Abstract
Recent findings suggested that the SUMO-interacting motifs (SIMs) present in the human TRIM5α (TRIM5αhu) protein play an important role in the ability of TRIM5αhu to restrict N-MLV. Here we explored the role of SIMs in the ability of rhesus TRIM5α (TRIM5αrh) to restrict HIV-1, and found that TRIM5αrh SIM mutants IL376KK (SIM1mut) and VI405KK (SIM2mut) completely lost their ability to block HIV-1 infection. Interestingly, these mutants also lost the recently described property of TRIM5αrh to shuttle into the nucleus. Analysis of these variants revealed that they are unable to interact with the HIV-1 core, which might explain the reason that these variants are not active against HIV-1. Furthermore, NMR titration experiments to assay the binding between the PRYSPRY domain of TRIM5αrh and the small ubiquitin-like modifier 1(SUMO-1) revealed no interaction. In addition, we examined the role of SUMOylation in restriction, and find out that inhibition of SUMOylation by the adenoviral protein Gam1 did not altered the retroviral restriction ability of TRIM5α. Overall, our results do not support a role for SIMs or SUMOylation in the antiviral properties of TRIM5α.
Keywords: TRIM5αrh, HIV-1, SUMOylation, restriction, SUMO-interacting motif
INTRODUCTION
Several newly discovered proteins endogenously expressed in primates show the ability to dominantly block retroviral infection and cross-species transmission by interfering with the early phase of viral replication (Kirmaier et al., 2010; Sayah et al., 2004; Stremlau et al., 2004). Of particular interest are members of the tripartite motif (TRIM) family of proteins (Reymond et al., 2001). The splicing variant alpha of TRIM5 from rhesus macaque (TRIM5αrh) is a ~53 kDa cytosolic protein that potently restricts HIV-1(Keckesova et al., 2004; Song et al., 2005a). TRIM5αrh blocks HIV-1 and certain other retroviruses soon after viral entry, but prior to reverse transcription (Keckesova et al., 2004; Stremlau et al., 2004). The retroviral capsid protein (CA) is the viral determinant for susceptibility to restriction by TRIM5α (Owens et al., 2003). Studies on the fate of the HIV-1 capsid in the cytosol of infected cells have correlated restriction with a decrease in the amount of cytosolic particulate capsid (Diaz-Griffero et al., 2007a; Diaz-Griffero et al., 2008; Perron et al., 2007; Roa et al., 2012; Stremlau et al., 2006), suggesting that TRIM5αrh acts by inducing premature uncoating in target cells.
TRIM5αrh is composed of four distinct domains: RING, B-box 2, coiled-coil and PRYSPRY (Reymond et al., 2001). The RING domain of TRIM5αrh is an E3 ubiquitin ligase (Diaz-Griffero et al., 2006a; Kar et al., 2008; Kim et al., 2011; Langelier et al., 2008; Lienlaf et al., 2011; Maegawa et al.; Yamauchi et al., 2008). The E3-ligase activity of TRIM5αrh correlates with the ability of TRIM5αrh to block HIV-1 (Lienlaf et al., 2011; Roa et al., 2012). The B-box 2 domain of TRIM5αrh and other TRIM proteins, such as TRIM63, self-associates into dimeric complexes that are important for TRIM5α higher-order self-association and capsid binding avidity; these B-box 2 domain functions are essential for full and potent restriction of HIV-1 (Diaz-Griffero et al., 2007b; Diaz-Griffero et al., 2009; Ganser-Pornillos et al.; Javanbakht et al., 2005; Mrosek et al., 2008; Perez-Caballero et al., 2005). The coiled-coil domain enables TRIM5αrh dimerization (Kar et al., 2008; Langelier et al., 2008), which is critical for interaction of the PRYSPRY domain with the HIV-1 capsid (Diaz-Griffero et al., 2006b; Sebastian and Luban, 2005; Stremlau et al., 2006). The PRYSPRY domain is the capsid recognition module that dictates the specificity of restriction (Biris et al., 2012; Nakayama et al., 2005; Sawyer et al., 2005; Song et al., 2005b; Stremlau et al., 2005; Yap et al., 2005).
Recent discoveries have demonstrated that TRIM5αrh has the ability to shuttle into the nucleus and then exported back into the cytoplasm by a CRM-1 dependent mechanism (Diaz-Griffero et al., 2011). This work demonstrated that the use of the CRM-1 inhibitor, Leptomycin B (LMB) (Fornerod et al., 1997; Kudo et al., 1998; Kuersten et al., 2001; Mohr et al., 2009; Watanabe et al., 1999; Wolff et al., 1997), induced the accumulation of TRIM5αrh in the nuclear compartment; however, the ability of TRIM5αrh to restrict HIV-1 infection was not altered by the use of LMB (Diaz-Griffero et al., 2011). Because the use of LMB leads to accumulation of TRIM5αrh in the nucleus while de novo synthesis of TRIM5αrh occurs in the cytoplasm, it is difficult to know whether the shuttling of TRIM5αrh through the nucleus is important for HIV-1 restriction. Therefore, the contribution of the nuclear shuttling properties of TRIM5αrh to HIV-1 restriction is not understood.
The addition of small ubiquitin-related modifiers (SUMO) to cellular proteins, a process known as SUMOylation(Johnson, 2004), is linked to the targeting of cytosolic proteins to the nuclear compartment (Wang et al., 2012). Recent work suggested that SUMO-interacting motifs (SIMs) in the human TRIM5α. TRIM5αhu protein are involved in the ability of TRIM5αhu to restrict N-tropic murine leukemia virus (N-MLV)(Arriagada et al., 2011). This work showed that mutation of the SIMs of TRIM5αhu inactivates its restriction capability (Arriagada et al., 2011). These studies, however, did not test whether TRIM5αhu bearing mutated SIMs lost its ability to shuttle into and out of the nucleus.
To understand the role of TRIM5αrh nuclear-shuttling and SUMOylation in HIV-1 restriction by TRIM5αrh, we performed the following studies: 1) Contribution of SU-MO-conjugating motifs (SCM) and SIM to the ability of TRIM5αrh to restrict HIV-1 was examined by independent mutagenesis of these motifs, 2) ability of TRIM5αrh SCM and SIM variants to shuttle into the nucleus by the use of LMB, 3) ability of TRIM5αrh variants to bind in vitro assembled HIV-1 capsid-nucleocapsid complexes and oligomerization, 4) ability of the SUMOylation inhibitor Gam1 to affect N-MLV restriction by cells that endogenously express TRIM5αhu, and 5) ability of the purified PRYSPRY domain of TRIM5αrh to interact with SUMO-1.
