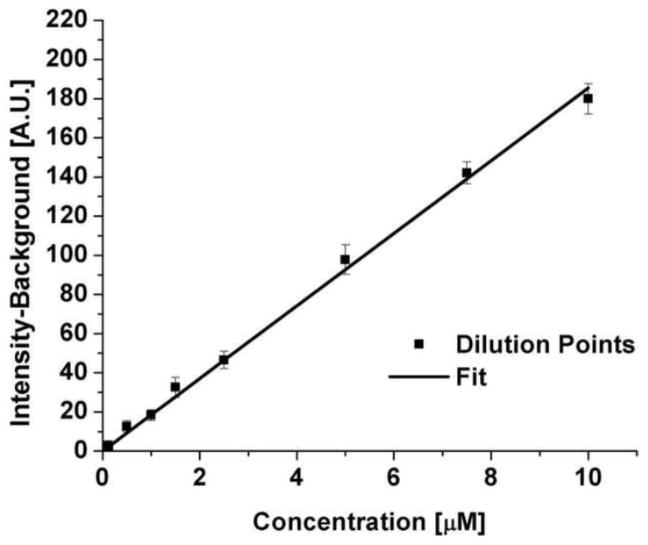
The red channel intensity recorded in the cells as a function of time was converted to concentration using a calibration curve of intensity versus Alexa Fluor 555-PAMAM concentration (Figure 2). The dilution curve was obtained across a range from zero to 10 μM of Alexa Fluor 555-PAMAM in complete medium without any cells present. The intensity was measured using a running average with a pixel size of 10 to smooth noise along a line randomly placed through the image. The background intensity was 15.74 ± 0.95 (fluorescence units), while the straight line fit to the data showed a slope of 18.55 ± 0.48 fluorescence units/μM.
Figure 2.
Dilution curve of Alexa Fluor 555-PAMAM G3 in complete medium.
2.5 Alexa Fluor 555-PAMAM uptake and efflux experiments
After incubation at 37°C for 15 minutes in the microscope without illumination, a suitable cluster of cells, 3 to 7 cells in the field of view that were not in a cluster, was selected and an image z-series was acquired. Next, the medium was removed and medium containing a given concentration of Alexa Fluor 555-PAMAM G3 was pipetted into the well. The chamber was covered with a microscope slide to minimize evaporation of the medium. During the first hour, z-series data was acquired at eight-minute intervals, followed by 36-minute intervals for up to three hours. The same procedure was followed for the efflux experiments, except the cells started in equilibrium with medium containing a given concentration of Alexa Fluor 555-PAMAM G3, which was replaced with plain medium.
2.6 Image and Data Analysis
The images were analyzed using the program Volocity (Release 3.1, Improvision). The cells were identified by setting the green channel threshold just above background intensity. Everything that was highlighted using this qualifier was deemed a cell and thus the volume and surface area of each cell was identified. All red Alexa Fluor 555 fluorescence inside the cell volume was considered to be Alexa Fluor 555-PAMAM present inside the cell. The average concentration of Alexa Fluor 555-PAMAM inside the cell was calculated from the average fluorescence measured throughout each cell, using the calibration curve in Figure 2.
The uptake of the Alexa Fluor 555-PAMAM into the cells was modeled as a first order mass transfer process, which over time will reach equilibrium (see Appendix 1 for derivation). The dendrimer concentration inside the cell ( ) is related to that in the buffer ( ) by:
| (1) |
is assumed to be constants as the reservoir is effectively infinite. Km is a mass transfer coefficient that includes all processes leading to cellular uptake and efflux, a is the cell surface area, and VII is the volume of the cell. M is a distribution coefficient defined as , where M is determined at long times when the bulk concentration and the concentration inside the cell are in equilibrium.
The same model with different integration limits ( , to , which is the final concentration in the reservoir at a value of zero in the efflux case) predicts the following efflux behavior:
| (2) |
where is the concentration of dendrimer bound inside the cell. The mass transfer coefficient is related to the commonly used rate constant, k, as shown in equation 3.
| (3) |
Note that the rate constant is proportional to the mass transfer coefficient (Km), but also includes the partition coefficient and ratio of cell area to volume. Through measurement of the cell area, volume and the partition coefficient, our method can isolate the fundamental uptake behavior that describes the effective overall process of cellular uptake. This can be used then, for example, in physiologically-based pharmacokinetic (PBPK) models [31]. In a typical PBPK model the various organs in the body are described by two compartments, a vascular and an extravascular compartment. The equation describing the extravascular compartment [39] has the same form as equation 1 above, further underlying how the parameter measured here can be used for PBPK modeling.
2.7 Correction of axial distortion
Some optical foreshortening will occur when recording the z-stack through cells due to the different refractive indices between the air, medium, cell and the glass coverslip making up the sample [40]. While distortions occur in the axial as well as the radial directions of the laser beam, the axial distortions are typically much more severe [40], and thus in this work only the non-linear geometrical correction for axial distortions developed by van Elburg et al. [41] will be used. The distortion in the z-axis can affect the surface area to volume ratio; therefore a correction factor for the surface area to volume ratio was calculated. The correction factor was determined by applying van Elburg’s focal shift to an ellipsoid of height 20 μm, and a major and minor axis of 19 μm and 8 μm, respectively, with details given in Appendix 2. The correction factor is calculated from equation 4, which gives a minimal correction of only Fz–axis = 0.996.
| (4) |
Although the effects of z-axis distortion on the volume (40%) and area (41%) are substantial, the ratio is largely unaffected. All surface area to volume ratios reported henceforth have been corrected for axial distortions using the correction factor, Fz–axis.
