Abstract
Vampire bats (Desmodus rotundus) are obligate blood feeders that have evolved specialized systems to suit their unique sanguinary lifestyle 1–3. Chief among such adaptations is the ability to detect infrared radiation as a means of locating hot spots on warm-blooded prey. Among vertebrates, only vampire bats, boas, pythons, and pit vipers are capable of detecting infrared radiation 1,4. In each case, infrared heat is detected by trigeminal nerve fibers that innervate specialized pit organs on the animal’s face 5–10. Thus, vampire bats and snakes have taken thermosensation to the extreme by developing specialized systems for detecting infrared radiation. As such, these creatures provide a window into the molecular and genetic mechanisms underlying evolutionary tuning of thermoreceptors in a species or cell type specific manner. Previously, we have shown that snakes co-opt a non-heat sensitive channel (vertebrate TRPA1) to produce an infrared detector 6. Here we show that vampire bats tune an already heat sensitive channel (TRPV1) by lowering its thermal activation threshold to ~30°C. This is achieved through alternative splicing of TRPV1 transcripts to produce a channel with a truncated C-terminal cytoplasmic domain. Remarkably, these splicing events occur exclusively in trigeminal ganglia (TG), and not dorsal root ganglia (DRG), thereby maintaining a role for TRPV1 as a detector of noxious heat in somatic afferents. This reflects a unique organization of the bat TRPV1 gene that we show to be characteristic of Laurasiatheria mammals (cows, dogs, and moles), supporting a close phylogenetic relationship with bats. These findings reveal a unique molecular mechanism for physiological tuning of thermosensory nerve fibers.
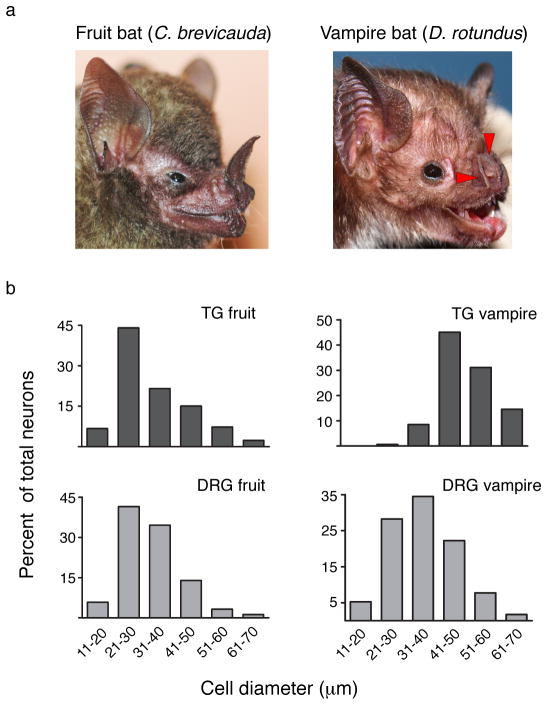
Vampire bats have three ‘leaf pits’ that surround the nose (Fig. 1a) and receive input from low-threshold heat-sensitive nerve fibers responding to stimuli >29°C (refs. 1,7,11,12). Closely related fruit bats (Carollia brevicauda) have a different nasal structure devoid of pit organs (Fig. 1a) and thus cannot detect infrared stimuli 3. Sensory ganglia of fruit bats showed a typical size distribution of neurons resembling that seen in other mammals (Fig. 1b). In contrast, TG from vampire bats showed marked skewing towards large diameter neurons, much like that observed in TG of pit bearing snakes 6, suggesting that anatomical specialization of TG in vampire bats has similarly evolved to suit a predominant role in infrared sensation. Consistent with this, vampire bat DRG showed a normal size distribution resembling sensory ganglia of fruit bats or other mammals (Fig. 1b). To our knowledge, vampire bats provide the only such example of TG specialization among mammalian species.
Figure 1. Anatomy of fruit bat and vampire bat sensory ganglia.
a, Facial anatomy of fruit bat (left) and vampire bat (right). Red arrowheads mark pit organs surrounding the vampire bat’s nose. b–e, Neuronal cell sizes determined from histological sections of fruit bat TG (n=300, 7 independent sections), fruit bat DRG (n=345, 9 sections), vampire bat TG (n=164, 6 sections), vampire bat DRG (n=400 neurons, 11 sections).
