Abstract
OBJECTIVES:
It has been demonstrated that circulating monocytes relocate to the intestinal mucosa during intestinal inflammation, but the phenotype and inflammatory mechanisms of these monocytes remain poorly understood. Here, we have investigated blood monocytes expressing high levels of HLA-DR and CCR9 in patients with inflammatory bowel disease (IBD).
METHODS:
Fifty-one patients with mild to severe ulcerative colitis (UC; n=31; UC-DAI 3–12) or Crohn's disease (CD; n=20; Harvey–Bradshaw indices (HBI) 2–16) were included together with 14 controls, during IBD therapy for four consecutive weeks. The frequency of CD14+HLA-DRhi monocytes was monitored weekly in peripheral blood, using flow cytometry. The surface phenotype and cytokine profile of these monocytes were established using flow cytometry and real-time PCR. Clinical parameters were assessed weekly in all patients.
RESULTS:
The frequency of circulating CD14+HLA-DRhi monocytes was significantly higher in IBD patients with moderate to severe disease compared with healthy controls (P<0.001). During treatment with corticosteroids and granulocyte/monocyte apheresis, the proportion of circulating CD14+HLA-DRhi monocytes was significantly reduced. CD14+HLA-DRhi monocytes produced high levels of inflammatory mediators, such as tumor necrosis factor (TNF)-α, and expressed the gut-homing receptor CCR9. Furthermore, we found that the CCR9 ligand, CCL25/TECK, was expressed at high levels in the colonic mucosa in IBD patients with active disease.
CONCLUSIONS:
CD14+HLA-DRhi blood monocytes were increased in patients with active IBD. These monocytes exhibit a pro-inflammatory, gut-homing phenotype with regard to their TNF-α production and expression of CCR9. Our results suggest that these monocytes are important in mediating intestinal inflammation, and provide potential therapeutic targets in IBD.
INTRODUCTION
Crohn's disease (CD) and ulcerative colitis (UC) (inflammatory bowel diseases, IBDs) are chronic, inflammatory disorders of the gastrointestinal tract resulting from a disrupted balance between the mucosal immune system and commensal flora. To date, the immunological pathophysiology behind IBD remains poorly understood. Traditionally, adaptive immunity was believed to have an important role in the onset of IBD. Studies in patients and animal models have shown that CD is driven by T-helper 1 signaling with interleukin (IL)-12 and interferon-γ production, whereas UC is characterized by T-helper 2 responses and IL-13.1 However, the Th1/Th2 paradigm has been questioned over the past decade.2 Since the discovery of the NOD2/CARD15 susceptibility locus that encodes a pattern recognition receptor mainly expressed on dendritic cells (DCs) and monocytes, the focus of IBD research has shifted toward innate immunity.3, 4 Currently, innate mechanisms are believed to be responsible for the onset of acute mucosal inflammation in genetically susceptible individuals, whereas the chronic state might be maintained by adaptive elements.5
Monocytes are bone marrow-derived leukocytes of the myeloid lineage that migrate to the tissue and differentiate into macrophages or DCs. Increased turnover rates and elevated levels of circulating monocytes have been demonstrated in IBD.6, 7 Furthermore, monocytes have the ability to migrate to the inflamed mucosa and mediate inflammation, but the phenotype of these monocytes as well as the mechanisms underlying this relocation remains to be elucidated.8, 9, 10 Currently, two main human monocyte subpopulations have been characterized. The CD14+CD16− cells have been shown to produce the regulatory cytokine IL-10 and are most commonly referred to as classical monocytes. The CD14loCD16+ subset is characterized by production of pro-inflammatory cytokines as well as high surface expression of inflammatory markers, such as CD43.11, 12, 13 However, a larger degree of heterogeneity among human myeloid cell populations with regard to their surface antigen expression as well as their functionality has lately been observed.14
Reports have established that myeloid-derived suppressor cells, characterized by their lack of expression of classical myeloid markers such as CD14 and HLA-DR, have the ability to significantly suppress antigen-specific T-cell responses in cancer patients.15, 16 It has also been shown that patients with IBD display elevated levels of functionally suppressive HLA-DR− myeloid-derived cells, reflecting the need for immunosuppression in the state of disease.17
It has been demonstrated that monocyte HLA-DR expression has an important role in conditions characterized by immune responses against bacterial agents.18 Although the CD14+CD16− subset has been reported to express HLA-DR, the specific contribution of CD14+HLA-DRhi monocytes to intestinal inflammation has not been studied. As it is well established that induction of colitis in human as well as in animal models requires the presence of bacteria, we set out to study CD14+HLA-DRhi monocytes in patients with chronic intestinal inflammation.20
MATERIALS AND METHODS
Patients
In total, 51 IBD patients (UC=31 and CD=20) were included in this study (Table 1). The patients were monitored during treatment with corticosteroids (n=16), the antitumor necrosis factor (TNF)-α antibodies infliximab or adalimumab (Remicade or Humira; n=17), or granulocyte/monocyte apheresis (GMA; Adacolumn; n=18). Four to six biopsies from affected rectum and sigmoid colon were collected together with blood samples before the start of treatment, followed by analysis of intestinal and blood specimens after 1–2 weeks and 4 weeks into treatment. Patients were clinically assessed using the UC-DAI (UC) and Harvey–Bradshaw (HBI; CD) indices. For glucocorticoid- and anti-TNF-treated patients, the response was evaluated at week 5, and GMA-treated patients were assessed at week 11 post treatment, owing to the delayed response observed in this treatment group. Clinical remission was defined as <3 for UC-DAI and <5 for HBI.8, 21 Fourteen controls without IBD were included in the study. All patients were enrolled through formal consent, and the study was approved by the regional ethics committee.
Table 1. Patient demography.
| Gender | Male | 32 |
| Female | 19 | |
| Age mean | 37.9 | |
| Diagnosis | Ulcerative colitis | 31 |
| Crohn's disease | 20 | |
| Extension | Extensive | 25 |
| Left-sided | 19 | |
| Proctitis | 5 | |
| Ileocekal | 2 | |
| Intervention | Corticosteroidsa,b | 16 |
| Anti-TNF-αc,d | 17 | |
| GMA apheresisd,e | 18 | |
| Azathioprine | Yes | 21 |
| No | 30 |
Abbreviations: GMA, granulocyte/monocyte apheresis; TNF, tumor necrosis factor.
Fifteen patients were introduced to 20–45 mg prednisone followed by tapering of 5 mg weekly.
One patient received topical corticosteroids for ulcerative proctosigmoiditis.
Anti-TNF-α treatment was administered either as infusions of 5 mg/kg infliximab week 0, 2, and 6 or subcutaneous injections of 80 mg Adalimumab week 0 followed by 40 mg every other week.
Some patients were receiving baseline corticosteroid medication.
In the GMA apheresis group, each patient received a total of 5–8 Adacolumn leukocytapheresis sessions 1–2 times weekly.