3. Results
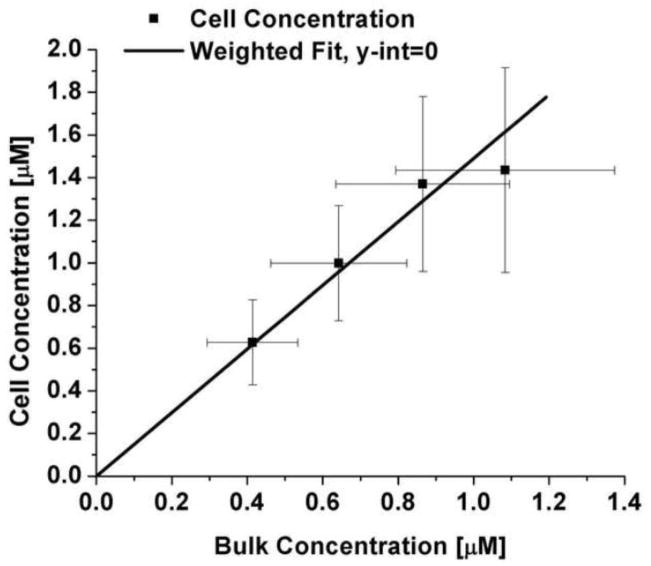
The mass transfer model as derived assumes that the final concentration of the probe inside the cell is proportional to the concentration in the reservoir. This assumption is reasonable as the cells will reach equilibrium with the reservoir outside the cell over time. This is shown to be valid by plotting the cell ( ) vs. bulk probe concentration ( ) (Figure 3), yielding a slope of 0.67 ± 0.10. This indicates that the dendrimer partitions favorably into the cell as compared to the surrounding medium.
Figure 3.
Final cell concentration as a function of bulk concentration.
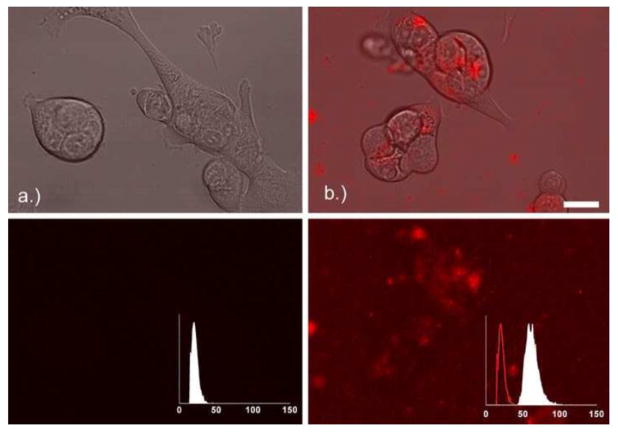
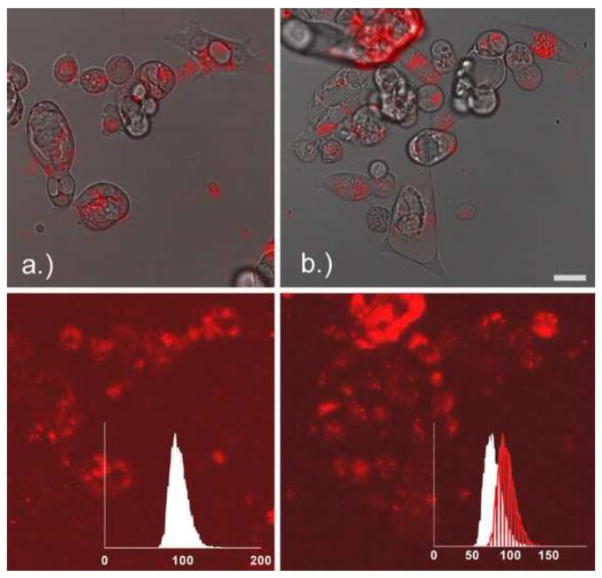
The red fluorescence inside the Capan-1 cells due to the Alexa Fluor 555-PAMAM G3 dendrimer was converted to concentration using the dilution curve shown in Figure 2. An example image showing the cells as well as the red fluorescence of the Alexa Fluor 555-PAMAM before and 3 hours after the addition of 1.1 μM of Alexa Fluor 555-PAMAM is shown in Figure 4.
Figure 4.
Capan-1 cells right before (a) and 3 hours after (b) the addition of 1.1 μM Alexa Fluor 555-PAMAM G3 dendrimer. Upper set of images shows visible and red channel, lower set of images is maximum intensity projection of red channel only. Inset is the normalized intensity distribution of the red channel solution surrounding the cells, where the line histogram in panel (b) is the intensity distribution at time zero (panel a). Scale bar indicates 20 μm.