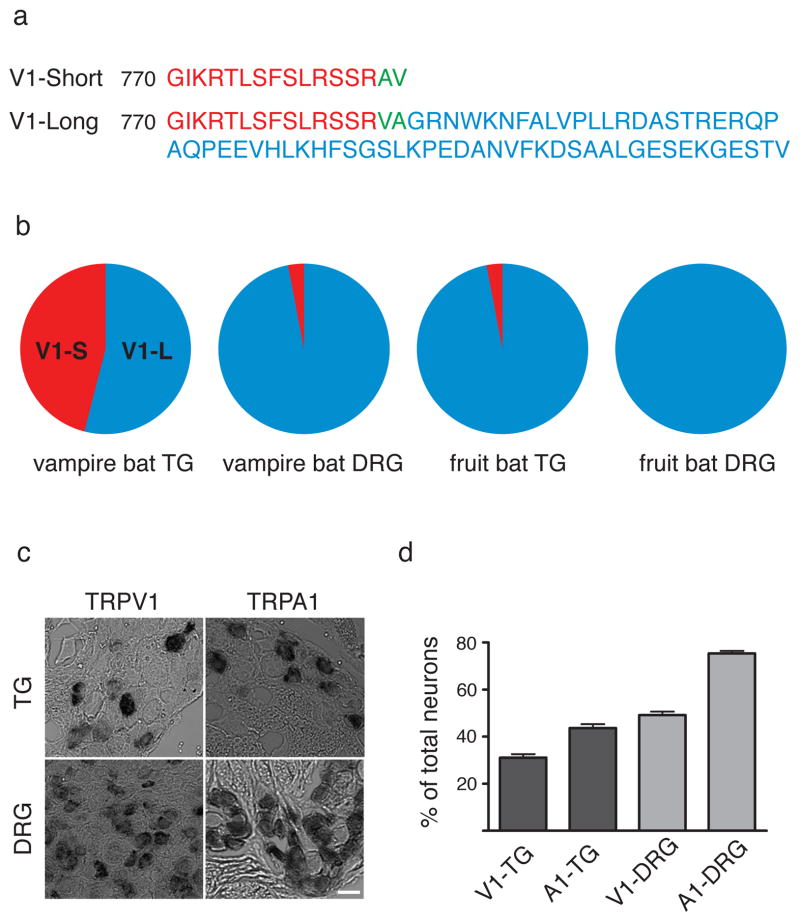
Given the distinct anatomy of vampire bat TG versus DRG, we asked whether these ganglia show differential patterns of gene expression that might highlight molecules specifically involved in infrared sensation, as in the case of infrared sensing snakes 6. Thus, we performed ‘deep sequencing’ of cDNAs from bat TG and DRG. Initially, transcriptomes from these ganglia appeared to be indistinguishable (SFig. 1). However, examination of cDNAs encoding candidate thermosensors revealed a novel short isoform of the capsaicin receptor, TRPV1, an excitatory ion channel that is activated by noxious heat (>43°C) (refs. 13–15). This novel isoform (TRPV1-S) lacks 62 amino acids from the C-terminus (Fig. 2a and SFig. 2) and is uniquely expressed in TG from vampire bat, constituting between 35–46% of all TRPV1 transcripts in TG, but <3% in DRG, as determined from transcriptome data or direct cloning and sequencing of RT-PCR products amplified from sensory ganglia (Fig. 2b). Moreover, TRPV1-S transcripts were barely detected (<6%) in fruit bat TG or DRG (Fig. 2b), and represented ≤1.8% of TRPV1 transcripts from TG of three other fruit, nectar, or insect feeding bat species (U. bilobatum, S. lilium, and A. cultrata). Taken together, these observations support the notion that TRPV1-S contributes to the specialized function of vampire bat TG, most notably infrared sensation.
Figure 2. Sequence and distribution of vampire bat TRPV1.
a, Deduced protein sequences for TRPV1 short (TRPV1-S) and long (TRPV1-L) isoforms from vampire bat sensory ganglia. b, Abundance of TRPV1 isoform transcripts in vampire and fruit bat sensory ganglia as determined by Solexa sequencing and/or direct sequencing of RT-PCR products (≥86 clones) amplified from sensory ganglia mRNA. c, In situ hybridization showing expression of TRPV1 and TRPA1 transcripts in histological sections from vampire bat TG or DRG (scale bar = 50 μm). d, Prevalence of TRPV1 or TRPA1 mRNA expression within vampire bat TG or DRG (mean ± s.e.m.; n = 554 neurons; 14 sections for V1-TG; 783 neurons; 17 sections for A1-TG; 1455 neurons; 15 sections for V1-DRG; 1030 neurons; 10 section for A1-DRG).
Discrete cellular expression of TRPV1 in vampire bat sensory ganglia was confirmed by in situ hybridization. Consistent with RNA-seq analysis, TRPV1 transcripts were found in similar percentages of TG and DRG neurons (31.1 ± 1.5% and 49.2 ± 1.5%, respectively) (Fig. 2c, d). Specificity of this pattern was confirmed by examining the distribution of TRPA1 transcripts, which were present in a larger percentage of TG and DRG neurons (43.6 ± 1.7 and 75.4 ± 1.0, respectively) compared to TRPV1 (Fig. 2c, d and SFig. 3).
