Abstract
Genetic factors may interact with lifestyle factors to modify obesity risk. FTO and PPARG2 are relevant obesogenes. Our aim was to explore the effect of Pro12Ala (rs1801282) of PPARG2 and rs9939609 of FTO on obesity risk and to examine their interaction with lifestyle factors in an elderly population. Subjects (n = 978; aged 69 ± 6) were recruited from the SUN (Seguimiento Universidad de Navarra) Project. DNA was obtained from saliva, and lifestyle and dietary data were collected by validated self-reported questionnaires. Genotyping was assessed by RT-PCR plus allele discrimination. Subjects carrying the Ala allele of PPARG2 gene had a significantly increased obesity risk compared to non-carrier (Pro12Pro) subjects (OR, 1.66; 95 % CI, 1.01–2.74; p = 0.045). Greater obesity risk was also found in inactive or high carbohydrate intake subjects with the Ala12 allele of PPARG2 gene. Interestingly, subjects carrying the Ala allele of the PPARG2 gene and with a high CHO (>246 g/day) intake had an increased obesity risk compared to Pro12Pro subjects (OR, 2.67; 95 % CI, 1.3–5.46; p = 0.007; p for [CHO × PPARG2] interaction = 0.046). Moreover, in subjects with a high CHO intake, the co-presence of the Ala allele of PPARG2 gene and one minor A allele (rs9939609) of FTO gene did increase obesity risk (OR, 3.26; 95 % CI, 1.19–8.89; p = 0.021) when compared to non-carrier (Pro12Pro/TT) subjects. In conclusion, it appears that lifestyle factors may act as effect modifiers for obesity risk linked to Ala12 allele of the PPARG2 gene and the minor A allele of FTO gene in an elderly population.
Keywords: PPARG2, Pro12ala, FTO, rs9939609, Obesity risk
Background
Obesity is a complex disease with genetic and environmental basis (Marti et al. 2008). FTO and PPARG2 gene variants for obesity risk have been widely studied (Razquin et al. 2011). Several meta-analyses showed an increased body mass index (BMI) in subjects with the Ala allele of the PPARG2 gene (Masud 2003; Tonjes et al. 2006). This observation was recently confirmed; carriers of the Ala allele of the PPARG2 gene had a significant higher BMI (+0.060 kg/m2) compared to non-carriers (Galbete et al., in press), with a total of 49,337 subjects.
As it is known, the FTO gene harbours the stronger association with adiposity in genome-wide association (GWA) studies, although the physiological function of FTO remains unclear (Tung 2011). In the large meta-analysis of GWAS, thus far performed with 123,865 individuals of European ancestry, the FTO locus was confirmed as one of the 32 variants associated with BMI with p-values <5 × 10−8 (Speliotes et al. 2010; Speakman et al. 2011). A significant association between rs9939609 SNP of FTO gene and obesity, with an overall odds ratio (OR) for obesity of 1.31 under per-allele comparison, was reported in another meta-analysis including 111,571 subjects (Peng et al. 2011).
Epidemiological studies have suggested that in addition to genetic factors, a variety of lifestyle factors (e.g. dietary composition, low level of physical activity (PA)) may contribute to the epidemic of obesity and interact with genetic factors to modify obesity risk (Chung 2008; Walley et al. 2009).
The interaction between lifestyle factors and these gene variants (Pro12Ala of PPARG2 (rs1801282) and rs9939609 of FTO) have been explored in different populations and cohorts. On one hand, some studies have reported an interaction between Ala allele of PPARG2 gene variant and carbohydrate (CHO) or fat intake, (Marti et al. 2002; Lamri et al. 2012) on obesity risk, whereas in others no association was found (Memisoglu et al. 2003; Nelson et al. 2007). A significant interaction between food intake and rs9939609 SNP of FTO gene on BMI was detected in some populations (Corella et al. 2011; Lappalainen et al. 2012; Moleres et al. 2012).
With regard to PA, recently, Kilpelainen et al. (2011) using data from 45 studies with a total of 218,166 adults conducted a meta-analysis. They reported a significant interaction between the minor A allele of rs9939609 and PA, the odds for obesity risk being 27 % smaller in active versus inactive subjects.
A cohort study is the best way to identify incidence and natural history of a disease and can be used to examine multiple outcomes after a single exposure (Grimes 2002). The SUN Project (Seguimiento Universidad de Navarra–University of Navarra Follow-up) is a multi-purpose prospective Mediterranean dynamic cohort designed to study the association of diet and other lifestyle factors with various health outcomes including cardiovascular disease, hypertension, diabetes or obesity (Martinez-Gonzalez et al. 2002; Segui-Gomez et al. 2006).