RESULTS
Role of SUMO-interacting and SUMO-conjugation motifs in HIV-1 restriction by TRIM5αrh
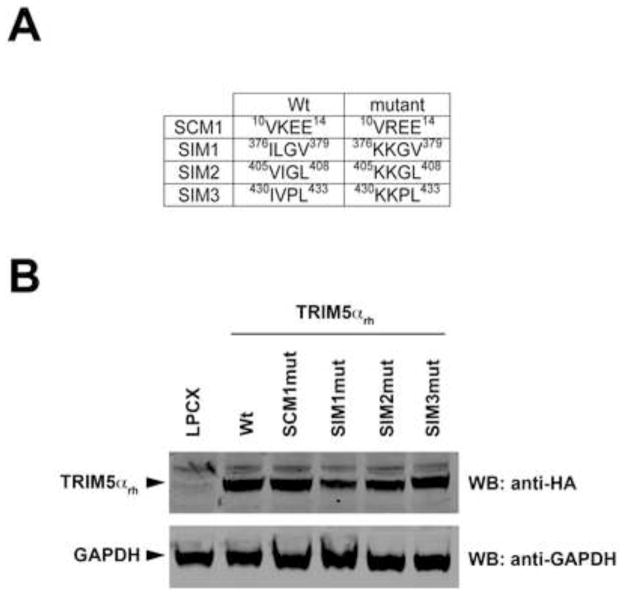
Previous work has reported that N-MLV restriction by TRIM5αhu requires the presence of intact SUMO-interacting motifs in the PRYSPRY domain of TRIM5αhu (Arriagada et al., 2011). To test the role of SUMO-interacting motifs (SIMs) and SUMO-conjugation motifs (SCMs) in the ability of TRIM5αrh to restrict HIV-1, we independently mutated the K10R (SCM1mut), IL376KK (SIM1mut), VI405KK (SIM2mut) and IV430KK (SIM3mut) (Figure 1A). The TRIM5αrh variants were stably expressed in dog Cf2Th cells (Figure 1B), and the ability of TRIM5αrh variants to restrict HIV-1 was tested (Figure 1C). In agreement with observations studying TRIM5αhu SCMmut and SIMmut motifs, we found that TRIM5αrh SIM1mut and SIM2mut did not restrict HIV-1 suggesting the involvement of SIMs in restriction (Figure 1C). By contrast, TRIM5αrh SCM1mut and SIM3mut blocked HIV-1 infection as potent as wild type. These results suggested that SIMs are important for the ability of TRIM5αrh to block HIV-1 infection.
Figure 1. Contribution of TRIM5αrh SUMO-conjugation and SUMO-interacting motifs(SIM) to restriction of HIV-1.
Dog Cf2Th cells were transduced with the LPCX vector expressing HA-tagged wild-type and mutant TRIM5αrh proteins. The TRIM5αrh SUMO-conjugation motif(SCM1) and the three SUMO-interacting motifs (SIM1, SIM2 and SIM3) variants are shown with their respective amino acid position (A). Stable cell lines were selected with 5 μg/ml puromycin, and the expression levels of mutant and wild-type TRIM5αrh proteins were assayed by Western blotting using anti-HA and anti-GAPDH antibodies (B). Stable cell lines were challenged with different amounts of HIV-1-GFP (C). The percentage of GFP-positive cells was measured 48 h later by FACS. The results of three independent experiments were similar; the results of a single experiment are shown.
Role of SIM1 and SIM2 in the ability of TRIM5αrh to shuttle into and out of the nucleus
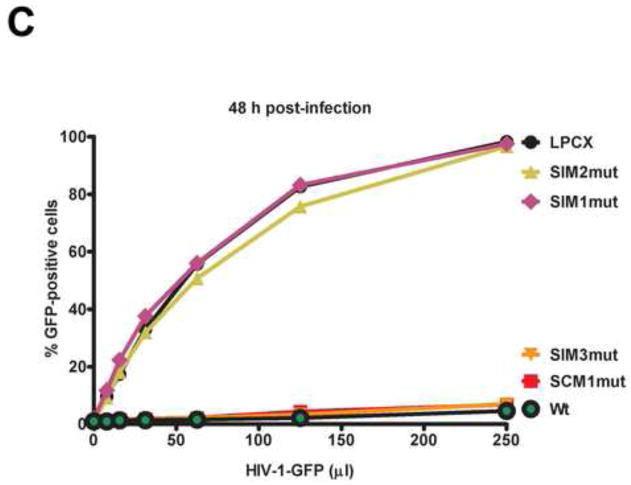
We have previously described the ability of TRIM5αrh to shuttle into and the nucleus (Diaz-Griffero et al., 2011). Because of SUMO’s involvement in the nuclear import of cellular proteins (Wang et al., 2012), we tested the role of SIMs in the ability of TRIM5αrh to shuttle into and out of the nucleus. For this purpose, we studied the localization of TRIM5αrh variants by immunofluorescence microscopy in the presence of leptomycin B (LMB), which is a specific inhibitor of nuclear export (Fornerod et al., 1997; Kudo et al., 1998; Kuersten et al., 2001; Mohr et al., 2009; Watanabe et al., 1999; Wolff et al., 1997). As shown in Figure 2, TRIM5αrh SIM1mut and SIM2mut were not retained in the nuclear compartment in the presence of LMB for 5 or 12 h when compared to wild type TRIM5αrh (supplemental Table 1). These experiments suggested that TRIM5μrh-SIM1mut and TRIM5αrh-SIM2mut have lost their ability to traffic into the nucleus. By contrast, TRIM5αrh SCM1mut and SIM3mut were retained in the nucleus similar to wild type TRIM5αrh (Figure 2 and Supplemental Table 1). These experiments suggested that SIM1 and SIM2 are involved in the translocation of TRIM5αrh into the nucleus.
Figure 2. Intracellular distribution of TRIM5αrh SIM mutant proteins in the presence of leptomycin B.
Human HeLa cells expressing the indicated TRIM5αrh variants were treated with 20 ng/ml of leptomycin B (LMB) or DMSO for either 5 or 12 h. Treated cells were were fixed and immunostained using anti-HA antibodies (green). Cellular nuclei were stained by using DAPI (blue). Representative figures are shown. The results of three independent experiments were similar (Supplemental Table 1); the result of a single experiment is shown.
Binding of TRIM5αrh variants to in vitro assembled HIV-1 capsid-nucleocapsid (CA-NC) complexes
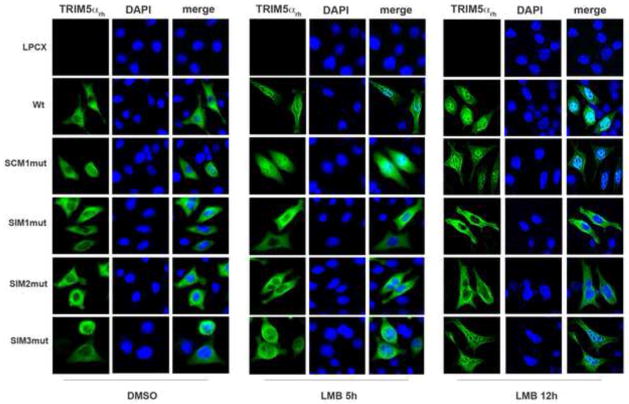
The PRYSPRY domain of TRIM5αrh mediates the interaction between TRIM5αrh and the HIV-1 core (Ohkura et al., 2006; Song et al., 2005c; Stremlau et al., 2006). Because TRIM5αrh SIM1mut and SIM2mut are located in the PRYSPRY domain, which could affect the ability of TRIM5αrh to bind the HIV-1 core (Stremlau et al., 2006); we examined the ability of the different TRIM5αrh variants to bind in vitro assembled HIV-1 CA-NC complexes (Figure 3), as previously described (Lienlaf et al., 2011; Roa et al., 2012; Stremlau et al., 2006). Interestingly, TRIM5αrh SIM1mut and SIM2mut lost their ability to bind the HIV-1 core when compared to wild type TRIM5αrh. These results explain the inability of TRIM5αrh SIM1mut and SIM2mut variants to block HIV-1 infection (Figure 1C). By contrast, TRIM5αrh SCM1mut and TRIM5αrh SIM3mut bound in vitro assembled HIV-1 CA-NC complexes similar to wild type TRIM5αrh. These results suggested that the inability of TRIM5αrh SIM1mut and SIM2mut to block HIV-1 infection is due to their failure to bind in vitro assembled HIV-1 CA-NC complexes.