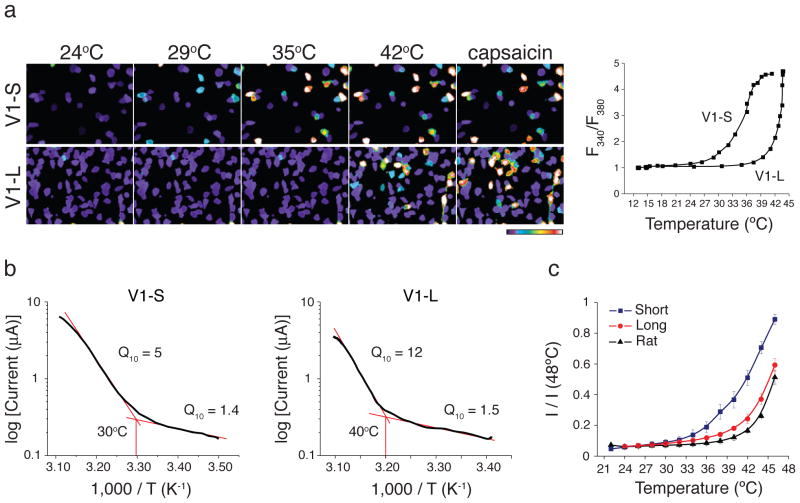
If TRPV1-S contributes to infrared detection in vampire bats, then this isoform should exhibit appropriate temperature sensitivity. We expressed and characterized TRPV1-S and TRPV1-L in HEK293 cells or Xenopus oocytes using calcium imaging or electrophysiological assays, respectively. Indeed, we observed a marked difference in temperature sensitivity such that the TRPV1-S isoform was activated at a substantially lower threshold compared to TRPV1-L (30.5 ± 0.7°C versus 39.6 ± 0.4°C in HEK293 cells and 31.2 ± 1.5°C versus 40.2 ± 0.7 in oocytes) (Fig. 3a–c and SFigs. 4, 5). Similar thresholds were observed for fruit bat TRPV1 isoforms (SFigs. 4, 6). This ~10°C threshold differential between TRPV1-S and TRPV1-L, together with unique expression of the short isoform in vampire bat TG, is consistent with a role for TRPV1-S in infrared detection. In contrast, heterologously expressed TRPA1 channels from either bat species were heat insensitive (SFig. 7).
Figure 3. Functional analyses of vampire bat TRPV1 isoforms.
a, HEK293 cells expressing vampire bat TRPV1 isoforms were analyzed for heat or capsaicin (10μM)-evoked responses using calcium imaging; color bar indicates relative change in fluorescence ratio, with purple and white denoting lowest and highest cytoplasmic calcium, respectively (n ≥ 141 cells per channel). Average temperature-response profiles for capsaicin-sensitive cells are shown at the right. b, Arrhenius plots show thermal thresholds and Q10 values for baseline and evoked responses for TRPV1-S and TRPV1-L (+80 mV, n=8). c, Relative heat response profiles of vampire bat TRPV1 isoforms compared with rat TRPV1 as measured electrophysiologically in oocytes (response at each temperature was normalized to maximal response at 48°C; VH = −80 mV, n ≥ 8).
Our histological analysis does not allow us to discriminate between TRPV1 isoforms. We therefore asked whether their co-expression would produce channels having lowered thermal thresholds compared to TRPV1-L alone. Injection of oocytes with equal amounts of TRPV1-S and -L cRNAs produced an intermediate temperature response curve with an activation threshold of 33.9 ± 1.2°C (SFig. 5), rather than biphasic thresholds, suggesting that short and long isoforms can produce functional heterotetrameric complexes. Oocytes expressing mostly one isoform (10S:1L or 1S:10L) showed a thermal threshold defined by the predominant species (SFig. 5). Thus, if individual TG neurons express equivalent levels of short and long isoforms, then they will have lower thermal thresholds compared to cells predominately expressing the long form.
The identification of C-terminal TRPV1 splice variants in vampire bats is somewhat surprising since, to our knowledge, such isoforms have not been previously reported for other species. Alignment between vampire bat TRPV1-S and -L cDNAs highlighted a 23-base pair insertion in the former (SFig. 8a) containing a stop codon that accounts for production of the short isoform. We used sequences flanking this insert to amplify and characterize the organization of the vampire bat TRPV1 locus in this vicinity. The resulting 5.5 kb fragment contained a tiny 23-bp exon (e14a) flanked by two introns (2.1 and 3.1 kb), each marked by canonical –GT/AG- donor-acceptor sites required for U2-dependent splicing, as well as obligate polypyrimidine tracts preceding potential splicing sites 16,17, allowing for formation of two splice variants that incorporate or bypass e14a to produce TRPV1-S or TRPV1-L, respectively (SFig. 9).
