Abstract
Cytochrome P450 (P450)-derived arachidonic acid (AA) metabolites serve pivotal physiological roles. Therefore, it is important to determine the dominant P450 AA monooxygenases in different organs. We investigated the P450 AA monooxygenases protein expression as well as regioselectivity, immunoinhibition, and kinetic profile of AA epoxygenation and hydroxylation in rat heart, lung, kidney, and liver. Thereafter, the predominant P450 epoxygenases and P450 hydroxylases in these organs were characterized. Microsomes from heart, lung, kidney, and liver were incubated with AA. The protein expression of CYP2B1/2, CYP2C11, CYP2C23, CYP2J3, CYP4A1/2/3, and CYP4Fs in the heart, lung, kidney, and liver were determined by Western blot analysis. The levels of AA metabolites were determined by liquid chromatography–electrospray ionization mass spectroscopy. This was followed by determination of regioselectivity, immunoinhibition effect, and the kinetic profile of AA metabolism. AA was metabolized to epoxyeicosatrienoic acids and 19- and 20-hydroxyeicosatetraenoic acid in the heart, lung, kidney, and liver but with varying metabolic activities and regioselectivity. Anti-P450 antibodies were found to differentially inhibit AA epoxygenation and hydroxylation in these organs. Our data suggest that the predominant epoxygenases are CYP2C11, CYP2B1, CYP2C23, and CYP2C11/CYP2C23 for the heart, lung, kidney, and liver, respectively. On the other hand, CYP4A1 is the major ω-hydroxylase in the heart and kidney; whereas CYP4A2 and/or CYP4F1/4 are probably the major hydroxlases in the lung and liver. These results provide important insights into the activities of P450 epoxygenases and P450 hydroxylases-mediated AA metabolism in different organs and their associated P450 protein levels.
Key words: arachidonic acid metabolism, cytochrome P450, kinetics, P450 epoxygenase activity, P450 hydroxylase activity, regioselectivity
INTRODUCTION
Arachidonic acid (AA) is an essential polyunsaturated fatty acid that is present as an esterified form in the membrane phospholipids. Upon release from the membrane phospholipids by phospholipase A2, AA is then metabolized by cyclooxygenases, lipoxygenases, or cytochrome P450 enzymes (P450) (1). AA is oxidized to epoxyeicosatrienoic acids (EETs) and 19- (19-HETE) and 20-hydroxyeicosatetraenoic acid (20-HETE) by P450 epoxygenases and P450 hydroxylases, respectively, in the presence of oxygen and nicotinamide adenine dinucleotide phosphate (NADPH) (2). EETs are quickly hydrolyzed by soluble epoxide hydrolase (sEH) to their corresponding, less biologically active, dihydroxyeicosatrienoic acid (DHET) metabolites (2).
Many studies have demonstrated the role of P450-dependent AA metabolites in several physiological functions. In the cardiovascular system, EETs exhibit potent vasodilatory, anti-platelet, anti-inflammatory, fibrinolytic, vascular smooth muscle anti-migratory, and angiogenic properties (3). In lungs however, EETs have been shown to exhibit both vasoconstrictor and vasodilator effects (4–6). In the kidney, EETs have a role in the regulation of glomerular filtration through activating the Na+/H+ exchanger (7). In the liver, EETs exhibit vasoconstrictor properties (8) and are involved in vasopressin-induced glycogenolysis (9).
On the other hand, 20-HETE is reported to be a potent vasoconstrictor (10) and has been shown to play a role in several pathological heart conditions, including cardiac hypertrophy (11–13). Renal 20-HETE levels are increased in the spontaneously hypertensive rat model of essential hypertension (14). Moreover, inhibition of 20-HETE increased urinary excretion of sodium, without altering renal hemodynamics in Ren-2 transgenic rats (15). In contrast to its vasoconstrictive effects in the peripheral vasculature, 20-HETE has been shown to be a potent vasodilator in human, rabbit, and canine pulmonary arteries and bronchial rings (16). Interestingly, 19-HETE is considered the endogenous antagonist for 20-HETE. It has been reported that 20-HETE could be competitively antagonized by 19(R)- (16) and 19(S)-HETE (17).
A few decades ago, P450 expressions were thought to be limited to the liver. However, many P450 genes have now been shown to be constitutively expressed in different organs (18,19). We have shown that nearly all important P450 involved in AA metabolism are expressed in rat liver as well as heart, lung, and kidney (18). CYP2Bs, CYP2C11, and CYP2J3 (AA P450 epoxygenases) and CYP2E1, CYP4As, and CYP4Fs (AA P450 hydroxylases) were found to have higher expression in the heart (18), while in the lung, CYP2B1, CYP2J3, CYP2E1, and CYP4Fs genes were found to be the predominant P450 epoxygenases and P450 hydroxylases expressed (18). As expected, the kidney and the liver did have high gene expression of all of P450 epoxygenases and hydroxylases (18).