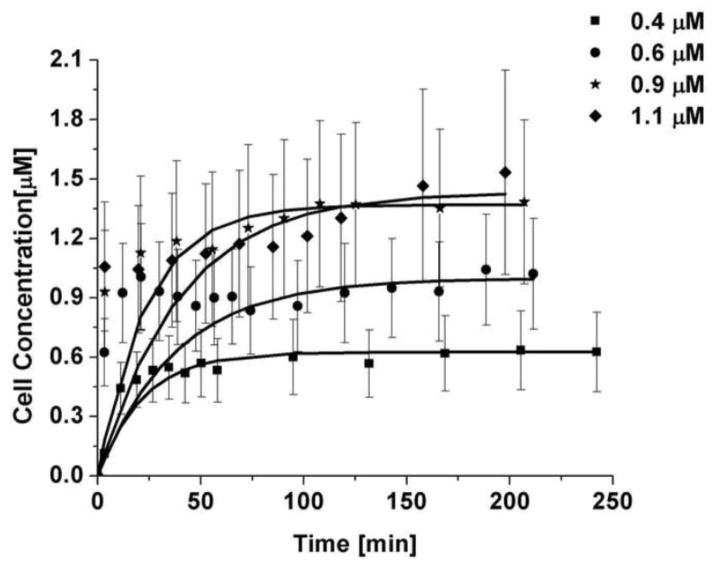
Four bulk concentrations were investigated, ranging from 0.4 to 1.1 μM. The concentration inside the cells was observed to rise rapidly within the first 30 to 50 minutes, then followed by a monotonic approach to a plateau. The resulting time course, along with a least square fit of the data to the linear form of eqn. 1, is shown in Figure 5. The error bars shown include both uncertainty propagated from the dilution curve along with the intensity uncertainty from the cellular intensity measurements. The fit parameters are shown in Table 1, while the resulting rate constants and mass transfer coefficients are shown in Table 2. From the fits an average mass transfer coefficient of 0.054 ± 0.043 μm/min was obtained.
Figure 5.
Fit uptake behavior of Alexa Fluor 555-PAMAM G3, each bulk concentration is an average of 5 cells. The error bars are a combination of the uncertainty from the fluorescence measurements in the cells and the error from the dilution curve.
Table 1.
Cell and bulk concentration of Alexa Fluor 555-PAMAM G3
| Nominal [μM] | [μM] | [μM] | M | AV [μm−1] |
|---|---|---|---|---|
| 0.4 | 0.41 ± 0.12 | 0.63 ± 0.20 | 0.66 ± 0.28 | 1.13 ± 0.06 |
| 0.6 | 0.64 ± 0.18 | 1.00 ± 0.27 | 0.64 ± 0.25 | 0.56 ± 0.02 |
| 0.9 | 0.87 ± 0.23 | 1.37 ± 0.41 | 0.63 ± 0.25 | 1.04 ± 0.07 |
| 1.1 | 1.08 ± 0.29 | 1.43 ± 0.48 | 0.76 ± 0.32 | 1.88 ± 0.14 |
-bulk concentration; -final concentration inside cells; M-distribution coefficient; AV -surface area to volume ratio
Table 2.
Fit results for uptake of Alexa Fluor 555-PAMAM G3
| Nominal [μM] | k [min−1] | k [h−1] | Km [μm/min] | Km[μm/h] | r2 |
|---|---|---|---|---|---|
| 0.4 | 0.046 ± 0.021 | 2.74 ± 1.27 | 0.062 ± 0.039 | 3.69 ± 2.32 | 0.89 |
| 0.6 | 0.026 ± 0.018 | 1.58 ± 1.08 | 0.073 ± 0.057 | 4.35 ± 3.42 | 0.46 |
| 0.9 | 0.043 ± 0.037 | 2.56 ± 2.23 | 0.065 ± 0.063 | 3.89 ± 3.75 | 0.77 |
| 1.1 | 0.025 ± 0.015 | 1.51 ± 0.91 | 0.018 ± 0.013 | 1.06 ± 0.79 | 0.63 |
-bulk concentration; k -rate constant; Km -mass transfer coefficient, r2-quality of fit
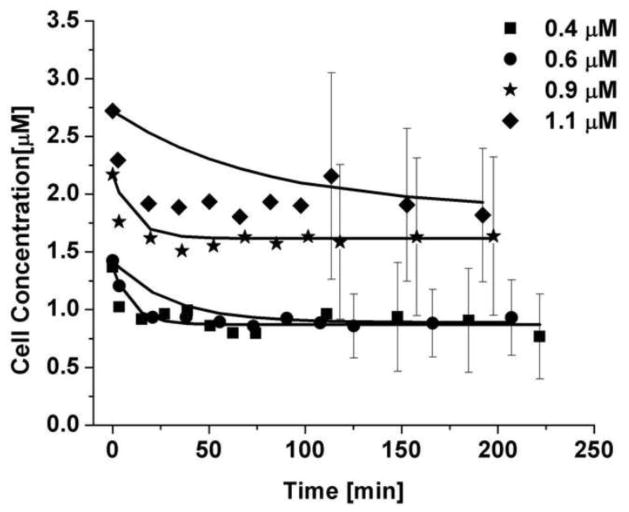
Using the protocol described above, the efflux of the labeled dendrimer from the live cells was measured by confocal fluorescence imaging. The efflux data were predicted using the mass transfer coefficient obtained from the uptake experiments, which is possible because we independently measured the cell area and volume during the efflux experiments. The only fit parameter is the amount of dendrimer that undergoes non-specific binding in the cell. The average fraction of Alexa Fluor 555-PAMAM G3 present in the cells at the end of the experiment was 0.67 ± 0.43. The parameters necessary to predict the efflux profile of the Alexa Fluor 555-PAMAM are shown in Table 3. The efflux data, as shown in Figure 6, exhibited a quick drop within the first 30 minutes and then leveled out to a steady plateau value. Sample images showing the cells as well as the red fluorescence of the Alexa Fluor 555-PAMAM before and 3 hours after the removal of 0.9 μM of Alexa Fluor 555-PAMAM are shown in Figure 7.