The aim of this study was to explore the effect of two gene variants, Pro12Ala of PPARG2 and rs9939609 of FTO, on obesity risk and to examine their interaction with lifestyle factors in an elderly population of the SUN study.
Subjects and methods
Sample population
This work has been conducted within the framework of the SUN Project (Martinez-Gonzalez et al. 2002). The SUN Project was initiated in December 1999 in Spain, and recruitment was permanently open. All participants were university graduates and about 50 % of them are health professionals themselves.
Lifestyle and dietary data were collected by self-reported biennially mailed questionnaires (Alonso et al. 2005; Bes-Rastrollo et al. 2005; Martinez-Gonzalez et al. 2005). Dietary intake was assessed using a semi-quantitative food frequency questionnaire (136 food items) included at baseline. Validity and reproducibility of this questionnaire has recently been re-evaluated (de la Fuente-Arrillaga et al. 2010). Nutrient intakes of 136 food items were calculated as frequency multiplied by nutrient composition of specified portion size for each food item, using an ad hoc computer program developed for this purpose. A trained dietician updated the nutrient databank using the latest available information from the food composition table for Spain. Baseline intake of macronutrients was analysed as quantitative variables (grams per day) (de la Fuente-Arrillaga et al. 2010; Fernandez-Ballart et al. 2010).
PA was ascertained through a baseline 17-item questionnaire. The index of metabolic equivalent task hours per week (METs-h/week) was computed using the time spent engaging in 17 activities and multiplying the time spent by the resting metabolic rate (MET-score) specific for each activity. The METs-h/week for all activities were combined to obtain a value of total METs-h/week, which adequately correlated with the objectively measured energy expenditure in a validation study in a subsample of the cohort (Martinez-Gonzalez et al. 2005).
For this research, elderly participants (more than 55 years old when the baseline questionnaire was completed) of the SUN Project were invited to participate in a genetic study in May 2008. Each participant received a kit designed to collect saliva, and 1085 participants agreed to participate but 986 kits were received back. Finally, 978 volunteers were correctly genotyped for the rs1801282 SNP (PPARG2), and 967 for the rs9939609 SNP (FTO). The mean age was 69 years (70 % male). Anthropometric data were collected from the baseline questionnaire. Self-reported information on BMI had been previously validated in a subsample of the SUN Project (Bes-Rastrollo et al. 2005). Specific written informed consent was required to participate in this study. The study protocol was performed in accordance with the ethical standards of the Declaration of Helsinki (as revised in Hong Kong in 1989, in Edinburgh in 2000 and in South Korea in 2008) and was approved by the Institutional Ethical Review Board of the University of Navarra.
Genotyping
Saliva samples were collected with specially designed kits (Oragene®ADN Self-Collection kit-OG250), and DNA was extracted according to the manufacturer’s instructions. The genotyping for the Pro12Ala SNP of PPARG2 gene (rs1801282) and for the rs9939609 SNP of the FTO gene were performed using TaqMan assays with allele-specific probes on the ABI Prism 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA, USA) according to standardized laboratory protocols.
Statistical analysis
Hardy–Weinberg equilibrium was tested using a χ2 test. This test was also used to analyse if there were differences on the genotype distribution according to obesity status.
The odds ratio (OR) for obesity associated with genotypes (dominant models) were fitted with an unconditional logistic regression model after adjustment for sex, age, PA and total energy intake as covariables. To address the combined effect of these two polymorphisms [Pro12Ala (rs1801282) of PPARG2 and rs9939609 of FTO gene (dominant model)], dummy variables were created. Non-carrier subjects (Pro12Pro and TT) were considered as the reference category. Three different categories according to the genotypes were considered: the presence of Ala allele (rs1801282) of PPARG2 gene, the presence of A allele (rs9939609) of FTO gene and the co-presence of the two risk alleles (Ala and A allele). The association between the different possible genotypes and BMI was analysed using linear regression models and analysis of covariance (ANCOVA), after adjusting for potential confounders (sex, age, PA and total energy intake). We also evaluated the interaction between the genetic variants Pro12Ala (rs1801282) of PPARG2 and rs9939609 of FTO and a high CHO intake or low PA practice (dichotomized at the median) on obesity risk. Indicated interactions were tested for obesity risk with the likelihood ratio test. Product terms between the SNPs and lifestyle factors were calculated firstly with the corresponding variables dichotomized at the median (model 1 and 3) and secondly as continuous variables (model 2 and 4). Interactions between the SNPs and lifestyle factors on BMI (as a continuous variable) were also tested.
Results
Anthropometrical and lifestyle characteristics of elderly subjects of the SUN cohort according to the two genotypes (Pro12Ala SNP (rs1801282) of PPARG2 and the rs9939609 SNP of FTO gene, dominant model) are shown in Table 1. The frequencies of these two SNPs did fulfil the Hardy–Weinberg equilibrium.