Figure 3. Binding of TRIM5αrh SIM mutant proteins to in vitro assembled HIV-1 capsid-nucleocapsid (CA-NC) complexes.
Human 293T cells were transfected with plasmids expressing the indicated wild-type and mutant HA-tagged TRIM5αrh proteins. Forty-eight h after transfection, cells were lysed. The lysates were incubated at room temperature for 1 h with in vitro assembled HIV-1 CA-NC complexes. The mixtures were applied to a 70% sucrose cushion and centrifuged. INPUT represents the lysates analyzed by Western blotting before being applied to the 70% cushion. The input mixtures were Western blotted using anti-HA antibodies. Similarly, the pellets from the 70% cushion (BOUND) were analyzed by Western blotting using anti-HA and anti-p24 antibodies. The results of three independent experiments were similar; the result of a single experiment is shown.
Oligomerization of TRIM5αrh variants
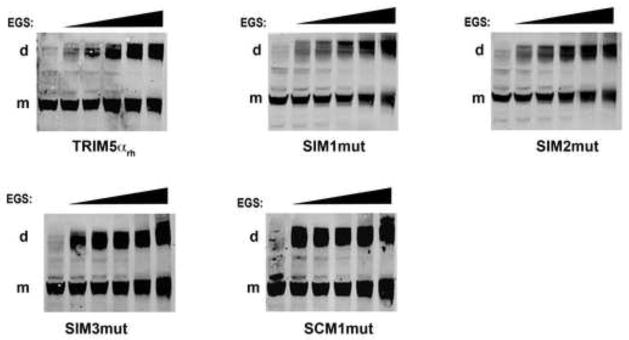
Next we examined the ability of TRIM5αrh variants to oligomerize by cross-linking using glycolbis-succinimidylsuccinate (EGS), as previously described (Diaz-Griffero et al., 2007b; Kar et al., 2008; Langelier et al., 2008). As shown in Figure 4, cross-linking experiments suggested that all TRIM5αrh variants studied here efficiently form oligomers. This observation is consistent with previous results demonstrating that the coiled-coil is the only domain required for oligomerization (Javanbakht et al., 2006).
Figure 4. Oligomerization of TRIM5αrh SIM mutant proteins.
Lysates from 293T cells expressing wild-type and mutant TRIM5αrh proteins were cross-linked with increasing concentrations of ethylene glycol-bis(succinimidyl succinate) (EGS), and were subjected to Western blotting using anti-HA antibodies, as described in Materials and Methods. “m” and “d” stands for monomer and dimer, respectively. Similar results were obtained in three independent experiments and a representative experiment is shown.
Role of SUMOylation in the ability of TRIM5αhu to restrict N-MLV
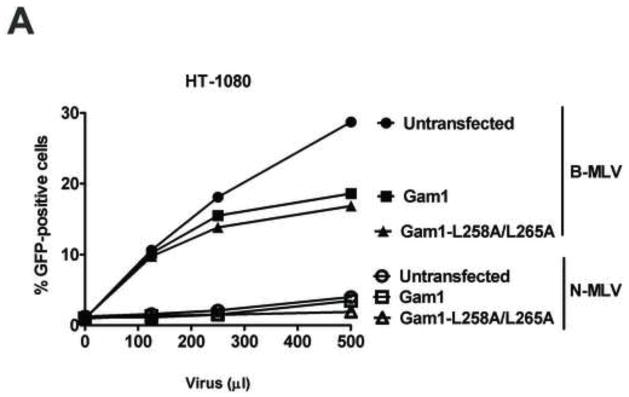
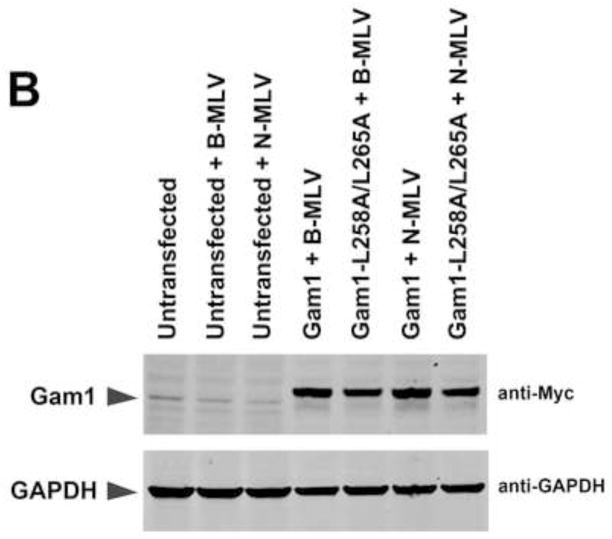
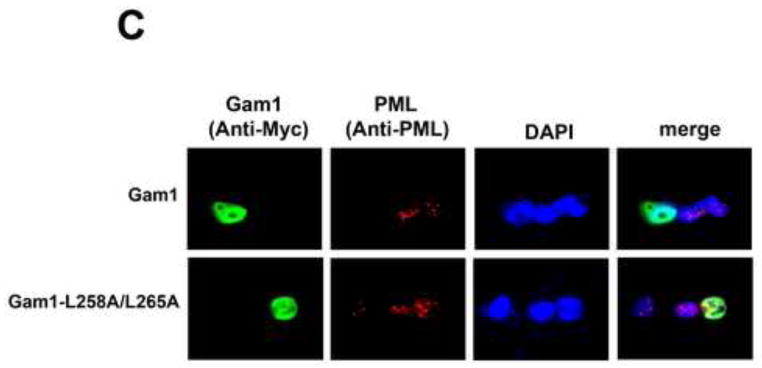
Our results suggested that TRIM5αrh SIM1mut and SIM2mut do not restrict HIV-1 because of their inability to bind the HIV-1 core; however, these results do not exclude the possibility that SUMOylation is a necessary process for the capacity of TRIM5α to restrict retroviruses, as previously suggested (Arriagada et al., 2011). To understand the role of SUMOylation in restriction, we took advantage of the avian adenoviral protein Gam1, which inhibits protein SUMOylation in human cells by degradation of SUMO E1 and E2 enzymes (Boggio et al., 2004; Boggio et al., 2007; Colombo et al., 2002). To understand the role of SUMOylation in N-MLV restriction by cells endogenously expressing TRIM5αhu, we challenged the fibrosarcoma human HT-1080 cell line with different amounts N-tropic murine leukemia virus bearing the green fluorescent protein as a reporter (N-MLV-GFP) in the presence of Gam1 (Figure 5A and B). Expression of the SUMOylation inhibitor Gam1 did not released the restriction imposed by TRIM5αhu to N-MLV-GFP infection (Figure 5A and B). As control, we performed similar infections in the presence of the mutant Gam1-L258A/L265A, which is unable to inhibit the SUMOylation pathway (Boggio et al., 2004; Chiocca et al., 1997; Colombo et al., 2002). To rule out the possibility that inhibition of SUMOylation is affecting N-MLV infection, we challenged HT-1080 cells with B-tropic MLV bearing a GFP reporter (B-MLV-GFP) in the presence of Gam1 (Figure 5A and B). The expression of Gam1 in HT-1080 cells did not affect the infection of B-MLV-GFP suggesting that Gam1 did not interfere with the process of infection (Figure 5A). Because inhibition of SUMOylation by expression of Gam1 destroys nuclear bodies that contain promyelocytic leukemia (PML) protein (Colombo et al., 2002), we tested whether expression of Gam1 in human HT-1080 cells displayed the same effect (Figure 5C). As expected, expression of Gam1 destroyed PML-containing nuclear bodies in HT-1080 cells when compared to Gam1-L258A/L265A (Figure 5C and supplemental Figure 1). Overall these results showed that inhibition of SUMOylation by expression of Gam1 does not affect the ability of TRIM5αhu to restrict N-MLV suggesting that TRIM5αhu does not require SUMOylation for restriction of N-MLV.