Assessing P450 distribution in organs via gene expression has an innate imperfection, as it assesses only effective gene transcription. In fact, actual P450 expression is a function of both effective transcription and translation. Previously, Thum and Borlak could not detect CYP3A4/5/7 mRNA in human heart. On the contrary, Minamiyama et al. reported significant CYP3A4 protein expression level in human heart (20,21). Furthermore, discrepancies between mRNA and protein levels of CYP2J2 and CYP2C8 have been previously reported (22). They reported that although CYP2J2 mRNA was about 1,000 times higher than CYP2C8, both enzymes had comparable protein levels in human heart tissue (22).
Since AA oxidation by P450 is dependent on other cofactors such as P450 reductase and cytochrome b5, correlation between P450 enzymes expression and activity is not always guaranteed. Marji et al. (23) described a lack of correlation between CYP4A enzymes expression and their activities due to insufficient P450 reductase in rat kidneys (23). Naturally, P450 are greatly varied in their catalytic activities (19). Therefore, a P450 enzyme of high activity, despite its low expression level, may dictate the levels of AA metabolites in a tissue more efficiently than a highly expressed P450 but with low activity.
Because P450-derived AA metabolites are important biologically, it is important to determine dominant P450 AA epoxygenases and hydroxylases in different organs. Therefore, the aims of the current study were to (1) determine different P450 monoxygenases protein expression and (2) define the kinetic profile, regioselectivity, and immunoinhibition characteristics of P450-mediated AA metabolism, in male Sprague Dawley rats heart, lung, kidney, and liver. Subsequently, the distribution of P450 that is significantly involved in AA epoxygenation and hydroxylation in different organs was identified.
MATERIALS AND METHODS
Materials
AA, NADPH, 4-hydroxybenzophenone, and sucrose were purchased from Sigma-Aldrich Chemical Co. (St. Louis, MO). AA metabolite standards: 5,6-epoxyeicosatrienoic acid (5,6-EET), 8,9-epoxyeicosatrienoic acid (8,9-EET), 11,12-epoxyeicosatrienoic acid (11,12-EET), 14,15-epoxyeicosatrienoic acid (14,15-EET), 5,6-dihydroxyeicosatrienoic acid (5,6-DHET), 8,9-dihydroxyeicosatrienoic acid (8,9-DHET), 11,12-dihydroxyeicosatrienoic acid (11,12-DHET), 14,15-dihydroxyeicosatrienoic acid (14,15-DHET), 19-HETE, and 20-HETE, were purchased from Cayman Chemical (Ann Arbor, MI). High-performance liquid chromatography grade reagents were used for liquid chromatography–electrospray ionization mass spectrometry (LC-ESI-MS). Acetonitrile, methanol, and ethyl acetate were purchased from EM Scientific (Gibbstawn, NJ). Potassium chloride, calcium chloride dihydrate, potassium dihydrogen orthophosphate, dipotassium hydrogen orthophosphate, and magnesium chloride hexahydrate were obtained from BDH (Toronto, ON, Canada). Acrylamide, N′N′-bis-methylene-acrylamide, ammonium persulphate, β-mercaptoethanol, glycine, nitrocellulose membrane (0.45 μm), and TEMED were purchased from Bio-Rad Laboratories (Hercules, CA). Chemiluminescence Western blotting detection reagents were purchased from GE Healthcare Life Sciences, Piscataway, NJ. CYP2C11, CYP2C23, and CYP4F2 primary antibodies were purchased from Abcam (Cambridge, UK). CYP2Js primary antibody was a generous gift from Dr. Darryl Zeldin (National Institute of Environmental Health Sciences, National Institutes of Health, Research Triangle Park, NC). CYP2B1/2, CYP4A1/2/3, secondary antibodies, and control IgG were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Other chemicals were purchased from Fisher Scientific Co. (Toronto, ON, Canada).
Animals
All rats were maintained and used in accordance with the animal protocol approved by the University of Alberta Health Sciences Animal Policy and Welfare Committee, Edmonton, AB, Canada. Male Sprague Dawley (SD) rats (Charles River Canada, St. Constant, QC, Canada) with mean weight of 450 g were used. All animals were allowed free access to food and water.
Microsomal Preparation
Animals were sacrificed under isoflurane anaesthesia. The heart, lung, kidney, and liver were excised, immediately frozen in liquid nitrogen and stored at −80°C until microsomal preparation. Microsomal fractions of the heart, lung, kidney, and liver of 5 SD rats pooled together were prepared by differential centrifugation of homogenized tissues as described previously (24). Briefly, organs were washed in ice-cold potassium chloride (1.15%, w/v). Subsequently, they were cut into pieces and homogenized in ice-cold 0.25 M sucrose solution sucrose (17%, w/v). After homogenizing, the tissues were separated by differential ultracentrifugation. The final pellet was re-suspended in cold sucrose and stored at −80°C. The microsomal protein concentration was determined by the Lowry method using bovine serum albumin as a standard (25).