Table 1.
Baseline characteristics according to genotype for elderly subjects from the SUN Project
| PPARG2 rs1801282 | FTO rs9939609 | |||||
|---|---|---|---|---|---|---|
| Pro12Pro (n = 814) |
Ala12 (n = 164) |
p-valuea | TT (n = 336) |
TA/AA (n = 631) |
p-valuea | |
| % male | 70 % | 74 % | 0.275 | 72 % | 70 % | 0.657 |
| Age (years) | 69 (6) | 70 (7) | 0.071 | 69 (6) | 69 (6) | 0.484 |
| BMI (kg/m2) | 25.7 (3.2) | 26.2 (3.2) | 0.091 | 25.6 (3.1) | 25.9 (3.2) | 0.173 |
| Total energy intake (kcal/day) | 2378 (903) | 2484 (1021) | 0.182 | 2412 (1038) | 2384 (862) | 0.654 |
| CHO intake (g/day) | 267 (129) | 281 (147) | 0.200 | 272 (147) | 267 (123) | 0.647 |
| Protein intake (g/day) | 107 (40) | 109 (37) | 0.686 | 109 (46) | 107 (36) | 0.449 |
| Fat intake (g/day) | 91 (39) | 93 (42) | 0.392 | 91 (41) | 91 (39) | 0.883 |
| Physical activity (METs-h/week) | 23.9 (20.7) | 24.3 (21.5) | 0.828 | 23.3 (20.1) | 24.5 (21.3) | 0.387 |
Values are expressed as mean (SD), unless otherwise stated
aContinuous variables were compared using student’s t-test. Categorical variables were compared using χ2 test
The ORs for obesity risk were calculated for each gene variant after adjustment for sex, age, PA and total energy intake. The presence of the Ala allele of PPARG2 gene significantly increased obesity risk in the adjusted models (including total population, subjects with high CHO intake and those with low PA practice). The obesity risk linked to the Ala12 allele of PPARG2 was 1.66 (95 % CI, 1.01–2.74; p = 0.045) in the total population (Table 2).
Table 2.
Odds ratios (OR) for obesity risk and linear regression coefficients for the association between the rs9939609 of FTO gene and Pro12Ala SNPs of the PPARG2 gene and BMI in elderly participants of the SUN Project
| OR (95 % CI) for obesity | p value | p for interactiona | B (95 % CI)b | p value | p for interactionc | ||
|---|---|---|---|---|---|---|---|
| PPARG2 (rs1801282) | |||||||
| Pro12Pro | 1 (ref) | 0 (ref) | |||||
| Ala12 | 1.66 (1.01–2.74) | 0.045 | 0.40 (−0.13–0.90) | 0.139 | |||
| FTO (rs9939609) | |||||||
| TT | 1 (ref) | 0 (ref) | |||||
| TA/AA | 1.03 (0.66–1.60) | 0.892 | 0.33 (−0.08–0.74) | 0.112 | |||
| Genotype | |||||||
| FTOe | PPARG2e | ||||||
| − | − | 1 (ref) | 0 (ref) | ||||
| − | + | 1.92 (0.86–4.27) | 0.111 | 0.46 (−0.36–1.30) | 0.287 | ||
| + | − | 1.10 (0.66–1.84) | 0.704 | 0.34 (−0.11–0.79) | 0.135 | ||
| + | + | 1.71 (0.84–3.48) | 0.138 | 0.79 (0.08–1.50) | 0.030 | ||
| High CHO intake (>246 g/day) | model 1/model 2 | ||||||
| PPARG2 (rs1801282) | 0.046/0.030 | 0.260 | |||||
| Pro12Pro | 1 (ref) | 0 (ref) | |||||
| Ala12 | 2.67 (1.30–5.46) | 0.007d | 0.49 (−0.21–1.20) | 0.169 | |||
| FTO (rs9939609) | 0.609/0.449 | 0.739 | |||||
| TT | 1 (ref) | 0 (ref) | |||||
| TA/AA | 1.15 (0.57–2.31) | 0.697 | 0.46 (−0.11–1.03) | 0.111 | |||
| Genotype | |||||||
| FTOe | PPARG2e | 0.814/0.844 | 0.973 | ||||
| − | − | 1 (ref) | 0 (ref) | ||||
| − | + | 1.92 (0.79–6.76) | 0.312 | 0.28 (−0.03–1.43) | 0.639 | ||
| + | − | 1.04 (0.60–2.41) | 0.924 | 0.41 (−0.22–1.03) | 0.204 | ||
| + | + | 3.26 (1.19–8.89) | 0.021 | 1.07 (0.10–2.03) | 0.031 | ||
| Low physical activity practice (< 18.6 METs-h/week) | model 3/model 4 | ||||||
| PPARG2 (rs1801282) | 0.266/0.243 | 0.741 | |||||
| Pro12Pro | 1 (ref) | 0 (ref) | |||||
| Ala12 | 2.14 (1.13–4.05) | 0.020 | 0.93 (0.16–1.69) | 0.017d | |||
| FTO (rs9939609) | 0.366/0.152 | 0.417 | |||||
| TT | 1 (ref) | 0 (ref) | |||||
| TA/AA | 1.14 (0.64–2.06) | 0.652 | 0.56 (−0.05–1.16) | 0.070 | |||
| Genotype | |||||||
| FTOe | PPARG2e | 0.346/0.230 | 0.360 | ||||
| − | − | 1 (ref) | 0 (ref) | ||||
| − | + | 2.31 (0.79–6.76) | 0.125 | 0.63 (−0.65–1.90) | 0.334 | ||
| + | − | 1.21 (0.60–2.41) | 0.594 | 0.48 (−0.18–1.14) | 0.156 | ||
| + | + | 2.51 (1.01–6.23) | 0.047 | 1.67 (0.63–2.71) | 0.002d | ||
Adjusted for gender, age, physical activity and total energy intake