Leukocyte isolation and activation
For flow cytometry studies, peripheral blood mononuclear cells (PBMCs) were obtained from heparinized whole blood by incubation in hypotonic buffer (160 mℳ NH4Cl, 10 mℳ Tris-HCl, pH=7.4). For PCR and CCL25 depletion experiments, PBMCs were obtained from anticoagulated healthy donor buffy coats by density gradient centrifugation using Ficoll-Paque (GE Healthcare, Amersham, UK). For PCR experiments, CD14+ monocytes were negatively isolated using Monocyte Isolation Kit II (Miltenyi Biotec, Bergisch Gladbach, Germany). Monocytes were subsequently activated with LPS (lipopolysaccharide; Sigma, St Louis, MO, USA) (200 ng/ml/106 cells) for 2 h (TNF-α PCR) or 6 h (PCR array) in RPMI medium (Thermo Scientific Hyclone, Waltham, MA, USA) supplemented with 10% fetal calf serum, 2 mℳℒ-glutamine, and 1% PEST (penicillin–streptomycin).
Flow cytometry
PBMCs were stained for flow cytometry analysis or sorting using combinations of the antibody conjugates described in Table 2. All stainings were carried out according to the instructions of the manufacturer of the respective antibody conjugate. Isotype- and fluorochrome-matched control antibodies were used to define chemokine receptor expression in Figures 5 and 6. Flow cytometry analyses and sorting experiments were carried out using a FACSAria cytometer and data were analyzed using FACSDiva software (BD Biosciences, San Jose, CA, USA).
Table 2. Flow cytometry antibodies used in the study.
| Marker | Conjugate | Clone | Manufacturer |
|---|---|---|---|
| CD4 | Pacific Blue | RPA-T4 | BD |
| CD14 | FITC | RMO52 | Beckman |
| CD16 | PE-Cy7 | 3G8 | BD |
| HLA-DR | APC-Cy7 | L243 | BD |
| CCR1 | Alexa Fluor 647 | TG4 | BioLegend |
| CCR2 | PerCP-Cy5.5 | TG5 | BioLegend |
| CCR3 | PE | 5E8 | BioLegend |
| CCR4 | PerCP-Cy5.5 | 1G1 | BD |
| CCR5 | PE | HM-CCR5 | BioLegend |
| CCR6 | PerCP-Cy5.5 | 11A9 | BD |
| CCR7 | PerCP-Cy5.5 | TG8 | BioLegend |
| CCR9 | APC | 112509 | R&D Systems |
| CCR10 | PE | 314305 | R&D Systems |
| CXCR1 | APC | 8F1 | BioLegend |
| CXCR2 | PE | 5E8 | BioLegend |
| CXCR3 | PerCP-Cy5.5 | TG1 | BioLegend |
| CXCR4 | APC | 12G5 | R&D Systems |
| CXCR5 | PerCP-Cy5.5 | TG2 | BioLegend |
| CXCR6 | PE | 56811 | R&D Systems |
| CXCR7 | PE | 8F11-M16 | BioLegend |
| CX3CR1 | APC | 2A9-1 | BioLegend |
| XCR1 | PE | polyclonal | R&D Systems |
| ChemR23 | APC | 84939 | R&D Systems |
PCR experiments
For TNF-α PCR experiments, RNA isolation was performed using TRIZOL reagent (Invitrogen, Carlsbad, CA, USA). For the CCL25 experiment, intestinal biopsies were collected through flexible sigmoidoscopy from UC patients and immediately submerged in RNAlater (Ambion, Austin, TX, USA). RNA was subsequently isolated using the RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. For the TNF-α and CCL25 experiments, 100 ng RNA per sample was included in a reverse transcriptase reaction using iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, USA). For PCR array analyses, RNA was isolated from sorted CD14+HLA-DRhi and CD14+HLA-DRlo populations using the RNeasy Mini Kit (Qiagen). For each of the analyzed populations, equal amounts of RNA from three independent donors were pooled, and complementary DNA was synthesized using RT2 First Strand Kit (SABiosciences, Germantown, MD, USA) from 150 ng of RNA. Subsequently, complementary DNA was put into a RT2 qPCR Master Mix (SABiosciences) reaction and loaded onto a Human Inflammatory Response and Autoimmunity 96-well PCR array plate according to the instructions of the manufacturer (SABiosciences). TNF-α PCR experiments were performed on an iCyclerIQ Optical System using 2 × IQ SYBR Green supermix and iCycler IQ Optical System Software v3.1 (Bio-Rad) for data retrieval. The CCL25 PCR was performed on a CFX96 PCR system (Bio-Rad) using Go-Taq hotstart polymerase (Promega, Madison, WI, USA). In the TNF-α and CCL25 experiments, expression levels were normalized to RNA polymerase II using the 2−ΔΔCt method. Primers used were TNF-α forward (5′-CTCTCTCCCCTGGAAAGGAC-3′), TNF-α reverse (5′-GCCAGAGGGCTGATTAGAGA-3′), CCL25 forward (5′-CCACACCCAAGGTGTCTTTGA-3′), CCL25 reverse (5′-GAGCACAGCCCACCCAAT-3′), RPII forward (5′-GCACCACGTCCAATGACAT-3′), RPII reverse (5′-GTGCGGCTGCTTCCATAA-3′), and CCL25 Taqman probe (5′-FAM-ACTGCTGCCTGGCCTACCACTACCC-TAMRA-3′ (Cybergene, Huddinge, Sweden). CCL25 primer sequences were adopted from Eksteen et al.22 For CCL25 PCR array analyses, expression levels were normalized to the arithmetic mean expression of the B2M, HPRT1, RPL13A, GAPDH, and ACTB housekeeping genes, using the 2−ΔΔCt method.
CCL25 depletion assay
Biotinylated CCL25 (Almac Sciences, Craigavon, UK) was bound to a solid support consisting of a streptavidin–sepharose matrix (GE Healthcare). PBMC from six healthy donors was perfused through the device, and CCR9 expression was analyzed before and after using flow cytometry.
Statistical analyses
All group analyses were carried out using two-tailed-dependent Student's t-test (Figures 4 and 6) or two-tailed-independent Student's t-test (Figures 2, 3, and 6). Regression analyses were performed using ordinal regression test for nonparametric data (Figures 2b and c). All calculations were carried out in GraphPad Prism v5 software (GraphPad Software, San Diego, CA, USA). Values of P≤0.05 were regarded as significant, and depicted as follows: P≤0.05*, P<0.01**, and P≤0.001***. In all figures, bars represent mean±s.d.
Ethical considerations
The study was approved by the Stockholm Regional Ethics Review Board in Stockholm, Sweden (http://www.epn.se). The ethical approval applies to all centers from which patients were recruited (South Hospital, Stockholm, Sweden; Karolinska Hospital, Stockholm, Sweden; and Danderyd Hospital, Stockholm, Sweden). All patients were enrolled through formal written consent.