Figure 5. Role of SUMOylation in the ability of TRIM5αhu to restrict N-MLV.
Human HT-1080 cells were transienly transfected with plasmids expressing Myc-tagged Gam1 or Gam1-L258A/L265A. After 24 h, cells were challenged with increasing amounts of N-MLV-GFP or B-MLV-GFP. Viruses expressing the reproter GFP were normilized in dog Cf2Th cells. Forthy-eight h post-transfection, infection was determined by counting the percentage of GFP-positive cells using FACS (A). The expression of Gam1 and Gam1-L258A/L265A were analized by Western blotting using anti-Myc and anti-GAPDH antibodies (B). The ability of Gam1 to destroy PML-containing nuclear bodies was assayed by transiently transfecting HT-1080 cells with plasmids expressing Gam1 or Gam1-L258A/L265A tagged with a Myc epitope. After 24 h, cells were fixed and and immunostained using anti-Myc (green) and anti-PML (red) antibodies (C). Cellular nuclei were stained by using DAPI (blue). Similar results were obtained in three independent experiments and a representative experiment is shown.
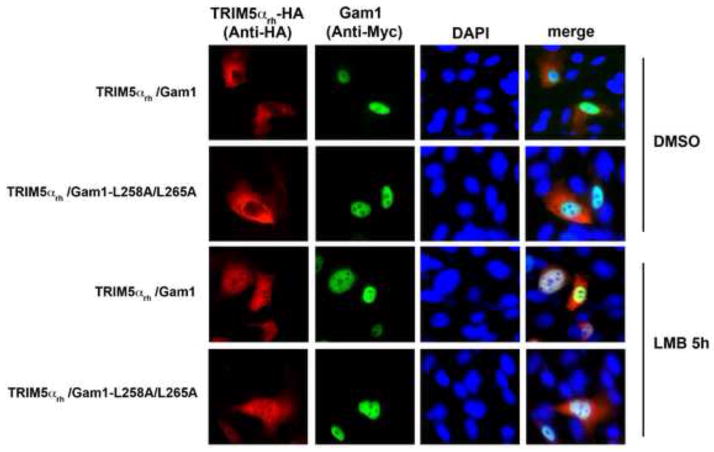
Inhibition of SUMOylation by Gam1 did not affect the ability of TRIM5αrh to traffic into the nucleus
Because SUMOylation is important for nuclear import of cellular proteins (Wang et al., 2012), we tested whether SUMOylation is necessary for the ability of TRIM5αrh to shuttle into the nucleus (Diaz-Griffero et al., 2011). For this purpose, we examined the subcellular localization of transiently transfected HT-1080 cells with HA-tagged TRIM5αrh (TRIM5αrh-HA) and Gam1 in the presence of LMB (Figure 6 and Supplemental Table 2). As previously shown (Diaz-Griffero et al., 2011), the treatment with LMB accumulated TRIM5αrh-HA proteins in the nuclear compartment; however, expression of Gam1 did not diminish or alter the accumulation of TRIM5αrh-HA proteins in the nuclear compartment. To show that TRIM5αrh does not interfere with the biological activity of Gam1, we demonstrated that Gam1 destroyed PML-nuclear bodies in the presence of TRIM5αrh (Supplemental Figure 2). These results suggested that SUMOylation is not involved in the ability of TRIM5αrh to shuttle into the nucleus.
Figure 6. Inhibition of SUMOylation by Gam1 does not affect the ability of TRIM5αrh to traffic into the nucleus.
Human HT-1080 cells were cotransfected with plasmids expressing Myc-tagged Gam1 and HA-tagged TRIM5αrh. After 24 h, cells were treated with 20 ng/ml of leptomycin B (LMB) or DMSO for 5 h, fixed and immunostained using antibodies anit-myc (green) and anti-HA (red) antibodies. Cellular nuclei were stained by using DAPI (blue). Similar results were obtained in three independent experients(Supplemental Table 2), and a representative experiment is shown.
The PRYSPRY domain of TRIM5αrh does not interact with SUMO-1
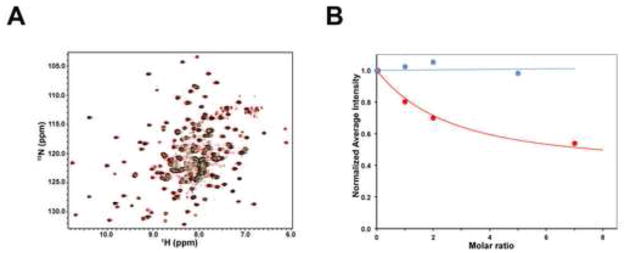
To test whether the protein SUMO-1 can interact with the PRYSPRY domain ofTRIM5αrh (residues 292–497), we performed NMR titrations of the 15N-labeled rhesus PRYSPRY domain with the unlabeled SUMO-1 protein. NMR titrations are sensitive to even very weak protein-protein interactions and can be used to map the interaction surfaces of the PRYSPRY domain (Biris et al., 2012). As shown in Figure 7A, the addition of a 5 fold excess of SUMO-1 to the PRYSPRY domain, did not produce observable shifts on the NMR signals in the 15N-HSQC spectrum. In some cases protein-protein interactions can produce attenuation rather than shifts of the peaks in the NMR spectra; however, Figure 7B showed that no signal attenuation was observed in NMR titrations of the PRYSPRY domain with SUMO-1. By contrast, signal attenuation is clearly visible in the titration of PRYSPRY with the N-terminal domain of the HIV-1 capsid, a protein that interacts with the PRYSPRY domain very weakly with a Kd~400 μM (Biris et al., 2012). Thus, no interaction between the PRYSPRY domain of TRIM5αrh and SUMO-1 was detected even at SUMO-1 concentration of up to 1.5 mM.
Figure 7. NMR titrations of the TRIM5αrh PRYSPRY domain with SUMO-1.
(A) The superposition of the 15N-HSQC spectra of the 0.3 mM 15N-labeled PRYSPRY sample in the presence of 1.5 mM SUMO-1 (green) or in the absence of SUMO-1 added (red) is shown. Overlapping signals appear black. (B) NMR titrations of the 15N-labeled PRYSPRY domain with SUMO-1 (blue) and the N-terminal domain of HIV-1 capsid (red). Addition of 5:1 molar excess of SUMO-1 does not show any detectable attenuation of the PRYSPRY NMR signals.