model 1: interaction term = genotype*CHO (dichotomized at the median); model 2: interaction term = genotype*CHO (continuous); model 3: interaction term = genotype*PA (dichotomized at the median); model 4: interaction term = genotype*PA (continuous)
ap value for likelihood ratio test for obesity risk
bAdjusted differences in average BMI (kg/m2) between genotypes
cp value for interaction for BMI (as continuous variable)
dp value <0.05 after correcting for Benjamini–Hochberg multiple comparisons
e(−) Non-carriers of the minor risk alleles (+). Subjects carrying the minor risk alleles, either Pro12Ala of PPARG2 gene or rs9939609 of FTO gene
Interestingly, as shown in Table 2, obesity risk was higher in subjects with a high CHO consumption (>246 g/day), carrying the Ala allele of the PPARG2 gene (OR, 2.67; 95 % CI, 1.30–5.46; p = 0.007). This p-value did remain statistically significant after applying the Benjamini–Hochberg multiple comparison correction. The interaction for obesity risk between CHO intake and PPARG2 gene was also statistically significant (p for [CHO × PPARG2] interaction = 0.046). Similar results for this interaction were also obtained when considering CHO as a continuous trait (p for [CHO × PPARG2] interaction = 0.030).
Furthermore, subjects with a high CHO intake and carriers of the Ala allele had an increased obesity risk by the co-presence of one minor A allele (rs9939609) of FTO gene (OR, 3.26; 95 % CI, 1.19–8.89; p = 0.021) compared to non-carriers of the two alleles (Pro12Pro and TT) in subjects with a high CHO intake.
The presence of the Ala12 allele (rs1801282) of PPARG2 gene increased obesity risk to 2.14 (95 % CI, 1.13–4.05; p = 0.020) in subjects with a sedentary lifestyle (<18.6 METs-h/week) compared to Pro12Pro subjects. However, there was no evidence of statistical interaction (p for interaction = 0.243). Furthermore, in inactive carrier subjects of the Ala12 allele of PPARG2, the co-presence of the minor A allele (rs9939609) of FTO gene had a further rise in obesity risk to 2.51 (95 % CI, 1.01–6.23; p = 0.047) compared to inactive non-carrier (Pro12Pro and TT) subjects. The interactions between the genetic variants and low PA practice for obesity risk were not statistically significant (Table 2).
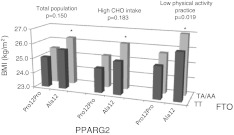
Linear regression models were also fitted to confirm the association between the co-presence of the two risk alleles (Ala of the PPARG2 and A allele of FTO gene) and BMI (as a continuous variable) in the three models undertaken: total population, high CHO intake and low PA practice (Table 2). Moreover, the same tendency was observed in the ANCOVA analysis (Fig. 1).
Fig. 1.
BMI differences according to genotype (dominant models for Pro12Ala and rs9939609 SNPs) for the three population groups (total population, only subjects with a high CHO intake or subjects with a low physical activity practice). Adjusted for sex, age, physical activity and total energy intake. *p < 0.05 between Ala12 + TA/AA and Pro12Pro + TT genotypes
Discussion
The main finding of this work is that a high CHO consumption seems to modify the obesity risk linked to the Pro12Ala SNPs of the PPARG2 gene in an elderly population. Some strengths of the SUN cohort deserve to be mentioned: the homogeneity of participants with regard to socio-economic status, which helps to better control confounding and the higher educational level of participants in the cohort that ensures a higher validity in self-reported information (Beunza et al. 2010; Sayon-Orea et al. 2011). A potential limitation in our study is the self-reported outcome; nevertheless, self-reported weight and BMI had been previously validated (Bes-Rastrollo et al. 2005). Another limitation is that identifying interactions between genetic variants and lifestyle factors may need much larger sample size (Smith 1984).
