Abstract
Sialic acids are a family of negatively charged monosaccharides which are commonly presented as the terminal residues in glycans of the glycoconjugates on eukaryotic cell surface or as components of capsular polysaccharides or lipooligosaccharides of some pathogenic bacteria. Due to their important biological and pathological functions, the biosynthesis, activation, transfer, breaking down, and recycle of sialic acids are attracting increasing attention. The understanding of the sialic acid metabolism in eukaryotes and bacteria leads to the development of metabolic engineering approaches for elucidating the important functions of sialic acid in mammalian systems and for large-scale production of sialosides using engineered bacterial cells. As the key enzymes in biosynthesis of sialylated structures, sialyltransferases have been continuously identified from various sources and characterized. Protein crystal structures of seven sialyltransferases have been reported. Wild-type sialyltransferases and their mutants have been applied with or without other sialoside biosynthetic enzymes for producing complex sialic acid-containing oligosaccharides and glycoconjugates. This mini-review focuses on current understanding and applications of sialic acid metabolism and sialyltransferases.
Keywords: carbohydrate, metabolism, sialic acid, sialoside, sialyltransferase
Introduction
Sialic acids are a family of α-keto acids with a nine-carbon backbone. More than 50 sialic acid forms have been found in nature including the most abundant N-acetylneuraminic acid (Neu5Ac), non-human N-glycolylneuraminic acid (Neu5Gc), 2-keto-3-deoxy-nonulosonic acid (or deaminoneuraminic acid) (Kdn), and their O-methyl, O-lactyl, O-sulfo, O-phospho-, single or multiple O-acetyl derivatives (Angata and Varki 2002; Schauer 2000; Chen and Varki 2010).
Sialic acids are commonly found as the terminal monosaccharides of the glycans presented in glycoconjugates (glycoproteins and glycolipids) on cell surfaces of vertebrates and higher invertebrates (Varki et al. 2011; Chen and Varki 2010). They are also components of lipooligosaccharides or capsular polysaccharides of some pathogenic bacteria including well-studied pathogens Escherichia coli K1, Haemophilus influenzae, Haemophilus ducreyi, Pasteurella multocida, Neisseria gonorrhoeae, Neisseria meningitidis, Campylobacter jejuni, and Streptococcus agalactiae (Almagro-Moreno and Boyd 2009; Vimr et al. 2004; Severi et al. 2007). Sialic acids play pivotal roles in many physiologically and pathologically important processes, including nervous system embryogenesis, cancer metastasis, immunological regulation, bacterial and viral infection, etc. (Angata and Varki 2002; Chen and Varki 2010).
Although sialic acid metabolism pathways differ in eukaryotes and bacteria, both involve the coordinated action of several enzymes that catalyze the biosynthesis, activation, and transfer of sialic acids for the formation of sialyl glycoconjugates, as well as modifications and degradation of sialyl glycoconjugates and sialic acids. Abnormal metabolism of sialic acid in human has been associated with various pathological conditions (Schwarzkopf et al. 2002). For example, mutation of human bifunctional enzyme GNE with both hydrolyzing uridine 5′-diphosphate-N-acetylglucosamine (UDP-GlcNAc) 2-epimerase and N-acetylmannosamine (ManNAc) kinase activities is related to two human disorders including sialuria (OMIM 269921) and hereditary inclusion body myopathy (HIBM, OMIM 600737) (Yardeni et al. 2011). Mutations of human lysosomal sialidase NEU1 have been related to the lysosomal storage disorder sialidosis (OMIM 256550) (Bonten et al. 1996; Pshezhetsky et al. 1997). In addition, normal metabolic incorporation of the non-human Neu5Gc from dietary sources (mainly red meat) to human tissues (mainly endothelia and epithelia) in the face of circulating anti-Neu5Gc antibodies led to chronic inflammation named xenosialitis (Varki et al. 2011).
Given the importance of sialic acids and the sequence and structural differences of human and bacterial enzymes involved in sialic acid metabolism, some enzymes are potential targets for drug development such as sialic acid synthases, CMP-sialic acid synthetases, sialyltransferases, sialidases, and sialic acid modification enzymes. In addition, although some human pathogenic bacteria (e.g. Vibrio cholerae, Anthrobacter ureafaciens, Clostridium perfringens, Salmonella typhimurium, and Streptococcus pneumoniae, etc.) (Chokhawala et al. 2007b), commensals (e.g. Bacteroides fragilis) (Thompson et al. 2009), probiotics (e.g. Bifidobacterium infantis) (Sela et al. 2011) and viruses (e.g. Newcastle disease virus and influenza virus) (von Itzstein 2007; Paulson et al. 1982) do not biosynthesize sialic acid or sialylglycoconjugates, they produce sialidases or neuraminidases (sialic acid-cleaving enzymes) which have been shown to be virulence factors, to provide nutrient, or to release the newly formed virons. Inhibitors against human influenza viruses generated by protein crystal structure-assisted rational drug design, such as Relenza (Zanamivir - ZMV) and Tamiflu (Oseltamivir - OTV), have been commercialized and used as effective anti-influenza virus drugs although recent emerging drug-resistant strains demand new anti-flu therapeutics (von Itzstein and Thomson 2009; Mitrasinovic 2010). A recent study showed that sialidase substrate specificity-based inhibitor design was also an effective approach for identification of selective inhibitors against certain sialidases (Li et al. 2011). Pathogenic bacterial sialidases have been shown to disrupt the repressive immune-regulation of sialic acid-based interaction and cause server damage of host tissues during bacterial sepsis. A cocktail of two bacterial sialidase inhibitors has been used to protect mice dying from sepsis in a Caecal Ligation and Puncture (CLP) model (Chen et al. 2011). Besides sialidases, the crystal structures of many other sialic acid metabolic enzymes have been reported. Nevertheless, a clear understanding of the significance of nature’s sialic acid structural diversity is still missing. This is mainly due to the analytical challenges in elucidating sialic acid-dependent interactions and synthetic difficulties in obtaining homogenous sialic acid-containing oligosaccharides and glycoconjugates, especially those contain diverse naturally occurring sialic acid modifications from natural sources (Yu and Chen 2007). Clearly, developing metabolic engineering approaches to characterize sialic acid-containing structures and sialic acid-binding proteins as well as establishing simple and efficient methods to synthesize sialylated structures in vitro are important to unravel the numerous biological roles of sialic acid and to assist drug sign.
This mini-review highlights current understanding of eukaryotic and bacterial sialic acid metabolic pathways and their applications in cell surface labeling and sialosides production via metabolic engineering. The natural functions of sialyltransferases and their applications in chemoenzymatic synthesis of various sialic acid-containing structures are also discussed.
Sialic acid metabolism
Among more than 50 different sialic acid forms that have been identified in nature, some are shared by bacteria, higher invertebrates, and vertebrates, while others have been identified exclusively in certain species. In addition, even for Neu5Ac, the most common sialic acid in nature, the metabolism pathways of bacteria and eukaryotes differ from each other, especially on the biosynthesis of sialic acids. In addition, the locations of enzymes involved in sialic acid activation and transfer as well as sialyl glycoconjugate degradation are different for bacteria and eukaryotes.
Sialic acid metabolism in eukaryotes
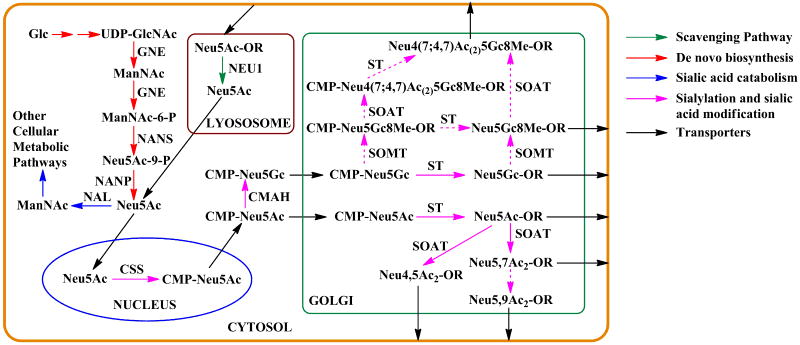
Sialic acid biosynthesis in vertebrates and higher invertebrates takes place in the cytosol involving three enzymes in a four-step process. The first two steps are catalyzed by a bifunctional enzyme called GNE with both hydrolyzing UDP-GlcNAc 2-epimerase and ManNAc kinase activities (Figure 1). The epimerase function of the GNE converts UDP-GlcNAc to ManNAc with removal of the UDP moiety and epimerization of the GlcNAc. The kinase function of GNE then phosphorylates ManNAc to form ManNAc-6-P. Neu5Ac is then produced by the condensation and dephosphorylation reactions catalyzed by Neu5Ac 9-phosphate synthase (NANS) and Neu5Ac-9-phosphate phosphatase (NANP), respectively (Varki and Schauer 2008).
Figure 1.
Sialic acid metabolism in eukaryotes. Abbreviations: Glc, glucose; UDP-GlcNAc, uridine 5′-diphospho-N-acetylglucosamine; ManNAc, N-acetylmannosamine; ManNAc-6-P, N-acetylmannosamine 6-phosphate; Neu5Ac-9-P, N-acetylneuraminic acid-9-phosphate; Neu5Ac, N-acetylneuraminic acid; CMP-Neu5Ac, cytidine 5′-monophospho-N-acetylneuraminic acid; CMP-Neu5Gc, cytidine 5′-monophospho-N-glycolylneuraminic acid; R, glycoprotein or glycolipid. Enzymes: GNE, hydrolyzing UDP-GlcNAc 2-epimerase/ManNAc-6-kinase; NANS, Neu5Ac-9-P synthetase; NANP, Neu5Ac-9-P phosphatase; NAL, N-acetylneuraminate lyase; CSS, CMP-sialic acid synthetase; CMAH, cytidine monophosphate-N-acetylneuraminic acid hydroxylase; ST, sialyltransferase; SOAT, sialate-O-acetyltransferase; SOMT, sialate-O-methyltransferase; NEU1, lysosomal sialidase.
Despite the differences in the biosynthesis of sialic acids in bacteria and eukaryotes, the sialic acid activation and transfer processes are conserved from bacteria through humans although the locations of enzymes differ. In eukaryotic cells, The Neu5Ac synthesized in the cytosol is transferred to nucleus and activated by cytosine 5′-monophosphate N-acetylneuraminic acid (CMP-Neu5Ac) synthetase (EC 2.7.7.43) to form CMP-Neu5Ac which then goes to Golgi to be used by sialyltransferases for the formation of glycoconjugates which are subsequently secreted or delivered to cell surface (Kean et al. 2004; Altheide et al. 2006).
Sialic acids on glycolipids and glycoproteins are released in lysosome by sialidases (e.g. human NEU1) as part of the overall degradation of these glycoconjugates and are pumped back into the cytosol, where they can go through another cycle of sialyl glycoconjugate production or be broken down by Neu5Ac lyase (or sialic acid aldolase) to form ManNAc and pyruvate (Verheijen et al. 1999). Multiple sialidases have been found in human cells. Other than the lysosomal sialidase NEU1 (Pshezhetsky et al. 1997), three additional human sialidases including the cytosolic sialidase NEU2 (Monti et al. 1999), the plasma membrane-associated sialidase NEU3 (Wada et al. 1999), and the lysosomal or mitochondrial membrane-associated sialidase NEU4 (Monti et al. 2004) have been identified. These four human sialidases showed different substrate specificities and physiological functions (Miyagi 2008b). Aberrant sialylation is closely associated with the malignant phenotype of cancer cells including metastatic potential and invasiveness (Miyagi 2008a; Miyagi et al. 2004).
The de novo biosynthesis CMP-Kdn is believed to start from mannose and follow a similar route as CMP-Neu5Ac from ManNAc although it is not as well elucidated (Angata and Varki 2002; Terada et al. 1993; Angata et al. 1994; Angata et al. 1999).
The biosynthesis of sialyl glycoconjugates, however, is complicated by the additional modifications on sialic acid either before or after the formation of sialyl linkages in the Golgi. One such example is the non-human Neu5Gc. In animals including nonhuman hominids (NHHs, previously great apes) such as chimpanzees, bonobos, gorillas, and orangutans, CMP-Neu5Ac is converted to CMP-Neu5Gc in the cytosol by Neu5Ac hydroxylase encoded by the Cmah gene. In contrast, humans do not synthesize CMP-Neu5Gc due to the inactivation of CMP-Neu5Ac hydroxylase by a frame-shift mutation of the CMAH gene. Nevertheless, non-human Neu5Gc can be metabolically incorporated from dietary sources (mainly red meat)to human tissues (mainly endothelia and epithelia) in the face of circulating anti-Neu5Gc antibodies (Varki et al. 2011; Taylor et al. 2010; Pham et al. 2009; Padler-Karavani et al. 2008). Neu5Gc-containing glycoconjugates have also been found in the sera of cancer patients, human cancerous tissues, cultured human cell lines, and recombinant therapeutic glycoproteins (Inoue et al. 2010; Ghaderi et al. 2010). Human anti-Neu5Gc antibodies against Neu5Gc-sialyl Tn antigen have been identified as novel serum biomarkers and immunotherapeutics in human cancer based on studies using a novel sialoside glycan microarray (Padler-Karavani et al. 2011).
Another common sialic acid modification is O-acetylation. It is one of the most frequent modifications of sialic acids in eukaryotes cells, and evidence has shown that sialic acid O-acetylation on glycoconjugates in the Golgi involves both an acetyl-CoA transporter and an intraluminal O-acetyltransferase (Varki and Diaz 1985; Diaz et al. 1989; Higa et al. 1989). C7- and C9-O-acetylation have been shown in bovine submandibular gland (Vandamme-Feldhaus and Schauer 1998; Lrhorfi et al. 2007) and C4-O-acetylation has been observed in microsomes from equine submandibular glands (Tiralongo et al. 2000). Human colon mucosa is also a rich source of O-acetylated sialic acids and the level of O-acetylation is reduced significantly in colorectal cancer (Corfield et al. 1999). Modifications of Neu5Gc have been shown in salmonid fish egg glycoproteins (Sato et al. 1993). Sialate-O-acetyltransferases (SOATs) and sialate-O-acetylesterases (SOAEs) are responsible for adding and removal of O-acetyl groups, respectively (Shen et al. 2004a; Srinivasan and Schauer 2009). In animals, the best characterized sialate-O-acetyltransferases (SOATs) are sialate-4-O-acetyltransferase in guinea pig liver (Iwersen et al. 1998; Iwersen et al. 2003) and sialate-7(9)-O-acetyltransferase studied in rat liver (Higa et al. 1989), human colon (Shen et al. 2004a), and bovine submandibular gland (Lrhorfi et al. 2007). The C9-O-acetyl sialic acid is believed to be formed by the migration of the C7-O-acetyl group on glycosidically bound sialic acids, most likely also catalyzed by an enzyme (Vandamme-Feldhaus and Schauer 1998) (Figure 1). The amount of O-acetylated sialic acid in a special tissue (e.g. human colon mucosa) or cell is believed to be dependent on the activities of both SOATs and SOAEs (Shen et al. 2004b). A positive correlation between the increased SOAT activity and the enhanced expression of Neu5,9Ac2-glycoconjugates is seen in the microsomes of lymphoblasts (Mandal et al. 2009). In addition, SOAE activity is decreased in both lysosomal and cytosolic fractions of acute lymphoblastic leukemia cell lines (Mandal et al. 2012). Human CasD1 gene, encoding a protein with a serine-glycine-asparagine-histidine hydrolase domain and a hydrophobic transmembrane domain, is believed to be involved in O-acetylation of α2–8-linked sialic acids (Arming et al. 2011).
Other than O-acetylation, O-methylation of sialic acids (mainly on lower invertebrates such as starfish) has also been well studied (Bergwerff et al. 1992; Zanetta et al. 2006; Kelm et al. 1998). In starfish Asterias rubens gonads, most Neu5Gc residues are 8-methylated in addition to C4- and/or C7-O-acetylation (Zanetta et al. 2006). Free Neu5Ac and Neu5Gc can be methylated, although those presented on oligosaccharides and glycoproteins are better substrates for enzymatic methylation (Kelm et al. 1998) (Figure 1).