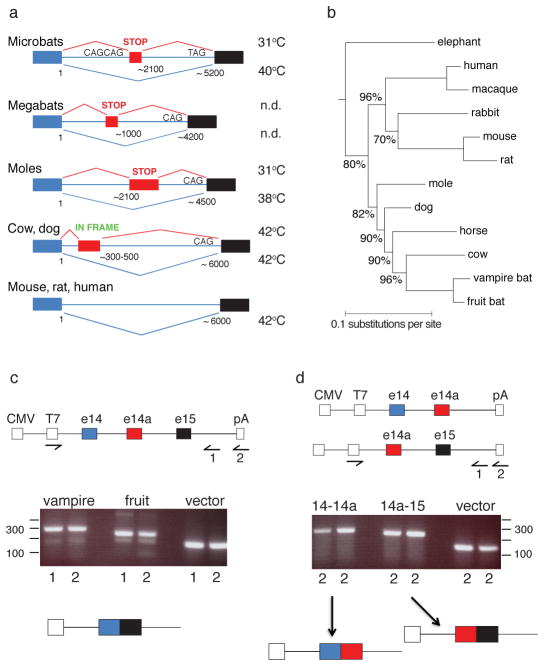
To determine whether exon 14a-based splicing is a hallmark of other Laurasiatheria orders, we carried out transcriptome and/or genomic analysis from Cetartiodactyla (cow, pig), Carnivora (dog), and Lipotyphla (Coast mole), as well as several additional bat species representing both Chiroptera suborders, including microbats and megabats 18–20. Two types of TRPV1 splicing events can occur (Fig. 4a): in microbats, megabats, and moles, e14a contains a premature stop codon accounting for production of a low threshold TRPV1-S isoform (SFigs. 4–6 and 10). In cows, pigs, and dogs e14a produces an in-frame insertion generating an extra-large isoform (TRPV1-XL). Thus, while members of the Laurasiatheria superorder exploit the same intronic region for TRPV1 modification, different suborders exhibit distinct e14a sequences and positions (relative to exons 14 and 15), indicative of independent evolutionary events leading to modification of this intronic region. Interestingly, the architecture of splice sites in microbats differs from that of other Laurasiatheria members in that the pyrimidine-rich tract is followed by a strong tandem splice site (CAGCAG), and a weak exon 15 acceptor site (TAG in microbats versus CAG in most other species) (Fig. 4a). Consistent with our failure to observe TRPV1-S-like isoforms in mouse sensory ganglia (not shown), neither rodent nor human TRPV1 gene contains an e14a equivalent, limiting splicing to a single isoform.
Figure 4. Genomic organization of mammalian TRPV1 locus.
a, Schematic of TRPV1 gene locus spanning putative exons 14 and 15 in animals of Laurasiatheria, Rodentia, and Primate groups. Splicing events between exons 14 (blue) and 15 (black) are shown, including those involving exon 14a (red). Thermal activation thresholds for resulting channel isoforms are shown at right (n.d., not determined). Lengths of intronic region and relative positions of exon 14a are indicated. b, Consensus phylogenetic tree from Bayesian estimation, with clade credibility values shown for branches with <100% support. c–d, Structures of mini-genes (top) used for in vivo splicing assay in HEK293 cells, showing location of CMV promoter and polyadenylation site (pA), defining start and end of transcribed unit. Reverse transcripts were generated with primers annealing to 3′ vector sequence (1) or oligo-dT (2), as indicated, and products amplified with T7 and primer (1) pair. Reaction products were resolved on agarose gel (middle) and major bands collected for characterization of splicing products (bottom).
Although cows and moles have the potential to produce TRPV1 splice variants, we found that <6% of TRPV1 transcripts corresponded to TRPV1-XL or TRPV1-S in TG of cow or mole, respectively, suggesting that this form of post-transcriptional regulation is not physiologically relevant in these animals. In cows, this is further underscored by the fact that TRPV1-L and TRPV1-XL isoforms are indistinguishable in regard to thermal response profiles (thresholds of 43.0 ± 0.8 and 42.7 ± 0.4°C, respectively) (SFigs. 4 and 11). Our analysis of TRPV1 gene organization in different orders (including Chiroptera, Cetartiodactyla, Carnivora, Lipotyphla, Afrotheria, and Rodentia) (Fig. 4b and SFigs. 9 and 12) is consistent with an independent molecular phylogeny of mammals 21,22 and supports the conclusion that bats are more closely related to cows, moles, and dogs 19,20, rather than to rodents as initially posited based on anatomical and morphological criteria23.
TRPV1 genomic sequences for vampire and other microbats in the vicinity of exon 14-14a-15 splice junctions are highly conserved (>90% identity). We therefore asked whether differential splicing was observed when mini-genes containing these genomic regions were introduced into HEK293 cells. Direct splicing of exon 14 to 15, with exclusion of exon 14a, was observed in all cases (Fig. 4c). In mini-gene constructs where direct splicing between exon 14 and 15 was not possible, splicing to or from exon 14a was readily observed (Fig. 4d), indicating that exon 14a is competent to engage in splicing, but disfavored by exon 14-15 competition. These results show that vampire and fruit bat genes exhibit the same default pattern of exon 14 to 15 splicing in this non-neural context, irrespective of small differences in gene structure. Indeed, this ‘constitutive’ splicing pattern predominates in vampire bat DRG, fruit bat TG and DRG, as well as cow and mole TG, suggesting that efficient splicing to exon 14a in vampire bat TG requires a specialized environment. Furthermore, we have used this assay to identify a putative exon 14a from megabat species, such as Pteropus vampyrus, where transcriptome data are not available (Fig. 4a and SFig. 13).