Western Blot Analysis
To determine the protein expression pattern of the main P450 epoxygenases, CYP2Bs, CYP2C11, CYP2C23, and CYP2J3, and the main P450 hydroxylases, CYP4As, and CYP4Fs for each organ, Western blot analysis was performed using a previously described method (26). Briefly, 5–40 μg of heart, lung, kidney, and liver microsomal preparations were separated by 10% sodium dodecyl sulfate-polyacrylamide gel (SDS-PAGE), and then electrophoretically transferred to nitrocellulose membrane. Protein blots were then blocked overnight at 4°C in blocking solution containing 0.15 M sodium chloride, 3 mM potassium chloride, 25 mM Tris-base, 5% skim milk, 2% bovine serum albumin, and 0.5% Tween-20. The blots were then incubated with a primary antibody: mouse anti-rat CYP2B1/2, rabbit anti-rat CYP2C11, rabbit anti-rat CYP2C23, rabbit anti-mouse CYP2Js, mouse anti-rat CYP4A1/2/3, rabbit anti-human CYP4F2, or rabbit anti-rat actin. After a 2-h incubation period (24 h for the mouse anti-rat CYP2B1/2 antibody), blots were incubated with a peroxidase-conjugated goat anti-rabbit or goat anti-mouse IgG secondary antibody for 1 h at room temperature. The bands were visualized using the enhanced chemiluminescence method according to the manufacturer’s instructions (GE Healthcare Life Sciences, Piscataway, NJ). The intensity of the protein bands were quantified relative to the signals obtained for actin, using ImageJ software (National Institutes of Health, Bethesda, MD, http://rsb.info.nih.gov/ij).
Microsomal Incubations
Preliminary incubations showed that the metabolism of AA was linear with respect to both incubation time and microsomal protein concentration used. The total microsomal protein concentrations used were 1,000 μg/mL for heart and lung and 500 μg/mL for kidney and liver. The total volume of microsomal incubates was 200 μL of incubation buffer (3 mM magnesium chloride hexahydrate dissolved in 100 mM potassium phosphate buffer, pH 7.4). AA was added at a final concentration ranging between 16 and 922 μM for heart and lung and 16–182 μM for kidney and liver. At each AA concentration, the formation of the metabolites was determined in triplicate. Incubations were conducted at 37°C in a shaking water bath (80 rpm). A pre-equilibration period of 5 min was performed. The reaction was initiated by the addition of the cofactor NADPH (final concentration, 2 mM) and terminated after 30 min for heart and lung, or 15 min for kidney and liver, by the addition of 600 μL ice-cold acetonitrile. AA metabolites were extracted twice by 1 mL ethyl acetate and dried using speed vacuum (Savant, Farmingdale, NY), then reconstituted in acetonitrile.
Immunoinhibition of P450-Mediated AA Epoxygenation and Hydroxylation Activity
The inhibitory effect of anti-CYP2B1/2, anti-CYP2C11, anti-CYP2C23, and anti-CYP4A1/2/3 antibodies on P450-mediated AA metabolism was studied in microsomal fraction of each organ; 15–25 μg of microsomal protein was incubated with 9–15 μg of antibody for 30 min on ice. Then AA was added to a concentration of 100 μM and final volume was 50 μL. The incubation time varied between 30 and 60 min according to organ used. GAPDH rat antibody was used as non-P450-specific antibody in control incubations. Initiation and termination of the reaction were performed as described under microsomal incubation experiment.
Analysis of AA Metabolites by LC-ESI-MS
AA metabolites were analyzed using a LC-ESI-MS (Waters Micromass ZQ 4000 spectrometer) method as described previously with minor modifications (27,28). The mass spectrometer runs in negative ionization mode with single ion recorder acquisition. The nebulizer gas was acquired from an in house high purity nitrogen source. The temperature of the source was set at 150°C, and the capillary and cone voltage were 3.51 kV and 25 V, respectively. The samples (10 μL) were separated on reverse phase C18 column (Kromasil, 250 × 3.2 mm) using the mobile phase water/acetonitrile with 0.005% acetic acid. The mobile phase was delivered using a linear gradient method at a flow rate of 0.2 mL/min as follows: 60% to 80% acetonitrile in 30 min, 80% to 100% acetonitrile in 5 min, and 100% for 5 min. The concentrations of the AA metabolites were calculated by comparing their corresponding analyte/reference response ratio based on peak area to calibration standards.
Data Analysis
To determine the enzyme kinetics of P450-mediated AA metabolism in the heart, lung, kidney, and liver microsomal preparations, the rate of AA metabolite formation versus AA concentration in each organ were fitted to three models of enzyme kinetics: one site saturation Michaelis–Menten sigmoidal model (Eq. 1), one site saturation + linear nonspecific component (Eq. 2), and two site saturation Michaelis–Menten model (Eq. 3). The nonlinear regressions were performed using SigmaPlot software, version 11.0.