Different from the de novo biosynthesis of sialic acid in most eukaryotes, some protozoa species, such as Trypanosoma cruzi (a causive agent of Chagas disease or American trypanosomiasis), use a surface α2–3-trans-sialidase (EC 2.4.1.-) to transfer α2–3-linked sialic acid residues directly from host sialyl glycoconjugates to the terminal β-galactose residues of the parasite mucins and form their own surface sialyl glycoconjugates. In comparison, a related American parasite Trypanosoma rangeli secretes a homologous sialidase but does not express trans-sialidase (Buschiazzo et al. 1997; Montagna et al. 2006). Two trans-sialidase (TS) forms (TS-1 and TS-2) have been purified from procyclic Trypanosoma congolense (a causive agent of animal African trypanosomiasis) cultures. The TS-1 form has higher TS activity and significantly less sialidase activity, whereas sialidase activity was predominately found in TS-2 form (Tiralongo et al. 2003). Two TS1 variants have been cloned and showed activity in sialylating asialofetuin (Koliwer-Brandl et al. 2011). Interestingly, the procyclic stage of Trypanosoma brucei (a human African trypanosome that causes sleeping sickness) in the insect vector expresses a surface α2–3-trans-sialidase (TbTS) and an α2–3-sialidase separately (Montagna et al. 2006). More recently, a second catalytically active α2–3-trans-sialidase has also been identified from T. brucei (Nakatani et al. 2011). The activity of trans-sialidase is crucial for the pathogenesis of the parasites (Eugenia Giorgi and de Lederkremer 2011). Together with majority of eukaryotic and bacterial exo-sialidases, all trans-sialidases have been grouped into the glycosidase hydrolase family GH33 in the Carbohydrate-Active enZymes (CAZy) database (URL: http://www.cazy.org) based on protein sequence homology (Henrissat 1991; Davies and Henrissat 1995). The most well studied trans-sialidase is TcTS which is a glycosylphosphatidylinositol-anchored protein that is also shed into the milieu (Sartor et al. 2010). Its crystal structures have been reported (Buschiazzo et al. 2002; Amaya et al. 2004). Like exo-sialidases, TcTS follows a double displacement mechanism with the formation of an enzyme-substrate intermediate and an overall retention of the anomeric carbon stereo-configuration of the sialic acid (Amaya et al. 2004; Damager et al. 2008; Watts et al. 2003). Tyr342 was identified as the nucleophile that attacks the anomeric carbon of the sialic acid and forms the enzyme-substrate intermediate (Watts et al. 2003). A Tyr342 to histidine mutation causes the inactivation of TcTSs and has been commonly observed for some T. cruzi strains (Cremona et al. 1996; Oppezzo et al. 2011). Due to its important roles in parasite infection and its functional difference from sialyltransferases or sialidases in human, TcTS is an attractive drug target (Sartor et al. 2010). Recently, a new generation of TcTS inhibitors have been designed (Buchini et al. 2008) and a highly specific high affinity (subnanomolar) TcTS neutralizing mouse monoclonal antibody (mAb 13G9) has been identified (Buschiazzo et al. 2012). TbTS has also been suggested as a target for DNA vaccine development (Silva et al. 2009). An antibody (mAb 7/23) specifically against T. congolense TS-1 form is also available (Tiralongo et al. 2003).
Metabolic engineering of vertebrate cell surface sialic acids
The understanding of sialic acid metabolic pathways in eukaryotes leads to the development of metabolic engineering approaches for labeling and functional studies of sialic acid-containing glycoconjugates and sialic acid-binding proteins. Sialic acids modified with a small chemical handle (e.g. ketone, azide, alkyne, diazirine or thiol) have been incorporated via metabolic engineering onto vertebrate cell surface in cell culture or in living animals and allow later detections using a bioorthogonal ligand conjugated to a fluorophore, a biotin molecule, or an antigen that can be recognized by specific antibodies. In addition to in vitro and in vivo imaging and characterization of sialic acid-containing glycoconjugates using N-levulinoylmannosamine and N-azidoacetylmannosamine (ManNAz) as sialic acid precursors for metabolic engineering (Mahal et al. 1997; Prescher et al. 2004), a sialic acid analog with an N-butanoyl group has been used to inhibit expression of α2–8-linked polysialic acids on cell surface (Mahal et al. 2001). Neu5Ac analogs with N-propionyl, N-iso-butanoyl, N-phenylacetyl derivatives of Neu5Ac have also been used to improve the immnuogenicity of glycan-based cancer vaccine (Krug et al. 2004; Chefalo et al. 2006; Wu and Guo 2006). Four processes have been used to metabolically incorporate sialic acid analogs onto cell surface. A less used approach is to start with GlcNAc analogs, which can form UDP-GlcNAc analogs to be used by sialic acid de novo synthetic pathway. It turns out that the strict substrate specificity of GNE limits the diversity of sialic acid analogs that can be generated using this approach (Yarema and Bertozzi 1998; Tanaka and Kohler 2008). A more frequently used approach is to start with ManNAc analogs, which are usually per-acetylated to help their admission into cells. For this approach, the Neu5Ac 9-phosphate synthase (NANS) is the bottleneck process and its substrate specificity excludes the use of long or branched N-acyl ManNAc (Jacobs et al. 2001; Viswanathan et al. 2003) or C6-modified ManNAc derivatives (Lawrence et al. 2000). Another frequently used approach is to start with per-acetylated sialic acid analogs to bypass the substrate limitations of GNE and NANS. A minor drawback is the relatively higher cost or increased difficulties of synthesizing sialic acid analogs comparing to GlcNAc or ManNAc analogs. The least used approach is to start with CMP-sialic acid analogs as the synthesis of such compounds is more time consuming and challenging. Besides the non-natural sialic acids, analogs of other common monosaccharides found in eukaryotes have also been successfully incorporated into the glycoconjugates on cell surface by metabolic glycoengineering methodologies. For more information on metabolic engineering of sialic acid and other sugars such as N-acetylgalactosamine (GalNAc), N-acetylglucosamine (GlcNAc), and fucose, readers are directed to two excellent reviews (Campbell et al. 2007; Du et al. 2009).
Bacterial sialic acid metabolism
Most bacteria are unable to synthesize sialic acid except for a limited number of pathogenic bacteria and commensals of which majority are related to human (Crocker and Varki 2001). Some pathogenic bacteria can coat themselves with sialic acid, protecting themselves from the detection of host immune system by regulating complement interaction as well as down regulate adaptive and innate immune responses (Carlin et al. 2009; Varki et al. 2011). Some bacteria use sialic acid as a nutrient (Severi et al. 2007).
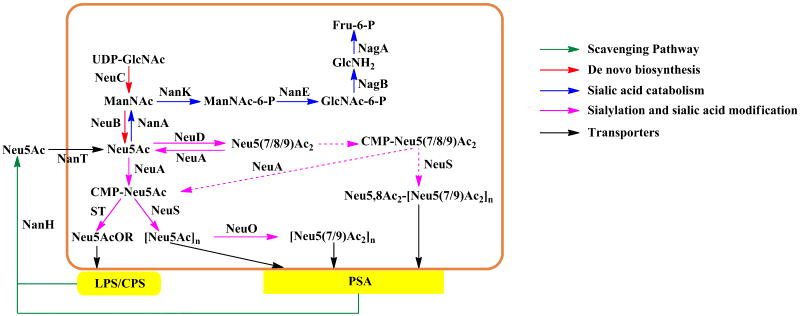
Bacterial pathogens have evolved two ways to obtain sialic acid: de novo and scavenging pathways. Similar to that of eukaryotes, the de novo pathway of bacteria (e.g. E coli K1, N. meningitidis, C jejuni, and S agalactiae) begins with the conversion of UDP-GlcNAc to ManNAc by hydrolyzing UDP-GlcNAc 2-epimerase (NeuC) (Figure 2). Different from eukaryotes, ManNAc produced by bacteria is directly used in the presence of phosphoenolpyruvate (PEP) by Neu5Ac synthase (NeuB) for the formation of Neu5Ac and inorganic phosphate without the involvement of a kinase or a phosphatase (Bravo et al. 2004; Angata and Varki 2002). The scavenging pathway involves two scenarios: donor scavenging (only found in Neisseria gonorrheae) in which CMP-sialic acid is scavenged from the hosts (Parsons et al. 1994), and precursor scavenging (e.g. H. influenzae, H. ducreyi, Haemophilus somnus, and P. multocida belonging to the Haemophilus-Actinobacillus-Pasteurella or HAP group) in which free sialic acid is obtained directly from the host (Steenbergen et al. 2005; Schilling et al. 2001).
Figure 2.
Bacterial sialic acid metabolism using E. coli K1 as a model system. Abbreviations: UDP-GlcNAc, uridine 5′-diphosphate-N-acetylglucosamine; ManNAc, N-acetylmannosamine; Neu5Ac, N-acetylneuraminic acid; CMP-Neu5Ac, cytidine 5′-monophospho-N-acetylneuraminic acid; GlcNAc, N-acetylglucosamine; Fru, fructose; LPS, lipopolysaccharide; CPS, capsule polysaccharide; PSA, polysialic acid; R, LPS or CPS. Enzymes: NeuC, hydrolyzing UDP-GlcNAc 2-epimerase; NeuB, sialic acid synthase; NanA, Sialic acid aldolase; NeuA, CMP-sialic acid synthetase (NeuA in E. coli K1 and Streptococcus agalactiae also possesses O-acetylesterase activity); NeuD, sialic acid O-acetyltransferase; ST, sialyltransferase; NeuS, polysialyltransferase; NeuO, polysialic acid O-acetyltransferase; NanK, ManNAc kinase; NanE, ManNAc-6-phosphate epimerase; NagB, GlcNAc-6-phosphate deacetylase; NagA, glucosamine-6-phosphate deaminase; NanH, sialidase; NanT, Neu5Ac transporter.
Similar to eukaryotes, sialylation in bacteria is mainly catalyzed by sialyltransferases (STs) and sialic acid catabolism is carried out by sialidases and Neu5Ac lyase (or sialic acid aldolases, NanA in E. coli K1 and K12) which catalyzes the breakdown of Neu5Ac to form ManNAc and pyruvate (Vimr et al. 2004).
In some bacteria (e.g. C. jejuni, E. coli K1, and S. agalactiae), sialic acid can be further modified, such as by O-acetylation. For example, the terminal sialic acid on the di-sialylated LPS in C. jejuni can be modified by an O-acetyltransferase identified recently (Houliston et al. 2006). Both E. coli K1 and S. agalactiae (or group B Streptococcus, GBS) have sialic acid O-acetyltransferases and the C-terminal sequence of their CMP-sialic acid synthetases has sialic acid O-acetylesterase activity (Steenbergen et al. 2006; Lewis et al. 2007). In vitro studies showed that GBS CMP-sialic acid synthetase (NeuA) de-O-acetylated sialic acid by two alternate pathways: de-O-acetylation of Neu5,9Ac2 followed by CMP activation of Neu5Ac, and activation of Neu5,9Ac2 followed by de-O-acetylation of CMP-Neu5,9Ac2 (Lewis et al. 2007). In comparison, N. meningitidis has a shorter CMP-sialic acid synthetase lacking the C-terminal sialic acid O-acetylesterase activity in E. coli K1 and GBS enzymes. Different from O-acetylation of sialic acids on oligosaccharide moieties of glycoconjugates, separate pathways are proposed for sialic acid-containing polymers. For example, O-acetylation at C-7 and C-9 of the sialic acid in polysialic acid (PSA) capsules is catalyzed by NeuO in E. coli K1 and OatC in N. meningitidis serogroup B. A minor NeuO-independent, NeuD-dependent, O-acetylation pathway with the involvement of NeuA and NeuS is also proposed (Steenbergen et al. 2006). For more information about sialic acid metabolism and function in bacterial pathogens, readers are referred to two excellent reviews (Vimr et al. 2004; Severi et al. 2007).
Production of sialosides by engineering bacterial sialic acid metabolic pathways
The understanding of sialic acid metabolic pathways in sialic acid-producing bacteria has helped to develop metabolic engineering approaches for large-scale production of sialosides using whole cell catalysts or by fermenting living cells (so called the living factory approach) (Chen and Varki 2010). Large-scale production of 3′-sialyllactose (Endo et al. 2000) and the carbohydrate portion of the sialyl-Tn epitope, Neu5Ac α2–6GalNAc (Endo et al. 2001), has been achieved using the whole cell catalysts strategy. This strategy uses a UTP/CDP-producing Corynebacterium ammoniagenes strain, and three E. coli strains harboring plasmids encoding a CTP synthetase, a CMP-Neu5Ac synthetase, and a suitable sialyltransferase, respectively. All cells were grown and collected separately. They were permeablized and combined in one-pot for sialoside production. In comparison, fermenting living cells engineered by adding suitable sialic acid biosynthetic genes and eliminating genes involved in divergent pathways seems to be more convenient and cost-effective. An earlier version of this approach was reported in 2002 for large-scale production of 3′-sialyllactose using a lacZ− E. coli strain engineered by inactivating the endogenous sialic acid aldolase gene (nanA−) and adding plasmids containing N. meningitidis CMP-Neu5Ac synthase gene (neuA) and N. meningitidis α2–3-sialyltransferase gene respectively. In this system, relatively expensive sialic acid was added exogenously and transported into the cells by endogenous permease NanT for the production of the target sialoside (Priem et al. 2002). A similar system with additional glycosyltransferase genes has been generated for large-scale synthesis of GM1 and GM2 oligosaccharides (Antoine et al. 2003; Fort et al. 2005). An improved more economic version of 3′-sialyllactose-production strain eliminates the need of adding exogenous sialic acid by knocking out the endogenous ManNAc kinase (nanK−) gene in lacZ− E. coli K12 strains devoid of sialic acid aldolase (nanA−) and introducing additional genes including neuC and neuB from C. jejuni in addition to the previously introduced neuA and α2–3-sialyltransferase genes. This strategy allows the bacterium to generate sialic acid from endogenous UDP-GlcNAc produced through its own metabolism (Fierfort and Samain 2008). Recently, this improved strategy has been used to produce 6′-sialyllactose, 6,6′-disialyllactose, and 6′-Kdo-lactose with metabolically engineered E. coli harboring a sialyltransferase from Photobacterium sp. JT-ISH-224 (Drouillard et al. 2010).
Sialyltransferases
As described above, sialyltransferases (STs) (EC 2.4.99.X) are key enzymes in the biosynthesis of sialic acid-containing oligosaccharides and glycoconjugates (Harduin-Lepers et al. 2005; Harduin-Lepers et al. 1995). They catalyze the reaction that transfers a sialic acid residue from its activated sugar nucleotide donor cytidine 5′-monophosphate sialic acid (CMP-Sia) to a variety of acceptor molecules, usually a structure terminated with a galactose (Gal), an N-acetylgalactosamine (GalNAc), or another sialic acid (Sia) residue (Chen and Varki 2010).
Classification of sialyltransferases
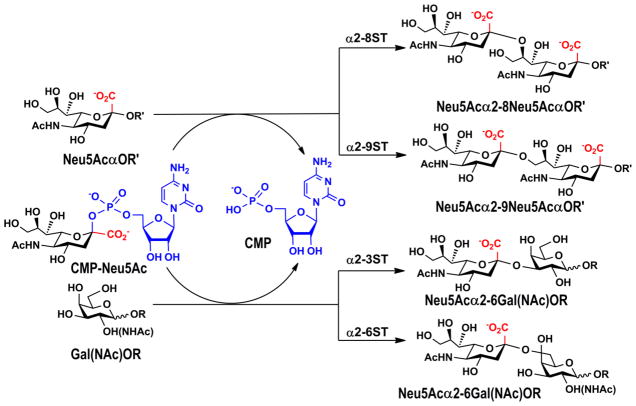
Based on the linkages that they form, common sialyltransferases have been classified to α2–3-, α2–6-, α2–8-sialyltransferases, and polysialyltransferases. In human and animals, α2–3- and α2–6-sialyltransferases can modify a numbers of core glycan structures on different proteins, while polysialyltransferases that catalyze the formation of α2–8-linked sialic acid homopolymers appear to be highly selective for their protein carriers. Other than autopolysialylation of ST8Sia IV and ST8Sia II, only four other protein carriers have been identified for ST8Sia IV and ST8Sia II: the neural cell adhesion molecule (NCAM, also called CD56), the α-subunit of voltage-gated sodium channel, CD36, and neuropilin (Drake et al. 2008). In bacteria, polysialyltransferases that catalyze the formation of α2–8- and/or α2–9-linked capsular polysaccharides of N. meningitidis serogroups B and C, E. coli K1 and K92 strains have been identified (Steenbergen and Vimr 2003). Examples of the products formed by different sialyltransferases are shown in Figure 3. In addition, capsular polysaccharide polymerases that contain both sialyltransferase and hexosyltransferase activities for the construction of sialic acid-containing heteropolymeric capsular polysaccharides of N. meningitidis serogroups W-135 and Y are also presented in nature (Claus et al. 2009).