Our findings suggest that variation within the TRPV1 C-terminus represents a genetic mechanism for tuning the thermal response profile of the channel in a species- or tissue-specific manner. We therefore asked whether more primitive and evolutionarily distant species use a similar strategy for thermosensory adaptation. Indeed, we found that zebrafish TRPV1 is activated with a threshold of 32.9 ± 1.2°C (SFig. 14a–d), consistent with physiological adaptation of the animal to its environment of 25–33°C (ref. 24). When compared to rat TRPV1, the zebrafish channel has a gap of 12 amino acids within the C-terminus corresponding precisely to the location of the splice junction in the vampire bat channel (SFig. 14e). This gap results from a polymorphism within exon 15 of the zebrafish TRPV1 gene, and thus zebrafish and vampire bats use different mechanisms to generate TRPV1 C-terminal variants.
RNA splicing extends the coding potential of the genome and enhances functionality of the proteome 25. The vampire bat uses this strategy to produce physiologically distinct channels, thereby generating a hypersensitive detector (TRPV1-S) within a specific thermosensory organ, without sacrificing somatic thermo-sensation and/or –nociception. Moreover, our results demonstrate that sequence variation within a specific region of the cytoplasmic C-terminus accounts for differential thermal response profiles of vampire bat TRPV1 isoforms. Indeed, we previously proposed that this region of the channel interacts with membrane phospholipids to modulate sensitivity to thermal and chemical stimuli 26.
In addition to illuminating mechanisms of thermosensation and sensory adaptation, the analysis of TRP channel gene structure provides a physiologically relevant marker for assessing phylogenetic relationships. Our findings support recent molecular classification in which bats are grouped together with horses, dogs, cows, moles, and dolphins (Laurasiatheria superorder), rather than with humans, monkeys, flying lemur, mouse, rat, and rabbits (Euarchontoglires superorder) as originally proposed on the basis of anatomical criteria.
Methods
Tissue collection
Vampire bats (Desmodus rotundus) and fruit/insect eating bats (Carollia brevicauda, Anoura cultrata, Uroderma bilobatum, and Sturnira lilium) were collected in pasture forests grazed by cattle at the “Hato Piñero” ranch, property of the “Centro Técnico de Producción Socialista Florentino”, located near to “El Baúl”, Cojedes State, Venezuela. Exact sampling points as listed in the table below. Genetic resource and animal sampling permits were obtained from the “Ministerio del Ambiente” (MINAMB). Global coordinates for animal collection are provided in Supplementary Figure 15.
Bovine trigeminal ganglia were collected from 12-day old calves for RNA extraction under supervision of C. Sehnert (Meat Sciences Laboratory, Dept. of Animal Sciences, UC Davis). Blood samples from megabats were generously provided by the Lubee Bat Conservatory (Gainesville, Florida). A summary of samples from all species examined is provided in Supplementary Figure 15.
Deep sequencing and analysis
Sequencing libraries were prepared from poly (A)+ RNA using the Illumina mRNA-Seq Sample Prep Kit according to the manufacturer’s instructions. Libraries were then sequenced on the Illumina Genome Analyser II using two 36-cycle sequencing kits to read 80 nucleotides of sequence from a single end of each insert, by standard protocols. Between 7.6 and 26.2 million inserts were sequenced for each sample.
Sequences were aligned to the cow RefSeq protein database (downloaded from NCBI on 25 August 2009) using the blastx tool from NCBI blast (version 2.2.21), which aligns a six- frame translation of each query against a protein database. The alignment was performed with a word size of four amino acids and a window size of five; a maximum E value of 1×10−5 was required. For each read that aligned to the cow proteome, a set of optimal hits was collected based on alignments whose bit score was within 0.2 of the highest bit score reported for that sequencing read. Each RefSeq alignment for a given sequencing read was converted to an Entrez Gene identifier and redundant alignments for a single read, which correspond to alignments against different isoforms of the same protein, were collapsed to a single count. The number of optimally aligning reads was then counted for each gene; in some cases a single read counted towards several genes.
Splice variants were quantified by aligning sequencing reads to the nucleotide sequence of the alternate TRPV1 isoforms using Bowtie v0.11.3. The alignments were manually inspected to ensure that they supported the splice junction with at least 24 nucleotides aligning on one side of the TRPV1 transcript and at least two nucleotides unambiguously distinguishing the isoform.