RESULTS
The frequency of CD14+HLA-DRhi monocytes is increased in active UC and CD
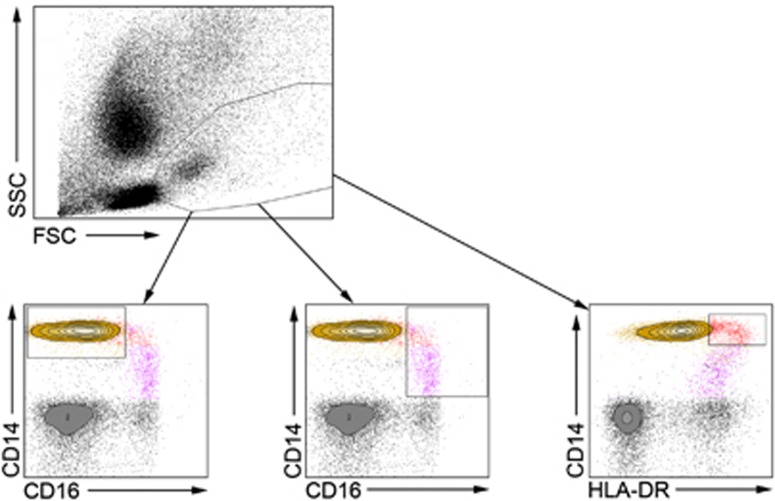
To investigate the role of CD14+HLA-DRhi monocytes in IBD patients, we used flow cytometry to identify the population in peripheral blood (Figure 1). To rule out any possibility that the CD14+HLA-DRhi population is in fact a mixed population mainly composed of CD16-expressing monocytes, we investigated the relative expression of CD16 and HLA-DR as well as side and forward scatter appearance in the CD14+HLA-DRhi, CD14loCD16+, and CD14+CD16− populations. This analysis revealed the HLA-DRhi subset to be clearly distinguished from the other established monocyte population with regard to its CD16 and HLA-DR expression pattern (data not shown).
Figure 1.
Flow cytometry gating strategies. Representative flow cytometry plots showing the gating strategies used throughout the study for the CD14+CD16− (yellow; lower left), CD14loCD16+ (purple; lower middle), and CD14+HLA-DRhi (red; lower right) monocyte populations. The presented plots are from one representative inflammatory bowel disease patient.
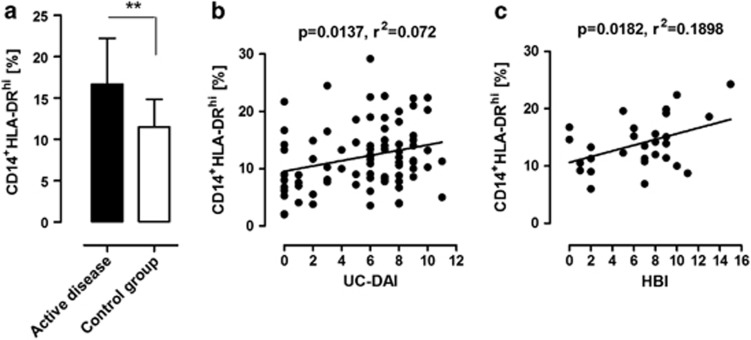
When analyzing blood from patients and controls, we found that active inflammation in the colon correlated to a significantly higher frequency of HLA-DRhi monocytes compared with the control group (Figure 2a, P=0.006). In addition, a correlation was observed with disease activity and the prevalence of HLA-DRhi monocytes in UC (UC-DAI) (Figure 2b; P=0.0137, r2=0.072) and CD (HBI) (Figure 2c; P=0.0182, r2=0.1898).
Figure 2.
CD14+HLA-DRhi monocytes are increased and correlate to disease activity in patients with ulcerative colitis (UC) and Crohn's disease (CD). (a) Frequency of CD14+HLA-DRhi monocytes in peripheral blood of inflammatory bowel disease patients compared with controls, as determined with flow cytometry. Bars represent mean values±s.d. from controls (n=11) and patients (n=31) with active UC (n=20; UC-DAI 6–12) or CD (n=11; Harvey–Bradshaw (HBI) 8–16) (P=0.006). (b) Regression analysis of CD14+HLA-DRhi monocytes and clinical disease activity in patients with UC (P=0.0137; r2=0.072). Data represent measurements (n=84) from 28 unique patients at different time points during treatment. (c) Regression analysis of CD14+HLA-DRhi monocytes and clinical disease activity in patients with CD (P=0.0182; r2=0.190). Data represent measurements (n=29) from 11 unique patients at different time points during treatment. Axes represent percentage CD14+HLA-DRhi of its parent CD14+ monocyte population.
CD14+HLA-DRhi monocytes are potential therapeutic targets and markers of inflammation in colitis
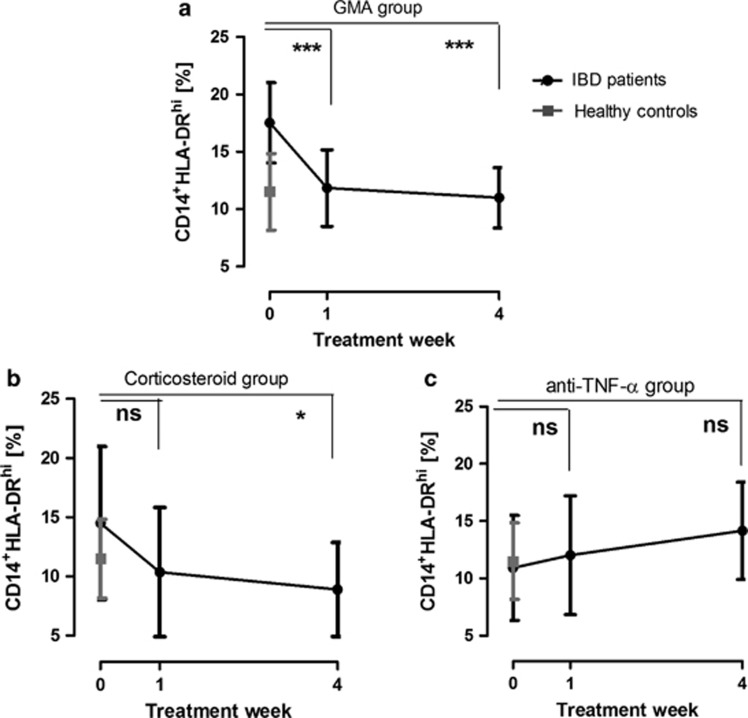
Next, we investigated whether the CD14+HLA-DRhi population was affected by conventional IBD therapy. Patients with active intestinal inflammation who received either corticosteroids or anti-TNF-α antibodies (infliximab or adalimumab) were monitored for 5 consecutive weeks. A patient group treated with GMA was included for comparison, considering the selective removal of monocytes associated with Adacolumn.23 When these treatment regimens were plotted separately, the patient group receiving GMA therapy accounted for the most prominent decrease (Figure 3a). The monocyte population was also attenuated after only 1 week of therapy among corticosteroid patients. The suppression was maintained, reaching levels well below those of healthy control patients at week 4 (Figure 3b, P<0.05). The decreased population of CD14+HLA-DRhi during treatment was not influenced by the diagnosis UC or CD, extension of the disease in the colon, concomitant azathioprine treatment, or gender (data not shown). The treatment groups were too small to allow for any comparison between responders and nonresponders. However, adding the patients of the GMA- and corticosteroid-treated groups together, the HLA-DRhi population was significantly decreased after 4 weeks of treatment (16.95±1.80–9.68±0.75; P<0.001) in those who achieved remission. This was not observed among the non-remission patients (15.36±1.66–11.85±1.73; not significant). Interestingly, biological therapy with antibodies against TNF-α did not significantly affect the proportion of CD14+HLA-DRhi monocytes (Figure 3c). Among these patients, CD14+HLA-DRhi never reached the reference level observed in the controls.