Figure 3.
Sialyltransferases catalyze the transfer of sialic acid from CMP-sialic acid to a suitable acceptor, commonly a Gal, GalNAc, or Sia terminated glycan or glycoconjugates. The most common sialic acid form, Neu5Ac, is shown as an example. R, glycans or glycoconjugates; R′, Neu5Ac- or Gal-terminated glycans or glycoconjugates.
Based on their protein sequence homology, all known sialyltransferase have been classified into six glycosyltransferase (GT) families (Thon et al. 2011) in the Carbohydrate-Active enZymes (CAZy) database (URL: http://www.cazy.org) (Campbell et al. 1997; Coutinho et al. 2003). All sialyltransferases and polysialyltransferases from eukaryotes and viruses are grouped into glycosyltransferase family 29 (GT 29). Bacterial STs have been grouped into GT4, GT38, GT42, GT52, and GT80 five CAZy GT families. More details are discussed below for eukaryotic and bacterial sialyltransferases respectively.
Eukaryotic sialyltransferases
Together with some viral sialyltransferases, all identified eukaryotic sialyltransferases share sequence homology and belong to CAZy GT29 family (Harduin-Lepers et al. 2001; Harduin-Lepers et al. 2005; Harduin-Lepers 2010). Due to the diversities of the sialyl linkages that they form and the acceptor substrates that they recognize, multiple sialyltransferases are produced by single species of eukaryotes. For example, twenty human sialyltransferases and polysialyltransferases have been identified and are classified into four groups according to the type of linkage formed and the nature of the sugar acceptor. These include six beta-galactoside α2–3-sialyltransferases (ST3Gal I-VI), two beta-galactoside α2–6-sialyltransferases (ST6Gal-I–II), six GalNAc α2–6-sialyltransferases (ST6GalNAc-I–VI), and six α2–8-sialyltransferases (ST8Sia-I–VI, among which ST8Sia-II and ST8Sia-IV are polysialyltransferases). The sialyltransferases in the same group share some overlapping but not identical acceptor substrate specificities. The presence of such a large number of sialyltransferases in human and other animals is another indication of the important roles of sialic acid-containing structures. Mammalian STs have quite strict acceptor substrate specificity but relatively more relaxed donor substrate specificity (Harduin-Lepers 2010; Datta 2009) although their donor substrate specificities have been less explored. CMP-Neu5Gc, CMP-Neu5,9Ac2 (Higa and Paulson 1985) and CMP-Kdn (Angata et al. 1998) have shown to be acceptable donor substrates by some mammalian sialyltransferases.
Like other vertebrate glycosyltransferases, mammalian sialyltransferases are type II membrane proteins. They are localized in Golgi and have an N-terminal cytoplasmic domain, a single transmembrane domain of 16–20 amino acid residues, a size-variable (20–300 amino acid residues) stem region and a soluble relatively well conserved C-terminal catalytic domain of 300±20 amino acid residues in Golgi lumen (Audry et al. 2011; Chen and Varki 2010). The catalytic domains of mammalian sialyltransferases and a viral sialyltransferase vST3Gal-I that can use fucose-containing Lewis × antigens as acceptors (Sugiarto et al. 2011b) have four conserved sialyl motifs including long (L), short (S), very small (VS) motifs, and motif 3 (Datta and Paulson 1995; Datta et al. 1998; Jeanneau et al. 2004). A disulfide bond stabilizing the L- and the S-motifs is also conserved among these sialyltransferases (Datta et al. 2001). Site-directed mutagenesis and structures of porcine ST3Gal-I show that the L-motif is involved in the donor binding, motifs 3 and VS contribute to the binding of the acceptor, and the S-motif participates in the binding of both donor and acceptor substrates (Datta 2009; Audry et al. 2011; Rao et al. 2009; Paulson and Rademacher 2009). A conserved histidine residue located in the VS-motif has been identified as the catalytic base from the x-ray crystal structures of porcine ST3Gal-I which have the GT-A fold with a single Rossmann domain (Rao et al. 2009).
Bacterial sialyltransferases
Bacterial sialyltransferases have been mainly identified and characterized from several pathogenic bacteria including N. meningitidis, N. gonorrheae, C. jejuni, H. influenzae and H. ducreyi, P. multocida, S.agalactiae, and some marine bacteria. Some of these bacteria including N. meningitidis, H. influenzae, H. ducreyi, P. multocida, and Photobacterium species JT-ISH-224 have multiple sialyltransferase genes. Different from mammalian sialyltransferases which are commonly monofunctional sialyltransferases, many bacterial sialyltransferases have multiple functions including sialyltransferase activities responsible for forming different sialyl linkages with or without additional sialidase and trans-sialidase activities (Chen and Varki 2010).
Unlike vertebrate and viral sialyltransferases which share protein sequence homology and all belong to CAZy GT29 family, bacterial sialyltransferases have less conserved protein sequences and are distributed into five CAZy GT families. The highly homologous capsular polysaccharide (CPS) polymerases (SiaDs) of N. meningitidis serogroups W135 and Y which have both hexosyltransferase (α1–4-galactosyltransferase activity for SiaDW135 and α1–4-glucosyltransferase activity for SiaDY) and sialyltransferase activities responsible for the synthesis of sialic acid-containing heteropolymeric CPSs [−6Gal/Glcα1–4Neu5Acα2-]n belong to GT4 along with other glycosyltransferases. In comparison, E. coli K1 α2–8-polysialyltransferase (NeuS), E. coli K92 alternating α2–8/9-polysialyltransferase (Vimr et al. 1992; Steenbergen et al. 1992; Shen et al. 1999), N. meningitidis serogroup B α2–8-polysialyltransferase (SiaD) encoded by siaD/synD, and N. meningitidis serogroup C α2–9-polysialyltransferase encoded by synE are grouped into GT38 (Peterson et al. 2011). GT42 includes α2–3-sialyltransferases (Cst-I and Cst-III) and a multifunctional α2–3/8-sialyltransferase (Cst-II) which also has α2–8-sialidase and α2–8-trans-sialidase activities (Cheng et al. 2008) from C. jejuni (Gilbert et al. 2002; Gilbert et al. 2000), a P. multocida α2–3-sialyltransferase (PmST3) encoded by Pm1174 gene (Thon et al. 2012), as well as an α2–3-sialyltransferase encoded by lic3A gene (Harrison et al. 2005) and a multifunctional α2–3/8-sialyltransferase (Lic3B) from H. influenzae (Fox et al. 2006). GT52 family contains characterized α2–3/6-sialyltransferases (Lsts) from N. meningitidis and N. gonorrhoeae (Gilbert et al. 1996), an H. influenzae α2–3-sialyltransferase (LsgB) (Jones et al. 2002), a H. ducreyi α2–3-sialyltransferase (Lst encoded by Hd0686 gene) (Bozue et al. 1999), and a recently reported P. multocida glycolipid α2–3-sialyltransferase (PmST2) encoded by Pm0508 gene (Thon et al. 2011). CpsK, another member of GT52 family and a homolog to the Lst of H. ducreyi, has also been identified as a putative α2–3-sialylatransferase for the synthesis of sialic acid-terminated capsular polysaccharide of S. agalactiae (Group B Streptococcus) (Chaffin et al. 2002). GT80 family contains bacterial α2–3- and/or α2–6-sialyltransferases including a multifunctional P. multocida α2–3/6-sialyltransferase (PmST1) encoded by Pm0188 gene which also has α2–3-sialidase and α2–3-trans-sialidase activities (Yu et al. 2005), an H. ducreyi α2–3-sialyltransferase (Hd2,3ST encoded by Hd0053 gene) (Li et al. 2007), and several marine bacterial sialyltransferases such as a Photobacterium damselae α2–6-sialyltransferase (Pd2,6ST or JT0160 Bst) (Yamamoto et al. 1998; Sun et al. 2008) which also has α2–6-sialidase and α2–6-trans-sialidase activity (Cheng et al. 2010), Photobacterium leiognathi α2–6-sialyltransferases with (Mine et al. 2010) or without additional α2–6-sialidase activity (Yamamoto et al. 2007), Photobacterium phosphoreum α2–3-sialyltransferase (Tsukamoto et al. 2007), an α2–3-sialyltransferase from Vibrio species (Takakura et al. 2007), as well as α2–3- and α2–6-sialyltransferases from Photobacterium species JT-ISH-224 (Tsukamoto et al. 2008).
Crystal structures of sialyltransferases
Despite the diversity of their primary protein sequences (grouped into 94 classified and one non-classified CAZy GT families), the tertiary structures of all glycosyltransferases characterized so far fall into only two structural folds: GT-A and GT-B folds. The GT-A fold consists of a single nucleotide-binding Rossmann domain with a typical α/β/α sandwich topology and a smaller fold. Although GT-A enzymes are commonly metal-ion-dependent and contain a conserved Asp-x-Asp (DxD) or equivalent motif that is crucial for catalysis, sialyltransferases usually do not require metal ion for catalysis and GT-A fold sialyltransferases have been identified as two variants that do not have the metal-binding DxD conserved motif (Chiu et al. 2004; Chiu et al. 2007). The GT-B fold consists of two separate Rossmann domains with a connecting linker region and a substrate binding site located in the cleft between the two domains. Although divalent cations may be required for full activity of some non-sialyltransferase GT-B enzymes, a bound metal ion associated with catalysis has not been seen in the GT-B fold glycosyltransferase structures characterized so far (Audry et al. 2011; Buschiazzo and Alzari 2008).
The crystal structures of seven sialyltransferases become available since the first report of the crystal structures of a bacterial multifunctional sialyltransferase Cst-II (a GT42 sialyltransferase) in 2004, ten years after the first report of the crystal structures of a glycosyltransferase (Vrielink et al. 1994). These sialyltransferases span four sialyltransferase GT families including GT29, GT42, GT52, and GT80. The crystal structures of capsular polysaccharide (CPS) polymerases (SiaDs) of N. meningitidis serogroups W135 and Y in GT4 family and any of the polysialyltransferases in GT38 family are still unknown.
The first crystal structure of the ST reported in 2004 for bacterial Cst-II from C. jejuni (GT42) belongs to the GT-A family (variant 1) (Chiu et al. 2004). Another C. jejuni sialyltransferase Cst-I (GT42) adopts a similar GT-A (variant 1) fold (Chiu et al. 2007). Both Cst-I and Cst-II are tetrameric. The recently reported crystal structures of porcine ST3Gal-I (GT29) adopts a second distinct GT-A variant (variant 2) (Rao et al. 2009). The multifunctional bacterial ST, PmST1 (GT80) from P. multocida was the second structure of a ST to be reported which belongs to the GT-B structural superfamily (Ni et al. 2006). In the same GT80 family, the crystal structures of two other STs sharing the GT-B fold, an α2–6ST from Photobacterium sp. JT-ISH-224 in complex with CMP and lactose (Kakuta et al. 2008) and an α2–3ST from P. phosphoreum in complex with CMP (Iwatani et al. 2009), have also been reported. Very recently, the first sialyltransferase structure of GT52 family for a membrane associated α2–3/6 lipooligosaccharide sialyltransferase from N. meningitidis serotype L1 (NST) adopting a GT-B fold, has been solved (Lin et al. 2011).
Despite their sequence and structural differences, all sialyltransferases characterized to-date are inverting glycosyltransferases which catalyze the formation of α-sialyl linkage in the product from β-linked sialic acid in CMP-sialic acid donor substrate. The sialyltransferase is believed to follow a single displacement mechanism that involves nucleophilic attack of the acceptor hydroxyl (activated by a catalytic base in the enzyme) to the C2 anomeric center of the sialic acid moiety in the donor substrate with inversion of stereochemistry. In STs of GT29 and GT42, a histidine residue [H319 in pST3Gal-I (Rao et al. 2009), H202 in Cst-I (Chiu et al. 2007), and H188 in Cst-II (Chan et al. 2009)] serves as a catalytic base to deprotonate the reactive oxygen of the acceptor, while the most likely candidate for the catalytic base is an aspartic acid residue in the STs of GT80 [D141 in Δ24PmST1 (Ni et al. 2007)] and GT52 [D258 in NST (Lin et al. 2011)].
The inverting reaction of glycosyltransferases follows a SN2-like mechanism (Lairson et al. 2008) and involves the formation of an oxocarbenium-like transition state with the concomitant departure of the nucleotide leaving group. In Cst-I and Cst-II, two conserved tyrosine residues [Y171 and Y177 in Cst-I (Chiu et al. 2007), Y156 and Y162 in Cst-II (Chiu et al. 2004)] are involved in the departure of the CMP group of CMP-Neu5Ac, while in GT29, GT52, and GT80 STs, a conserved histidine residue [H302 in pST3GalI (Rao et al. 2009), H280 in NST (Lin et al. 2011), and H311 in Δ24PmST1 (Ni et al. 2007)] is involved in stabilizing the phosphate of the departing CMP.
Sialyltransferase mutants by crystal structure-based design or directed evolution
The structures of PmST1, a multifunctional highly active α2–3-sialyltransferase (pH 6.0–10.0) with α2–3-sialidase (pH = 5.0–5.5), α2–3-trans-sialidase (pH = 5.5–6.5), and α2–6-sialyltransferase (pH =4.5–7.0) activities, in the presence or the absence of CMP, CMP-3F(axial)Neu5Ac, CMP-3F(equatorial)Neu5Ac and/or lactose provide a good understanding of the mechanism and the key residues involved in the sialyltransferase and the sialidase activities of the enzyme and allow the rational design of a PmST1 double mutant E271F/R313Y with significantly decreased α2–3-sialidase activity without affecting its α2–3-sialyltransferase activity (Sugiarto et al. 2011a). Besides using the site-directed mutagenesis methodology to engineer sialyltransferases for desired properties, a fluorescence-based high-throughput screening (HTS) method was developed for screening sialyltransferase mutants generated via error-prone polymerase chain reactions (PCRs). Using this directed evolution approach, a library of > 105 sialyltransferase mutants were screened and a variant with up to 400-fold higher catalytic efficiency using a fluorescently labeled acceptor was obtained (Aharoni et al. 2006). Recently, this method was improved by the introduction of a two-color screening protocol to minimize the possibility of false positive mutants (Yang et al. 2010).
Sialyltransferase-catalyzed enzymatic and chemoenzymatic syntheses of sialosides
Chemical sialylation has been considered one of the most challenging glycosylation reactions due to the hindered tertiary anomeric center and the lack of a neighboring participating group in sialic acids (Boons and Demchenko 2000). Sialyltransferase-catalyzed reaction has been used as a preferred alternative approach (Izumi et al. 2001).
Vertebrate sialyltransferases are usually mono-functional although they have certain tolerance towards acceptors with some variations and are more promiscuous towards donor substrates (Harduin-Lepers 2010; Datta 2009). Before the identification and cloning of sialyltransferases from bacterial sources, mammalian sialyltransferases have been used in enzymatic and chemoenzymatic syntheses but only a few reactions were carried out in preparative scales mainly due to the limited access of a large amount of sialyltransferases and the high cost of sugar nucleotide donor CMP-sialic acid (Blixt et al. 2002). Several sialyltransferases such as rat recombinant α2–3-(N)-sialyltransferase, rat recombinant α2–3-(O)-sialyltransferase, and human recombinant α2–6-(N)-sialyltransferase, were commercially available from CalBioChem (now EMD). Due to their limited expression levels in insect cells such as Spodoptera frugiperda, challenges in adapting to E. coli expression systems, and less flexible substrate tolerance, vertebrate sialyltransferases have found less application in synthesis, especially in preparative and large-scale preparation, of sialosides. In comparison, bacterial sialyltransferases have been increasingly used in enzymatic synthesis of sialosides since the reports of cloning α2–3-sialyltransferases from N. meningitidis and N. gonorrhoeae in 1996 (Gilbert et al. 1996) and an α2–6-sialyltransferase from P. damselae (Yamamoto et al. 1998).