The TRPV1 protein sequences were taken from this study, Ensembl (finch, macaque, horse, and elephant), and RefSeq (all others) and aligned using t_coffee [Notredame, Higgins, and Heringa, JMB, 302: 205–217) (2000)] in “accurate” mode with the blosum80mt distance matrix. The protein multiple sequence alignment was used as input to MrBayes [Ronquist and Huelsenbeck MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574 (2003)] and the phylogeny was estimated using gamma-distributed substitution rates under the Blosum62 distance matrix combined with a fraction of invariant sites. The phylogeny distribution was estimated across 10 runs, each of 6,000,000 MCMC cycles, with samples ever 100,000 cycles following a 1,000,000 cycle burn-in. The chicken and finch sequences were used to root the mammalian tree and are not shown.
Cloning
Functional cDNAs were amplified from first strand cDNA generated by reverse transcription using following primers:
Vampire bat TRPV1 (forward 5′-3′: GCAAGGATGAAGAAACGGG; reverse 5′-3′: CCTCACTTTTCCCCTAAAGC)
Vampire bat TRPA1 (forward 5′-3′: GGCCTGCGTAACATCAGAAGC; reverse 5′-3′: TACTGCTAAGGTCACTGCATTTTGG)
Carollia brevicauda TRPV1 (forward 5′-3′: CTTGGGCAAAGATGAAGAAACG; reverse 5′-3′: GTGTGCTAGTCTAAGGCCCA).
Carollia brevicauda TRPA1 (forward 5′-3′: GCATCCAGGGTAGGATCCAT; reverse 5′-3′: CAAGAAACGTGTGTGTGGGA)
Zebrafish TRPV1 (forward 5′-3: CCTCAAGCCAAGTTACTCAC; reverse 5′-3′: TCGAAGGACACCTTGTAGAC)
Cow TRPV1 (forward 5′-3: ATGAAGAAATGGGGGAGCTCAG; reverse 5′-3′: TCACTTCTCCCCTAAAGCCAC)
Coast Mole TRPV1 (forward 5′-3: GGAGCTGGCAAGGATGAAG; reverse 5′-3′: ATCCTAAGGCCCAACAGAGT)
Genomic DNA was extracted from vampire (Desmondus rotundus), fruit bat (Carollia brevicauda, Sturnira billobatum) brain or coast mole TG using TRIzol Reagent (Invitrogen); genomic DNA of Megabats was extracted from blood using QIAmp DNA Blood mini kit (Qiagen). Genomic region between exon 14 and 15 was amplified using following primers: Vampirebat, Megabats (forward 5′-3′: CAGGCAATTGTGAAGGCATC; reverse 5′-3′: CCAGGGCAAAGTTCTTCCAG); Carollia brevicauda, Sturnira billobatum (forward 5′-3′: CGGGCAATTGTGAAGGCAT; reverse 5′-3′: CCAGGGCAAAGTTCTTCCAG); Coast Mole (forward 5′-3′: CTTCTCCCTGCGGTCAAG; reverse 5′-3′: CTCGAGTGCTTGCATCTCTTAA).
Oocyte electrophysiology
Surgically extracted oocytes from Xenopus laevis (Nasco) were cultured, injected with 5 ng of RNA, and analyzed 4–5 days post-injection by TEVC using a Geneclamp 500 amplifier (Axon Instruments, Inc) as described 47. Microelectrodes were pulled from borosilicate glass capillary tubes to obtain resistances of 0.3–.07 MΩ. Bath solution contained 10 mM Hepes, 120 mM NaCl, 2 mM KCl, 1 mM EGTA, and 2 mM MgCl2 buffered to a final pH of 7.4 with NaOH. Data were analyzed using pCLAMP10 software.
Determination of thermal threshold and temperature coefficients (Q10)
Temperature thresholds represent the point of intersection between linear fits to baseline and the steepest component of the Arrhenius profile. Values are derived from averages of individual curves; n ≥ 8. Arrhenius curve were obtained by plotting the current (+80 mV) on a log-scale against the reciprocal of the absolute temperature. Q10 was used to characterize the temperature dependence of the ionic current as calculated using the following equation:
where R2 is the current at the higher temperature T2 and R1 is the current at the lower temperature T1 (ref. 38).
Calcium imaging
All tested channels were transiently expressed in HEK293 cells with the use of Lipofectamin 2000 (Invitrogen). Calcium imaging of HEK293 cells using Fura-2/AM was performed on coverslips coated with Matrigel (BD). Fluorecent images were acquired with Metaflour software (Molecular Device) and analyzed using Graph Pad Prizm 4.
In situ hybridization histochemistry
TG and DRG tissue were dissected and fixed in 4% PFA in PBS for 2 days. Cryostat sections (12–15 μm thick) were processed and probed with a digoxigenin-labeled cRNA. Probes were generated by T7/T3 in vitro transcription reactions using a full length of TRPV1 cDNA and 3 kb fragment of TRPA1 cDNA. Signal was developed with alkaline phosphatase-conjugated anti-digoxigenin Fab fragments according to the manufacturer’s instructions.