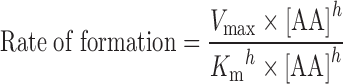 |
1 |
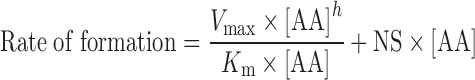 |
2 |
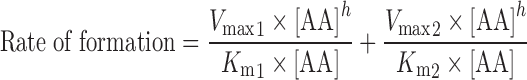 |
3 |
Where Vmax is the maximal rate of formation, Km is the affinity constant, [AA] is the concentration of AA, h is the shape factor of a value of “1” or other values to fit the data, and NS represents the nonspecific activity. In the two binding sites model, two Km (Km1 and Km2) and two Vmax (Vmax1 and Vmax2) were used.
In the heart, lung, kidney, and liver microsomes, the intrinsic clearance (Clint) for EETs as well as 19- and 20-HETE formation was calculated by determining the Vmax to Km ratio. The rate of formation of the four EETs was calculated as the sum of the rate of formation of each EET + the corresponding DHETs, due to the rapid metabolism of some EETs to their corresponding DHETs by sEH enzyme.
The optimal enzyme kinetics model was determined by the Akaike information criterion as a measure of the goodness of fit. The model that demonstrated the best goodness of fit was the one site saturation Michaelis–Menten sigmoidal model (Eq. 1). Applying a constraint of “h = 1” (simple Michaelis–Menten model) or no constraints (sigmoidal model) was also evaluated by Akaike information criterion.
Statistical Analysis
Data are presented as mean ± SE One-way analysis of variance was used to compare kinetic parameters or protein expression between the four organs followed by a Tukey’s post hoc test. A result was considered statistically significant where p < 0.05.
RESULTS
Protein Expression of P450 Epoxygenases and P450 Hydroxylases in the Heart, Lung, Kidney, and Liver
The P450 protein expression in the microsomal fractions of the heart, lung, kidney, and liver was determined by Western blot analysis. The four organs expressed all P450 enzymes investigated, albeit in different extent. The protein expression profile of the main P450 epoxygenase enzymes (CYP2B1/2, CYP2C11, CYP2C23, and CYP2J3) and P450 ω-hydroxylase (CYP4A1/2/3 and CYP4Fs) was found to be organ specific. Figure 1 depicts this profile for the heart, lung, and kidney compared with the liver. It is apparent that CYP2C11 and CYP2J3 were the main epoxygenases to be expressed in the heart. On the other hand, CYP2B1/2, CYP2C11, and CYP2J3 were expressed in the lung.
Fig. 1.
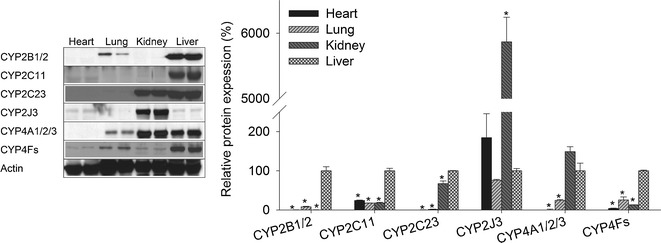
P450 protein expression in heart, lung, kidney, and liver. Microsomal protein was isolated from the heart, lung, kidney, and liver and separated on a 10% SDS-PAGE. CYP2B1/2, CYP2C11, CYP2C23, CYP2J3, CYP4A1/2/3, CYP4Fs, and actin proteins were detected by the enhance chemiluminescence method. The graph represents the amount of protein normalized to the loading control (mean ± SEM, n = 3), and the results are expressed as a percentage of the liver protein expression value (*p < 0.05) compared with the liver
For the kidney, CYP2B1/2 was negligibly expressed compared with the liver or the lung. Kidney was the organ with the highest expression of CYP2J3. CYP2C23 has a predominant expression in the kidney compared with CYP2C11. Except for CYP2J3, the liver was found to be the organ with the highest expression of all the tested epoxygenases. Interestingly, the expression level of CYP2J3 was higher in the heart and kidney than in the liver.
Regarding the hydroxylase activity, ω-hydroxylation of AA has been attributed to CYP4A1/2/3 and CYP4Fs (29). We found that the protein levels of CYP4A1/2/3 as well as CYP4Fs in the lung were ∼25% of their levels in the liver (Fig. 1). CYP4A1/2/3 were highly expressed in the kidney compared with the other organs (Fig. 1). CYP4Fs were the highest in the liver followed by the lung and then the kidney (Fig. 1). Regarding the heart, CYP4A1/2/3 and CYP4Fs expression was significantly lower compared with other organs (Fig. 1).