To avoid the use of expensive CMP-Neu5Ac for sialyltransferase-catalyzed synthesis of sialosides, an in situ CMP-Neu5Ac regeneration system was developed by the Wong group (Ichikawa et al. 1991b; Ichikawa et al. 1991a; Ichikawa et al. 1992). In this system, The CMP formed from sialyltransferase-catalyzed reaction was reacted with ATP to form CDP and ADP by nucleoside monophosphate kinase. A pyruvate kinase in the presence of phosphoenolpyruvate (PEP) was used to regenerate CTP and ATP from CDP and ADP, respectively. The CTP formed reacted with Neu5Ac to form CMP-Neu5Ac and pyrophosphate by CMP-Neu5Ac synthetase. An inorganic phosphorylase was used to breakdown the pyrophosphate to drive the reaction towards completion. The inputs of this system are metal cofactor(s), stoichiometric amounts of a sialyltransferase acceptor and Neu5Ac, at least two equivalents of PEP, and catalytic amount of ATP and CMP. Neu5Ac can also be replaced by excess ManNAc and pyruvate using a sialic acid aldolase-catalyzed reaction (Ichikawa et al. 1991a).
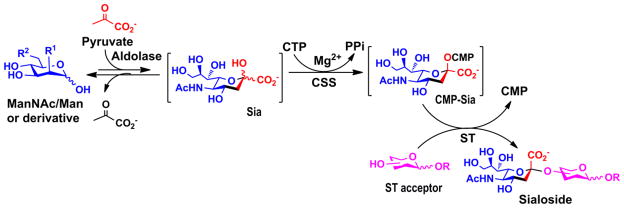
Earlier applications of mammalian or bacterial sialyltransferases and other sialic acid-biosynthetic enzymes in preparative-scale or large-scale synthesis of sialosides with or without in situ cofactor regeneration have been mainly focused on Neu5Ac-containing sialosides (Ichikawa et al. 1991b; Ichikawa et al. 1991a; Ichikawa et al. 1992; Ito 1993; Gilbert et al. 1998; Johnson 1999). More recently, systematic synthesis of a large library of sialosides containing diverse sialic acid forms linked to various underlying glycans with α2–3-, α2–6-, or α2–8- sialyl linkages in preparative-scale has been achieved efficiently from ManNAc, mannose, and their derivative as precursors for naturally existing and non-natural sialic acid forms using highly reactive and substrate promiscuous recombinant bacterial sialyltransferases in the presence of a sialic acid aldolase and a CMP-sialic acid synthetase in a one-pot three-enzyme system (Figure 4) (Yu et al. 2006a).
Figure 4.
One-pot three-enzyme synthesis of sialosides. R, glycans or glycoconjugates; R1 [-NHAc, -OH, -NHC(O)CH2OH, -OAc, -OMe, -NHC(O)CH2OAc, -NHC(O)CH2OMe, -NHC(O)CH2NHCbz, -N3, -NHC(O)CH2N3, -NHC(O)OCH2C≡CH, -NHC(O)CH2F, -H, -F] and R2 [-OH, -OAc, -OC(O)CH(CH3)OH, -N3], various substituents.
Sialic acid aldolase (N-acetylneuraminate lyase, NAL; EC 4.1.3.3) is a class I aldolase that catalyzes the cleavage of sialic acid to pyruvate and ManNAc with an equilibrium that favors Neu5Ac cleavage. It has been found in pathogenic as well as non-pathogenic bacteria (Aisaka et al. 1991) and in mammalian tissues. Both C2- and C5-modified N-acetylmannosamine (ManNAc) or mannose derivatives can be used as substrates by E. coli K-12 sialic acid aldolase (EcNanA) and P. multocida P-1059 sialic acid aldolase (PmNanA) (Li et al. 2008; Cao et al. 2009b). Nevertheless, PmNanA is a more efficient enzyme than EcNanA for synthesizing C8-modi ed sialic acids, especially for 5-O-methyl ManNAc, 5-O-methyl N-glycolylmannosamine (ManNGc5OMe), 5-O-methyl mannose, and 5-deoxy-mannose (Yu et al. 2011; Li et al. 2008). For activating sialic acid to form CMP-sialic acid, the donor of sialyltransferase, CMP-sialic acid synthetase (CSS, or sialic acid cytidylyltransferase, EC 2.7.7.43) from N. meningitidis (NmCSS) was the first bacterial CSS that was characterized in detail (Warren and Blacklow 1962; Yu et al. 2004). The crystal structures of NmCSS (Horsfall et al. 2010) and N-terminal catalytically active domain of murine CSS (Krapp et al. 2003) have been reported. The enzyme has also been cloned from E. coli K1 (Yu et al. 2004), S. agalactiae or GBS (Yu et al. 2006c), H. ducreyi (Tullius et al. 1996), Pasteurella haemolytica A2 (Bravo et al. 2001), Clostridium thermocellum (Mizanur and Pohl 2007) as summarized in a recent review (Mizanur and Pohl 2008) and more recently from P. multocida (Li et al. 2012). The substrate promiscuity of NmCSS enables its application in catalyzing the formation of diverse CMP-sialic acids (Morley and Withers 2010; Yu et al. 2004; Rauvolfova et al. 2008; Hartlieb et al. 2008; Gilbert et al. 1998). A recent study has shown that the substrate promiscuity of NmCSS can be further improved by site-directed mutagenesis (e.g. NmCSS_S81R and NmCSS_Q163A) (Li et al. 2012).
Using the one-pot three-enzyme sialylation system containing a multifunctional P. multocida sialyltransferase PmST1 encoded by Pm0188 gene, an E. coli or a P. multocida sialic acid aldolase (EcNanA or PmNanA), and NmCSS, α2–3-linked structurally diverse sialosides containing C5-, C8-, C9-, and other modified sialic acids and/or various acceptors including mono- and oligosaccharides, were synthesized in preparative-scale at 37°C, pH 8.5 (for sialosides that do not have a base labile O-acetyl or O-lactyl group) or pH 7.5 (for sialosides with an O-acetyl or O-lactyl group) (Yu et al. 2005; Yu et al. 2011; Cao et al. 2008; Lau et al. 2011; Yu et al. 2006a). Typical yields for preparative-scale (>20 mg) synthesis were higher than 60%, many reactions were achieved with more than 90% yields. Sialosides containing an azido group on the sialic acids were readily obtained and the azido group can be easily converted to an amino group and used as a handle for chemical acylation to generate a series of sialosides containing various sialic acid forms (Cao et al. 2009b). The tumor-associated sialyl T-antigens and derivatives were also synthesized using PmST1 in an efficient sequential two-step multienzyme approach (Lau et al. 2011). Recently, both PmST1 and Cst-I, an α2–3-sialyltransferase from C. jejuni, have been used synthesizing sialosides containing C8-modified sialic acid (Yu et al. 2011; Morley and Withers 2010).
Quite interesting, unlike PmST1 encoded by Pm0188 gene homolog in Pm strain P-1059 which is a multifunctional α2–3-sialyltransferase that prefers Gal-terminated oligosaccharides as acceptors, PmST2 encoded by Pm0508 gene in the same P. multocida strain is a monofunctional α2–3-sialyltransferase that prefers β1–4-linked galactosyl glycolipids as acceptors (Thon et al. 2011). In comparison, PmST3 encoded by Pm1174 gene in Pm70 strain is absent from Pm strains P-1059 and P-934. It is a monofunctional α2–3-sialyltransferase that can use both β1–4-linked galactosyl oligosaccharides and glycolipids as acceptors (Thon et al. 2012).
The one-pot three-enzyme system (Figure 4) has also been used for synthesizing α2–6-linked sialosides. The extremely flexible substrate specificity of the P. damselae α2–6 sialyltransferase (Pd2,6ST) enables its application in highly efficient chemoenzymatic synthesis of naturally occurring and non-naturally α2–6-linked sialosides (Yu et al. 2006b; Yu et al. 2011; Yu et al. 2006a; Muthana et al. 2007). Size-defined polysaccharide analogues were also chemoenzymatically synthesis by Pd2,6ST-catalyzed block transfer of di- or tetra-oligosaccharides from their CMP-activated forms (Muthana et al. 2007). These polysaccharides were used to produce novel macrocyclic structures (Muthana et al. 2009). For synthesizing sialyl Tn antigens (Siaα2–6GalNAca1-O-Ser/Thr) and derivatives, the recombinant marine bacterial α2,6-sialyltransferase cloned from Photobacterium sp. JT-ISH-224 (Psp2,6ST) (Tsukamoto et al. 2008) was shown to be a more efficient catalyst than Pd2,6ST (Yu et al. 2007; Yu et al. 2006b).
CMP-sialic acids and α2–3- and α2–6-linked sialosides containing 3F(axial)-sialic acid or 3F(equatorial)-sialic acid residue have also been obtained using a one-pot two-enzyme system containing a sialyltransferase and NmCSS from purified 3F(axial)-sialic acid or 3F(equatorial)-sialic acid synthesized by an E. coli sialic acid-catalyzed reaction (Chokhawala et al. 2007a).
In addition to α2–3- and α2–6-linked sialosides, a recombinant Cst-II mutant, Cst-IIΔ32I53S, in which the predicted C-terminal membrane associated domain of 32 amino acids was removed and an I53S mutation was introduced to enhance its stability and α2–8-sialyltransferase activity (Gilbert et al. 2002), was used in an efficient chemoenzymatic approach for the synthesis of a series of ganglioside oligosaccharides including GM3 (Neu5Acα2–3Lac), GD3 (Neu5Acα2–8Neu5Acα2–3Lac), GT3 (Neu5Acα2–8Neu5Ac α2–8Neu5Ac α2–3Lac), and other disialyl glycans containing a terminal Sia α2–8Sia component with different natural and non-natural sialic acids (Yu et al. 2009; Blixt et al. 2005).
Three types of bacterial polysialyltransferases (polySTs) that catalyze the sialic acid polymerization have been cloned and characterized. The polySTs encoded by neuS gene in E. coli K1 (Vimr et al. 1992; Steenbergen et al. 1992) and synD gene in N. meningitidis serogroup B (Steenbergen and Vimr 2003) catalyze the synthesis of α2–8-linked polysialic acid homopolymers. The polyST encoded by synE gene in N. meningitidis serogroup C catalyzes the formation α2–9-linked polysialic acid (Steenbergen and Vimr 2003). The polyST encoded by the E. coli K92 neuS gene can synthesize polysialic acid capsules with alternating sialyl α2–8 and α2–9 linkages (Vimr et al. 1992; Steenbergen et al. 1992; McGowen et al. 2001). Neither E. coli nor N. meningitidis can initiate polysialic acid synthesis de novo. They require oligosialic acids or endogenous acceptors (Ferrero and Aparicio 2010). All polysialyltransferases are cytoplasmic membrane associated enzymes. Therefore, earlier attempts of purification of polySTs failed and resulted in the inactivation of the enzymes. Success in expressing soluble N. meningitidis serogroup B polyST was achieved by introducing a maltose binding protein (MBP) as a fusion protein partner (Freiberger et al. 2007; Willis et al. 2008). Recently, the characterization of several soluble chimeras of the N. meningitidis serogroup C polySTs clearly demonstrated that only a single protein is required for elongation of polysialic acid acceptors (Peterson et al. 2011). Several vaccines based on the N. meningococcal capsular polysialic acids have been licensed (Tan et al. 2010). The polysialyltransferase genes have been used for the diagnosis of N. meningitidis infection by polymerase chain reaction (PCR)-based assays (Lewis et al. 2003).
Sialosides synthesized by the one-pot multienzyme system have been used to probe the interactions of sialic acid-binding proteins. Many sialosides synthesized have an alkyl azido aglycon which can be conveniently reduced to an amido group for efficient conjugation to proteins (Yu et al. 2007) for ELISA-based studies. It has also been used to produce biotinylated sialosides for surface plasmon resonance (SPR) imaging (Linman et al. 2008; Linman et al. 2009, 2012) and glycan array studies (Fei et al. 2011) to probe the interaction of sialosides and sialic acid-binding proteins. Some have a para-nitrophenyl aglycon for high-throughput substrate specificity studies of sialidases (Chokhawala et al. 2007b; Cao et al. 2009a; Li et al. 2011). The propyl azide aglycon on a library of α2–3/6/8-linked sialosides has also been reduced to propyl amine for sialyl glycan array studies (Padler-Karavani et al. 2011). The one-pot three-enzyme system has also been used to sialylate fluorophore-derivatized complex glycans such as 2-amino-N-(2-aminoethyl)-benzamide (AEAB)-derivatized lactose, lacto-N-tetraose, lacto-N-neotetraose, and complex-type biantennary N-glycan with diverse natural occurring and non-natural sialic acid forms for sialyl glycan microarray studies (Song et al. 2011; Bradley et al. 2011).
Although the one-pot multi-enzyme chemoenzymatic synthesis is an efficient approach to obtain structurally defined sialosides, the product purification steps are still tedious and time-consuming for generating a large library of sialosides. To avoid the tedious product purification process, a combinatorial chemoenzymatic sialylation approach has been developed. In this strategy, sialyltransferase acceptors were linked to a biotin molecule through a hexa-ethylene glycol linker to minimize non-specific binding in the protein binding assays. One-pot multienzyme sialylation using the biotinylated acceptors followed by the ELISA-type protein binding analysis using NeutrAvidin-coated plates allows high-throughput screening of sialic acid-binding proteins without product purification (Chokhawala et al. 2008).
Prospective
Despite a good understanding of general sialic acid metabolic processes for eukaryotes and bacteria, the details and significance of diverse sialic acid post-glycosylational modifications are still not fully elucidated and required further investigation. Due to the importance of sialyl glycoconjugates in cellular recognition, cell signaling, immune regulation, as well as bacterial and viral infection, key enzymes in the metabolism of sialyl glycoconjugates, such as sialyltransferases and sialidases are attractive targets for drug design. In spite of the characterization of an increasing number of sialyltransferases, CMP-sialic acid synthetases, and sialic acid aldolase for chemoenzymatic synthesis of diverse sialyl oligosaccharides and glycoconjugates, the substrate specificity of the sialoside biosynthetic enzymes limits the diversity of sialoside products. Cloning, characterizing, and exploring substrate specificity of additional wild-type bacterial enzymes, protein crystal structure-based tailor design of enzyme mutants with altered substrate specificity, and directed evolution-based screening of enzyme mutagenesis with broader substrate promiscuity will contribute greatly to increase the size of obtainable sialic acid-containing structures. Obtaining structurally diverse sialosides including those with modified underlying sugars (e.g. O-sulfated Gal or GlcNAc in O-sulfated sialyl Lewis x structures) and various sialic acid forms such as Neu5Ac, Neu5Gc, Kdn, and their natural existing O-acetylated and other post-glycosylationally modified forms is critical for unraveling the important functions of sialic acid-containing molecules.
Acknowledgments
The authors are grateful for the financial supports from NSF grant CHE1012511, NIH grant R01HD065122, the Camille Dreyfus Teacher-Scholarship, and the UC-Davis Chancellor’s Fellowship.