Figure 3.
CD14+HLA-DRhi monocyte levels are decreased during inflammatory bowel disease (IBD) therapy. CD14+HLA-DRhi monocyte levels in IBD patients during treatment with (a) granulocyte/monocyte apheresis (GMA) apheresis (n=18), (b) corticosteroids (n=16), or (c) anti-tumor necrosis factor (TNF)-α biological therapy (n=14). Control patient reference levels (n=11) are included in all graphs. Error bars represent group mean values±s.d. Axes represent percentage CD14+HLA-DRhi of its parent CD14+ monocyte population. NS, not significant.
CD14+HLA-DRhi monocytes produce high levels of inflammatory mediators
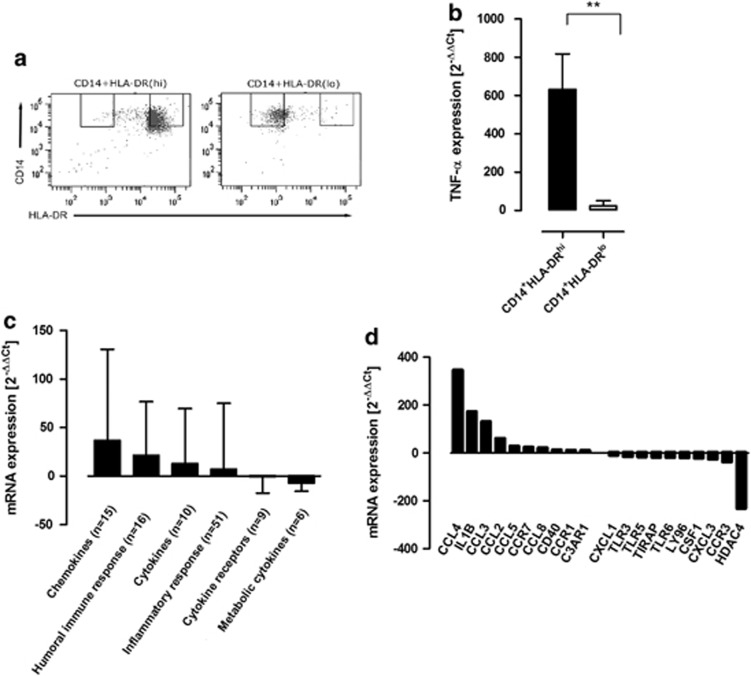
With the purpose of investigating the capacity of CD14+HLA-DRhi monocytes to produce inflammatory mediators, monocytes from healthy blood donors were cultured in the presence of LPS. The HLA-DRhi population and its CD14+HLA-DRlo counterpart were subsequently sorted using flow cytometry (Figure 4a). The two cell populations were investigated with regard to the production of the pro-inflammatory cytokine TNF-α. Interestingly, the CD14+HLA-DRhi population produced 500-fold increased levels of TNF-α transcripts upon LPS stimulation compared with the CD14+HLA-DRlo cells (Figure 4b). Furthermore, PCR array analyses were carried out on sorted CD14+HLA-DRhi monocytes from three independent donors after activation with LPS in order to establish the distinctive phenotype of the population. In accordance with our hypothesis, several gene transcripts described as being involved in monocyte-mediated immune responses were upregulated in the CD14+HLA-DRhi monocytes. Increased gene expression was mainly found among chemotactic cytokines and genes involved in the humoral immune response (Figure 4c; Supplementary Table). The most prominent fold-change difference between the CD14+HLA-DRhi and the CD14+HLA-DRlo monocytes was observed for the chemotactic cytokine CCL4. The transcript with the most apparent downregulation among HLA-DRhi monocytes was the HDAC4 gene that encodes a histone deacetylase that functions as a transcriptional repressor.24 Together, these data show that CD14+HLA-DRhi monocytes have strong pro-inflammatory potential.
Figure 4.
CD14+HLA-DRhi monocytes produce high levels of inflammatory mediators in response to lipopolysaccharide (LPS). (a) Representative flow cytometry plots depicting CD14+HLA-DRhi and CD14+HLA-DRlo purity after flow cytometry sorting. (b) PCR analysis of tumor necrosis factor (TNF)-α in CD14+HLA-DRhi monocytes after LPS activation for 2 h (n=4, P=0.0047). Graph shows analysis of duplicate samples using the 2−ΔΔCt method and RNA polymerase II as housekeeping gene. (c) Target transcripts from PCR array analysis of CD14+HLA-DRhi monocytes from three independent healthy donors after LPS activation for 6 h, grouped into functional categories. (d) The 20 target transcripts that represented the strongest up- and downregulation in PCR array analyses of CD14+HLA-DRhi monocytes following LPS stimulation for 6 h. Fold changes range from 347.3 to 10.9 and −10.3 to −232.3, respectively. Axes in (c) and (d) represent transcript fold changes in the CD14+HLA-DRhi subset using CD14+HLA-DRlo as a reference population, and the arithmetic mean of the B2M, HPRT1, RPL13A, GAPDH, and ACTB transcript levels as housekeeping genes using the 2−ΔΔCt method. Panels (c) and (d) represent data from equal amounts of pooled input RNA isolated from three independent healthy blood donors. Error bars represent group mean values±s.d. mRNA, messenger RNA.
CD14+HLA-DRhi monocytes express the gut-homing chemokine receptor CCR9
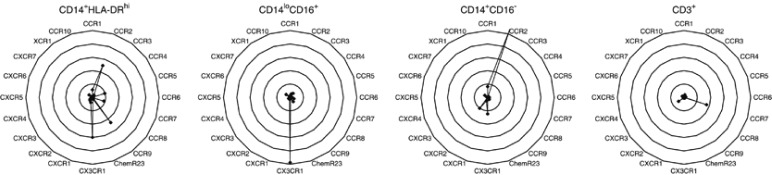
Next, we studied the surface expression of various chemokine receptors on CD14+HLA-DRhi monocytes in relation to the CD14+CD16− and CD14loCD16+ subsets. Although we could observe significant overlap between many of these markers in terms of their expression in the respective subsets, the CD14+HLA-DRhi population was clearly distinguished by its expression of CCR7 and CCR9 (Figure 5; Supplementary Figure 1). CCR7 is mainly described as a lymph-node-homing receptor for DCs and T-helper cells, and has previously not been reported to be expressed on circulating monocytes.25 CCR9 has been shown to be important in lymphocyte homing to the gut through interactions with its ligand CCL25, expressed in the intestinal mucosa.26, 27 Although CCR9–CCL25 interactions have been well characterized in T-helper cells, their role in monocytes is unclear. The general CD14+ monocyte population exhibited clearly higher CCR9 expression compared with CD3+ T lymphocytes, which has been described as the main CCR9-expressing cell type (Figure 6a).28
Figure 5.