Table 3.
Parameters used to predict the Alexa Fluor 555-PAMAM G3 efflux behavior
| Nominal [μM] | [μM] | [μM] | Km,uptake [μm/min] | AVrelease [μm−1] | Fraction Remaining | r2 |
|---|---|---|---|---|---|---|
| 0.4 | 1.37 ± 0.75 | 0.87 ± 0.43 | 0.062 ± 0.039 | 2.52 ± 0.16 | 0.64 ± 0.47 | 0.70 |
| 0.6 | 1.42 ± 0.69 | 0.89 ± 0.30 | 0.073 ± 0.057 | 0.74 ± 0.11 | 0.62 ± 0.37 | 0.84 |
| 0.9 | 2.17 ± 0.87 | 1.62 ± 0.68 | 0.065 ± 0.063 | 2.40 ± 0.18 | 0.75 ± 0.43 | 0.82 |
| 1.1 | 2.72 ± 1.47 | 1.86 ± 0.71 | 0.018 ± 0.013 | 0.99 ± 0.15 | 0.68 ± 0.45 | 0.55 |
-bulk concentration; -initial concentration inside cells; -final concentration inside cells; Km,uptake -mass transfer coefficient; AVrelease-surface area to volume ratio, r2-quality of fit
Figure 6.
Prediction of the efflux of Alexa Fluor 555-PAMAM G3 dendrimers from Capan-1 cells. The error bars at early times are similar to those seen for the uptake and have thus been excluded for clarity. Each bulk concentration is an average of 5 cells. The error bars are a combination of the uncertainty from the fluorescence measurements in the cells and the error from the dilution curve.
Figure 7.
Capan-1 cells right before (a) and 3 hours after (b) removing the 0.9 μM bulk solution of Alexa Fluor 555-PAMAM G3 dendrimer. Upper set of images shows visible and red channel, lower set of images is maximum intensity projection of red channel only. Inset is the normalized intensity distribution of the red channel solution surrounding the cells, where the line histogram in panel (b) is the intensity distribution at time zero (panel a). Scale bar indicates 20 μm.
4. Discussion
The protocol for quantitative measurement of the rate of cellular uptake and efflux is demonstrated to give consistent results across a range of dendrimer concentrations. Using a simple, lumped mass transfer model, the effective rate of mass transfer, Km, for uptake was also found to describe the subsequent efflux. All previous studies of cellular uptake of a compound reported rate constants, without reduction to the more fundamental mass transfer coefficient. To put the rate constants obtained by our method in perspective, the rate constants for the uptake of a variety of biologically active molecules, along with the rate constant calculated in this work, are compared in Table 4. Comparing the average rate constant obtained for the Alexa Fluor 555-PAMAM G3 dendrimer, 0.035 min−1, to molecules taken up commonly by cells in vitro, the results obtained here are comparable to those for other compounds taken up by endocytosis. It should be noted in particular that antibodies and the IGF-1 peptide hormone are internalized by the cell through receptor-mediated endocytosis. While it is not known to which cell surface macromolecule Alexa Fluor 555-PAMAM binds in order to be internalized, it is known that the cell carries a negative charge, while the Alexa Fluor 555-PAMAM at approximately neutral pH is positively charged. This promotes attachment of the Alexa Fluor 555-PAMAM to the cell surface. Earlier studies discovered that the PAMAM dendrimer is taken up via endocytosis and accumulates in multivesicular bodies [19, 20, 22, 23], which is consistent with our results.
Table 4.
Rate constants of uptake of different molecules into cells
Though no full kinetic or mass transfer analysis has been reported previously for PAMAM dendrimers, there have been studies that have recorded the accumulation of fluorescently labeled PAMAM G4 dendrimers carrying a steroid [15], ibuprofen [16], or fluorescent label [23] into mammalian cells. In each case, relatively rapid uptake occurred within the first 30 to 50 minutes, which was then followed by a slower increase, reaching a plateau. That behavior is comparable to the uptake curves measured in this work. The uptake of an intrinsically fluorescent dendrimer into mammalian cells has also been reported [29]. Although the time sequence of the uptake was not reported, the authors noted that after 5 minutes the dendrimer was present in the cytoplasm, and after 35 minutes the dendrimer had reached the nucleus, indicating that it had diffused throughout most of the cell. This agrees roughly with the time course seen in the uptake data from this work.
The model parameters obtained from the uptake experiment were able to represent the efflux data, although a considerable fraction was observed to be immobilized. This is most likely due to nonspecific binding inside the cells resulting from the high number of positively charged amines on the unmodified end groups of the dendrimer. Also, it has been shown that fluorescent dyes can bind non-specifically to cells [47, 48]. In efflux experiments of Oregon green-PAMAM dendrimers from murine melanoma cells, no efflux was recorded, which might indicate a difference in how different cell types internalize and efflux PAMAM dendrimers [20].