References
- Aharoni A, Thieme K, Chiu CP, Buchini S, Lairson LL, Chen H, Strynadka NC, Wakarchuk WW, Withers SG. High-throughput screening methodology for the directed evolution of glycosyltransferases. Nat Methods. 2006;3:609–614. doi: 10.1038/nmeth899. [DOI] [PubMed] [Google Scholar]
- Aisaka K, Igarashi A, Yamaguchi K, Uwajima T. Purification, crystallization and characterization of N-acetylneuraminate lyase from Escherichia coli. Biochem J. 1991;276:541–546. doi: 10.1042/bj2760541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almagro-Moreno S, Boyd EF. Insights into the evolution of sialic acid catabolism among bacteria. BMC Evol Biol. 2009;9:118. doi: 10.1186/1471-2148-9-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altheide TK, Hayakawa T, Mikkelsen TS, Diaz S, Varki N, Varki A. System-wide genomic and biochemical comparisons of sialic acid biology among primates and rodents: Evidence for two modes of rapid evolution. J Biol Chem. 2006;281:25689–25702. doi: 10.1074/jbc.M604221200. [DOI] [PubMed] [Google Scholar]
- Amaya MF, Watts AG, Damager I, Wehenkel A, Nguyen T, Buschiazzo A, Paris G, Frasch AC, Withers SG, Alzari PM. Structural insights into the catalytic mechanism of Trypanosoma cruzi trans-sialidase. Structure. 2004;12:775–784. doi: 10.1016/j.str.2004.02.036. [DOI] [PubMed] [Google Scholar]
- Angata T, Kitazume S, Terada T, Kitajima K, Inoue S, Troy FA, 2nd, Inoue Y. Identification, characterization, and developmental expression of a novel alpha 2→8-KDN-transferase which terminates elongation of alpha 2→8-linked oligo-polysialic acid chain synthesis in trout egg polysialoglycoproteins. Glycoconj J. 1994;11:493–499. doi: 10.1007/BF00731286. [DOI] [PubMed] [Google Scholar]
- Angata T, Matsuda T, Kitajima K. Synthesis of neoglycoconjugates containing deaminated neuraminic acid (KDN) using rat liver alpha2,6-sialyltransferase. Glycobiology. 1998;8:277–284. doi: 10.1093/glycob/8.3.277. [DOI] [PubMed] [Google Scholar]
- Angata T, Nakata D, Matsuda T, Kitajima K, Troy FA., 2nd Biosynthesis of KDN (2-keto-3-deoxy-D-glycero-D-galacto-nononic acid). Identification and characterization of a KDN-9-phosphate synthetase activity from trout testis. J Biol Chem. 1999;274:22949–22956. doi: 10.1074/jbc.274.33.22949. [DOI] [PubMed] [Google Scholar]
- Angata T, Varki A. Chemical diversity in the sialic acids and related alpha-keto acids: an evolutionary perspective. Chem Rev. 2002;102:439–469. doi: 10.1021/cr000407m. [DOI] [PubMed] [Google Scholar]
- Antoine T, Priem B, Heyraud A, Greffe L, Gilbert M, Wakarchuk WW, Lam JS, Samain E. Large-scale in vivo synthesis of the carbohydrate moieties of gangliosides GM1 and GM2 by metabolically engineered Escherichia coli. Chembiochem. 2003;4:406–412. doi: 10.1002/cbic.200200540. [DOI] [PubMed] [Google Scholar]
- Arming S, Wipfler D, Mayr J, Merling A, Vilas U, Schauer R, Schwartz-Albiez R, Vlasak R. The human CAS1 protein: a sialic acid-specific O-acetyltransferase? Glycobiology. 2011;21:553–564. doi: 10.1093/glycob/cwq153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audry M, Jeanneau C, Imberty A, Harduin-Lepers A, Delannoy P, Breton C. Current trends in the structure-activity relationships of sialyltransferases. Glycobiology. 2011;21:716–726. doi: 10.1093/glycob/cwq189. [DOI] [PubMed] [Google Scholar]
- Bergwerff AA, Hulleman SH, Kamerling JP, Vliegenthart JF, Shaw L, Reuter G, Schauer R. Nature and biosynthesis of sialic acids in the starfish Asterias rubens. Identification of sialo-oligomers and detection of S-adenosyl-L-methionine: N-acylneuraminate 8-O-methyltransferase and CMP-N-acetylneuraminate monooxygenase activities. Biochimie. 1992;74:25–37. doi: 10.1016/0300-9084(92)90181-d. [DOI] [PubMed] [Google Scholar]
- Blixt O, Allin K, Pereira L, Datta A, Paulson JC. Efficient chemoenzymatic synthesis of O-linked sialyl oligosaccharides. J Am Chem Soc. 2002;124:5739–5746. doi: 10.1021/ja017881+. [DOI] [PubMed] [Google Scholar]
- Blixt O, Vasiliu D, Allin K, Jacobsen N, Warnock D, Razi N, Paulson JC, Bernatchez S, Gilbert M, Wakarchuk W. Chemoenzymatic synthesis of 2-azidoethyl-ganglio-oligosaccharides GD3, GT3, GM2, GD2, GT2, GM1, and GD1a. Carbohydr Res. 2005;340 (12):1963–1972. doi: 10.1016/j.carres.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Bonten E, van der Spoel A, Fornerod M, Grosveld G, d’Azzo A. Characterization of human lysosomal neuraminidase defines the molecular basis of the metabolic storage disorder sialidosis. Genes Dev. 1996;10:3156–3169. doi: 10.1101/gad.10.24.3156. [DOI] [PubMed] [Google Scholar]
- Boons GJ, Demchenko AV. Recent advances in O-sialylation. Chem Rev. 2000;100:4539–4566. doi: 10.1021/cr990313g. [DOI] [PubMed] [Google Scholar]
- Bozue JA, Tullius MV, Wang J, Gibson BW, Munson RS., Jr Haemophilus ducreyi produces a novel sialyltransferase. Identification of the sialyltransferase gene and construction of mutants deficient in the production of the sialic acid-containing glycoform of the lipooligosaccharide. J Biol Chem. 1999;274:4106–4114. doi: 10.1074/jbc.274.7.4106. [DOI] [PubMed] [Google Scholar]
- Bradley KC, Galloway SE, Lasanajak Y, Song X, Heimburg-Molinaro J, Yu H, Chen X, Talekar GR, Smith DF, Cummings RD, Steinhauer DA. Analysis of influenza virus hemagglutinin receptor binding mutants with limited receptor recognition properties and conditional replication characteristics. J Virol. 2011;85:12387–12398. doi: 10.1128/JVI.05570-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo IG, Barrallo S, Ferrero MA, Rodriguez-Aparicio LB, Martinez-Blanco H, Reglero A. Kinetic properties of the acylneuraminate cytidylyltransferase from Pasteurella haemolytica A2. Biochem J. 2001;358:585–598. doi: 10.1042/bj3580585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo IG, Garcia-Vallve S, Romeu A, Reglero A. Prokaryotic origin of cytidylyltransferases and alpha-ketoacid synthases. Trends Microbiol. 2004;12:120–128. doi: 10.1016/j.tim.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Buchini S, Buschiazzo A, Withers SG. A new generation of specific Trypanosoma cruzi trans-sialidase inhibitors. Angew Chem Int Ed Engl. 2008;47:2700–2703. doi: 10.1002/anie.200705435. [DOI] [PubMed] [Google Scholar]
- Buschiazzo A, Alzari PM. Structural insights into sialic acid enzymology. Curr Opin Chem Biol. 2008;12:565–572. doi: 10.1016/j.cbpa.2008.06.017. [DOI] [PubMed] [Google Scholar]
- Buschiazzo A, Amaya MF, Cremona ML, Frasch AC, Alzari PM. The crystal structure and mode of action of trans-sialidase, a key enzyme in Trypanosoma cruzi pathogenesis. Mol Cell. 2002;10:757–768. doi: 10.1016/s1097-2765(02)00680-9. [DOI] [PubMed] [Google Scholar]
- Buschiazzo A, Campetella O, Frasch AC. Trypanosoma rangeli sialidase: cloning, expression and similarity to T. cruzi trans-sialidase. Glycobiology. 1997;7:1167–1173. doi: 10.1093/glycob/7.8.1167. [DOI] [PubMed] [Google Scholar]
- Buschiazzo A, Muia R, Larrieux N, Pitcovsky T, Mucci J, Campetella O. Trypanosoma cruzi trans-sialidase in complex with a neutralizing antibody: Structure/function studies towards the rational design of inhibitors. PLoS Pathog. 2012;8:e1002474. doi: 10.1371/journal.ppat.1002474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell CT, Sampathkumar SG, Yarema KJ. Metabolic oligosaccharide engineering: perspectives, applications, and future directions. Mol Biosyst. 2007;3:187–194. doi: 10.1039/b614939c. [DOI] [PubMed] [Google Scholar]
- Campbell JA, Davies GJ, Bulone V, Henrissat B. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem J. 1997;326:929–939. doi: 10.1042/bj3260929u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Huang S, Cheng J, Li Y, Muthana S, Son B, Chen X. Chemical preparation of sialyl Lewis x using an enzymatically synthesized sialoside building block. Carbohydr Res. 2008;343:2863–2869. doi: 10.1016/j.carres.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Li Y, Lau K, Muthana S, Yu H, Cheng J, Chokhawala HA, Sugiarto G, Zhang L, Chen X. Sialidase substrate specificity studies using chemoenzymatically synthesized sialosides containing C5-modified sialic acids. Org Biomol Chem. 2009a;7:5137–5145. doi: 10.1039/b916305k. [DOI] [PubMed] [Google Scholar]
- Cao H, Muthana S, Li Y, Cheng J, Chen X. Parallel chemoenzymatic synthesis of sialosides containing a C5-diversified sialic acid. Bioorg Med Chem Lett. 2009b;19:5869–5871. doi: 10.1016/j.bmcl.2009.08.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlin AF, Uchiyama S, Chang YC, Lewis AL, Nizet V, Varki A. Molecular mimicry of host sialylated glycans allows a bacterial pathogen to engage neutrophil Siglec-9 and dampen the innate immune response. Blood. 2009;113:3333–3336. doi: 10.1182/blood-2008-11-187302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffin DO, McKinnon K, Rubens CE. CpsK of Streptococcus agalactiae exhibits alpha2,3-sialyltransferase activity in Haemophilus ducreyi. Mol Microbiol. 2002;45:109–122. doi: 10.1046/j.1365-2958.2002.02988.x. [DOI] [PubMed] [Google Scholar]
- Chan PH, Lairson LL, Lee HJ, Wakarchuk WW, Strynadka NC, Withers SG, McIntosh LP. NMR spectroscopic characterization of the sialyltransferase CstII from Campylobacter jejuni: histidine 188 is the general base. Biochemistry. 2009;48:11220–11230. doi: 10.1021/bi901606n. [DOI] [PubMed] [Google Scholar]
- Chefalo P, Pan Y, Nagy N, Guo Z, Harding CV. Efficient metabolic engineering of GM3 on tumor cells by N-phenylacetyl-D-mannosamine. Biochemistry. 2006;45:3733–3739. doi: 10.1021/bi052161r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GY, Chen X, King S, Cavassani KA, Cheng J, Zheng X, Cao H, Yu H, Qu J, Fang D, Wu W, Bai XF, Liu JQ, Woodiga SA, Chen C, Sun L, Hogaboam CM, Kunkel SL, Zheng P, Liu Y. Amelioration of sepsis by inhibiting sialidase-mediated disruption of the CD24-SiglecG interaction. Nat Biotechnol. 2011;29:428–435. doi: 10.1038/nbt.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Varki A. Advances in the biology and chemistry of sialic acids. ACS Chem Biol. 2010;5:163–176. doi: 10.1021/cb900266r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Huang S, Yu H, Li Y, Lau K, Chen X. Trans-sialidase activity of Photobacterium damsela alpha2,6-sialyltransferase and its application in the synthesis of sialosides. Glycobiology. 2010;20:260–268. doi: 10.1093/glycob/cwp172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Yu H, Lau K, Huang S, Chokhawala HA, Li Y, Tiwari VK, Chen X. Multifunctionality of Campylobacter jejuni sialyltransferase CstII: characterization of GD3/GT3 oligosaccharide synthase, GD3 oligosaccharide sialidase, and trans-sialidase activities. Glycobiology. 2008;18:686–697. doi: 10.1093/glycob/cwn047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu CP, Lairson LL, Gilbert M, Wakarchuk WW, Withers SG, Strynadka NC. Structural analysis of the alpha-2,3-sialyltransferase Cst-I from Campylobacter jejuni in apo and substrate-analogue bound forms. Biochemistry. 2007;46:7196–7204. doi: 10.1021/bi602543d. [DOI] [PubMed] [Google Scholar]
- Chiu CP, Watts AG, Lairson LL, Gilbert M, Lim D, Wakarchuk WW, Withers SG, Strynadka NC. Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analog. Nat Struct Mol Biol. 2004;11:163–170. doi: 10.1038/nsmb720. [DOI] [PubMed] [Google Scholar]
- Chokhawala HA, Cao H, Yu H, Chen X. Enzymatic synthesis of fluorinated mechanistic probes for sialidases and sialyltransferases. J Am Chem Soc. 2007a;129:10630–10631. doi: 10.1021/ja072687u. [DOI] [PubMed] [Google Scholar]
- Chokhawala HA, Huang S, Lau K, Yu H, Cheng J, Thon V, Hurtado-Ziola N, Guerrero JA, Varki A, Chen X. Combinatorial chemoenzymatic synthesis and high-throughput screening of sialosides. ACS Chem Biol. 2008;3:567–576. doi: 10.1021/cb800127n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chokhawala HA, Yu H, Chen X. High-throughput substrate specificity studies of sialidases by using chemoenzymatically synthesized sialoside libraries. Chembiochem. 2007b;8:194–201. doi: 10.1002/cbic.200600410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claus H, Stummeyer K, Batzilla J, Muhlenhoff M, Vogel U. Amino acid 310 determines the donor substrate specificity of serogroup W-135 and Y capsule polymerases of Neisseria meningitidis. Mol Microbiol. 2009;71:960–971. doi: 10.1111/j.1365-2958.2008.06580.x. [DOI] [PubMed] [Google Scholar]
- Corfield AP, Myerscough N, Warren BF, Durdey P, Paraskeva C, Schauer R. Reduction of sialic acid O-acetylation in human colonic mucins in the adenoma-carcinoma sequence. Glycoconj J. 1999;16 (6):307–317. doi: 10.1023/a:1007026314792. [DOI] [PubMed] [Google Scholar]
- Coutinho PM, Deleury E, Davies GJ, Henrissat B. An evolving hierarchical family classification for glycosyltransferases. J Mol Biol. 2003;328:307–317. doi: 10.1016/s0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- Cremona ML, Pollevick GD, Frasch AC, Campetella O. Effect of primary structure modifications in Trypanosoma cruzi neuraminidase/trans-sialidase activities. Cell Mol Biol. 1996;42:697–702. [PubMed] [Google Scholar]
- Crocker PR, Varki A. Siglecs, sialic acids and innate immunity. Trends Immunol. 2001;22:337–342. doi: 10.1016/s1471-4906(01)01930-5. [DOI] [PubMed] [Google Scholar]
- Damager I, Buchini S, Amaya MF, Buschiazzo A, Alzari P, Frasch AC, Watts A, Withers SG. Kinetic and mechanistic analysis of Trypanosoma cruzi trans-sialidase reveals a classical ping-pong mechanism with acid/base catalysis. Biochemistry. 2008;47:3507–3512. doi: 10.1021/bi7024832. [DOI] [PubMed] [Google Scholar]
- Datta AK. Comparative sequence analysis in the sialyltransferase protein family: analysis of motifs. Curr Drug Targets. 2009;10:483–498. doi: 10.2174/138945009788488422. [DOI] [PubMed] [Google Scholar]
- Datta AK, Chammas R, Paulson JC. Conserved cysteines in the sialyltransferase sialylmotifs form an essential disulfide bond. J Biol Chem. 2001;276:15200–15207. doi: 10.1074/jbc.M010542200. [DOI] [PubMed] [Google Scholar]
- Datta AK, Paulson JC. The sialyltransferase “sialylmotif” participates in binding the donor substrate CMP-NeuAc. J Biol Chem. 1995;270:1497–1500. doi: 10.1074/jbc.270.4.1497. [DOI] [PubMed] [Google Scholar]
- Datta AK, Sinha A, Paulson JC. Mutation of the sialyltransferase S-sialylmotif alters the kinetics of the donor and acceptor substrates. J Biol Chem. 1998;273:9608–9614. doi: 10.1074/jbc.273.16.9608. [DOI] [PubMed] [Google Scholar]