Determination of the Kinetic Parameters of P450-Mediated AA Metabolism in the Heart, Lung, Kidney, and Liver
The AA concentration ranges needed for the full kinetic profile were up to 922 μM for heart and lung and up to 182 μM for kidney and liver, as shown in Figs. 2 and 3. In comparison, the reported endogenous AA concentration was up to 100 μM (30). Moreover, the kinetic model that provided the best fit was the simple Michaelis–Menten models rather than the more complex models, for all metabolites in all organs. This indicates that the formation of a metabolite was either controlled by a single P450 enzyme, or by more than one P450 enzymes of similar Km and Vmax values. The enzyme kinetics parameters values, Vmax, and Km are shown in Table I. Again, the Km values of the EETs formation in the heart in addition to the 14,15-EET formation in the lung are substantially higher than the endogenous AA concentration (30). 19-HETE was identified by its retention time (14.6 min) which was consistent with the authentic standard. Linearity for 19-HETE was in the range between 0.01 and 4 μg/mL and the lower limit of detection was 0.001 μg/mL.
Fig. 2.
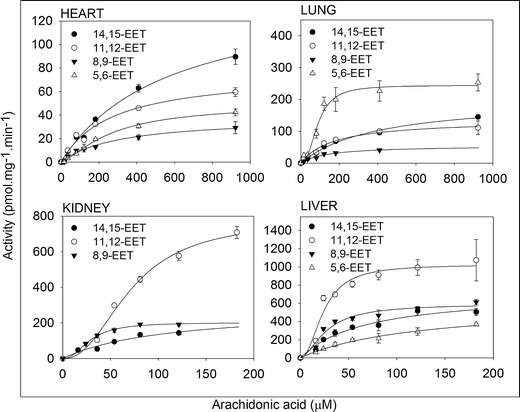
Kinetic profile of arachidonic acid metabolism by P450 epoxygenases in the heart, lung, kidney, and liver microsomal incubates. In a 200-μL total volume, 100 (heart and lung) and 200 μg (kidney and liver) of microsomal protein pooled from five rats were incubated with arachidonic acid for 30 min for heart and lung and 15 min for kidney and liver. The experimental values for arachidonic acid metabolism were expressed as mean ± SEM. Each point was measured in triplicate
Fig. 3.
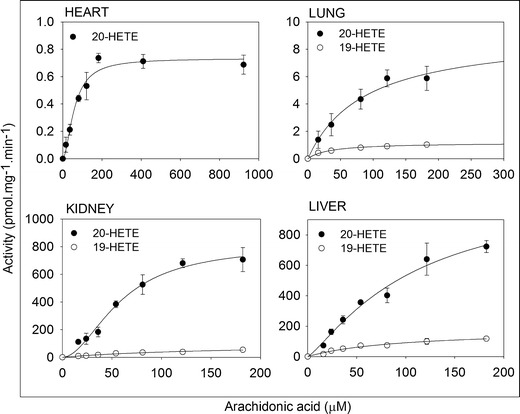
Kinetic profile of arachidonic acid metabolism by P450 hydroxylases in the heart, lung, kidney, and liver microsomal incubates. In a 200-μL total volume, 100 (heart and lung) and 200 μg (kidney and liver) of microsomal protein pooled from five rats were incubated with arachidonic acid for 30 min for heart and lung and 15 min for kidney and liver. The experimental values for arachidonic acid metabolism were expressed as mean ± SEM. Each point was measured in triplicate
Table I.
Enzyme Kinetic Parameters for the Formation of EETs and HETEs in Rat Heart, Lung, Kidney, and Liver Microsomes
| Parameters | Heart | Lung | Kidney | Liver | |
|---|---|---|---|---|---|
| 5,6-EET | V max | 49.3 ± 3.4 | 244 ± 11 | – | 596 ± 71 |
| h | 1.40 ± 0.13 | 2.41 ± 0.45 | – | – | |
| K m | 268 ± 35 | 89.2 ± 7.1 | – | 124 ± 26 | |
| Clint | 0.18 | 2.74 | – | 4.83 | |
| R 2 | 0.99 | 0.95 | – | 0.95 | |
| 8,9-EET | V max | 38.2 ± 2.7 | 55.1 ± 3.1 | 202 ± 8 | 592 ± 35 |
| h | – | – | 2.25 ± 0.33 | 1.71 ± 0.33 | |
| K m | 310 ± 50 | 141 ± 18 | 28.3 ± 1.4 | 26.9 ± 2.8 | |
| Clint | 0.12 | 0.39 | 7.15 | 22 | |
| R 2 | 0.96 | 0.98 | 0.99 | 0.95 | |
| 11,12-EET | V max | 76.6 ± 4 | 136 ± 7 | 775 ± 40 | 1,024 ± 57 |
| h | – | – | 2.30 ± 0.23 | 2.13 ± 0.50 | |
| K m | 270 ± 33 | 172 ± 24 | 71.3 ± 4.5 | 24.2 ± 2.4 | |
| Clint | 0.28 | 0.79 | 10.9 | 42.4 | |
| R 2 | 0.97 | 0.96 | 0.99 | 0.92 | |
| 14,15-EET | V max | 146 ± 8 | 204 ± 8 | 283 ± 44 | 729 ± 60 |
| h | – | – | – | – | |
| K m | 570 ± 60 | 401 ± 32 | 109 ± 29 | 68.4 ± 12.6 | |
| Clint | 0.26 | 0.51 | 2.59 | 10.7 | |
| R 2 | 0.99 | 0.99 | 0.96 | 0.94 | |
| 19-HETE | V max | – | 1.18 ± 0.03 | 100 ± 19 | 177 ± 18 |
| h | – | – | – | – | |