The relative chemokine receptor expression in the CD14+HLA-DRhi, CD14loCD16+, and CD14+CD16− monocyte populations, as well as CD3+ lymphocytes, as determined by flow cytometry. Each monocyte subset is represented by one spider web chart. Data points indicate isotype control-subtracted expression for each chemokine receptor. Expression levels are defined as median fluorescence intensity channel numbers (MFC). Data presented are mean values from five healthy control patients.
Figure 6.
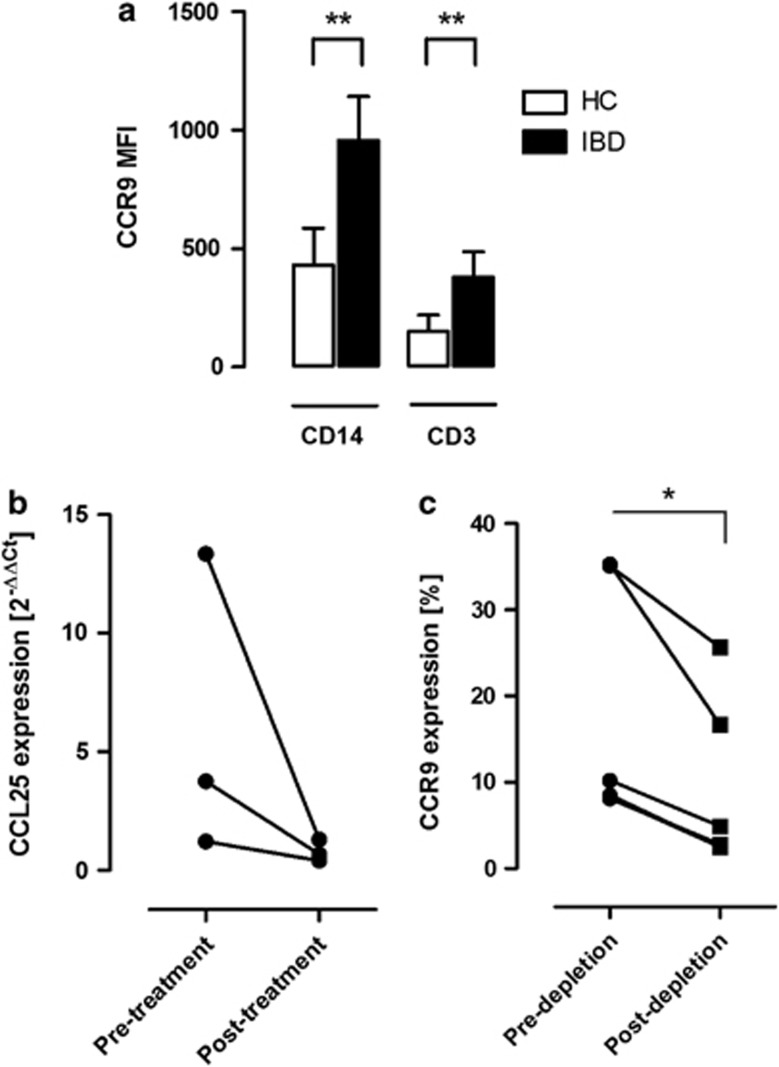
The CCR9–CCL25 axis has a role in monocytes contributing to chronic intestinal inflammation. (a) CCR9 expression levels in CD14+ monocytes and CD3+ lymphocytes are significantly increased in peripheral blood in inflammatory bowel disease (IBD) patients with active intestinal inflammation (n=5) compared with healthy controls (HC) (n=5), as determined by flow cytometry. Expression levels are defined as median fluorescence intensity channel numbers (MFC). Error bars represent group mean values±s.d. (b) The graph shows CCL25 messenger RNA transcript levels from colonic biopsy specimens from IBD patients (n=3) before and after 4 weeks of corticosteroid therapy. Graph shows analysis of duplicate samples using the 2−ΔΔCt method and RNA polymerase II as housekeeping gene. (c) Depletion of peripheral blood CCR9-expressing CD14+HLA-DRhi monocytes from IBD patients using CCL25-coated beads (n=5; P=0.011).
When comparing CCR9 expression in CD14+HLA-DRhi with CD14loCD16+ and CD14+CD16− monocytes, the HLA-DRhi subset displayed markedly increased levels (Figure 5; Supplementary Figure 1). In contrast, the expression of CCR2, a chemokine receptor responsible for general monocyte migration, was not increased on the HLA-DRhi monocytes, indicating that gut-homing phenotype constitutes a specific feature of the monocytes, rather than reflecting general immunological activation (Figure 5; Supplementary Figure 1).29 In conclusion, pro-inflammatory CD14+HLA-DRhi monocytes express high levels of the gut-homing chemokine receptor CCR9, which directs them to the site of mucosal inflammation.
CCR9 expression on CD14+ monocytes is significantly increased during active intestinal inflammation
After establishing that CCR9 is indeed expressed on human monocytes, we investigated monocytic CCR9 expression levels during active colonic inflammation. In patients with active IBD, we observed significantly increased CCR9 expression levels on CD14+ monocytes compared with healthy controls (Figure 6a; P<0.01). The same pattern was observed on CD3+ lymphocytes, although the levels were generally lower (P<0.01).
We also analyzed colonic mucosal tissue biopsies with real-time PCR and found that the CCR9 ligand, CCL25/TECK, was expressed in the colon (Figure 6b). To establish a functional interaction between CCL25 and CCR9, we carried out depletion experiments by perfusing peripheral blood cells through a solid support containing CCL25-coated sepharose beads. The frequency of CCR9-positive CD14+HLA-DRhi monocytes was significantly reduced after encounter with the CCL25-coated sepharose beads, showing that CCR9 on CD14+HLA-DRhi monocytes could bind CCL25 and be removed from the blood (Figure 6c; P<0.05). These results indicate that CCR9 expressed on monocytes may functionally interact with colonic CCL25/TECK and has a role in human colonic inflammation.