- Davies G, Henrissat B. Structures and mechanisms of glycosyl hydrolases. Structure. 1995;3:853–859. doi: 10.1016/S0969-2126(01)00220-9. [DOI] [PubMed] [Google Scholar]
- Diaz S, Higa HH, Hayes BK, Varki A. O-Acetylation and de-O-acetylation of sialic acids. 7- and 9-O-acetylation of alpha 2,6-linked sialic acids on endogenous N-linked glycans in rat liver Golgi vesicles. J Biol Chem. 1989;264:19416–19426. [PubMed] [Google Scholar]
- Drake PM, Nathan JK, Stock CM, Chang PV, Muench MO, Nakata D, Reader JR, Gip P, Golden KP, Weinhold B, Gerardy-Schahn R, Troy FA, 2nd, Bertozzi CR. Polysialic acid, a glycan with highly restricted expression, is found on human and murine leukocytes and modulates immune responses. J Immunol. 2008;181:6850–6858. doi: 10.4049/jimmunol.181.10.6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouillard S, Mine T, Kajiwara H, Yamamoto T, Samain E. Efficient synthesis of 6′-sialyllactose, 6,6′-disialyllactose, and 6′-KDO-lactose by metabolically engineered E. coli expressing a multifunctional sialyltransferase from the Photobacterium sp. JT-ISH-224. Carbohydr Res. 2010;345:1394–1399. doi: 10.1016/j.carres.2010.02.018. [DOI] [PubMed] [Google Scholar]
- Du J, Meledeo MA, Wang Z, Khanna HS, Paruchuri VD, Yarema KJ. Metabolic glycoengineering: sialic acid and beyond. Glycobiology. 2009;19:1382–1401. doi: 10.1093/glycob/cwp115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T, Koizumi S, Tabata K, Kakita S, Ozaki A. Large-scale production of the carbohydrate portion of the sialyl-Tn epitope, alpha-Neup5Ac-(2→6)-D-GalpNAc, through bacterial coupling. Carbohydr Res. 2001;330:439–443. doi: 10.1016/s0008-6215(01)00007-6. [DOI] [PubMed] [Google Scholar]
- Endo T, Koizumi S, Tabata K, Ozaki A. Large-scale production of CMP-NeuAc and sialylated oligosaccharides through bacterial coupling. Appl Microbiol Biotechnol. 2000;53:257–261. doi: 10.1007/s002530050017. [DOI] [PubMed] [Google Scholar]
- Eugenia Giorgi M, de Lederkremer RM. Trans-sialidase and mucins of Trypanosoma cruzi: an important interplay for the parasite. Carbohydr Res. 2011;346:1389–1393. doi: 10.1016/j.carres.2011.04.006. [DOI] [PubMed] [Google Scholar]
- Fei Y, Sun YS, Li Y, Lau K, Yu H, Chokhawala HA, Huang S, Landry JP, Chen X, Zhu X. Fluorescent labeling agents change binding profiles of glycan-binding proteins. Mol Biosyst. 2011;7:3343–3352. doi: 10.1039/c1mb05332a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrero MA, Aparicio LR. Biosynthesis and production of polysialic acids in bacteria. Appl Microbiol Biotechnol. 2010;86:1621–1635. doi: 10.1007/s00253-010-2531-5. [DOI] [PubMed] [Google Scholar]
- Fierfort N, Samain E. Genetic engineering of Escherichia coli for the economical production of sialylated oligosaccharides. J Biotechnol. 2008;134:261–265. doi: 10.1016/j.jbiotec.2008.02.010. [DOI] [PubMed] [Google Scholar]
- Fort S, Birikaki L, Dubois MP, Antoine T, Samain E, Driguez H. Biosynthesis of conjugatable saccharidic moieties of GM2 and GM3 gangliosides by engineered E. coli. Chem Commun (Camb) 2005:2558–2560. doi: 10.1039/b500686d. [DOI] [PubMed] [Google Scholar]
- Fox KL, Cox AD, Gilbert M, Wakarchuk WW, Li J, Makepeace K, Richards JC, Moxon ER, Hood DW. Identification of a bifunctional lipopolysaccharide sialyltransferase in Haemophilus influenzae: incorporation of disialic acid. J Biol Chem. 2006;281:40024–40032. doi: 10.1074/jbc.M602314200. [DOI] [PubMed] [Google Scholar]
- Freiberger F, Claus H, Gunzel A, Oltmann-Norden I, Vionnet J, Muhlenhoff M, Vogel U, Vann WF, Gerardy-Schahn R, Stummeyer K. Biochemical characterization of a Neisseria meningitidis polysialyltransferase reveals novel functional motifs in bacterial sialyltransferases. Mol Microbiol. 2007;65:1258–1275. doi: 10.1111/j.1365-2958.2007.05862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaderi D, Taylor RE, Padler-Karavani V, Diaz S, Varki A. Implications of the presence of N-glycolylneuraminic acid in recombinant therapeutic glycoproteins. Nat Biotechnol. 2010;28:863–867. doi: 10.1038/nbt.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert M, Bayer R, Cunningham AM, DeFrees S, Gao Y, Watson DC, Young NM, Wakarchuk WW. The synthesis of sialylated oligosaccharides using a CMP-Neu5Ac synthetase/sialyltransferase fusion. Nat Biotechnol. 1998;16:769–772. doi: 10.1038/nbt0898-769. [DOI] [PubMed] [Google Scholar]
- Gilbert M, Brisson JR, Karwaski MF, Michniewicz J, Cunningham AM, Wu Y, Young NM, Wakarchuk WW. Biosynthesis of ganglioside mimics in Campylobacter jejuni OH4384. Identification of the glycosyltransferase genes, enzymatic synthesis of model compounds, and characterization of nanomole amounts by 600-MHz (1)H and (13)C NMR analysis. J Biol Chem. 2000;275:3896–3906. doi: 10.1074/jbc.275.6.3896. [DOI] [PubMed] [Google Scholar]
- Gilbert M, Karwaski MF, Bernatchez S, Young NM, Taboada E, Michniewicz J, Cunningham AM, Wakarchuk WW. The genetic bases for the variation in the lipo-oligosaccharide of the mucosal pathogen, Campylobacter jejuni. Biosynthesis of sialylated ganglioside mimics in the core oligosaccharide. J Biol Chem. 2002;277:327–337. doi: 10.1074/jbc.M108452200. [DOI] [PubMed] [Google Scholar]
- Gilbert M, Watson DC, Cunningham AM, Jennings MP, Young NM, Wakarchuk WW. Cloning of the lipooligosaccharide alpha-2,3-sialyltransferase from the bacterial pathogens Neisseria meningitidis and Neisseria gonorrhoeae. J Biol Chem. 1996;271:28271–28276. doi: 10.1074/jbc.271.45.28271. [DOI] [PubMed] [Google Scholar]
- Harduin-Lepers A. Comprehensive analysis of sialyltransferases in vertebrate genomes. Glycobiol Insights. 2010;2:29–61. [Google Scholar]
- Harduin-Lepers A, Mollicone R, Delannoy P, Oriol R. The animal sialyltransferases and sialyltransferase-related genes: a phylogenetic approach. Glycobiology. 2005;15:805–817. doi: 10.1093/glycob/cwi063. [DOI] [PubMed] [Google Scholar]
- Harduin-Lepers A, Recchi MA, Delannoy P. 1994, the year of sialyltransferases. Glycobiology. 1995;5:741–758. doi: 10.1093/glycob/5.8.741. [DOI] [PubMed] [Google Scholar]
- Harduin-Lepers A, Vallejo-Ruiz V, Krzewinski-Recchi MA, Samyn-Petit B, Julien S, Delannoy P. The human sialyltransferase family. Biochimie. 2001;83:727–737. doi: 10.1016/s0300-9084(01)01301-3. [DOI] [PubMed] [Google Scholar]
- Harrison A, Dyer DW, Gillaspy A, Ray WC, Mungur R, Carson MB, Zhong H, Gipson J, Gipson M, Johnson LS, Lewis L, Bakaletz LO, Munson RS., Jr Genomic sequence of an otitis media isolate of nontypeable Haemophilus influenzae: comparative study with H. influenzae serotype D, strain KW20. J Bacteriol. 2005;187:4627–4636. doi: 10.1128/JB.187.13.4627-4636.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartlieb S, Gunzel A, Gerardy-Schahn R, Munster-Kuhnel AK, Kirschning A, Drager G. Chemoenzymatic synthesis of CMP-N-acetyl-7-fluoro-7-deoxy-neuraminic acid. Carbohydr Res. 2008;343:2075–2082. doi: 10.1016/j.carres.2008.02.003. [DOI] [PubMed] [Google Scholar]
- Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1991;280:309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higa HH, Butor C, Diaz S, Varki A. O-Acetylation and de-O-acetylation of sialic acids. O-acetylation of sialic acids in the rat liver Golgi apparatus involves an acetyl intermediate and essential histidine and lysine residues - a transmembrane reaction? J Biol Chem. 1989;264:19427–19434. [PubMed] [Google Scholar]
- Higa HH, Paulson JC. Sialylation of glycoprotein oligosaccharides with N-acetyl-, N-glycolyl-, and N-O-diacetylneuraminic acids. J Biol Chem. 1985;260:8838–8849. [PubMed] [Google Scholar]
- Horsfall LE, Nelson A, Berry A. Identification and characterization of important residues in the catalytic mechanism of CMP-Neu5Ac synthetase from Neisseria meningitidis. FEBS J. 2010;277:2779–2790. doi: 10.1111/j.1742-4658.2010.07696.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houliston RS, Endtz HP, Yuki N, Li J, Jarrell HC, Koga M, van Belkum A, Karwaski MF, Wakarchuk WW, Gilbert M. Identification of a sialate O-acetyltransferase from Campylobacter jejuni: demonstration of direct transfer to the C-9 position of terminalalpha-2, 8-linked sialic acid. J Biol Chem. 2006;281:11480–11486. doi: 10.1074/jbc.M512183200. [DOI] [PubMed] [Google Scholar]
- Ichikawa Y, Lin Y-C, Dumas DP, Shen G-J, Garcia-Junceda E, Williams MA, Bayer R, Ketcham C, Walker LE, Paulson JC, Wong C-H. Chemical-enzymatic synthesis and conformational analysis of sialyl Lewis x and derivatives. J Am Chem Soc. 1992;114:9283–9298. [Google Scholar]
- Ichikawa Y, Liu JLC, Shen GJ, Wong CH. A highly efficient multienzyme system for the one-step synthesis of a sialyl trisaccharide - in situ generation of sialic acid and N-acetyllactosamine coupled with regeneration of UDP-glucose, UDP-galactose, and CMP-sialic acid. J Am Chem Soc. 1991a;113:6300–6302. [Google Scholar]
- Ichikawa Y, Shen GJ, Wong CH. Enzyme-catalyzed synthesis of sialyl oligosaccharide with in situ regeneration of CMP-sialic acid. J Am Chem Soc. 1991b;113:4698–4700. [Google Scholar]
- Inoue S, Sato C, Kitajima K. Extensive enrichment of N-glycolylneuraminic acid in extracellular sialoglycoproteins abundantly synthesized and secreted by human cancer cells. Glycobiology. 2010;20:752–762. doi: 10.1093/glycob/cwq030. [DOI] [PubMed] [Google Scholar]
- Ito YP, JC Combined use of trans-sialidase and sialyltransferase for enzymatic synthesis of alphaNeuAc2-3-beta-Gal-OR. J Am Chem Soc. 1993;115:7862–6863. [Google Scholar]
- Iwatani T, Okino N, Sakakura M, Kajiwara H, Takakura Y, Kimura M, Ito M, Yamamoto T, Kakuta Y. Crystal structure of alpha/beta-galactoside alpha2,3-sialyltransferase from a luminous marine bacterium, Photobacterium phosphoreum. FEBS Lett. 2009;583:2083–2087. doi: 10.1016/j.febslet.2009.05.032. [DOI] [PubMed] [Google Scholar]
- Iwersen M, Dora H, Kohla G, Gasa S, Schauer R. Solubilisation and properties of the sialate-4-O-acetyltransferase from guinea pig liver. Biol Chem. 2003;384:1035–1047. doi: 10.1515/BC.2003.116. [DOI] [PubMed] [Google Scholar]
- Iwersen M, Vandamme-Feldhaus V, Schauer R. Enzymatic 4-O-acetylation of N-acetylneuraminic acid in guinea-pig liver. Glycoconj J. 1998;15:895–904. doi: 10.1023/a:1006911100081. [DOI] [PubMed] [Google Scholar]
- Izumi M, Shen GJ, Wacowich-Sgarbi S, Nakatani T, Plettenburg O, Wong CH. Microbial glycosyltransferases for carbohydrate synthesis: alpha-2,3-sialyltransferase from Neisseria gonorrheae. J Am Chem Soc. 2001;123:10909–10918. doi: 10.1021/ja011382r. [DOI] [PubMed] [Google Scholar]
- Jacobs CL, Goon S, Yarema KJ, Hinderlich S, Hang HC, Chai DH, Bertozzi CR. Substrate specificity of the sialic acid biosynthetic pathway. Biochemistry. 2001;40:12864–12874. doi: 10.1021/bi010862s. [DOI] [PubMed] [Google Scholar]
- Jeanneau C, Chazalet V, Auge C, Soumpasis DM, Harduin-Lepers A, Delannoy P, Imberty A, Breton C. Structure-function analysis of the human sialyltransferase ST3Gal I: role of N-glycosylation and a novel conserved sialylmotif. J Biol Chem. 2004;279:13461–13468. doi: 10.1074/jbc.M311764200. [DOI] [PubMed] [Google Scholar]
- Johnson KF. Synthesis of oligosaccharides by bacterial enzymes. Glycoconj J. 1999;16:141–146. doi: 10.1023/a:1026440509859. [DOI] [PubMed] [Google Scholar]
- Jones PA, Samuels NM, Phillips NJ, Munson RS, Jr, Bozue JA, Arseneau JA, Nichols WA, Zaleski A, Gibson BW, Apicella MA. Haemophilus influenzae type b strain A2 has multiple sialyltransferases involved in lipooligosaccharide sialylation. J Biol Chem. 2002;277:14598–14611. doi: 10.1074/jbc.M110986200. [DOI] [PubMed] [Google Scholar]
- Kakuta Y, Okino N, Kajiwara H, Ichikawa M, Takakura Y, Ito M, Yamamoto T. Crystal structure of Vibrionaceae Photobacterium sp. JT-ISH-224 alpha2,6-sialyltransferase in a ternary complex with donor product CMP and acceptor substrate lactose: catalytic mechanism and substrate recognition. Glycobiology. 2008;18:66–73. doi: 10.1093/glycob/cwm119. [DOI] [PubMed] [Google Scholar]
- Kean EL, Munster-Kuhnel AK, Gerardy-Schahn R. CMP-sialic acid synthetase of the nucleus. Biochim Biophys Acta. 2004;1673:56–65. doi: 10.1016/j.bbagen.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Kelm A, Shaw L, Schauer R, Reuter G. The biosynthesis of 8-O-methylated sialic acids in the starfish Asterias rubens--isolation and characterisation of S-adenosyl-L-methionine: sialate-8-O-methyltransferase. Eur J Biochem. 1998;251:874–884. doi: 10.1046/j.1432-1327.1998.2510874.x. [DOI] [PubMed] [Google Scholar]
- Koliwer-Brandl H, Gbem TT, Waespy M, Reichert O, Mandel P, Drebitz E, Dietz F, Kelm S. Biochemical characterization of trans-sialidase TS1 variants from Trypanosoma congolense. BMC Biochem. 2011;12:39. doi: 10.1186/1471-2091-12-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp S, Munster-Kuhnel AK, Kaiser JT, Huber R, Tiralongo J, Gerardy-Schahn R, Jacob U. The crystal structure of murine CMP-5-N-acetylneuraminic acid synthetase. J Mol Biol. 2003;334:625–637. doi: 10.1016/j.jmb.2003.09.080. [DOI] [PubMed] [Google Scholar]
- Krug LM, Ragupathi G, Ng KK, Hood C, Jennings HJ, Guo Z, Kris MG, Miller V, Pizzo B, Tyson L, Baez V, Livingston PO. Vaccination of small cell lung cancer patients with polysialic acid or N-propionylated polysialic acid conjugated to keyhole limpet hemocyanin. Clin Cancer Res. 2004;10:916–923. doi: 10.1158/1078-0432.ccr-03-0101. [DOI] [PubMed] [Google Scholar]
- Lairson LL, Henrissat B, Davies GJ, Withers SG. Glycosyltransferases: structures, functions, and mechanisms. Annu Rev Biochem. 2008;77:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- Lau K, Yu H, Thon V, Khedri Z, Leon ME, Tran BK, Chen X. Sequential two-step multienzyme synthesis of tumor-associated sialyl T-antigens and derivatives. Org Biomol Chem. 2011;9:2784–2789. doi: 10.1039/c0ob01269f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence SM, Huddleston KA, Pitts LR, Nguyen N, Lee YC, Vann WF, Coleman TA, Betenbaugh MJ. Cloning and expression of the human N-acetylneuraminic acid phosphate synthase gene with 2-keto-3-deoxy-D-glycero-D-galacto-nononic acid biosynthetic ability. J Biol Chem. 2000;275:17869–17877. doi: 10.1074/jbc.M000217200. [DOI] [PubMed] [Google Scholar]
- Lewis AL, Cao H, Patel SK, Diaz S, Ryan W, Carlin AF, Thon V, Lewis WG, Varki A, Chen X, Nizet V. NeuA sialic acid O-acetylesterase activity modulates O-acetylation of capsular polysaccharide in group B Streptococcus. J Biol Chem. 2007;282:27562–27571. doi: 10.1074/jbc.M700340200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis C, Diggle MA, Clarke SC. Nucleotide sequence analysis of the sialyltransferase genes of meningococcal serogroups B, C, Y and W135. J Mol Microbiol Biotechnol. 2003;5:82–86. doi: 10.1159/000069978. [DOI] [PubMed] [Google Scholar]