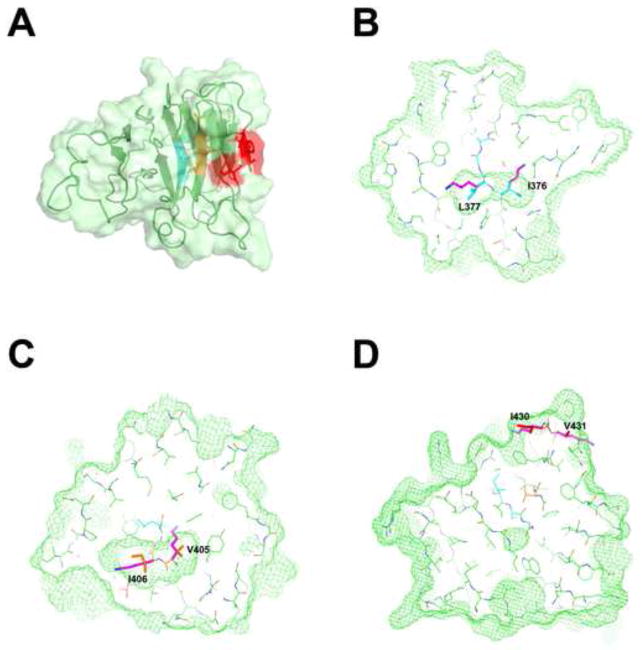
Mapping of SIM residues on the PRYSPRY domain of TRIM5αrh
We have recently solved the structure of the PRYSPRY domain of TRIM5αrh (Biris et al., 2012). SIM1, SIM2 and SIM3 motifs were predicted based on the consensus amino acid sequence, but these hydrophobic amino acid motifs have to be exposed on the protein surface in order to mediate protein-protein interactions. Inspection of the TRIM5αrh PRYSPRY structure revealed that out of the three predicted SIM motifs, only SIM3 contains surface-exposed residues, while SIM1 and SIM2 are buried in the hydrophobic core of the PRYSPRY structure; therefore, SIM1 and SIM2 cannot mediate protein-protein interactions (Figure 8).
Figure 8. Putative SUMO interaction motifs (SIM) mapped onto the TRIM5αrh PRYSPRY structure.
(A)SIM1, SIM2 and SIM3 mapped onto the TRIM5αrh PRYSPRY structure. The SPRY domain is shown as a cartoon with the protein surface colored in light green. The segments of the protein identified as SUMO interacting motifs SIM-1, SIM-2 and SIM-3 are colored in cyan, orange and red respectively. Only the SIM-3 segment contains surface-exposed residues, while SIM-1 and SIM-2 are buried in the hydrophobic core of the protein. (B) Slice through the TRIM5αrh PRYSPRY structure showing the volume (green mesh) occupied by residues I376 and L377 of SIM1 (cyan). The two residues are completely buried within the hydrophobic core and the lysine residues of the SIM1 mutant (magenta) can not be accomodated within that space. The resulting steric clash and buried positive charge are incompatible with the stably-folded native structure. (C) The same is true for SIM2. (D) In contrast, SIM3 is exposed on the protein surface, therefore SIM3mut is not expected to disrupt the overall PRYSPRY structure.
DISCUSSION
The recent discovery of the ability of TRIM5αrh and TRIM5αhu to shuttle into and out of the nucleus suggested the exiting possibility that the nuclear shuttling ability of TRIM5α is playing a role in restriction (Diaz-Griffero et al., 2011). Since SUMOylation is important for the ability of some cellular proteins to shuttle into the nucleus and recent findings suggested a role for SIM in restriction (Arriagada et al., 2011), here we explored the role of SCM, SIM and SUMOylation in the ability of TRIM5α to restrict retroviral infection.
In agreement with previous observations (Arriagada et al., 2011), we found that changes TRIM5αrh SIM1mut and SIM2mut lost their ability to restrict HIV-1. Interestingly, both TRIM5αrh variants also lost their ability to shuttle into the nucleus. However, these variants were unable to bind in vitro assembled HIV-1 CA-NC complexes, which suggest that these TRIM5αrh variants are unable to restrict HIV-1 because of their impaired binding to the HIV-1 core. The most likely explanation is that TRIM5αrh SIM1mut and SIM2mut contain a misfolded PRYSPRY domain. In agreement, analysis of SIM1 and SIM2 motifs in the structure of the PRYSPRY domain revealed that these motifs are buried in the hydrophobic core of the protein, and their mutation to charged residues is likely to disrupt the structural integrity of the PRYSPRY domain (Figure 8). Taken together, we would like to suggest that TRIM5αrh SIM1mut and SIM2mut are proteins that contain a misfolded PRYSPRY domain, and this is the reason that they are unable to restrict retroviral infection.
Although our results do not support a role for SUMOylation in the ability of TRIM5αrh to restrict HIV-1, this work does not discard the possibility that TRIM5αrh is SUMOylated. We have attempted to detect TRIM5αrh SUMOylation by using a tagged SUMO protein without any success, as tested by other groups (Arriagada et al., 2011). The development of antibodies against the endogenous TRIM5αrh will allow the exploration of this interesting possibility.
To directly test the ability of the PRYSPRY domain of TRIM5αrh to interact with SUMO protein, we tested the ability of recombinant PRYSPRY domain from TRIM5αrh to interact with SUMO-1 by NMR spectroscopy. However, we were unable to detect any interaction suggesting that TRIM5αrh does not interact with SUMO-1. An alternative possibility is that the interaction with SUMO-1 requires the full-length TRIM5αrh, which is likely to contain two PRYSPRY domains in close proximity, as previously suggested (Arriagada et al., 2011).
We tested the role of SUMOylation in the restriction ability of TRIM5αhu by using the SUMOylation inhibitor Gam1(Boggio et al., 2004; Boggio et al., 2007; Colombo et al., 2002). For this purpose, we used the human fibrosarcoma cell line HT-1080 that expresses TRIM5αhu and is able to potently block infection of N-MLV in a TRIM5αhu-dependent manner. Even though Gam1 destroyed the formation of PML nuclear bodies (Figure 5C), it did not affect the ability of TRIM5αhu to restrict N-MLV suggesting that SUMOylation is not involved the ability of TRIM5αhu to restrict N-MLV. Altogether these results suggested that the process of SUMOylation is not involved in the ability of TRIM5αhu to restrict N-MLV; however, the use of Gam1 doesn’t affect the levels of SUMO in the cell, which could be important for restriction.
Interestingly, increasing the levels of SUMO-1 in human 293T or TE671 cells increases the restriction imposed to N-MLV by TRIM5αhu (Arriagada et al., 2011), arguing for a role of SUMO-1 in restriction. One possibility is that the SUMOylation of capsid described for Moloney murine leukemia virus is important for the ability of TRIM5αhu to bind N-MLV capsid and restrict infection (Yueh et al., 2006).
Taken altogether these findings do not support the role of SCM, SIMs or SUMOylation in the ability of TRIM5α to restrict retroviral infection. However, the contribution of the ability of TRIM5α to shuttle into and out of the nucleus still remains unknown. The future finding of TRIM5α variants that do not shuttle into and out the nucleus and bind the HIV-1 core will be essential to uncover a role for nuclear shuttling on retroviral restriction by TRIM5α.
MATERIALS AND METHODS
Plasmid construction
The pLPCX-TRIM5α-HA plasmid, specific primers and a Quik-Change site-directed mutagenesis kit (Stratagene) were used according to the manufacturer’s specifications to generate recombinant plasmids that expressed TRIM5α-HA variants K10R, IL376KK, VI405KK and IV430KK. The plasmids expressing Gam1 and Gam1-L258A/L265A proteins tagged with Myc epitope, was a generous gift from Dr. Susanna Chiocca, and has been described previously (Boggio et al., 2004).