| K m | – | 33.8 ± 3.5 | 166 ± 52 | 94.6 ± 19 | |
| Clint | – | 0.03 | 0.60 | 1.87 | |
| R 2 | – | 0.97 | 0.91 | 0.94 | |
| 20-HETE | V max | 0.73 ± 0.03 | 9.19 ± 1.28 | 812 ± 62 | 1,118 ± 264 |
| h | 1.77 ± 0.27 | – | 1.91 ± 0.26 | 1.23 ± 0.22 | |
| K m | 59.6 ± 6.1 | 87.8 ± 27.1 | 58.7 ± 6.7 | 108 ± 45 | |
| Clint | 0.01 | 0.10 | 13.9 | 10.4 | |
| R 2 | 0.95 | 0.93 | 0.97 | 0.96 |
Data are the mean ± SEE. V max (in picomoles per minute per milligram of protein), K m (in micromolars), and h were determined as per simple Michaelis–Menten or sigmoidal model (Eq. 1). Clint (in microliters per minute per milligram of protein) was calculated as V max/K m
In the heart, P450-mediated metabolism of AA was mainly through epoxygenation (Fig. 2), whereas, 20-HETE was formed at a significantly lower rate (Fig. 3). Heart microsomal fraction formed minor amounts of 19-HETE which was below the limit of quantification. Clint values were 0.18, 0.12, 0.28, and 0.26 μL min−1 mg−1 protein for 5,6-, 8,9-, 11,12-, and 14,15-EET, respectively, and 0.01 μL min−1 mg−1 protein for 20-HETE (Table I). The Km values were 268, 310, 271, and 570 μM for 5,6-, 8,9-, 11,12-, and 14,15-EET, respectively (Table I). While the Vmax values were 49.3, 38.2, 76.6, and 146 pmol min−1 mg−1 protein by the same order (Table I). The Vmax and Km values for the 20-HETE formation were 0.73 pmol min−1 mg−1 protein and 59.6 μM, respectively (Table I).
Similar to the heart, the epoxygenase activity in the lung was higher than that of the hydroxylase (Figs. 2 and 3). Clint values were 2.74, 0.39, 0.79, and 0.51 μL min−1 mg−1 protein for 5,6-, 8,9-, 11,12-, and 14,15-EET and 0.03 and 0.1 μL min−1 mg−1 protein for 19- and 20-HETE, respectively (Table I). The Km values were 89.2, 141, 172, and 401 μM, and the Vmax values were 244, 55.1, 136, and 204 pmol min−1 mg−1 protein for 5,6-, 8,9-, 11,12-, and 14,15-EET, respectively (Table I). The Vmax and Km values for the 19-HETE formation were 1.18 pmol min−1 mg−1 protein and 33.8 μM and for 20-HETE were 9.19 pmol min−1 mg−1 protein and 87.8 μM, respectively (Table I).
AA hydroxylation in the kidney and liver was substantially higher than those of the heart or the lung (Fig. 3). For the kidney, 5,6-EET formation was below the limit of quantification in the kidney but not in the other three organs. Clint values were 7.15, 10.9, and 2.59 μL min−1 mg−1 protein for 8,9-, 11,12-, and 14,15-EET and 0.6 and 13.9 μL min−1 mg−1 protein for 19- and 20-HETE, respectively (Table I). The Km values were 28.3, 71.3, and 109 μM, and the Vmax values were 202, 775, and 283 pmol min−1 mg−1 protein for 8,9-, 11,12-, and 14,15-EET, respectively (Table I). The Vmax and Km values for the 19-HETE formation were 100 pmol min−1 mg−1 protein and 166 μM and for 20-HETE were 812 pmol min−1 mg−1 protein and 58.7 μM, respectively (Table I).
For the liver, Clint values were 4.83, 22, 42.4, and 10.7 μL min−1 mg−1 protein for 5,6-, 8,9-, 11,12-, and 14,15-EET and 1.87 and 10.4 μL min−1 mg−1 protein for 19- and 20-HETE, respectively (Table I). The Km values were 124, 26.9, 24.2, and 68.4 μM and the Vmax values were 596, 592, 1,024, and 729 pmol min−1 mg−1 protein for 5,6-, 8,9-, 11,12-, and 14,15-EET, respectively (Table I). The Vmax and Km values for the 19-HETE formation were 177 pmol min−1 mg−1 protein and 94.6 μM and for 20-HETE were 1,118 pmol min−1 mg−1 protein and 108 μM, respectively (Table I).