The present work shows that the co-presence of these two risk alleles in PPARG2 and FTO gene increases obesity predisposition, but novel studies are needed to elucidate the potential mechanisms. Pro12Ala variant of PPARG2 gene is one of the most studied genes as potentially linked to obesity phenotypes (Razquin et al. 2011). Previous meta-analysis had associated the Ala12 minor allele with a higher BMI (Masud 2003; Tonjes et al. 2006; Galbete et al. in press), and this study confirmed in a larger sample of aged subject the association of the Ala12 allele with obesity risk.
Depending on the genotype, the response of individuals to a dietary component or components could be different. The Pro12Ala genetic variant is probably the most studied mutation in relation to the interaction with dietary components on adiposity features. Fatty acids are natural agonists of PPARG transcription factor; consequently, most of the studies have been directed to analyse the interaction between Pro12Ala and fat intake. However, this study replicated an earlier association of this PPARG2 genetic variant with obesity risk linked to a high CHO intake (Marti et al. 2002). Notably, in our study, the interaction between CHO consumption and this Ala12 allele of PPARG2 for obesity risk was statistically significant although confirmation is needed in larger sample studies.
The PPARG Pro12Ala genotype seems to be associated with obesity, type 2 diabetes and CHD risk (Dallongeville et al. 2009). This variant is a diet-dependent sensor, and in the presence of a positive energy balance, the adipogenic capacity of the Ala allele exceeds that of the Pro12 genotype, being partially attributed to diet-dependent effects of the PPARG2 Pro12Ala genotype on adiponectin signalling and on the interaction of PPARG2 with several transcriptional coregulators (Anderson et al. 2010). From a mechanistic point of view, it is shown that the Ala12 allele alters ligand interaction between PPARG2 and its cofactors (Pgc1alfa, SRC1, Ncor), leading to an effect beyond decreased DNA binding efficiency (Heikkinen et al. 2009). The enhancement in obesity risk linked to a high CHO intake may be partly explained by the fact that CHO are not able to activate the PPARG protein and could worsen the action of the Ala12 substitution on the receptor activity.
The impact of this rs9939609 SNP of FTO gene on human body weight is mainly through energy intake; however, some results are contradictory (Berentzen et al. 2008; Do et al. 2008; Speakman et al. 2008, Goossens et al. 2009; Haupt et al. 2009). In our elderly population, no effect of FTO on obesity was found. This observation agrees with former findings in mature subjects. Hardy et al. (2010) described a weak association between FTO and BMI at the age of 50. Jacobsson et al. (2011) suggested that the effect of FTO on corporal adiposity may decrease by age. Our limited sample size could also impair our ability to find significant results.
With regard to PA, it is well known that there is an inverse relationship with obesity (Levine et al. 1999; Levine et al. 2005; Kuliczkowska et al. 2008). Previous studies had reported that a high PA practice was linked to a lower fasting insulin level in Pro12 homozygous subjects of PPARG2 gene (Franks et al. 2004), but no studies were found on interactions between Pro12Ala polymorphism and inactivity on obesity risk. Nevertheless, our results suggested an association of this genetic variant with obesity risk linked to a low PA practice.
To our knowledge, we assessed for the first time the joint association of PPARG2 and FTO gene variants on obesity risk when modulated by lifestyle factors. Previous studies have found a higher obesity risk associated with the combined effect of several polymorphisms. Some research work which reported the combined effect of PPARG and ADRB3 or ACE I/D gene variants for increasing BMI (Huang et al. 2011; Passaro et al. 2011) stated that the combined effect of FTO and MC4R genetic variants was strongly associated with obesity risk and BMI. Similarly, Cauchi et al. (2009) observed that these two genetic variants increased obesity risk by 24 %, and low PA levels did accentuate this effect. Our study showed that the effect of PPARG2 (Ala12 allele) and FTO (rs9939609) gene variants on obesity effect might depend on high CHO intake.
In summary, it seems that lifestyle factors may act as effect modifiers for obesity risk linked to Ala12 allele of the PPARG2 gene and the minor A allele of FTO gene in an elderly population.