Antibodies
Monoclonal antibodies against the HA epitope were from Sigma. Anti-GAPDH rabbit polyclonal antibodies, anti-mouse-Cy2, anti-rabbit-Cy3, anti-mouse Alexa 488, and anti-goat Alexa 594 were obtained from Sigma. Anti-p24 HIV-1 capsid was obtained from Immunodiagnostics. Anti-Myc monoclonal and anti-PML goat polyclonal antibodies were purchased from Santa Cruz Biotechnology.
Protein analysis
Cellular proteins were extracted with radioimmunoprecipitation assay (RIPA) buffer, as previously described(Lienlaf et al., 2011). Detection of proteins by Western blotting was performed using anti-HA (Sigma), anti-GAPDH (Sigma), anti-P24 (Sigma), anti-Myc (Santa Cruz Biotechnology) and anti-rabbit and anti-mouse antibodies conjugated to Alexa Fluor 680 (Li-Cor). Bands were detected by scanning blots with the Li-Cor Odyssey Imaging System using the 700 channel.
Binding of TRIM5αrh variants to in vitro assembled HIV-1 capsid-nucleocapsid (CA-NC) complexes
The HIV-1 CA-NC protein was expressed, purified, and assembled as previously described (Lienlaf et al., 2011). HIV-1 CA-NC particles were assembled in vitro by diluting the CA-NC protein to a concentration of 0.3 mM in 50 mM Tris-HCl (pH 8.0), 0.5 M NaCl and 2 mg/ml DNA oligo (TG)50. The mixture was incubated at 4°C overnight and centrifuged at 8,600xg for 5 min. These CA-NC complexes were used for the binding assay as follows. 293T cells were transfected with plasmids expressing wild-type or mutant TRIM5αrh proteins. Forty-eight h after transfection, cell lysates were prepared as follows: washed cells were resuspended in hypotonic lysis buffer (10mMTris [pH 7.4], 1.5mM MgCl2, 10mM KCl, 0.5mM DTT). The cell suspension was frozen and thawed and then incubated on ice for 10 min. Next, the lysate was centrifuged at full speed in a refrigerated Eppendorf microcentrifuge (14,000xg) for 5 min. To test binding, 5 μl of CA-NC complexes assembled in vitro were incubated with 200 μl of cell lysate at room temperatura for 1 h. A fraction of this mixture was stored (input). The mixture was spun through a 70% sucrose cushion (70% sucrose, 1X PBS, and 0.5 mM DTT) at 100,000 xg in an SW55 rotor (Beckman) for 1 h at 4°C. After centrifugation, the supernatant was carefully removed and the pellet was resuspended in 1X SDS-PAGE loading buffer (pellet). The level of TRIM5αrh proteins was determined by Western blotting using anti-HA antibodies (Sigma). The level of HIV-1 CA-NC protein in the pellet was assessed by Western blotting with an anti-p24 capsid antibody (Immunodiagnostics).
Immunofluorescence Microscopy
Transfections of cell monolayers were performed using Lipofectamine Plus reagent (Invitrogen), according to the manufacturer’s instructions. Transfected cells were incubated at 37 °C for 24 h, unless otherwise stated. For indirect immunofluorescence microscopy, cell monolayers grown on coverslips were transfected. Monolayers were washed twice with PBS and fixed for 15 min with 4% paraformaldehyde in PBS. Paraformaldehyde-fixed cells were washed twice with PBS, incubated for 4 min in permeabilizing buffer (0.5% Triton X-100 in PBS), and then blocked using PBS containing 2% bovine serum albumin for 1 h at room temperature. Subsequently, cells were incubated for 1 h at room temperature with primary antibodies in blocking buffer, as previously described (Diaz-Griffero et al., 2002). After three washes with PBS, the cells were incubated for 30 min with secondary antibodies. Cellular nuclei were labeled with 1 μg/ml of DAPI (49, 69-diamidino-2-phenylindole) in PBS. Subsequently, samples were mounted for fluorescence microscopy by using the ProLong Anti-fade Kit (Molecular Probes, Eugene, OR). Images were obtained with a Zeiss Observer.Z1 microscope using a 63X objective, and deconvolution was performed using the software AxioVision V4.8.1.0 (Carl Zeiss Imaging Solutions).
Generation of cells stably expressing TRIM5αrh variants
Retroviral vectors encoding wild-type or mutant TRIM5αrh were created using the pLPCX vector (Clontech). Recombinant viruses were produced in 293T cells by cotransfecting the pLPCX plasmids with the pVPack-GP and pVPack-VSV-G packaging plasmids (Stratagene). The pVPack-VSV-G plasmid encodes the vesicular stomatitis virus G envelope glycoprotein, which allows efficient entry into a wide range of vertebrate cells (Yee et al., 1994). Transduced Cf2Th cells were selected in 5 μg/ml of puromycin.
Infection with viruses expressing green fluorescent protein (GFP)
Recombinant HIV-1 expressing GFP was prepared as described (Diaz-Griffero et al., 2008). The virus was pseudotyped with the VSV-G glycoprotein. For infections, 5 × 104 cells seeded in 24-well plates were incubated at 37°C with virus for 24 h. Cells were washed and returned to culture for 48 h unless otherwise stated, and the percentage of GFP positive cells was determined by flow cytometry (Becton Dickinson). Viral stocks were titrated by serial dilution on Cf2Th cells to determine the concentration of infectious viruses.
Cross-linking of TRIM5αrh
The HA-tagged TRIM5αrh variants were expressed transiently in 293T cells. Cells were washed in phosphate-buffered saline (PBS) and lysed in NP40 lysis buffer (0.5% Nonidet P40 (NP40), 1X protease inhibitor (complete EDTA-free, Roche Diagnostics) in PBS) for 45 min at 4 °C. Lysates were centrifuged at 14,000 xg for 15 min at 4 °C. The cleared lysates were not stored or frozen, but rather were directly cross-linked. Approximately 100–200 μl of cleared lysates were diluted with PBS/1 mM EDTA to a final volume of 400 μl. Lysates were cross-linked with varying concentrations (up to 2 mM) of ethylene glycol-bis(succinimidyl succinate) (EGS) for 30 min at room temperature and centrifuged briefly in a tabletop centrifuge. The reaction mix was quenched with 0.1 M Tris– HCl, pH 7.5 and briefly centrifuged. The cleared, cross-linked lysates were analyzed by Western blotting using antibodies against HA epitope (Sigma).
Recombinant proteins preparation
The pET28b expression vector encoding the SUMO1 with a C-terminal 6His tag was a generous gift from Dr. Christopher Lima. Transformed BL21 Rosetta 2 competent cells (Novagen) were grown at 37°C until cell density reached A600 ~ 0.6. The temperature was then lowered to 30°C and protein production was induced with 0.75 mM isopropyl-β-d-thiogalactopyranoside (IPTG). After 4 h cells were harvested by centrifugation (6,000 x g) and lysed by sonication. The purification of the C-terminally His-tagged SUMO-1 was carried out by metal affinity followed by size-exclusion chromatography on a Superdex 75 (GE life sciences). The cloning, expression and purification of PRYSPRY domain from TRIM5αrh has been described previously (Biris et al., 2012). All NMR samples were prepared in 50 mM sodium phosphate pH 7.0, 50 mM NaCl and 10 mM dithiothreitol buffer.