It should be noted, however, that the long duration of the uptake and efflux experiments can adversely affect the cells. Upon introducing fresh medium to observe the dendrimer efflux, not all the cells remained attached to the coverslip, which may indicate that those cells were not healthy anymore. Great care was taken to select healthy cells for the efflux experiments, which were chosen by visual inspection. While dead cells, indicated by very high Alexa Fluor 555-PAMAM concentration inside the cell ( ), were excluded from the analysis, some cell damage due to the primary amines of the PAMAM dendrimer is possible, due to the nature of PAMAM dendrimers [22, 26, 27]. Phototoxicity due to light from the mercury arc lamp used to select a cluster of cells for the efflux experiment, combined with the reported cytotoxicity of amine-terminated PAMAM dendrimers [22, 26], cannot be ruled out. Commercially available live/dead dyes were not used in order to avoid interference from the live/dead dye with the CMFDA and Alexa Fluor 555 dye signals used to quantify the uptake behavior. While the potential for cell death needs to be considered, the consistency of the data shown in Figures 5 and 6 suggest that any cell wall damage did not lead to non-specific uptake or efflux, as the overall behavior was captured with the mass transfer model. Additionally, the well-documented interaction between PAMAM dendrimers [22, 26, 27] and mammalian cells, and the resulting disruption of the cell wall, would be observed to some degree in all uptake experiments of this nature. Therefore these effects are part of the uptake or efflux behavior of PAMAM dendrimers, and not a shortcoming of the method introduced here.
5. Conclusions
A method for quantitatively studying the uptake of fluorescently labeled macromolecules into mammalian cells was developed and demonstrated to determine the rate of uptake and efflux of fluorescently labeled Alexa Fluor 555-PAMAM G3. A simple mass transfer model was able to describe the overall observed behavior. The dendrimer was found to partition preferentially into the cells (M=0.67 ± 0.10) with a mass transfer coefficient of 0.054 ± 0.043 μm/min. The corresponding effective rate constant is comparable to previous studies of other biologically active compounds that enter cells by endocytosis. While the results shown here are only applicable to the particular cell line and dendrimer studied here, the technique introduced with these results holds great promise for quantitative study of in vitro uptake and efflux behavior of fluorescently labeled macromolecules in mammalian cells.
The protocol established here has advantages over other techniques currently used. Most importantly, the concentration of a compound can be studied over time in the same cluster of cells, while knowing the exact number of cells being investigated. Furthermore, the surface area and volume of each cell can be determined, which enables determination of the effective mass transfer coefficient. This is a more fundamental measure of the effective rate of cellular transfection that could potentially be used in PBPK models [49]. The mass transfer model used here draws close parallels to the equations typically used to describe the behavior of organs in PBPK models. Therefore the method described here can potentially be used to measure parameters for a PBPK model in vitro before having to do a full tissue distribution study.
A limitation with using high laser intensity for increased signal is photobleaching and even cell death. Naturally, reducing laser light intensity for fluorophore excitation will result in a lower signal-to-noise ratio. Noise can be reduced by decreasing the scan speed or increased averaging, yet the total scan time is limited by the need to capture the uptake and efflux kinetics. For this technique to be applied successfully, the scan time for the necessary number of z-slices through the sample should be two orders of magnitude shorter than the actual time of uptake. As shown here, this is necessary to ensure that the initial rise in concentration inside the cell is accurately captured. This guideline can be relaxed if a fast scanning confocal microscope is available (i.e., line scanning or spinning disk).
Overall, this work has demonstrated a novel technique to study the uptake of fluorescently labeled macromolecules into mammalian cells. This will prove to be useful in the study of our genetic imaging nanoparticles, as they have been shown effective using MRI [50], PET [33], and SPECT [51] imaging techniques, while current work is investigating fluorescent labels [52]. The technique discussed in this work will be used next to study the behavior of genetic imaging nanoparticles with cancer cells in vitro.
Supplementary Material
Highlights for.
Developed method to quantitatively determine uptake of fluorescent molecules into mammalian cells
Mass transfer model can describe uptake behavior.