In vivo splicing assay
Mini-gene constructs
Genomic region containing vampire bat or Carollia brevicauda splice junction between exon 14-15, exon 14-14a, exon14a-15 was topo-cloned, then subcloned into pcDNA3(+) vector (Invitrogen) using BamH1/Xho1 restriction sites.
Transfections
HEK293 cells were cultured in DMEM medium supplemented with 10% fetal bovine serum in 6-well plates. At 50% confluent cells were transiently transfected with 1 μg of mini-gene plasmid or empty pcDNA3 (+) vector by use of Lipofectamin method. Cells were harvested 48h after transfection using TRizol reagent (Invitrogen).
cDNA synthesis and RT-PCR
RNA was extracted using Trizol reagent. 10 μg of RNA was used as template for cDNA synthesis using SuperScriptIII first strand synthesis system (Invitrogen). Synthesis was primed with oligo (dT) or by use of primer annealing to 3′-untranslated sequences in pcDNA3 (CAGTCGAGGCTGATCAGCGAGCT). 5% of cDNA was used in 36-cycle PCR using forward 5′-3′: TAATACGACTCACTATAGGG; reverse 5′-3′: CAGTCGAGGCTGATCAGCGAGCT primers. Reaction products were resolved by electrophoresis on 1.5% agarose gel. PCR products were isolated from the gel, cloned into Strataclone vector and sequenced.
Supplementary Material
Acknowledgments
We thank Y. Kelly and J. Poblete for expert technical assistance, C. Sehnert (UC Davis) for help with bovine tissue collection, M. Suzawa and H. Ingraham for providing zebrafish mRNA, and A. Walsh (Lubee Bat Conservancy) for providing megabat blood samples. This work was supported by a Ruth L. Kirschstein National Research Service Award (GM080853) (N.T.I.), a Pathway to Independence Fellowship from the UCSF CVRI (E.O.G.), the Howard Hughes Medical Institute (J.S.W.), and grants from NIH, including P01 AG010770 (J.S.W.) and NS047723 and NS055299 (D.J.).
Footnotes
Author contributions E.O.G., J.F.C-M. and N.T.I. designed and performed experiments and analyzed data. N.T.I. and J.S.W. developed analytical tools and analyzed data. J.A.G-C., C.I.A., and C.M. collected bat species and obtained tissues for analysis. E.O.G., J.F.C-M. and D.J. wrote the manuscript with discussion and contributions from all authors. J.S.W. and D.J. provided advice and guidance throughout.
Deep sequencing data are archived under GEO accession number GSE28243. GenBank accession numbers are JN006855 (Desmodus rotundus TRPV1-S), JN006856 (Desmodus rotundus TRPV1-L), JN006857 (Desmodus rotundus TRPA1), JN006858 (Carollia brevicauda TRPA1), JN006859 (Carollia brevicauda TRPV1-L), JN006860 (Carollia brevicauda TRPV1-S), JN006861 (Scapanus orarius TRPV1-L), JN006862 (Scapanus orarius TRPV1-S), JN006863 (Pteropus rodricensis intron), JN006864 (Desmodus rotundus intron), JN006865 (Carollia brevicauda intron), JN006866 (Pteropus vampyrus intron), JN006867 Rousettus aegyptiacus intron), JN006868 (Scapanus orarius intron).
The authors declare no competing financial interests.
References
- 1.Kurten L, Schmidt U. Thermoperception in the common vampire bat (Desmodus rotundus) J Comp Physiol. 1982;146:223–228. [Google Scholar]
- 2.Schutt B. Dark Banquet: Blood and the Curious Lives of Blood-Feeding Creatures. Three Rivers Press; 2008. [Google Scholar]
- 3.Tellgren-Roth A, et al. Keeping the blood flowing-plasminogen activator genes and feeding behavior in vampire bats. Naturwissenschaften. 2009;96:39–47. doi: 10.1007/s00114-008-0446-0. [DOI] [PubMed] [Google Scholar]
- 4.Campbell AL, Naik RR, Sowards L, Stone MO. Biological infrared imaging and sensing. Micron. 2002;33:211–225. doi: 10.1016/s0968-4328(01)00010-5. [DOI] [PubMed] [Google Scholar]
- 5.Bullock TH, Cowles RB. Physiology of an Infrared Receptor: The Facial Pit of Pit Vipers. Science. 1952;115:541–543. doi: 10.1126/science.115.2994.541-a. [DOI] [PubMed] [Google Scholar]
- 6.Gracheva EO, et al. Molecular basis of infrared detection by snakes. Nature. 2010;464:1006–1011. doi: 10.1038/nature08943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kurten L, Schmidt U, Schafer K. Warm and cold receptors in the nose of the vampire bat Desmodus rotundus. Naturwissenschaften. 1984;71:327–328. doi: 10.1007/BF00396621. [DOI] [PubMed] [Google Scholar]