Acknowledgments
The SUN study has received funding from the Spanish Government (Grants PI01/0619, PI030678, PI040233, PI042241, PI050976, PI070240, PI070312, PI081943, PI080819, PI1002658, PI1002293, RD06/0045, G03/140 and 87/2010), the Navarra Regional Government (36/2001, 43/2002, 41/2005 and 36/2008) and the University of Navarra, Línea Especial, Nutrición y Obesidad (University of Navarra), Carlos III Health Institute (CIBER project, CB06/03/1017) and RETICS network. The scholarship to C. Galbete from the Asociación de Amigos de la Universidad de Navarra is fully acknowledged.
Conflict of interest
The authors have no competing interests.
References
- Alonso A, Beunza JJ, Delgado-Rodriguez M, Martinez-Gonzalez MA. Validation of self reported diagnosis of hypertension in a cohort of university graduates in Spain. BMC Public Health. 2005;5:94. doi: 10.1186/1471-2458-5-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson AL, Harris TB, Houston DK, Tylavsky FA, Lee JS, Sellmeyer DE, Sahyoun NR. Relationships of dietary patterns with body composition in older adults differ by gender and PPAR-gamma Pro12Ala genotype. Eur J Nutr. 2010;49:385–394. doi: 10.1007/s00394-010-0096-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berentzen T, Kring SI, Holst C, et al. Lack of association of fatness-related FTO gene variants with energy expenditure or physical activity. J Clin Endocrinol Metab. 2008;93:2904–2908. doi: 10.1210/jc.2008-0007. [DOI] [PubMed] [Google Scholar]
- Bes-Rastrollo M, Perez Valdivieso JR, Sánchez-Villegas A, Alonso A, Martinez-Gonzalez MA. Validación del peso e indice de masa corporal auto-declarados de los participantes de una cohorte de graduados universitarios. Rev Esp Obes. 2005;3:183–189. [Google Scholar]
- Beunza JJ, Toledo E, Hu FB, et al. Adherence to the Mediterranean diet, long-term weight change, and incident overweight or obesity: the Seguimiento Universidad de Navarra (SUN) cohort. Am J Clin Nutr. 2010;92:1484–1493. doi: 10.3945/ajcn.2010.29764. [DOI] [PubMed] [Google Scholar]
- Cauchi S, Stutzmann F, Cavalcanti-Proenca C, et al. Combined effects of MC4R and FTO common genetic variants on obesity in European general populations. J Mol Med (Berl) 2009;87:537–546. doi: 10.1007/s00109-009-0451-6. [DOI] [PubMed] [Google Scholar]
- Chung WK, Leibel RL. Considerations regarding the genetics of obesity. Obesity (Silver Spring) 2008;16(Suppl 3):S33–S39. doi: 10.1038/oby.2008.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corella D, Arnett DK, Tucker KL, et al. A high intake of saturated fatty acids strengthens the association between the fat mass and obesity-associated gene and BMI. J Nutr. 2011;141:2219–2225. doi: 10.3945/jn.111.143826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallongeville J, Iribarren C, Ferrieres J et al (2009) Peroxisome proliferator-activated receptor gamma polymorphisms and coronary heart disease. PPAR Res. doi:10.1155/2009/543746 [DOI] [PMC free article] [PubMed]
- Fuente-Arrillaga C, Ruiz ZV, Bes-Rastrollo M, Sampson L, Martinez-Gonzalez MA. Reproducibility of an FFQ validated in Spain. Public Health Nutr. 2010;13:1364–1372. doi: 10.1017/S1368980009993065. [DOI] [PubMed] [Google Scholar]
- Do R, Bailey SD, Desbiens K, et al. Genetic variants of FTO influence adiposity, insulin sensitivity, leptin levels, and resting metabolic rate in the Quebec Family Study. Diabetes. 2008;57:1147–1150. doi: 10.2337/db07-1267. [DOI] [PubMed] [Google Scholar]
- Fernandez-Ballart JD, Pinol JL, Zazpe I, et al. Relative validity of a semi-quantitative food-frequency questionnaire in an elderly Mediterranean population of Spain. Br J Nutr. 2010;103:1808–1816. doi: 10.1017/S0007114509993837. [DOI] [PubMed] [Google Scholar]
- Franks PW, Luan J, Browne PO, Harding AH, O’Rahilly S, Chatterjee VK, Wareham NJ. Does peroxisome proliferator-activated receptor gamma genotype (Pro12ala) modify the association of physical activity and dietary fat with fasting insulin level? Metabolism. 2004;53:11–16. doi: 10.1016/j.metabol.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Goossens GH, Petersen L, Blaak EE, et al. Several obesity- and nutrient-related gene polymorphisms but not FTO and UCP variants modulate postabsorptive resting energy expenditure and fat-induced thermogenesis in obese individuals: the NUGENOB study. Int J Obes (Lond) 2009;33:669–679. doi: 10.1038/ijo.2009.59. [DOI] [PubMed] [Google Scholar]