Efflux behavior can be described with mass transfer model when accounting for non-specific binding inside cell.
Acknowledgments
The authors would like to thank Dr. Nariman Amirkhanov for his expert help in synthesizing the Alexa Fluor 555-PAMAM dendrimers, and Dr. Kim Roberts for help with the cell culture work necessary for these experiments. Microscopy performed at the Delaware Biotechnology Institute Bio-Imaging Center was supported by funding to N. J. Wagner and A. W. Opitz from Grant Number 2 P20 RR016472-08 under the INBRE Program of the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH). This work was supported in part by NCI contract N01 CO27175 to Eric Wickstrom.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Xu ZP, Hua Zeng Q, Lu GQ, Bing Yu A. Inorganic nanoparticles as carriers for efficient cellular delivery. Chemical Engineering Science. 2006;61:1027–1040. [Google Scholar]
- 2.Morgan MT, Nakanishi Y, Kroll DJ, Griset AP, Carnahan MA, Wathier M, Oberlies NH, Manikumar G, Wani MC, Grinstaff MW. Dendrimer-encapsulated camptothecins: Increased solubility, cellular uptake, and cellular retention affords enhanced anticancer activity in vitro. Cancer Research. 2006;66:11913–11921. doi: 10.1158/0008-5472.CAN-06-2066. [DOI] [PubMed] [Google Scholar]
- 3.Kocisova E, Praus P, Mojzes P, Sureau F, Stepanek J, Seksek O, Turpin RY. Cellular uptake of phosphorothioate oligonucleotide facilitated by cationic porphyrin: A microfluorescence study. Biopolymers. 2006;82:325–328. doi: 10.1002/bip.20495. [DOI] [PubMed] [Google Scholar]
- 4.Vives E. Cellular uptake of the Tat peptide: an endocytosis mechanism following ionic interactions. Journal of Molecular Recognition. 2003;16:265–271. doi: 10.1002/jmr.636. [DOI] [PubMed] [Google Scholar]
- 5.Oehlke J, Birth P, Klauschenz E, Wiesner B, Beyermann M, Oksche A, Bienert M. Cellular uptake of antisense oligonucleotides after complexing or conjugation with cell-penetrating model peptides. European Journal of Biochemistry. 2002;269:4025–4032. doi: 10.1046/j.1432-1033.2002.03093.x. [DOI] [PubMed] [Google Scholar]
- 6.Parimi S, Barnes TJ, Callen DF, Prestidge CA. Mechanistic Insight into Cell Growth, Internalization, and Cytotoxicity of PAMAM Dendrimers. Biomacromolecules. 2010;11:382–389. doi: 10.1021/bm9010134. [DOI] [PubMed] [Google Scholar]
- 7.Tomalia DA, Baker H, Dewald J, Hall M, Kallos G, Martin S, Roeck J, Ryder J, Smith P. A New Class of Polymers - Starburst-Dendritic Macromolecules. Polymer Journal. 1985;17:117–132. [Google Scholar]
- 8.Chen CZS, Cooper SL. Interactions between dendrimer biocides and bacterial membranes. Biomaterials. 2002;23:3359–3368. doi: 10.1016/s0142-9612(02)00036-4. [DOI] [PubMed] [Google Scholar]
- 9.DeLong RK, Nolting A, Fisher M, Chen Q, Wickstrom E, Kligshteyn M, Demirdji S, Caruthers M, Juliano RL. Comparative pharmacokinetics, tissue distribution, and tumor accumulation of phosphorothioate, phosphorodithioate, and methylphosphonate oligonucleotides in nude mice. Antisense & Nucleic Acid Drug Development. 1997;7:71–77. doi: 10.1089/oli.1.1997.7.71. [DOI] [PubMed] [Google Scholar]
- 10.Bielinska A, KukowskaLatallo JF, Johnson J, Tomalia DA, Baker JR. Regulation of in vitro gene expression using antisense oligonucleotides or antisense expression plasmids transfected using starburst PAMAM dendrimers. Nucleic Acids Research. 1996;24:2176–2182. doi: 10.1093/nar/24.11.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kukowska-Latallo JF, Bielinska AU, Johnson J, Spindler R, Tomalia DA, Baker JR. Efficient transfer of genetic material into mammalian cells using Starburst polyamidoamine dendrimers. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:4897–4902. doi: 10.1073/pnas.93.10.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chavanpatil MD, Khdair A, Panyam J. Nanoparticles for cellular drug delivery: Mechanisms and factors influencing delivery. Journal of Nanoscience and Nanotechnology. 2006;6:2651–2663. doi: 10.1166/jnn.2006.443. [DOI] [PubMed] [Google Scholar]
- 13.Shieh MJ, Peng CL, Lou PJ, Chiu CH, Tsai TY, Hsu CY, Yeh CY, Lai PS. Non-toxic phototriggered gene transfection by PAMAM-porphyrin conjugates. Journal of Controlled Release. 2008;129:200–206. doi: 10.1016/j.jconrel.2008.03.024. [DOI] [PubMed] [Google Scholar]
- 14.Chandrasekar D, Sistla R, Ahmad FJ, Khar RK, Diwan PV. The development of folate-PAMAM dendrimer conjugates for targeted delivery of anti-arthritic drugs and their pharmacokinetics and biodistribution in arthritic rats. Biomaterials. 2007;28:504–512. doi: 10.1016/j.biomaterials.2006.07.046. [DOI] [PubMed] [Google Scholar]
- 15.Khandare J, Kolhe P, Pillai O, Kannan S, Lieh-Lai M, Kannan RM. Synthesis, cellular transport, and activity of polyamidoamine dendrimer-methylprednisolone conjugates. Bioconjugate Chemistry. 2005;16:330–337. doi: 10.1021/bc0498018. [DOI] [PubMed] [Google Scholar]