DISCUSSION
In this study, we identify CCR9-expressing CD14+HLA-DRhi blood monocytes as an important factor in intestinal inflammation. The expression of HLA-DR on monocytes is vital to the inflammatory response and has been shown to determine the efficacy of antigen presentation to T-helper cells.30, 31 Monocytes with high expression of HLA-DR have also been shown to infiltrate the joints of patients with rheumatoid arthritis, an inflammatory disease successfully treated with TNF-α antibodies.32 In addition, carrying the class II HLA-DRB1*0103 allele correlates with an increased risk for developing UC.33
Several studies have suggested that monocytes per se are targeted by conventional IBD therapy.6, 34, 35 Our results suggest that specific downregulation of the HLA-DRhi subpopulation may be an important mechanism behind resolution of the inflammation. In this study, patients treated with GMA were added for comparison, as Adacolumn is the only treatment specifically targeting circulating monocytes. These cells are removed through Fcγ receptor binding to the cellulose acetate beads in the column, leaving circulating T cells unaffected.23 Corticosteroid therapy mediates a decrease in the number of circulating CD14+HLA-DRhi monocytes that is comparable to GMA (Figure 3b). Surprisingly, patients subjected to biological treatment did not display a decrease of pro-inflammatory monocytes (Figure 3c). We speculate that by removing TNF-α, one of the main products of these monocytes, autocrine feedback mechanisms leading to cellular activation might be induced. The observation underscores the difference in mode of action between anti-TNF-α antibodies and corticosteroids, and should be further investigated, as anti-TNF-α failure may partly depend on the monocytes' production of additional pro-inflammatory chemokines, cytokines, and integrin receptors counterbalancing TNF-α suppression.
Classically, leukocyte populations have been defined through their capability to produce inflammatory mediators such as cytokines and chemokines. In order to gain a functional understanding of how CD14+HLA-DRhi monocytes mediate inflammation, we investigated their pro-inflammatory potential at the messenger RNA level compared with their HLA-DRlo-expressing counterpart. In this context, the HLA-DRhi subset produced markedly elevated levels of gene transcripts associated with activation and pro-inflammatory phenotype. The population displayed a 500-fold increase of TNF-α transcript levels, which establishes the HLA-DRhi subset as one of the most important producers of this cytokine. Other genes were also investigated by PCR array analysis, revealing the highest upregulation in CCL4, CCL3, and IL-1β, all cytokines previously described as being involved in the recruitment of inflammatory cells to the intestinal mucosa in IBD (Figure 4d).36, 37, 38, 39, 40 The most prominent difference was observed in the CCL4/MIP-1β gene, with upregulated transcript levels of more than 300-fold. The inflammatory role of CCL4 was reported by Bystry et al.,36 who demonstrated that activated T-helper cells efficiently migrate toward a CCL4 tissue gradient through CCR5 interaction. Thus, our finding that CCL4 transcripts were produced in high levels by CD14+HLA-DRhi monocytes supports their inflammatory potential.41 The transcript with the most apparent downregulation in HLA-DRhi monocytes was the HDAC4 gene that encodes a histone deacetylase that functions as a transcriptional repressor.24 Together, these data indicate that CD14+HLA-DRhi is a transcriptionally active subset that readily expresses genes important for mediating mucosal immune responses.
On the surface level, CD14+HLA-DRhi monocytes only partially express CD16, suggesting that the population constitutes a separate subset that is not included in its entirety when defining pro-inflammatory monocytes through their expression of CD14loCD16+ (Figure 1). The population is further defined by its expression of CCR7 and the gut-homing receptor CCR9, which clearly distinguishes HLA-DRhi monocytes from the CD14+CD16− and CD14loCD16+ subsets (Figure 5; Supplementary Figure 1).
In the context of intestinal inflammation, interactions between CCR9-expressing T cells and CCL25 (TECK) expressed in the gut epithelium have been implicated as an important mechanism for recruiting circulating lymphocytes to the intestinal mucosa.26 However, whether this mechanism also applies to the extensive infiltration of blood monocytes to the intestinal mucosa observed during inflammation has never been studied.42 It was recently shown that CCR9-expressing monocytes are increased in the peripheral blood of patients with rheumatoid arthritis. In accordance with the results from this study, we show that the CD14+CD16− and CD14loCD16+ subsets express similar levels of CCR9.43 Those levels were notably superseded by the CCR9 expression on CD14+HLA-DRhi monocytes (Figure 5; Supplementary Figure 1). Interestingly, this expression was considerably higher than that observed on T cells, which are considered to be the main CCR9-carrying cell type (Figure 6a).28 In contrast, the expression of CCR2, another chemokine receptor important for monocyte relocation in several disease groups, was not increased on HLA-DRhi monocytes (Figure 5; Supplementary Figure 1).29 This suggests that the specific increase in CCR9 expression among CD14+HLA-DRhi may reflect a gut-specific phenotype, rather than a generally activated subset. CCL25, the ligand for CCR9, was found to be expressed in mucosal tissue by QT-PCR analysis, which is supported by other reports identifying CCL25 in the colon in mice (Figure 6b).27, 44 Interestingly, colonic CCL25 messenger RNA levels were consistently higher in pretreatment samples (UC-DAI 7–8, HBI 9; n=3) compared with samples from the same patients after 4 weeks of corticosteroid therapy (UC-DAI 1–2, HBI 2) (Figure 6b).
Our data also show that CCR9 expression levels on circulating blood monocytes are significantly increased in patients with active colonic inflammation (Figure 6a). In conclusion, these data suggest that monocytes in general and CD14+HLA-DRhi in particular possess the ability to relocate to the intestinal mucosa through CCR9–CCL25 interactions, particularly in patients with active disease.
T cells have been shown to acquire their CCR9 expression through retinoic acid-dependent imprinting by mesenteric lymph node DCs.45 The issue of whether CCR9 expression on monocytes is acquired through similar mechanisms seems controversial, especially as blood monocytes are not known to traffic lymph nodes to the same extent as T cells. Here, we show that CD14+HLA-DRhi monocytes are defined by their high expression of CCR7, a marker mainly described as a lymph-node-homing receptor for DCs and T-helper cells (Figure 5; Supplementary Figure 1).25 Therefore, it is tempting to speculate that CCR7-expressing CD14+HLA-DRhi monocytes traffic the lymph nodes to a higher extent than has previously been known, and that CCR9 imprinting in these monocytes may occur through mechanisms similar to those reported in T cells. Being beyond the scope of this study, the mechanisms behind CCR9 induction in monocytes, as well as the functional role of their CCR7 expression, need to be addressed in the future.
In this study, we have shown that CD14+HLA-DRhi blood monocytes are increased in patients with active intestinal inflammation. This subset is distinguished by its capability to produce pro-inflammatory cytokines and its expression of CCR9 that may direct the monocytes toward CCL25 gradients produced in the inflamed colon during IBD. In summary, these findings indicate that CD14+HLA-DRhi blood monocytes have an important role in IBD and that future studies evaluating these monocytes as specific targets for IBD therapy are highly indicated.