- Grimes DA, Schulz KF. Cohort studies: marching towards outcomes. Lancet. 2002;359:341–345. doi: 10.1016/S0140-6736(02)07500-1. [DOI] [PubMed] [Google Scholar]
- Hardy R, Wills AK, Wong A, et al. Life course variations in the associations between FTO and MC4R gene variants and body size. Hum Mol Genet. 2010;19:545–552. doi: 10.1093/hmg/ddp504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt A, Thamer C, Staiger H, et al. Variation in the FTO gene influences food intake but not energy expenditure. Exp Clin Endocrinol Diabetes. 2009;117:194–197. doi: 10.1055/s-0028-1087176. [DOI] [PubMed] [Google Scholar]
- Heikkinen S, Argmann C, Feige JN, et al. The Pro12Ala PPARgamma2 variant determines metabolism at the gene-environment interface. Cell Metab. 2009;9:88–98. doi: 10.1016/j.cmet.2008.11.007. [DOI] [PubMed] [Google Scholar]
- Huang W, Sun Y, Sun J. Combined effects of FTO rs9939609 and MC4R rs17782313 on obesity and BMI in Chinese Han populations. Endocrine. 2011;39:69–74. doi: 10.1007/s12020-010-9413-6. [DOI] [PubMed] [Google Scholar]
- Jacobsson JA, Almen MS, Benedict C, et al. Detailed analysis of variants in FTO in association with body composition in a cohort of 70-year-olds suggests a weakened effect among elderly. PLoS ONE. 2011;6:e20158. doi: 10.1371/journal.pone.0020158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpelainen TO, Qi L, Brage S, et al. Physical activity attenuates the influence of FTO variants on obesity risk: a meta-analysis of 218,166 adults and 19,268 children. PLoS Med. 2011;8:e1001116. doi: 10.1371/journal.pmed.1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuliczkowska J, Filus A, Trzmiel A, Tworowska U, Demissie M, Jedrzejuk D, Milewicz A. PPAR-gamma2 Pro12Ala polymorphism in the population of obese and non-obese men of the city of Wroclaw. Endokrynol Pol. 2008;59:312–315. [PubMed] [Google Scholar]
- Lamri A, Abi Khalil C, Jaziri R, et al. Dietary fat intake and polymorphisms at the PPARG locus modulate BMI and type 2 diabetes risk in the D.E.S.I.R. prospective study. Int J Obes (Lond) 2012;36:218–224. doi: 10.1038/ijo.2011.91. [DOI] [PubMed] [Google Scholar]
- Lappalainen T, Lindstrom J, Paananen J, Eriksson JG, Karhunen L, Tuomilehto J, Uusitupa M. Association of the fat mass and obesity-associated (FTO) gene variant (rs9939609) with dietary intake in the finnish diabetes prevention study. Br J Nutr. 2012;23:1–7. doi: 10.1017/S0007114511007410. [DOI] [PubMed] [Google Scholar]
- Levine JA, Eberhardt NL, Jensen MD. Role of nonexercise activity thermogenesis in resistance to fat gain in humans. Science. 1999;283:212–214. doi: 10.1126/science.283.5399.212. [DOI] [PubMed] [Google Scholar]
- Levine JA, Lanningham-Foster LM, McCrady SK, et al. Interindividual variation in posture allocation: possible role in human obesity. Science. 2005;307:584–586. doi: 10.1126/science.1106561. [DOI] [PubMed] [Google Scholar]
- Marti A, Corbalan MS, Martinez-Gonzalez MA, Forga L, Martinez JA. CHO intake alters obesity risk associated with Pro12Ala polymorphism of PPARgamma gene. J Physiol Biochem. 2002;58:219–220. doi: 10.1007/BF03179859. [DOI] [PubMed] [Google Scholar]
- Marti A, Martinez-Gonzalez MA, Martinez JA. Interaction between genes and lifestyle factors on obesity. Proc Nutr Soc. 2008;67:1–8. doi: 10.1017/S002966510800596X. [DOI] [PubMed] [Google Scholar]
- Martinez-Gonzalez MA, Sanchez-Villegas A, Irala J, Marti A, Martinez JA. Mediterranean diet and stroke: objectives and design of the SUN project. Seguimiento Universidad de Navarra. Nutr Neurosci. 2002;5:65–73. doi: 10.1080/10284150290007047. [DOI] [PubMed] [Google Scholar]
- Martinez-Gonzalez MA, Lopez-Fontana C, Varo JJ, Sanchez-Villegas A, Martinez JA. Validation of the Spanish version of the physical activity questionnaire used in the nurses’ health study and the health professionals’ follow-up study. Public Health Nutr. 2005;8:920–927. doi: 10.1079/PHN2005745. [DOI] [PubMed] [Google Scholar]