Interestingly, the Km values for all metabolites were significantly different between the organs except in three cases (Table I), viz. the Km values of 8,9-EET formation between liver and kidney and the Km values of 20-HETE formation between the heart and kidney and those of the lung and liver.
Determination of the Regioselectivity of AA Hydroxylation and Epoxygenation in the Heart, Lung, Kidney and Liver
The regioselectivity for each organ was determined for different AA concentrations. The concentrations used were ranged between 16 and 81 μM to reflect the endogenous AA level. Figure 4 depicts the relative formation of four EET regioisomers, as well as the 19- and 20-HETE formation rate, represented as a percentage of the total measured AA metabolites formation. In the heart, 20-HETE formation represented 0.7% of total metabolites. There were two main EET regioisomers, 14,15- and 11,12-EETs. Their formation was 34.2% and 37.1% of the total EETs. Then the 8,9- and 5,6-EET which contributed to 17.1% and 11.6%, respectively. Only minute amount of 19-HETE was observed but could not be accurately quantified, while the lung formed 19- and 20-HETE at a rate of 0.66% and 2.72% of the total measured AA metabolites, respectively. Interestingly, the main EET was 5,6-EET which represented 45.4% of the total EETs in the lung. 11,12- and 14,15-EETs represented 24% and 18.7% of the total EETs, respectively. The least EET to be formed was 8,9-EET which represented 11.9% of total EETs in the lung.
Fig. 4.
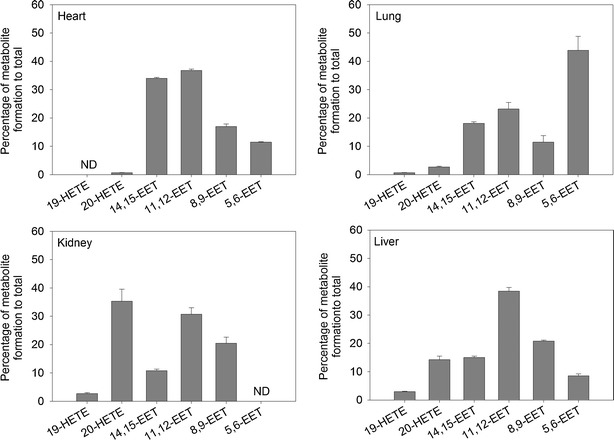
Regioselectivity of arachidonic acid epoxygenation and hydroxylation by the heart, lung, kidney, and liver microsomal fraction. The regioselectivity was determined using different AA concentrations ranging between 16 and 81 μM. Data were expressed as mean ± SEM. The y-axis indicates the percentage of metabolite formation to total investigated P450-derived arachidonic acid metabolites formation
Regarding the kidneys, quantification of 5,6-EET was problematic due to its low rate of formation. The main EET formed in the kidney was 11,12-EET followed by 8,9-EET and lastly 14,15-EET, and they represented 49.6%, 33%, and 17.4% of the total EETs formation, respectively. 20-HETE formation represented 35.3% compared with total metabolites formed, while 19-HETE represented 2.73%. Similar pattern was observed for the liver; the EET regioisomers formation was: 10.3%, 25.2%, 46.5%, and 18.1% for 5,6-, 8,9-, 11,12-, and 14,15-EET, respectively. 19- and 20-HETE represented 2.98% and 14.3% of the total measured AA metabolites formation.
Immunoinhibition of P450-Mediated AA Epoxygenation and Hydroxylation Activity
Anti-CYP2C11 antibody efficiently inhibited AA epoxygenation in the heart and liver by 89% and 88.5%, respectively (Table II). Anti-CYP2B1/2 antibody caused predominant inhibition of AA epoxygenation in the lung by 46% (Table II). For anti-CYP2C23 antibody, it inhibited AA epoxygenation in the kidney by 72.5% and the liver by 62.6% (Table II). Anti-CYP4A1/2/3 inhibited AA hydroxylation in the four organs. The percentage of inhibition was 76%, 23.9%, 71.5%, and 35.1% for the heart, lung, kidney, and liver, respectively (Table II).
Table II.