NMR titrations
NMR titrations of 15N-labeled PRYSPRY domain of TRIM5αrh with unlabeled His-tagged SUMO1 and unlabeled N-terminal domain of the HIV capsid (residues 1–147) were carried out on a Bruker Avance 700 MHz spectrometer at 298 K by recording a series of 1H-15N TROSY-HSQC spectra containing molar ratios from 1:0 up to 1:5 for SUMO1 and from 1:0 up to 1:7 for the N-terminal domain of HIV-1 capsid.
Supplementary Material
Acknowledgments
We thank Andre Rosowsky for critical reading of the manuscript. We thank Dr. Andrei Tomashevski for his help with recombinant protein purification. We thank Susanna Chiocca for providing reagents. This work was funded by an NIH R01 AI087390 to F.D.-G, CPRIT Scholar Award to D.I. and a K99/R00 Pathway to Independence Award to F.D.-G. from the National Institutes of Health 4R00MH086162-02.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arriagada G, Muntean LN, Goff SP. SUMO-interacting motifs of human TRIM5alpha are important for antiviral activity. PLoS Pathog. 2011;7:e1002019. doi: 10.1371/journal.ppat.1002019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biris N, Yang Y, Taylor AB, Tomashevski A, Guo M, Hart PJ, Diaz-Griffero F, Ivanov DN. Structure of the rhesus monkey TRIM5alpha PRYSPRY domain, the HIV capsid recognition module. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1203536109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggio R, Colombo R, Hay RT, Draetta GF, Chiocca S. A mechanism for inhibiting the SUMO pathway. Mol Cell. 2004;16:549–561. doi: 10.1016/j.molcel.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Boggio R, Passafaro A, Chiocca S. Targeting SUMO E1 to ubiquitin ligases: a viral strategy to counteract sumoylation. J Biol Chem. 2007;282:15376–15382. doi: 10.1074/jbc.M700889200. [DOI] [PubMed] [Google Scholar]
- Chiocca S, Baker A, Cotten M. Identification of a novel antiapoptotic protein, GAM-1, encoded by the CELO adenovirus. J Virol. 1997;71:3168–3177. doi: 10.1128/jvi.71.4.3168-3177.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo R, Boggio R, Seiser C, Draetta GF, Chiocca S. The adenovirus protein Gam1 interferes with sumoylation of histone deacetylase 1. EMBO Rep. 2002;3:1062–1068. doi: 10.1093/embo-reports/kvf213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Griffero F, Gallo DE, Hope TJ, Sodroski J. Trafficking of some old world primate TRIM5alpha proteins through the nucleus. Retrovirology. 2011;8:38. doi: 10.1186/1742-4690-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Griffero F, Hoschander SA, Brojatsch J. Endocytosis is a critical step in entry of subgroup B avian leukosis viruses. J Virol. 2002;76:12866–12876. doi: 10.1128/JVI.76.24.12866-12876.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Griffero F, Kar A, Lee M, Stremlau M, Poeschla E, Sodroski J. Comparative requirements for the restriction of retrovirus infection by TRIM5alpha and TRIMCyp. Virology. 2007a doi: 10.1016/j.virol.2007.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Griffero F, Kar A, Perron M, Xiang SH, Javanbakht H, Li X, Sodroski J. Modulation of retroviral restriction and proteasome inhibitor-resistant turnover by changes in the TRIM5alpha B-box 2 domain. J Virol. 2007b;81:10362–10378. doi: 10.1128/JVI.00703-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Griffero F, Li X, Javanbakht H, Song B, Welikala S, Stremlau M, Sodroski J. Rapid turnover and polyubiquitylation of the retroviral restriction factor TRIM5. Virology. 2006a;349:300–315. doi: 10.1016/j.virol.2005.12.040. [DOI] [PubMed] [Google Scholar]
- Diaz-Griffero F, Perron M, McGee-Estrada K, Hanna R, Maillard PV, Trono D, Sodroski J. A human TRIM5alpha B30.2/SPRY domain mutant gains the ability to restrict and prematurely uncoat B-tropic murine leukemia virus. Virology. 2008;378:233–242. doi: 10.1016/j.virol.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Griffero F, Qin XR, Hayashi F, Kigawa T, Finzi A, Sarnak Z, Lienlaf M, Yokoyama S, Sodroski J. A B-box 2 surface patch important for TRIM5alpha self-association, capsid binding avidity, and retrovirus restriction. J Virol. 2009;83:10737–10751. doi: 10.1128/JVI.01307-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Griffero F, Vandegraaff N, Li Y, McGee-Estrada K, Stremlau M, Welikala S, Si Z, Engelman A, Sodroski J. Requirements for capsid-binding and an effector function in TRIMCyp-mediated restriction of HIV-1. Virology. 2006b;351:404–419. doi: 10.1016/j.virol.2006.03.023. [DOI] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Ganser-Pornillos BK, Chandrasekaran V, Pornillos O, Sodroski JG, Sundquist WI, Yeager M. Hexagonal assembly of a restricting TRIM5alpha protein. Proc Natl Acad Sci U S A. 2011;108:534–539. doi: 10.1073/pnas.1013426108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javanbakht H, Diaz-Griffero F, Stremlau M, Si Z, Sodroski J. The contribution of RING and B-box 2 domains to retroviral restriction mediated by monkey TRIM5alpha. J Biol Chem. 2005;280:26933–26940. doi: 10.1074/jbc.M502145200. [DOI] [PubMed] [Google Scholar]
- Javanbakht H, Yuan W, Yeung DF, Song B, Diaz-Griffero F, Li Y, Li X, Stremlau M, Sodroski J. Characterization of TRIM5alpha trimerization and its contribution to human immunodeficiency virus capsid binding. Virology. 2006;353:234–246. doi: 10.1016/j.virol.2006.05.017. [DOI] [PubMed] [Google Scholar]
- Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- Kar AK, Diaz-Griffero F, Li Y, Li X, Sodroski J. Biochemical and biophysical characterization of a chimeric TRIM21-TRIM5alpha protein. J Virol. 2008;82:11669–11681. doi: 10.1128/JVI.01559-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keckesova Z, Ylinen LM, Towers GJ. The human and African green monkey TRIM5alpha genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc Natl Acad Sci U S A. 2004;101:10780–10785. doi: 10.1073/pnas.0402474101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Tipper C, Sodroski J. Role of TRIM5{alpha} RING Domain E3 Ubiquitin Ligase Activity in Capsid Disassembly, Reverse Transcription Blockade, and Restriction of Simian Immunodeficiency Virus. J Virol. 2011;85:8116–8132. doi: 10.1128/JVI.00341-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirmaier A, Wu F, Newman RM, Hall LR, Morgan JS, O’Connor S, Marx PA, Meythaler M, Goldstein S, Buckler-White A, Kaur A, Hirsch VM, Johnson WE. TRIM5 suppresses cross-species transmission of a primate immunodeficiency virus and selects for emergence of resistant variants in the new species. PLoS Biol. 2010:8. doi: 10.1371/journal.pbio.1000462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo N, Wolff B, Sekimoto T, Schreiner EP, Yoneda Y, Yanagida M, Horinouchi S, Yoshida M. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp Cell Res. 1998;242:540–547. doi: 10.1006/excr.1998.4136. [DOI] [PubMed] [Google Scholar]
- Kuersten S, Ohno M, Mattaj IW. Nucleocytoplasmic transport: Ran, beta and beyond. Trends Cell Biol. 2001;11:497–503. doi: 10.1016/s0962-8924(01)02144-4. [DOI] [PubMed] [Google Scholar]