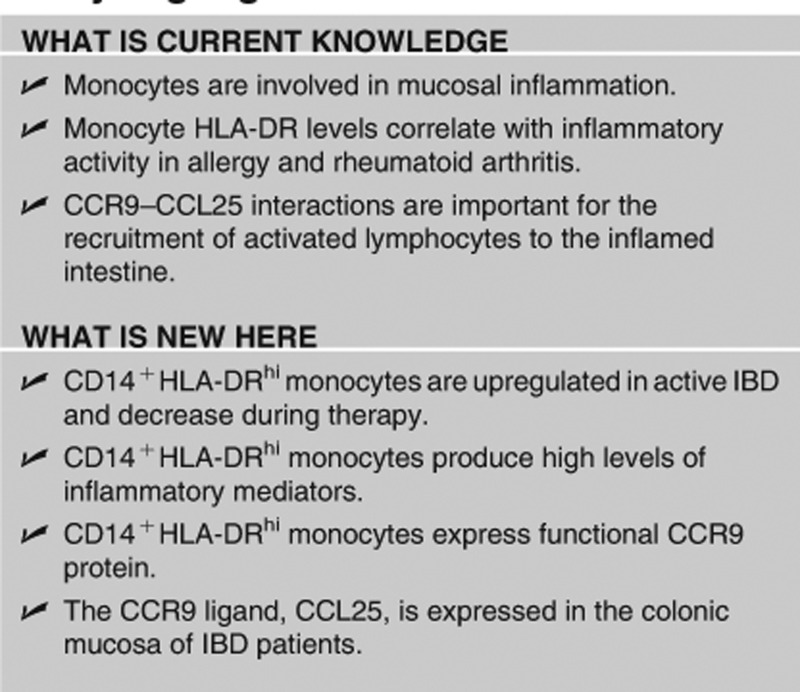
Study Highlights
Acknowledgments
We gratefully acknowledge Martina Jones and Petra Jones for valuable input and excellent technical assistance.
Guarantor of the article: Michael Eberhardson, MD, PhD.
Specific author contributions: Study design: ME and OW; data collection: LL, EH, and JG; data analysis and interpretation: LL, MK, OW, EL, and ME; manuscript drafting: LL, EL, and ME; critical revision of the manuscript: OW, PK, and HG; statistical analysis: MK and LL; obtained funding: OW, HG, and ME; and study supervision and patient inclusion: AL, PK, IJ, and RB.
Financial support: The investigators have received financial support from the Swedish Medical Society, Stockholm, Sweden; ALF-support, Stockholm, Sweden; Immune Therapy Holdings AB, Stockholm, Sweden; Schering-Plough AB, Stockholm, Sweden; and Otsuka Pharma Scandinavia AB, Stockholm, Sweden. The study was designed and performed independent of any financial source.
Potential competing interests: None.
Footnotes
Supplementary Information accompanies this paper on the Clinical and TranslationalGastroenterology website (http://www.nature.com/ctg)
Supplementary Material
References
- Bouma G, Strober W. The immunological and genetic basis of inflammatory bowel disease. Nat Rev Immunol. 2003;3:521–533. doi: 10.1038/nri1132. [DOI] [PubMed] [Google Scholar]
- Desreumaux P, Brandt E, Gambiez L, et al. Distinct cytokine patterns in early and chronic ileal lesions of Crohn's disease. Gastroenterology. 1997;113:118–126. doi: 10.1016/s0016-5085(97)70116-1. [DOI] [PubMed] [Google Scholar]
- Hugot JP, Laurent-Puig P, Gower-Rousseau C, et al. Mapping of a susceptibility locus for Crohn's disease on chromosome 16. Nature. 1996;379:821–823. doi: 10.1038/379821a0. [DOI] [PubMed] [Google Scholar]
- Hugot JP, Chamaillard M, Zouali H, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- Arseneau KO, Tamagawa H, Pizarro TT, et al. Innate and adaptive immune responses related to IBD pathogenesis. Curr Gastroenterol Rep. 2007;9:508–512. doi: 10.1007/s11894-007-0067-3. [DOI] [PubMed] [Google Scholar]
- Hanai H, Iida T, Takeuchi K, et al. Adsorptive depletion of elevated proinflammatory CD14+CD16+DR++ monocytes in patients with inflammatory bowel disease. Am J Gastroenterol. 2008;103:1210–1216. doi: 10.1111/j.1572-0241.2007.01714.x. [DOI] [PubMed] [Google Scholar]
- Mahida YR, Wu KC, Jewell DP. Respiratory burst activity of intestinal macrophages in normal and inflammatory bowel disease. Gut. 1989;30:1362–1370. doi: 10.1136/gut.30.10.1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Haens G, Sandborn WJ, Feagan BG, et al. A review of activity indices and efficacy end points for clinical trials of medical therapy in adults with ulcerative colitis. Gastroenterology. 2007;132:763–786. doi: 10.1053/j.gastro.2006.12.038. [DOI] [PubMed] [Google Scholar]
- Rugtveit J, Brandtzaeg P, Halstensen TS, et al. Increased macrophage subset in inflammatory bowel disease: apparent recruitment from peripheral blood monocytes. Gut. 1994;35:669–674. doi: 10.1136/gut.35.5.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smythies LE, Maheshwari A, Clements R, et al. Mucosal IL-8 and TGF-beta recruit blood monocytes: evidence for cross-talk between the lamina propria stroma and myeloid cells. J Leukoc Biol. 2006;80:492–499. doi: 10.1189/jlb.1005566. [DOI] [PubMed] [Google Scholar]
- Feng G, Lu J, Gross J. Generation of transgenic mice. Methods Mol Med. 2004;99:255–267. doi: 10.1385/1-59259-770-X:255. [DOI] [PubMed] [Google Scholar]
- Frankenberger M, Sternsdorf T, Pechumer H, et al. Differential cytokine expression in human blood monocyte subpopulations: a polymerase chain reaction analysis. Blood. 1996;87:373–377. [PubMed] [Google Scholar]
- Belge KU, Dayyani F, Horelt A, et al. The proinflammatory CD14+CD16+DR++ monocytes are a major source of TNF. J Immunol. 2002;168:3536–3542. doi: 10.4049/jimmunol.168.7.3536. [DOI] [PubMed] [Google Scholar]
- Tallone T, Turconi G, Soldati G, et al. Heterogeneity of human monocytes: an optimized four-color flow cytometry protocol for analysis of monocyte subsets. J Cardiovasc Transl Res. 2011;4:211–219. doi: 10.1007/s12265-011-9256-4. [DOI] [PubMed] [Google Scholar]
- Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9:162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almand B, Clark JI, Nikitina E, et al. Increased production of immature myeloid cells in cancer patients: a mechanism of immunosuppression in cancer. J Immunol. 2001;166:678–689. doi: 10.4049/jimmunol.166.1.678. [DOI] [PubMed] [Google Scholar]
- Haile LA, von Wasielewski R, Gamrekelashvili J, et al. Myeloid-derived suppressor cells in inflammatory bowel disease: a new immunoregulatory pathway Gastroenterology 2008135871–881.881 e1–e5. [DOI] [PubMed] [Google Scholar]
- Genel F, Atlihan F, Ozsu E, et al. Monocyte HLA-DR expression as predictor of poor outcome in neonates with late onset neonatal sepsis. J Infect. 2010;60:224–228. doi: 10.1016/j.jinf.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Fu Q, Cui N, Yu W, et al. Percentages of CD4+ T regulatory cells and HLA-DR expressing monocytes in severe intra-abdominal infections. Scand J Infect Dis. 2010;42:475–478. doi: 10.3109/00365541003660021. [DOI] [PubMed] [Google Scholar]
- Blumberg RS, Saubermann LJ, Strober W. Animal models of mucosal inflammation and their relation to human inflammatory bowel disease. Curr Opin Immunol. 1999;11:648–656. doi: 10.1016/s0952-7915(99)00032-1. [DOI] [PubMed] [Google Scholar]