- Masud S, Ye S. Effect of the peroxisome proliferator activated receptor-gamma gene Pro12Ala variant on body mass index: a meta-analysis. J Med Genet. 2003;40:773–780. doi: 10.1136/jmg.40.10.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memisoglu A, Hu FB, Hankinson SE, Manson JE, Vivo I, Willett WC, Hunter DJ. Interaction between a peroxisome proliferator-activated receptor gamma gene polymorphism and dietary fat intake in relation to body mass. Hum Mol Genet. 2003;12:2923–2929. doi: 10.1093/hmg/ddg318. [DOI] [PubMed] [Google Scholar]
- Moleres A, Ochoa MC, Rendo-Urteaga T, Martinez-Gonzalez MA, Azcona San Julian MC, Martinez JA, Marti A. Dietary fatty acid distribution modifies obesity risk linked to the rs9939609 polymorphism of the fat mass and obesity-associated gene in a Spanish case-control study of children. Br J Nutr. 2012;107:533–538. doi: 10.1017/S0007114511003424. [DOI] [PubMed] [Google Scholar]
- Nelson TL, Fingerlin TE, Moss L, Barmada MM, Ferrell RE, Norris JM. The PPARgamma Pro12Ala polymorphism is not associated with body mass index or waist circumference among hispanics from Colorado. Ann Nutr Metab. 2007;51:252–257. doi: 10.1159/000104145. [DOI] [PubMed] [Google Scholar]
- Passaro A, Dalla Nora E, Marcello C, et al. PPAR-gamma Pro12Ala and ACE I/D polymorphisms are associated with BMI and fat distribution, but not metabolic syndrome. Cardiovasc Diabetol. 2011;10:112. doi: 10.1186/1475-2840-10-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S, Zhu Y, Xu F, Ren X, Li X, Lai M. FTO gene polymorphisms and obesity risk: a meta-analysis. BMC Med. 2011;9:71. doi: 10.1186/1741-7015-9-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razquin C, Marti A, Martinez JA. Evidences on three relevant obesogenes: MC4R, FTO and PPARgamma. Approaches for personalized nutrition. Mol Nutr Food Res. 2011;55:136–149. doi: 10.1002/mnfr.201000445. [DOI] [PubMed] [Google Scholar]
- Sayon-Orea C, Bes-Rastrollo M, Basterra-Gortari FJ, Beunza JJ, Guallar-Castillon P, Fuente-Arrillaga C, Martinez-Gonzalez MA. Consumption of fried foods and weight gain in a Mediterranean cohort: The SUN project. Nutr Metab Cardiovasc Dis. 2011 doi: 10.1016/j.numecd.2011.03.014. [DOI] [PubMed] [Google Scholar]
- Segui-Gomez M, Fuente C, Vazquez Z, Irala J, Martinez-Gonzalez MA. Cohort profile: the ‘Seguimiento Universidad de Navarra’ (SUN) study. Int J Epidemiol. 2006;35:1417–1422. doi: 10.1093/ije/dyl223. [DOI] [PubMed] [Google Scholar]
- Smith PG, Day NE. The design of case-control studies: the influence of confounding and interaction effects. Int J Epidemiol. 1984;13:356–365. doi: 10.1093/ije/13.3.356. [DOI] [PubMed] [Google Scholar]
- Speakman JR, Rance KA, Johnstone AM. Polymorphisms of the FTO gene are associated with variation in energy intake, but not energy expenditure. Obesity (Silver Spring) 2008;16:1961–1965. doi: 10.1038/oby.2008.318. [DOI] [PubMed] [Google Scholar]
- Speakman JR, Levitsky DA, Allison DB, et al. Set points, settling points and some alternative models: theoretical options to understand how genes and environments combine to regulate body adiposity. Dis Model Mech. 2011;4:733–745. doi: 10.1242/dmm.008698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speliotes EK, Willer CJ, Berndt SI, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonjes A, Scholz M, Loeffler M, Stumvoll M. Association of Pro12Ala polymorphism in peroxisome proliferator-activated receptor gamma with Pre-diabetic phenotypes: meta-analysis of 57 studies on nondiabetic individuals. Diabetes Care. 2006;29:2489–2497. doi: 10.2337/dc06-0513. [DOI] [PubMed] [Google Scholar]
- Tung YC, Yeo GS. From GWAS to biology: lessons from FTO. Ann N Y Acad Sci. 2011;1220:162–171. doi: 10.1111/j.1749-6632.2010.05903.x. [DOI] [PubMed] [Google Scholar]
- Walley AJ, Asher JE, Froguel P. The genetic contribution to non-syndromic human obesity. Nat Rev Genet. 2009;10:431–442. doi: 10.1038/nrg2594. [DOI] [PubMed] [Google Scholar]