- 8.Molenaar GJ. The sensory trigeminal system of a snake in the possession of infrared receptors. II. The central projections of the trigeminal nerve. J Comp Neurol. 1978;179:137–151. doi: 10.1002/cne.901790109. [DOI] [PubMed] [Google Scholar]
- 9.Bakken GS, Krochmal AR. The imaging properties and sensitivity of the facial pits of pitvipers as determined by optical and heat-transfer analysis. J Exp Biol. 2007;210:2801–2810. doi: 10.1242/jeb.006965. [DOI] [PubMed] [Google Scholar]
- 10.Safer AB, Grace MS. Infrared imaging in vipers: differential responses of crotaline and viperine snakes to paired thermal targets. Behav Brain Res. 2004;154:55–61. doi: 10.1016/j.bbr.2004.01.020. [DOI] [PubMed] [Google Scholar]
- 11.Kishida R, Goris RC, Terashima S, Dubbeldam JL. A suspected infrared-recipient nucleus in the brainstem of the vampire bat, Desmodus rotundus. Brain Res. 1984;322:351–355. doi: 10.1016/0006-8993(84)90132-x. [DOI] [PubMed] [Google Scholar]
- 12.Schafer K, Braun HA, Kurten L. Analysis of cold and warm receptor activity in vampire bats and mice. Pflugers Arch. 1988;412:188–194. doi: 10.1007/BF00583749. [DOI] [PubMed] [Google Scholar]
- 13.Caterina MJ, et al. The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature. 1997;389:816–824. doi: 10.1038/39807. [DOI] [PubMed] [Google Scholar]
- 14.Jordt SE, McKemy DD, Julius D. Lessons from peppers and peppermint: the molecular logic of thermosensation. Curr Opin Neurobiol. 2003;13:487–492. doi: 10.1016/s0959-4388(03)00101-6. [DOI] [PubMed] [Google Scholar]
- 15.Ramsey IS, Delling M, Clapham DE. An introduction to TRP channels. Annu Rev Physiol. 2006;68:619–647. doi: 10.1146/annurev.physiol.68.040204.100431. [DOI] [PubMed] [Google Scholar]
- 16.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 17.Will CL, Luhrmann R. Splicing of a rare class of introns by the U12-dependent spliceosome. Biol Chem. 2005;386:713–724. doi: 10.1515/BC.2005.084. [DOI] [PubMed] [Google Scholar]
- 18.Murphy WJ, et al. Molecular phylogenetics and the origins of placental mammals. Nature. 2001;409:614–618. doi: 10.1038/35054550. [DOI] [PubMed] [Google Scholar]
- 19.Murphy WJ, et al. Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science. 2001;294:2348–2351. doi: 10.1126/science.1067179. [DOI] [PubMed] [Google Scholar]
- 20.Nishihara H, Hasegawa M, Okada N. Pegasoferae, an unexpected mammalian clade revealed by tracking ancient retroposon insertions. Proc Natl Acad Sci U S A. 2006;103:9929–9934. doi: 10.1073/pnas.0603797103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Asher RJ, Bennett N, Lehmann T. The new framework for understanding placental mammal evolution. Bioessays. 2009;31:853–864. doi: 10.1002/bies.200900053. [DOI] [PubMed] [Google Scholar]
- 22.Prasad AB, Allard MW, Green ED. Confirming the phylogeny of mammals by use of large comparative sequence data sets. Mol Biol Evol. 2008;25:1795–1808. doi: 10.1093/molbev/msn104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pettigrew JD, et al. Phylogenetic relations between microbats, megabats and primates (Mammalia: Chiroptera and Primates) Philos Trans R Soc Lond B Biol Sci. 1989;325:489–559. doi: 10.1098/rstb.1989.0102. [DOI] [PubMed] [Google Scholar]
- 24.Spence R, Gerlach G, Lawrence C, Smith C. The behaviour and ecology of the zebrafish, Danio rerio. Biol Rev Camb Philos Soc. 2008;83:13–34. doi: 10.1111/j.1469-185X.2007.00030.x. [DOI] [PubMed] [Google Scholar]
- 25.Grabowski PJ, Black DL. Alternative RNA splicing in the nervous system. Prog Neurobiol. 2001;65:289–308. doi: 10.1016/s0301-0082(01)00007-7. [DOI] [PubMed] [Google Scholar]
- 26.Prescott ED, Julius D. A modular PIP2 binding site as a determinant of capsaicin receptor sensitivity. Science. 2003;300:1284–1288. doi: 10.1126/science.1083646. [DOI] [PubMed] [Google Scholar]
- 27.Chuang HH, Neuhausser WM, Julius D. The super-cooling agent icilin reveals a mechanism of coincidence detection by a temperature-sensitive TRP channel. Neuron. 2004;43:859–869. doi: 10.1016/j.neuron.2004.08.038. [DOI] [PubMed] [Google Scholar]
- 28.DeCoursey TE, Cherny VV. Temperature dependence of voltage-gated H+ currents in human neutrophils, rat alveolar epithelial cells, and mammalian phagocytes. J Gen Physiol. 1998;112:503–522. doi: 10.1085/jgp.112.4.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.