- Vermeire S, Schreiber S, Sandborn WJ, et al. Correlation between the Crohn's disease activity and Harvey-Bradshaw indices in assessing Crohn's disease severity. Clin Gastroenterol Hepatol. 2010;8:357–363. doi: 10.1016/j.cgh.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Eksteen B, Grant AJ, Miles A, et al. Hepatic endothelial CCL25 mediates the recruitment of CCR9+ gut-homing lymphocytes to the liver in primary sclerosing cholangitis. J Exp Med. 2004;200:1511–1517. doi: 10.1084/jem.20041035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saniabadi AR, Hanai H, Takeuchi K, et al. Adacolumn, an adsorptive carrier based granulocyte and monocyte apheresis device for the treatment of inflammatory and refractory diseases associated with leukocytes. Ther Apher Dial. 2003;7:48–59. doi: 10.1046/j.1526-0968.2003.00012.x. [DOI] [PubMed] [Google Scholar]
- Bertos NR, Wang AH, Yang XJ. Class II histone deacetylases: structure, function, and regulation. Biochem Cell Biol. 2001;79:243–252. [PubMed] [Google Scholar]
- Forster R, Davalos-Misslitz AC, Rot A. CCR7 and its ligands: balancing immunity and tolerance. Nat Rev Immunol. 2008;8:362–371. doi: 10.1038/nri2297. [DOI] [PubMed] [Google Scholar]
- Johansson-Lindbom B, Agace WW. Generation of gut-homing T cells and their localization to the small intestinal mucosa. Immunol Rev. 2007;215:226–242. doi: 10.1111/j.1600-065X.2006.00482.x. [DOI] [PubMed] [Google Scholar]
- Wurbel MA, Philippe JM, Nguyen C, et al. The chemokine TECK is expressed by thymic and intestinal epithelial cells and attracts double- and single-positive thymocytes expressing the TECK receptor CCR9. Eur J Immunol. 2000;30:262–271. doi: 10.1002/1521-4141(200001)30:1<262::AID-IMMU262>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Zabel BA, Agace WW, Campbell JJ, et al. Human G protein-coupled receptor GPR-9–6/CC chemokine receptor 9 is selectively expressed on intestinal homing T lymphocytes, mucosal lymphocytes, and thymocytes and is required for thymus-expressed chemokine-mediated chemotaxis. J Exp Med. 1999;190:1241–1256. doi: 10.1084/jem.190.9.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmane SL, Kremlev S, Amini S, et al. Monocyte chemoattractant protein-1 (MCP-1): an overview. J Interferon Cytokine Res. 2009;29:313–326. doi: 10.1089/jir.2008.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Don Porto Carero A, Hoet PH, Nemery B, et al. Increased HLA-DR expression after exposure of human monocytic cells to air particulates. Clin Exp Allergy. 2002;32:296–300. doi: 10.1046/j.1365-2222.2002.01266.x. [DOI] [PubMed] [Google Scholar]
- Portillo G, Turner M, Chantry D, et al. Effect of cytokines on HLA-DR and IL-1 production by a monocytic tumour, THP-1. Immunology. 1989;66:170–175. [PMC free article] [PubMed] [Google Scholar]
- Ridley MG, Kingsley G, Pitzalis C, et al. Monocyte activation in rheumatoid arthritis: evidence for in situ activation and differentiation in joints. Br J Rheumatol. 1990;29:84–88. doi: 10.1093/rheumatology/29.2.84. [DOI] [PubMed] [Google Scholar]
- Anderson CA, Massey DC, Barrett JC, et al. Investigation of Crohn's disease risk loci in ulcerative colitis further defines their molecular relationship Gastroenterology 2009136523–529.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingerle-Rowson G, Angstwurm M, Andreesen R, et al. Selective depletion of CD14+ CD16+ monocytes by glucocorticoid therapy. Clin Exp Immunol. 1998;112:501–506. doi: 10.1046/j.1365-2249.1998.00617.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch S, Kucharzik T, Heidemann J, et al. Investigating the role of proinflammatory CD16+ monocytes in the pathogenesis of inflammatory bowel disease. Clin Exp Immunol. 2010;161:332–341. doi: 10.1111/j.1365-2249.2010.04177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystry RS, Aluvihare V, Welch KA, et al. B cells and professional APCs recruit regulatory T cells via CCL4. Nat Immunol. 2001;2:1126–1132. doi: 10.1038/ni735. [DOI] [PubMed] [Google Scholar]
- Ottonello L, Montecucco F, Bertolotto M, et al. CCL3 (MIP-1alpha) induces in vitro migration of GM-CSF-primed human neutrophils via CCR5-dependent activation of ERK 1/2. Cell Signal. 2005;17:355–363. doi: 10.1016/j.cellsig.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Ramos CD, Canetti C, Souto JT, et al. MIP-1alpha[CCL3] acting on the CCR1 receptor mediates neutrophil migration in immune inflammation via sequential release of TNF-alpha and LTB4. J Leukoc Biol. 2005;78:167–177. doi: 10.1189/jlb.0404237. [DOI] [PubMed] [Google Scholar]
- Uguccioni M, Gionchetti P, Robbiani DF, et al. Increased expression of IP-10, IL-8, MCP-1, and MCP-3 in ulcerative colitis. Am J Pathol. 1999;155:331–336. doi: 10.1016/S0002-9440(10)65128-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm MC, Elsbury SK, Pavli P, et al. Enhanced expression and production of monocyte chemoattractant protein-1 in inflammatory bowel disease mucosa. J Leukoc Biol. 1996;59:804–812. doi: 10.1002/jlb.59.6.804. [DOI] [PubMed] [Google Scholar]
- Zhou L, Braat H, Faber KN, et al. Monocytes and their pathophysiological role in Crohn's disease. Cell Mol Life Sci. 2009;66:192–202. doi: 10.1007/s00018-008-8308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm MC, Pullman WE, Bennett GM, et al. Direct evidence of monocyte recruitment to inflammatory bowel disease mucosa. J Gastroenterol Hepatol. 1995;10:387–395. doi: 10.1111/j.1440-1746.1995.tb01589.x. [DOI] [PubMed] [Google Scholar]
- Schmutz C, Cartwright A, Williams H, et al. Monocytes/macrophages express chemokine receptor CCR9 in rheumatoid arthritis and CCL25 stimulates their differentiation. Arthritis Res Ther. 2010;12:R161. doi: 10.1186/ar3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurbel MA, McIntire MG, Dwyer P, et al. CCL25/CCR9 interactions regulate large intestinal inflammation in a murine model of acute colitis. PLoS One. 2011;6:e16442. doi: 10.1371/journal.pone.0016442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata M, Hirakiyama A, Eshima Y, et al. Retinoic acid imprints gut-homing specificity on T cells. Immunity. 2004;21:527–538. doi: 10.1016/j.immuni.2004.08.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.