- Li Y, Cao H, Yu H, Chen Y, Lau K, Qu J, Thon V, Sugiarto G, Chen X. Identifying selective inhibitors against the human cytosolic sialidase NEU2 by substrate specificity studies. Mol Biosyst. 2011;7:1060–1072. doi: 10.1039/c0mb00244e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Sun M, Huang S, Yu H, Chokhawala HA, Thon V, Chen X. The Hd0053 gene of Haemophilus ducreyi encodes an alpha2,3-sialyltransferase. Biochem Biophys Res Commun. 2007;361:555–560. doi: 10.1016/j.bbrc.2007.07.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Yu H, Cao H, Lau K, Muthana S, Tiwari VK, Son B, Chen X. Pasteurella multocida sialic acid aldolase: a promising biocatalyst. Appl Microbiol Biotechnol. 2008;79:963–970. doi: 10.1007/s00253-008-1506-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Yu H, Cao H, Muthana S, Chen X. Pasteurella multocida CMP-sialic acid synthetase and mutants of Neisseria meningitidis CMP-sialic acid synthetase with improved substrate promiscuity. Appl Microbiol Biotechnol. 2012;93:2411–2423. doi: 10.1007/s00253-011-3579-6. [DOI] [PubMed] [Google Scholar]
- Lin LY, Rakic B, Chiu CP, Lameignere E, Wakarchuk WW, Withers SG, Strynadka NC. Structure and mechanism of the lipooligosaccharide sialyltransferase from Neisseria meningitidis. J Biol Chem. 2011;286:37237–37248. doi: 10.1074/jbc.M111.249920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linman MJ, Taylor JD, Yu H, Chen X, Cheng Q. Surface plasmon resonance study of protein-carbohydrate interactions using biotinylated sialosides. Anal Chem. 2008;80:4007–4013. doi: 10.1021/ac702566e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linman MJ, Yu H, Chen X, Cheng Q. Fabrication and characterization of a sialoside-based carbohydrate microarray biointerface for protein binding analysis with surface plasmon resonance imaging. ACS Appl Mater Interfaces. 2009;1:1755–1762. doi: 10.1021/am900290g. [DOI] [PubMed] [Google Scholar]
- Linman MJ, Yu H, Chen X, Cheng Q. Surface plasmon resonance imaging analysis of protein binding to a sialoside-based carbohydrate microarray. Methods Mol Biol. 2012;808:183–194. doi: 10.1007/978-1-61779-373-8_13. [DOI] [PubMed] [Google Scholar]
- Lrhorfi LA, Srinivasan GV, Schauer R. Properties and partial purification of sialate-O-acetyltransferase from bovine submandibular glands. Biol Chem. 2007;388:297–306. doi: 10.1515/BC.2007.033. [DOI] [PubMed] [Google Scholar]
- Mahal LK, Charter NW, Angata K, Fukuda M, Koshland DE, Jr, Bertozzi CR. A small-molecule modulator of poly-alpha 2,8-sialic acid expression on cultured neurons and tumor cells. Science. 2001;294:380–381. doi: 10.1126/science.1062192. [DOI] [PubMed] [Google Scholar]
- Mahal LK, Yarema KJ, Bertozzi CR. Engineering chemical reactivity on cell surfaces through oligosaccharide biosynthesis. Science. 1997;276:1125–1128. doi: 10.1126/science.276.5315.1125. [DOI] [PubMed] [Google Scholar]
- Mandal C, Chandra S, Schauer R. Regulation of O-acetylation of sialic acids by sialate-O-acetyltransferase and sialate-O-acetylesterase activities in childhood acute lymphoblastic leukemia. Glycobiology. 2012;22:70–83. doi: 10.1093/glycob/cwr106. [DOI] [PubMed] [Google Scholar]
- Mandal C, Srinivasan GV, Chowdhury S, Chandra S, Schauer R. High level of sialate-O-acetyltransferase activity in lymphoblasts of childhood acute lymphoblastic leukaemia (ALL): enzyme characterization and correlation with disease status. Glycoconj J. 2009;26:57–73. doi: 10.1007/s10719-008-9163-3. [DOI] [PubMed] [Google Scholar]
- McGowen MM, Vionnet J, Vann WF. Elongation of alternating alpha 2,8/2,9 polysialic acid by the Escherichia coli K92 polysialyltransferase. Glycobiology. 2001;11:613–620. doi: 10.1093/glycob/11.8.613. [DOI] [PubMed] [Google Scholar]
- Mine T, Katayama S, Kajiwara H, Tsunashima M, Tsukamoto H, Takakura Y, Yamamoto T. An alpha2,6-sialyltransferase cloned from Photobacterium leiognathi strain JT-SHIZ-119 shows both sialyltransferase and neuraminidase activity. Glycobiology. 2010;20:158–165. doi: 10.1093/glycob/cwp157. [DOI] [PubMed] [Google Scholar]
- Mitrasinovic PM. Advances in the structure-based design of the influenza A neuraminidase inhibitors. Curr Drug Targets. 2010;11:315–326. doi: 10.2174/138945010790711932. [DOI] [PubMed] [Google Scholar]
- Miyagi T. Aberrant expression of sialidase and cancer progression. Proc Jpn Acad Ser B Phys Biol Sci. 2008a;84:407–418. doi: 10.2183/pjab/84.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagi T. Physiological and pathological roles of mammalian sialidases. Seikagaku. 2008b;80:13–23. [PubMed] [Google Scholar]
- Miyagi T, Wada T, Yamaguchi K, Hata K. Sialidase and malignancy: a minireview. Glycoconj J. 2004;20:189–198. doi: 10.1023/B:GLYC.0000024250.48506.bf. [DOI] [PubMed] [Google Scholar]
- Mizanur RM, Pohl NL. Cloning and characterization of a heat-stable CMP-N-acylneuraminic acid synthetase from Clostridium thermocellum. Appl Microbiol Biotechnol. 2007;76:827–834. doi: 10.1007/s00253-007-1053-2. [DOI] [PubMed] [Google Scholar]
- Mizanur RM, Pohl NL. Bacterial CMP-sialic acid synthetases: production, properties, and applications. Appl Microbiol Biotechnol. 2008;80:757–765. doi: 10.1007/s00253-008-1643-7. [DOI] [PubMed] [Google Scholar]
- Montagna GN, Donelson JE, Frasch AC. Procyclic Trypanosoma brucei expresses separate sialidase and trans-sialidase enzymes on its surface membrane. J Biol Chem. 2006;281:33949–33958. doi: 10.1074/jbc.M604951200. [DOI] [PubMed] [Google Scholar]
- Monti E, Bassi MT, Bresciani R, Civini S, Croci GL, Papini N, Riboni M, Zanchetti G, Ballabio A, Preti A, Tettamanti G, Venerando B, Borsani G. Molecular cloning and characterization of NEU4, the fourth member of the human sialidase gene family. Genomics. 2004;83:445–453. doi: 10.1016/j.ygeno.2003.08.019. [DOI] [PubMed] [Google Scholar]
- Monti E, Preti A, Rossi E, Ballabio A, Borsani G. Cloning and characterization of NEU2, a human gene homologous to rodent soluble sialidases. Genomics. 1999;57:137–143. doi: 10.1006/geno.1999.5749. [DOI] [PubMed] [Google Scholar]
- Morley TJ, Withers SG. Chemoenzymatic synthesis and enzymatic analysis of 8-modified cytidine monophosphate-sialic acid and sialyl lactose derivatives. J Am Chem Soc. 2010;132:9430–9437. doi: 10.1021/ja102644a. [DOI] [PubMed] [Google Scholar]
- Muthana S, Yu H, Cao H, Cheng J, Chen X. Chemoenzymatic synthesis of a new class of macrocyclic oligosaccharides. J Org Chem. 2009;74:2928–2936. doi: 10.1021/jo8027856. [DOI] [PubMed] [Google Scholar]
- Muthana S, Yu H, Huang S, Chen X. Chemoenzymatic synthesis of size-defined polysaccharides by sialyltransferase-catalyzed block transfer of oligosaccharides. J Am Chem Soc. 2007;129:11918–11919. doi: 10.1021/ja075736b. [DOI] [PubMed] [Google Scholar]
- Nakatani F, Morita YS, Ashida H, Nagamune K, Maeda Y, Kinoshita T. Identification of a second catalytically active trans-sialidase in Trypanosoma brucei. Biochem Biophys Res Commun. 2011;415:421–425. doi: 10.1016/j.bbrc.2011.10.085. [DOI] [PubMed] [Google Scholar]
- Ni L, Chokhawala HA, Cao H, Henning R, Ng L, Huang S, Yu H, Chen X, Fisher AJ. Crystal structures of Pasteurella multocida sialyltransferase complexes with acceptor and donor analogues reveal substrate binding sites and catalytic mechanism. Biochemistry. 2007;46:6288–6298. doi: 10.1021/bi700346w. [DOI] [PubMed] [Google Scholar]
- Ni L, Sun M, Yu H, Chokhawala H, Chen X, Fisher AJ. Cytidine 5′-monophosphate (CMP)-induced structural changes in a multifunctional sialyltransferase from Pasteurella multocida. Biochemistry. 2006;45:2139–2148. doi: 10.1021/bi0524013. [DOI] [PubMed] [Google Scholar]
- Oppezzo P, Obal G, Baraibar MA, Pritsch O, Alzari PM, Buschiazzo A. Crystal structure of an enzymatically inactive trans-sialidase-like lectin from Trypanosoma cruzi: the carbohydrate binding mechanism involves residual sialidase activity. Biochim Biophys Acta. 2011;1814:1154–1161. doi: 10.1016/j.bbapap.2011.04.012. [DOI] [PubMed] [Google Scholar]
- Padler-Karavani V, Hurtado-Ziola N, Pu M, Yu H, Huang S, Muthana S, Chokhawala HA, Cao H, Secrest P, Friedmann-Morvinski D, Singer O, Ghaderi D, Verma IM, Liu YT, Messer K, Chen X, Varki A, Schwab R. Human xeno-autoantibodies against a non-human sialic acid serve as novel serum biomarkers and immunotherapeutics in cancer. Cancer Res. 2011;71:3352–3363. doi: 10.1158/0008-5472.CAN-10-4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padler-Karavani V, Yu H, Cao H, Chokhawala H, Karp F, Varki N, Chen X, Varki A. Diversity in specificity, abundance, and composition of anti-Neu5Gc antibodies in normal humans: potential implications for disease. Glycobiology. 2008;18:818–830. doi: 10.1093/glycob/cwn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons NJ, Constantinidou C, Cole JA, Smith H. Sialylation of lipopolysaccharide by CMP-NANA in viable gonococci is enhanced by low Mr material released from blood cell extracts but not by some UDP sugars. Microb Pathog. 1994;16:413–421. doi: 10.1006/mpat.1994.1041. [DOI] [PubMed] [Google Scholar]
- Paulson JC, Rademacher C. Glycan terminator. Nat Struct Mol Biol. 2009;16:1121–1122. doi: 10.1038/nsmb1109-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson JC, Weinstein J, Dorland L, van Halbeek H, Vliegenthart JF. Newcastle disease virus contains a linkage-specific glycoprotein sialidase. Application to the localization of sialic acid residues in N-linked oligosaccharides of alpha 1-acid glycoprotein. J Biol Chem. 1982;257:12734–12738. [PubMed] [Google Scholar]
- Peterson DC, Arakere G, Vionnet J, McCarthy PC, Vann WF. Characterization and acceptor preference of a soluble meningococcal group C polysialyltransferase. J Bacteriol. 2011;193:1576–1582. doi: 10.1128/JB.00924-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham T, Gregg CJ, Karp F, Chow R, Padler-Karavani V, Cao H, Chen X, Witztum JL, Varki NM, Varki A. Evidence for a novel human-specific xeno-auto-antibody response against vascular endothelium. Blood. 2009;114:5225–5235. doi: 10.1182/blood-2009-05-220400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescher JA, Dube DH, Bertozzi CR. Chemical remodelling of cell surfaces in living animals. Nature. 2004;430:873–877. doi: 10.1038/nature02791. [DOI] [PubMed] [Google Scholar]
- Priem B, Gilbert M, Wakarchuk WW, Heyraud A, Samain E. A new fermentation process allows large-scale production of human milk oligosaccharides by metabolically engineered bacteria. Glycobiology. 2002;12:235–240. doi: 10.1093/glycob/12.4.235. [DOI] [PubMed] [Google Scholar]
- Pshezhetsky AV, Richard C, Michaud L, Igdoura S, Wang S, Elsliger MA, Qu J, Leclerc D, Gravel R, Dallaire L, Potier M. Cloning, expression and chromosomal mapping of human lysosomal sialidase and characterization of mutations in sialidosis. Nat Genet. 1997;15:316–320. doi: 10.1038/ng0397-316. [DOI] [PubMed] [Google Scholar]
- Rao FV, Rich JR, Rakic B, Buddai S, Schwartz MF, Johnson K, Bowe C, Wakarchuk WW, Defrees S, Withers SG, Strynadka NC. Structural insight into mammalian sialyltransferases. Nat Struct Mol Biol. 2009;16:1186–1188. doi: 10.1038/nsmb.1685. [DOI] [PubMed] [Google Scholar]
- Rauvolfova J, Venot A, Boons GJ. Chemo-enzymatic synthesis of C-9 acetylated sialosides. Carbohydr Res. 2008;343:1605–1611. doi: 10.1016/j.carres.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartor PA, Agusti R, Leguizamon MS, Campetella O, de Lederkremer RM. Continuous nonradioactive method for screening trypanosomal trans-sialidase activity and its inhibitors. Glycobiology. 2010;20:982–990. doi: 10.1093/glycob/cwq056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato C, Kitajima K, Tazawa I, Inoue Y, Inoue S, Troy FA., 2nd Structural diversity in the alpha 2→8-linked polysialic acid chains in salmonid fish egg glycoproteins. Occurrence of poly(Neu5Ac), poly(Neu5Gc), poly(Neu5Ac, Neu5Gc), poly(KDN), and their partially acetylated forms. J Biol Chem. 1993;268:23675–23684. [PubMed] [Google Scholar]
- Schauer R. Achievements and challenges of sialic acid research. Glycoconj J. 2000;17:485–499. doi: 10.1023/A:1011062223612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling B, Goon S, Samuels NM, Gaucher SP, Leary JA, Bertozzi CR, Gibson BW. Biosynthesis of sialylated lipooligosaccharides in Haemophilus ducreyi is dependent on exogenous sialic acid and not mannosamine. Incorporation studies using N-acylmannosamine analogues, N-glycolylneuraminic acid, and 13C-labeled N-acetylneuraminic acid. Biochemistry. 2001;40:12666–12677. doi: 10.1021/bi0107849. [DOI] [PubMed] [Google Scholar]
- Schwarzkopf M, Knobeloch KP, Rohde E, Hinderlich S, Wiechens N, Lucka L, Horak I, Reutter W, Horstkorte R. Sialylation is essential for early development in mice. Proc Natl Acad Sci USA. 2002;99:5267–5270. doi: 10.1073/pnas.072066199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela DA, Li Y, Lerno L, Wu S, Marcobal AM, German JB, Chen X, Lebrilla CB, Mills DA. An infant-associated bacterial commensal utilizes breast milk sialyloligosaccharides. J Biol Chem. 2011;286:11909–11918. doi: 10.1074/jbc.M110.193359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severi E, Hood DW, Thomas GH. Sialic acid utilization by bacterial pathogens. Microbiology. 2007;153:2817–2822. doi: 10.1099/mic.0.2007/009480-0. [DOI] [PubMed] [Google Scholar]
- Shen GJ, Datta AK, Izumi M, Koeller KM, Wong CH. Expression of alpha 2,8/2,9-polysialyltransferase from Escherichia coli K92 - Characterization of the enzyme and its reaction products. J Biol Chem. 1999;274:35139–35146. doi: 10.1074/jbc.274.49.35139. [DOI] [PubMed] [Google Scholar]
- Shen Y, Kohla G, Lrhorfi AL, Sipos B, Kalthoff H, Gerwig GJ, Kamerling JP, Schauer R, Tiralongo J. O-Acetylation and de-O-acetylation of sialic acids in human colorectal carcinoma. Eur J Biochem. 2004a;271:281–290. doi: 10.1046/j.1432-1033.2003.03927.x. [DOI] [PubMed] [Google Scholar]
- Shen Y, Tiralongo J, Kohla G, Schauer R. Regulation of sialic acid O-acetylation in human colon mucosa. Biol Chem. 2004b;385:145–152. doi: 10.1515/BC.2004.033. [DOI] [PubMed] [Google Scholar]