- Langelier CR, Sandrin V, Eckert DM, Christensen DE, Chandrasekaran V, Alam SL, Aiken C, Olsen JC, Kar AK, Sodroski JG, Sundquist WI. Biochemical characterization of a recombinant TRIM5alpha protein that restricts human immunodeficiency virus type 1 replication. J Virol. 2008;82:11682–11694. doi: 10.1128/JVI.01562-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienlaf M, Hayashi F, Di Nunzio F, Tochio N, Kigawa T, Yokoyama S, Diaz-Griffero F. Contribution of E3-ubiquitin ligase activity to HIV-1 restriction by TRIM5alpha(rh): structure of the RING domain of TRIM5alpha. J Virol. 2011;85:8725–8737. doi: 10.1128/JVI.00497-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maegawa H, Miyamoto T, Sakuragi J, Shioda T, Nakayama EE. Contribution of RING domain to retrovirus restriction by TRIM5alpha depends on combination of host and virus. Virology. 399:212–220. doi: 10.1016/j.virol.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Mohr D, Frey S, Fischer T, Guttler T, Gorlich D. Characterisation of the passive permeability barrier of nuclear pore complexes. Embo J. 2009;28:2541–2553. doi: 10.1038/emboj.2009.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrosek M, Meier S, Ucurum-Fotiadis Z, von Castelmur E, Hedbom E, Lustig A, Grzesiek S, Labeit D, Labeit S, Mayans O. Structural analysis of B-Box 2 from MuRF1: identification of a novel self-association pattern in a RING-like fold. Biochemistry. 2008;47:10722–10730. doi: 10.1021/bi800733z. [DOI] [PubMed] [Google Scholar]
- Nakayama EE, Miyoshi H, Nagai Y, Shioda T. A specific region of 37 amino acid residues in the SPRY (B30.2) domain of African green monkey TRIM5alpha determines species-specific restriction of simian immunodeficiency virus SIVmac infection. J Virol. 2005;79:8870–8877. doi: 10.1128/JVI.79.14.8870-8877.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura S, Yap MW, Sheldon T, Stoye JP. All three variable regions of the TRIM5alpha B30.2 domain can contribute to the specificity of retrovirus restriction. J Virol. 2006;80:8554–8565. doi: 10.1128/JVI.00688-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens CM, Yang PC, Gottlinger H, Sodroski J. Human and simian immunodeficiency virus capsid proteins are major viral determinants of early, postentry replication blocks in simian cells. J Virol. 2003;77:726–731. doi: 10.1128/JVI.77.1.726-731.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Caballero D, Hatziioannou T, Yang A, Cowan S, Bieniasz PD. Human tripartite motif 5alpha domains responsible for retrovirus restriction activity and specificity. J Virol. 2005;79:8969–8978. doi: 10.1128/JVI.79.14.8969-8978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perron MJ, Stremlau M, Lee M, Javanbakht H, Song B, Sodroski J. The human TRIM5alpha restriction factor mediates accelerated uncoating of the N-tropic murine leukemia virus capsid. J Virol. 2007;81:2138–2148. doi: 10.1128/JVI.02318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, Riganelli D, Zanaria E, Messali S, Cainarca S, Guffanti A, Minucci S, Pelicci PG, Ballabio A. The tripartite motif family identifies cell compartments. Embo J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roa A, Hayashi F, Yang Y, Lienlaf M, Zhou J, Shi J, Watanabe S, Kigawa T, Yokoyama S, Aiken C, Diaz-Griffero F. RING domain mutations uncouple TRIM5alpha restriction of HIV-1 from inhibition of reverse transcription and acceleration of uncoating. J Virol. 2012;86:1717–1727. doi: 10.1128/JVI.05811-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SL, Wu LI, Emerman M, Malik HS. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc Natl Acad Sci U S A. 2005;102:2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayah DM, Sokolskaja E, Berthoux L, Luban J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature. 2004;430:569–573. doi: 10.1038/nature02777. [DOI] [PubMed] [Google Scholar]
- Sebastian S, Luban J. TRIM5alpha selectively binds a restriction-sensitive retroviral capsid. Retrovirology. 2005;2:40. doi: 10.1186/1742-4690-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B, Diaz-Griffero F, Park DH, Rogers T, Stremlau M, Sodroski J. TRIM5alpha association with cytoplasmic bodies is not required for antiretroviral activity. Virology. 2005a doi: 10.1016/j.virol.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Song B, Gold B, O’Huigin C, Javanbakht H, Li X, Stremlau M, Winkler C, Dean M, Sodroski J. The B30.2(SPRY) domain of the retroviral restriction factor TRIM5alpha exhibits lineage-specific length and sequence variation in primates. J Virol. 2005b;79:6111–6121. doi: 10.1128/JVI.79.10.6111-6121.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B, Javanbakht H, Perron M, Park DH, Stremlau M, Sodroski J. Retrovirus restriction by TRIM5alpha variants from Old World and New World primates. J Virol. 2005c;79:3930–3937. doi: 10.1128/JVI.79.7.3930-3937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- Stremlau M, Perron M, Lee M, Li Y, Song B, Javanbakht H, Diaz-Griffero F, Anderson DJ, Sundquist WI, Sodroski J. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc Natl Acad Sci U S A. 2006;103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stremlau M, Perron M, Welikala S, Sodroski J. Species-specific variation in the B30.2(SPRY) domain of TRIM5alpha determines the potency of human immunodeficiency virus restriction. J Virol. 2005;79:3139–3145. doi: 10.1128/JVI.79.5.3139-3145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YE, Pernet O, Lee B. Regulation of the nucleocytoplasmic trafficking of viral and cellular proteins by ubiquitin and small ubiquitin-related modifiers. Biol Cell. 2012;104:121–138. doi: 10.1111/boc.201100105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Fukuda M, Yoshida M, Yanagida M, Nishida E. Involvement of CRM1, a nuclear export receptor, in mRNA export in mammalian cells and fission yeast. Genes Cells. 1999;4:291–297. doi: 10.1046/j.1365-2443.1999.00259.x. [DOI] [PubMed] [Google Scholar]
- Wolff B, Sanglier JJ, Wang Y. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem Biol. 1997;4:139–147. doi: 10.1016/s1074-5521(97)90257-x. [DOI] [PubMed] [Google Scholar]
- Yamauchi K, Wada K, Tanji K, Tanaka M, Kamitani T. Ubiquitination of E3 ubiquitin ligase TRIM5 alpha and its potential role. FEBS J. 2008;275:1540–1555. doi: 10.1111/j.1742-4658.2008.06313.x. [DOI] [PubMed] [Google Scholar]
- Yap MW, Nisole S, Stoye JP. A single amino acid change in the SPRY domain of human Trim5alpha leads to HIV-1 restriction. Curr Biol. 2005;15:73–78. doi: 10.1016/j.cub.2004.12.042. [DOI] [PubMed] [Google Scholar]
- Yee JK, Friedmann T, Burns JC. Generation of high-titer pseudotyped retroviral vectors with very broad host range. Methods Cell Biol. 1994;43(Pt A):99–112. doi: 10.1016/s0091-679x(08)60600-7. [DOI] [PubMed] [Google Scholar]
- Yueh A, Leung J, Bhattacharyya S, Perrone LA, de los Santos K, Pu SY, Goff SP. Interaction of moloney murine leukemia virus capsid with Ubc9 and PIASy mediates SUMO-1 addition required early in infection. J Virol. 2006;80:342–352. doi: 10.1128/JVI.80.1.342-352.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.