- 16.Kolhe P, Khandare J, Pillai O, Kannan S, Lieh-Lal M, Kannan RM. Preparation, cellular transport, and activity of polyamidoamine-based dendritic nanodevices with a high drug payload. Biomaterials. 2006;27:660–669. doi: 10.1016/j.biomaterials.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 17.Patri AK, Kukowska-Latallo JF, Baker JR. Targeted drug delivery with dendrimers: Comparison of the release kinetics of covalently conjugated drug and non-covalent drug inclusion complex. Advanced Drug Delivery Reviews. 2005;57:2203–2214. doi: 10.1016/j.addr.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 18.Wiwattanapatapee R, Carreno-Gomez B, Malik N, Duncan R. Anionic PAMAM dendrimers rapidly cross adult rat intestine in vitro: A potential oral delivery system? Pharmaceutical Research. 2000;17:991–998. doi: 10.1023/a:1007587523543. [DOI] [PubMed] [Google Scholar]
- 19.Jevprasesphant R, Penny J, Attwood D, D’Emanuele A. Transport of dendrimer nanocarriers through epithelial cells via the transcellular route. Journal of Controlled Release. 2004;97:259–267. doi: 10.1016/j.jconrel.2004.03.022. [DOI] [PubMed] [Google Scholar]
- 20.Seib FP, Jones AT, Duncan R. Comparison of the endocytic properties of linear and branched PEIs, and cationic PAMAM dendrimers in B16f10 melanoma cells. Journal of Controlled Release. 2007;117:291–300. doi: 10.1016/j.jconrel.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 21.Manunta M, Tan PH, Sagoo P, Kashefi K, George AJT. Gene delivery by dendrimers operates via a cholesterol dependent pathway. Nucleic Acids Research. 2004;32:2730–2739. doi: 10.1093/nar/gkh595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kitchens KM, Kolhatkar RB, Swaan PW, Eddington ND, Ghandehari H. Transport of poly(amidoamine) dendrimers across Caco-2 cell monolayers: Influence of size, charge and fluorescent labeling. Pharmaceutical Research. 2006;23:2818–2826. doi: 10.1007/s11095-006-9122-2. [DOI] [PubMed] [Google Scholar]
- 23.Perumal OP, Inapagolla R, Kannan S, Kannan RM. The effect of surface functionality on cellular trafficking of dendrimers. Biomaterials. 2008;29:3469–3476. doi: 10.1016/j.biomaterials.2008.04.038. [DOI] [PubMed] [Google Scholar]
- 24.Tiriveedhi V, Kitchens KM, Nevels KJ, Ghandehari H, Butko P. Kinetic analysis of the interaction between poly(amidoamine) dendrimers and model lipid membranes. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2011;1808:209–218. doi: 10.1016/j.bbamem.2010.08.017. [DOI] [PubMed] [Google Scholar]
- 25.Jevprasesphant R, Penny J, Attwood D, McKeown NB, D’Emanuele A. Engineering of dendrimer surfaces to enhance transepithelial transport and reduce cytotoxicity. Pharmaceutical Research. 2003;20:1543–1550. doi: 10.1023/a:1026166729873. [DOI] [PubMed] [Google Scholar]
- 26.Jevprasesphant R, Penny J, Jalal R, Attwood D, McKeown NB, D’Emanuele A. The influence of surface modification on the cytotoxicity of PAMAM dendrimers. International Journal of Pharmaceutics. 2003;252:263–266. doi: 10.1016/s0378-5173(02)00623-3. [DOI] [PubMed] [Google Scholar]
- 27.Hong SP, Bielinska AU, Mecke A, Keszler B, Beals JL, Shi XY, Balogh L, Orr BG, Baker JR, Holl MMB. Interaction of poly(amidoamine) dendrimers with supported lipid bilayers and cells: Hole formation and the relation to transport. Bioconjugate Chemistry. 2004;15:774–782. doi: 10.1021/bc049962b. [DOI] [PubMed] [Google Scholar]
- 28.Mecke A, Majoros IJ, Patri AK, Baker JR, Holl MMB, Orr BG. Lipid bilayer disruption by polycationic polymers: The roles of size and chemical functional group. Langmuir. 2005;21:10348–10354. doi: 10.1021/la050629l. [DOI] [PubMed] [Google Scholar]
- 29.Al-Jamal KT, Ruenraroengsak P, Hartell N, Florence AT. An intrinsically fluorescent dendrimer as a nanoprobe of cell transport. Journal of Drug Targeting. 2006;14:405–412. doi: 10.1080/10611860600834441. [DOI] [PubMed] [Google Scholar]
- 30.Lai JC, Yuan CL, Thomas JL. Single-cell measurements of polyamidoamine dendrimer binding. Annals of Biomedical Engineering. 2002;30:409–416. doi: 10.1114/1.1470180. [DOI] [PubMed] [Google Scholar]
- 31.Opitz AW, Thakur ML, Wickstrom E, Wagner NJ. Physiologically based pharmacokinetics of molecular imaging nanoparticles for mRNA detection determined in tumor-bearing mice. Oligonucleotides. 2010;20:117–125. doi: 10.1089/oli.2009.0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tian X, Chakrabarti A, Amirkhanov N, Aruva MR, Zhang K, Cardi CA, Lai S, Thakur ML, Wickstrom E. Receptor-mediated internalization of chelator-PNA-peptide hybridization probes for radioimaging or magnetic resonance imaging of oncogene mRNAs in tumours. Biochemical Society Transactions. 2007;35:72–76. doi: 10.1042/BST0350072. [DOI] [PubMed] [Google Scholar]
- 33.Chakrabarti A, Zhang K, Aruva MR, Cardi CA, Opitz AW, Wagner NJ, Thakur ML, Wickstrom E. Radiohybridization PET Imaging of KRAS G12D mRNA Expression in Human Pancreas Cancer Xenografts with [Cu-64]DO3A-Peptide Nucleic Acid-Peptide Nanoparticles. Cancer Biology & Therapy. 2007;6:948–956. doi: 10.4161/cbt.6.6.4191. [DOI] [PubMed] [Google Scholar]