- Silva MS, Prazeres DM, Lanca A, Atouguia J, Monteiro GA. Trans-sialidase from Trypanosoma brucei as a potential target for DNA vaccine development against African trypanosomiasis. Parasitology research. 2009;105:1223–1229. doi: 10.1007/s00436-009-1542-6. [DOI] [PubMed] [Google Scholar]
- Song X, Yu H, Chen X, Lasanajak Y, Tappert MM, Air GM, Tiwari VK, Cao H, Chokhawala HA, Zheng H, Cummings RD, Smith DF. A sialylated glycan microarray reveals novel interactions of modified sialic acids with proteins and viruses. J Biol Chem. 2011;286:31610–31622. doi: 10.1074/jbc.M111.274217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan GV, Schauer R. Assays of sialate-O-acetyltransferases and sialate-O-acetylesterases. Glycoconj J. 2009;26:935–944. doi: 10.1007/s10719-008-9131-y. [DOI] [PubMed] [Google Scholar]
- Steenbergen SM, Lee YC, Vann WF, Vionnet J, Wright LF, Vimr ER. Separate pathways for O-acetylation of polymeric and monomeric sialic acids and identification of sialyl O-acetyl esterase in Escherichia coli K1. J Bacteriol. 2006;188:6195–6206. doi: 10.1128/JB.00466-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenbergen SM, Lichtensteiger CA, Caughlan R, Garfinkle J, Fuller TE, Vimr ER. Sialic Acid metabolism and systemic pasteurellosis. Infect Immun. 2005;73:1284–1294. doi: 10.1128/IAI.73.3.1284-1294.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenbergen SM, Vimr ER. Functional relationships of the sialyltransferases involved in expression of the polysialic acid capsules of Escherichia coli K1 and K92 and Neisseria meningitidis groups B or C. J Biol Chem. 2003;278:15349–15359. doi: 10.1074/jbc.M208837200. [DOI] [PubMed] [Google Scholar]
- Steenbergen SM, Wrona TJ, Vimr ER. Functional analysis of the sialyltransferase complexes in Escherichia coli K1 and K92. J Bacteriol. 1992;174:1099–1108. doi: 10.1128/jb.174.4.1099-1108.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiarto G, Lau K, Li Y, Khedri Z, Yu H, Le D-T, Chen X. Decreasing the sialidase activity of multifunctional Pasteurella multocida α2–3-sialyltransferase 1 (PmST1) by site-directed mutagenesis. Mol Biosyst. 2011a;7:3021–3027. doi: 10.1039/c1mb05182b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiarto G, Lau K, Yu H, Vuong S, Thon V, Li Y, Huang S, Chen X. Cloning and characterization of a viral α2–3-sialyltransferase (vST3Gal-I) for the synthesis of sialyl Lewisx. Glycobiology. 2011b;21:387–396. doi: 10.1093/glycob/cwq172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Li Y, Chokhawala HA, Henning R, Chen X. N-Terminal 112 amino acid residues are not required for the sialyltransferase activity of Photobacterium damsela alpha2,6-sialyltransferase. Biotechnol Lett. 2008;30:671–676. doi: 10.1007/s10529-007-9588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takakura Y, Tsukamoto H, Yamamoto T. Molecular cloning, expression and properties of an alpha/beta-galactoside alpha2,3-sialyltransferase from Vibrio sp. JT-FAJ-16. J Biochem. 2007;142:403–412. doi: 10.1093/jb/mvm147. [DOI] [PubMed] [Google Scholar]
- Tan LK, Carlone GM, Borrow R. Advances in the development of vaccines against Neisseria meningitidis. N Engl J Med. 2010;362:1511–1520. doi: 10.1056/NEJMra0906357. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Kohler JJ. Photoactivatable crosslinking sugars for capturing glycoprotein interactions. J Am Chem Soc. 2008;130:3278–3279. doi: 10.1021/ja7109772. [DOI] [PubMed] [Google Scholar]
- Taylor RE, Gregg CJ, Padler-Karavani V, Ghaderi D, Yu H, Huang S, Sorensen RU, Chen X, Inostroza J, Nizet V, Varki A. Novel mechanism for the generation of human xeno-autoantibodies against the nonhuman sialic acid N-glycolylneuraminic acid. J Exp Med. 2010;207:1637–1646. doi: 10.1084/jem.20100575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada T, Kitazume S, Kitajima K, Inoue S, Ito F, Troy FA, Inoue Y. Synthesis of CMP-deaminoneuraminic acid (CMP-KDN) using the CTP:CMP-3-deoxynonulosonate cytidylyltransferase from rainbow trout testis. Identification and characterization of a CMP-KDN synthetase. J Biol Chem. 1993;268:2640–2648. [PubMed] [Google Scholar]
- Thompson H, Homer KA, Rao S, Booth V, Hosie AH. An orthologue of Bacteroides fragilis NanH is the principal sialidase in Tannerella forsythia. J Bacteriol. 2009;191:3623–3628. doi: 10.1128/JB.01618-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thon V, Lau K, Yu H, Tran BK, Chen X. PmST2: A novel Pasteurella multocida glycolipid α2–3-sialyltransferase. Glycobiology. 2011;21:1206–1216. doi: 10.1093/glycob/cwr054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thon V, Li Y, Yu H, Lau K, Chen X. PmST3 from Pasteurella multocida encoded by Pm1174 gene is a monofunctional alpha2–3-sialyltransferase. Appl Microbiol Biotechnol. 2012 doi: 10.1007/s00253-011-3676-6. [DOI] [PubMed] [Google Scholar]
- Tiralongo E, Schrader S, Lange H, Lemke H, Tiralongo J, Schauer R. Two trans-sialidase forms with different sialic acid transfer and sialidase activities from Trypanosoma congolense. J Biol Chem. 2003;278:23301–23310. doi: 10.1074/jbc.M212909200. [DOI] [PubMed] [Google Scholar]
- Tiralongo J, Schmid H, Thun R, Iwersen M, Schauer R. Characterisation of the enzymatic 4-O-acetylation of sialic acids in microsomes from equine submandibular glands. Glycoconj J. 2000;17:849–858. doi: 10.1023/a:1010965128335. [DOI] [PubMed] [Google Scholar]
- Tsukamoto H, Takakura Y, Mine T, Yamamoto T. Photobacterium sp. JT-ISH-224 produces two sialyltransferases, alpha-/beta-galactoside alpha2,3-sialyltransferase and beta-galactoside alpha2,6-sialyltransferase. J Biochem. 2008;143:187–197. doi: 10.1093/jb/mvm208. [DOI] [PubMed] [Google Scholar]
- Tsukamoto H, Takakura Y, Yamamoto T. Purification, cloning, and expression of an alpha/beta-galactoside alpha-2,3-sialyltransferase from a luminous marine bacterium, Photobacterium phosphoreum. J Biol Chem. 2007;282:29794–29802. doi: 10.1074/jbc.M701907200. [DOI] [PubMed] [Google Scholar]
- Tullius MV, Munson RS, Jr, Wang J, Gibson BW. Purification, cloning, and expression of a cytidine 5′-monophosphate N-acetylneuraminic acid synthetase from Haemophilus ducreyi. J Biol Chem. 1996;271:15373–15380. doi: 10.1074/jbc.271.26.15373. [DOI] [PubMed] [Google Scholar]
- Vandamme-Feldhaus V, Schauer R. Characterization of the enzymatic 7-O-acetylation of sialic acids and evidence for enzymatic O-acetyl migration from C-7 to C-9 in bovine submandibular gland. J Biochem. 1998;124:111–121. doi: 10.1093/oxfordjournals.jbchem.a022069. [DOI] [PubMed] [Google Scholar]
- Varki A, Diaz S. The transport and utilization of acetyl coenzyme A by rat liver Golgi vesicles. O-acetylated sialic acids are a major product. J Biol Chem. 1985;260:6600–6608. [PubMed] [Google Scholar]
- Varki A, Schauer R. Sialic acid. In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of Glycobiology. 2. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2008. pp. 199–217. [Google Scholar]
- Varki NM, Strobert E, Dick EJ, Jr, Benirschke K, Varki A. Biomedical differences between human and nonhuman hominids: potential roles for uniquely human aspects of sialic acid biology. Annu Rev Pathol. 2011;6:365–393. doi: 10.1146/annurev-pathol-011110-130315. [DOI] [PubMed] [Google Scholar]
- Verheijen FW, Verbeek E, Aula N, Beerens CE, Havelaar AC, Joosse M, Peltonen L, Aula P, Galjaard H, van der Spek PJ, Mancini GM. A new gene, encoding an anion transporter, is mutated in sialic acid storage diseases. Nat Genet. 1999;23:462–465. doi: 10.1038/70585. [DOI] [PubMed] [Google Scholar]
- Vimr ER, Bergstrom R, Steenbergen SM, Boulnois G, Roberts I. Homology among Escherichia coli K1 and K92 polysialytransferases. J Bacteriol. 1992;174:5127–5131. doi: 10.1128/jb.174.15.5127-5131.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vimr ER, Kalivoda KA, Deszo EL, Steenbergen SM. Diversity of microbial sialic acid metabolism. Microbiol Mol Biol Rev. 2004;68:132–153. doi: 10.1128/MMBR.68.1.132-153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan K, Lawrence S, Hinderlich S, Yarema KJ, Lee YC, Betenbaugh MJ. Engineering sialic acid synthetic ability into insect cells: identifying metabolic bottlenecks and devising strategies to overcome them. Biochemistry. 2003;42:15215–15225. doi: 10.1021/bi034994s. [DOI] [PubMed] [Google Scholar]
- von Itzstein M. The war against influenza: discovery and development of sialidase inhibitors. Nat Rev Drug Discov. 2007;6:967–974. doi: 10.1038/nrd2400. [DOI] [PubMed] [Google Scholar]
- von Itzstein M, Thomson R. Anti-influenza drugs: the development of sialidase inhibitors. Handb Exp Pharmacol. 2009:111–154. doi: 10.1007/978-3-540-79086-0_5. [DOI] [PubMed] [Google Scholar]
- Vrielink A, Ruger W, Driessen HP, Freemont PS. Crystal structure of the DNA modifying enzyme beta-glucosyltransferase in the presence and absence of the substrate uridine diphosphoglucose. EMBO J. 1994;13:3413–3422. doi: 10.1002/j.1460-2075.1994.tb06646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Yoshikawa Y, Tokuyama S, Kuwabara M, Akita H, Miyagi T. Cloning, expression, and chromosomal mapping of a human ganglioside sialidase. Biochem Biophys Res Commun. 1999;261:21–27. doi: 10.1006/bbrc.1999.0973. [DOI] [PubMed] [Google Scholar]
- Warren L, Blacklow RS. The biosynthesis of cytidine 5′-monophospho-N-acetylneuraminic acid by an enzyme from Neisseria meningitidis. J Biol Chem. 1962;237:3527–3534. [PubMed] [Google Scholar]
- Watts AG, Damager I, Amaya ML, Buschiazzo A, Alzari P, Frasch AC, Withers SG. Trypanosoma cruzi trans-sialidase operates through a covalent sialyl-enzyme intermediate: tyrosine is the catalytic nucleophile. J Am Chem Soc. 2003;125:7532–7533. doi: 10.1021/ja0344967. [DOI] [PubMed] [Google Scholar]
- Willis LM, Gilbert M, Karwaski MF, Blanchard MC, Wakarchuk WW. Characterization of the alpha-2,8-polysialyltransferase from Neisseria meningitidis with synthetic acceptors, and the development of a self-priming polysialyltransferase fusion enzyme. Glycobiology. 2008;18:177–186. doi: 10.1093/glycob/cwm126. [DOI] [PubMed] [Google Scholar]
- Wu J, Guo Z. Improving the antigenicity of sTn antigen by modification of its sialic acid residue for development of glycoconjugate cancer vaccines. Bioconjug Chem. 2006;17:1537–1544. doi: 10.1021/bc060103s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Hamada Y, Ichikawa M, Kajiwara H, Mine T, Tsukamoto H, Takakura Y. A beta-galactoside alpha2,6-sialyltransferase produced by a marine bacterium, Photobacterium leiognathi JT-SHIZ-145, is active at pH 8. Glycobiology. 2007;17:1167–1174. doi: 10.1093/glycob/cwm086. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Nakashizuka M, Terada I. Cloning and expression of a marine bacterial beta-galactoside alpha2,6-sialyltransferase gene from Photobacterium damsela JT0160. J Biochem. 1998;123:94–100. doi: 10.1093/oxfordjournals.jbchem.a021921. [DOI] [PubMed] [Google Scholar]
- Yang G, Rich JR, Gilbert M, Wakarchuk WW, Feng Y, Withers SG. Fluorescence activated cell sorting as a general ultra-high-throughput screening method for directed evolution of glycosyltransferases. J Am Chem Soc. 2010;132:10570–10577. doi: 10.1021/ja104167y. [DOI] [PubMed] [Google Scholar]
- Yardeni T, Choekyi T, Jacobs K, Ciccone C, Patzel K, Anikster Y, Gahl WA, Kurochkina N, Huizing M. Identification, tissue distribution, and molecular modeling of novel human isoforms of the key enzyme in sialic acid synthesis, UDP-GlcNAc 2-epimerase/ManNAc kinase. Biochemistry. 2011;50:8914–8925. doi: 10.1021/bi201050u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarema KJ, Bertozzi CR. Chemical approaches to glycobiology and emerging carbohydrate-based therapeutic agents. Curr Opin Chem Biol. 1998;2:49–61. doi: 10.1016/s1367-5931(98)80035-5. [DOI] [PubMed] [Google Scholar]
- Yu H, Cao H, Tiwari VK, Li Y, Chen X. Chemoenzymatic synthesis of C8-modified sialic acids and related alpha2–3- and alpha2–6-linked sialosides. Bioorg Med Chem Lett. 2011;21:5037–5040. doi: 10.1016/j.bmcl.2011.04.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Chen X. Carbohydrate post-glycosylational modifications. Org Biomol Chem. 2007;5:865–872. doi: 10.1039/b700034k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Cheng J, Ding L, Khedri Z, Chen Y, Chin S, Lau K, Tiwari VK, Chen X. Chemoenzymatic synthesis of GD3 oligosaccharides and other disialyl glycans containing natural and non-natural sialic acids. J Am Chem Soc. 2009;131:18467–18477. doi: 10.1021/ja907750r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Chokhawala H, Karpel R, Wu B, Zhang J, Zhang Y, Jia Q, Chen X. A multifunctional Pasteurella multocida sialyltransferase: a powerful tool for the synthesis of sialoside libraries. J Am Chem Soc. 2005;127:17618–17619. doi: 10.1021/ja0561690. [DOI] [PubMed] [Google Scholar]
- Yu H, Chokhawala HA, Huang S, Chen X. One-pot three-enzyme chemoenzymatic approach to the synthesis of sialosides containing natural and non-natural functionalities. Nat Protoc. 2006a;1:2485–2492. doi: 10.1038/nprot.2006.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Chokhawala HA, Varki A, Chen X. Efficient chemoenzymatic synthesis of biotinylated human serum albumin-sialoglycoside conjugates containing O-acetylated sialic acids. Org Biomol Chem. 2007;5:2458–2463. doi: 10.1039/b706507h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Huang S, Chokhawala H, Sun M, Zheng H, Chen X. Highly efficient chemoenzymatic synthesis of naturally occurring and non-natural alpha-2,6-linked sialosides: a P. damsela alpha-2,6-sialyltransferase with extremely flexible donor-substrate specificity. Angew Chem Int Ed Engl. 2006b;45:3938–3944. doi: 10.1002/anie.200600572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Ryan W, Yu H, Chen X. Characterization of a bifunctional cytidine 5′-monophosphate N-acetylneuraminic acid synthetase cloned from Streptococcus agalactiae. Biotechnol Lett. 2006c;28:107–113. doi: 10.1007/s10529-005-4955-z. [DOI] [PubMed] [Google Scholar]
- Yu H, Yu H, Karpel R, Chen X. Chemoenzymatic synthesis of CMP-sialic acid derivatives by a one-pot two-enzyme system: comparison of substrate flexibility of three microbial CMP-sialic acid synthetases. Bioorg Med Chem. 2004;12:6427–6435. doi: 10.1016/j.bmc.2004.09.030. [DOI] [PubMed] [Google Scholar]
- Zanetta JP, Srinivasan V, Schauer R. Analysis of monosaccharides, fatty constituents and rare O-acetylated sialic acids from gonads of the starfish Asterias rubens. Biochimie. 2006;88:171–178. doi: 10.1016/j.biochi.2005.07.010. [DOI] [PubMed] [Google Scholar]