- 34.Chakrabarti A, Aruva MR, Sajankila SP, Thakur ML, Wickstrom E. Synthesis of novel peptide nucleic acid-peptide chimera for non-invasive imaging of cancer. Nucleosides Nucleotides & Nucleic Acids. 2005;24:409–414. doi: 10.1081/ncn-200061865. [DOI] [PubMed] [Google Scholar]
- 35.Amirkhanov NV, Zhang K, Aruva MR, Thakur ML, Wickstrom E. Scintigraphic imaging of KRAS2 mRNA in human pancreas cancer xenografts with (111In-DO3A)n-poly(diamidopropanoate)m-PNA-D(Cys-Ser-Lys-Cys) hybridization probes. Bioconjug Chem. 2010;21 doi: 10.1021/bc900523c. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alexa Fluor® 555 Protein Labeling Kit. Molecular Probes; Eugene, OR: 2001. [Google Scholar]
- 37.Sakai K, Sadayama S, Yoshimura T, Esumi K. Direct force measurements between adlayers consisting of poly(amidoamine) dendrimers with primary amino groups or quaternary ammonium groups. Journal of Colloid and Interface Science. 2002;254:406–409. doi: 10.1006/jcis.2002.8601. [DOI] [PubMed] [Google Scholar]
- 38.CellTracker (TM) Probes for Long-Term Tracing of Living Cells. Vol. 2005. Molecular Probes; Eugene, OR: 2005. [Google Scholar]
- 39.Peng B, Andrews J, Nestorov I, Brennan B, Nicklin P, Rowland M. Tissue distribution and physiologically based pharmacokinetics of antisense phosphorothioate oligonucleotide ISIS 1082 in rat. Antisense & Nucleic Acid Drug Development. 2001;11:15–27. doi: 10.1089/108729001750072092. [DOI] [PubMed] [Google Scholar]
- 40.Hell S, Reiner G, Cremer C, Stelzer EHK. Aberrations in Confocal Fluorescence Microscopy Induced by Mismatches in Refractive-Index. Journal of Microscopy-Oxford. 1993;169:391–405. [Google Scholar]
- 41.Van Elburg HJ, Kuypers LC, Decraemer WF, Dirckx JJJ. Improved correction of axial geometrical distortion in index-mismatched fluorescent confocal microscopic images using high-aperture objective lenses. Journal of Microscopy-Oxford. 2007;228:45–54. doi: 10.1111/j.1365-2818.2007.01822.x. [DOI] [PubMed] [Google Scholar]
- 42.Shih LB, Lu HHZ, Xuan H, Goldenberg DM. Internalization and Intracellular Processing of an Anti B-Cell Lymphoma Monoclonal-Antibody, Ll2. International Journal of Cancer. 1994;56:538–545. doi: 10.1002/ijc.2910560413. [DOI] [PubMed] [Google Scholar]
- 43.Shih LB, Xuan H, Aninipot R, Stein R, Goldenberg DM. In-Vitro and in-Vivo Reactivity of an Internalizing Antibody, Rs7, with Human Breast-Cancer. Cancer Research. 1995;55:S5857–S5863. [PubMed] [Google Scholar]
- 44.Stein R, Chen S, Sharkey RM, Goldenberg DM. Murine Monoclonal-Antibodies Raised against Human Non-Small Cell-Carcinoma of the Lung - Specificity and Tumor Targeting. Cancer Research. 1990;50:1330–1336. [PubMed] [Google Scholar]
- 45.Paye JMD, Forsten-Williams K. Regulation of insulin-like growth factor-I (IGF-I) delivery by IGF binding proteins and receptors. Annals of Biomedical Engineering. 2006;34:618–632. doi: 10.1007/s10439-005-9064-6. [DOI] [PubMed] [Google Scholar]
- 46.Gupta N, Hoffman RP, Veng-Pedersen P. Pharmacokinetic/pharmacodynamic differentiation of pancreatic responsiveness in obese and lean children. Biopharmaceutics & Drug Disposition. 2005;26:287–294. doi: 10.1002/bdd.461. [DOI] [PubMed] [Google Scholar]
- 47.Mahmudi-Azer S, Lacy P, Bablitz B, Moqbel R. Inhibition of nonspecific binding of fluorescent-labelled antibodies to human eosinophils. Journal of Immunological Methods. 1998;217:113–119. doi: 10.1016/s0022-1759(98)00105-7. [DOI] [PubMed] [Google Scholar]
- 48.Panchuk-Voloshina N, Haugland RP, Bishop-Stewart J, Bhalgat MK, Millard PJ, Mao F, Leung WY, Haugland RP. Alexa dyes, a series of new fluorescent dyes that yield exceptionally bright, photostable conjugates. Journal of Histochemistry & Cytochemistry. 1999;47:1179–1188. doi: 10.1177/002215549904700910. [DOI] [PubMed] [Google Scholar]
- 49.Opitz AW. Structural and functional investigations of a molecular imaging nanoparticle for magnetic resonance imaging of oncogene expression in the pancreas. University of Delaware; Newark, DE: 2008. p. 457. Vol. PhD. [Google Scholar]
- 50.Amirkhanov NV, Dimitrov I, Opitz AW, Zhang K, Lackey JP, Cardi CA, Lai S, Wagner NJ, Thakur ML, Wickstrom E. Design of (Gd-DO3A)n-Polydiamidopropanoyl-Peptide Nucleic Acid-D(Cys-Ser-Lys-Cys) Magnetic Resonance Contrast Agents. Biopolymers. 2008;89:1061–1076. doi: 10.1002/bip.21059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Amirkhanov NV, Zhang K, Aruva MR, Thakur ML, Wickstrom E. Scintigraphic imaging of KRAS2 mRNA in human pancreas cancer xenografts with (111In-DO3A)n-poly(diamidopropanoate)m-PNA-D(Cys-Ser-Lys-Cys) hybridization probes. Bioconjugate Chemistry. 2010;21:731–740. doi: 10.1021/bc900523c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Amirkhanov NV, Wickstrom E. Twin fluorophore near infrared KRAS peptide nucleic acid molecular beacons. A7. American Association for Cancer Research; Philadelphia PA: 2008. p. 29. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.