Immunoinhibition of P450-Mediated AA Epoxygenation and Hydroxylation in Rat Heart, Lung, Kidney, and Liver Microsomes
| Antibody | % inhibition | |||||||
|---|---|---|---|---|---|---|---|---|
| Heart | Lung | Kidney | Liver | |||||
| Epoxygenation | Hydroxylation | Epoxygenation | Hydroxylation | Epoxygenation | Hydroxylation | Epoxygenation | Hydroxylation | |
| Anti-CYP2B1/2 | 4.48 ± 3.63 | 8.20 ± 4.17 | 46 ± 14 | 20.5 ± 3.9 | 11.2 ± 5.5 | 21.5 ± 11.6 | 14.5 ± 9.8 | 8.00 ± 9.43 |
| Anti-CYP2C11 | 89 ± 25 | 11 ± 5 | 17.5 ± 6.6 | 15.2 ± 6.8 | 58.1 ± 22.8 | 25.4 ± 21.9 | 88.5 ± 26.5 | 30.2 ± 7.1 |
| Anti-CYP2C23 | 5.02 ± 3.66 | 8.87 ± 4.18 | 9.51 ± 6.02 | 3.02 ± 2.38 | 72.5 ± 16.9 | 4.73 ± 6.93 | 62.6 ± 28 | 27.3 ± 8.4 |
| Anti-CYP4A1/2/3 | 17.5 ± 4.2 | 76 ± 28 | 23 ± 10 | 23.9 ± 15.8 | 27.7 ± 23.7 | 71.5 ± 27.3 | 21.4 ± 6.1 | 35.1 ± 10.1 |
Data are the mean of at least three determinations and SEM
DISCUSSION
P450-derived AA metabolites, most notably EETs and 20-HETE, play pivotal physiological roles in various organs (3,17). AA is metabolized to 5,6-, 8,9-, 11,12-, and 14,15-EET by P450 epoxygenases (31). In several tissues of rats, CYP2B, CYP2C, and CYP2J3 isoforms each serve as major P450 epoxygenases (32). On the other hand, P450 hydroxylation has been generally attributed to CYP4A and CYP 4F isoforms (29,32). Other P450s, such as CYP1As, CYP1B1, CYP2As, and CYP2E1, also contribute to AA monoxygenation. CYP1A1 as well as CYP1B1 were reported to possess inconsequential activity towards AA metabolism. They catalyze the formation of HETEs other than the 20-HETE, especially mid-chain HETEs for CYP1B1 (32,33). CYP2A isoforms were also observed to have a very low epoxygenase activity (19). CYP2E1 has a substantial contribution to ω-1-hydroxylation activity in the liver but not in the kidney of rats and rabbits; however, this was reported only after its induction and not in the constitutive state (32,34,35). Despite the documented physiological and pathophysiological role of P450-derived AA metabolites, the kinetic properties of the P450 epoxygenases and hydroxylases activities have not been investigated at the organ level.
Our results have demonstrated that AA is metabolized to EETs and HETEs in all organs tested, albeit with different levels of metabolic activity. In the heart, the P450 epoxygenase activity was 84 times the P450 ω-hydroxylase activity based on Clint. This is consistent with the relatively low cardiac expressions of CYP4A1/2/3 and CYP4Fs compared with those of CYP2C11 and CYP2J3. In agreement with our results, it has been previously reported that P450 epoxygenase activity was greater than P450 ω-hydroxylase activity in the heart microsomal fraction of mice (36).
Similar to our observations, it has been previously reported that P450 epoxygenase activity was higher than P450 hydroxylase activity in the lung of mice (36). CYP4A1/2/3 and CYP4Fs exhibited moderate expressions in the lung, representing ∼25% of their relative expression in liver. Consequently, the pulmonary 20-HETE formation rate was expected to be about 25% of that in the liver at saturating concentration of AA, given the linearity between metabolite formation rate and enzyme concentration. However, the observed hydroxylase activity was lower than expected based on protein expression. Not all CYP4A and CYP4F isoforms exhibit activity towards AA metabolism; CYP4A3, CYP4F5, and CYP4F6 have no or very low activity (29,32,37). The unexpected results could be explained by lung microsomes possessing relatively greater concentrations of these inactive isoforms compared with the liver.
In the kidney, as reported elsewhere, HETEs and EETs are formed in considerable amounts (38). Our results have demonstrated that the kidney has P450 hydroxylase activity comparable to that of the epoxygenase activity, based on Clint. This is attributable to the high expressions of CYP2C23, CYP2J3, and CYP4A1/2/3 in the kidney. Similar to what was seen in the heart and kidney, enzyme activities in the liver correlated well with protein expression. Based on Clint, P450 epoxygensae activity was 6.5 times the hydroxylase activity. In agreement with previously published results (8), P450 expoxygenases (CYP2B1/2, CYP2C11, CYP2C23, and CYP2J3) and P450 hydroxylases (CYP4A1/2/3 and CYP4Fs) were both highly expressed in the liver.
Another objective of the current study was to characterize specific P450 epoxygenases and hydroxylases that contribute primarily to the formation of EETs and HETEs by heart, lung, kidney, and liver microsomes. Our strategy was to exclude P450 enzymes that showed relatively low expression in each organ. Then, the P450 that plays the predominant role was identified, based on the published AA metabolism regioselectivity for different P450 enzymes. The identity of these P450 enzymes was further confirmed by immunoinhibition studies and kinetic profile of AA metabolism.
In the heart, almost no epoxygenases other than CYP2C11 and CYP2J3 were expressed. It has been previously demonstrated that recombinant CYP2J3 produces EETs as a mixture of regioisomers with relative ratios of 1.5:1:1 of 14,15-, 11,12-, and 8,9-EET, respectively (39); while for CYP2C11, the corresponding ratios were 1:1:0.5, respectively (40). Compared with the regioselectivity of the microsomal incubates, we can conclude that CYP2C11 dominates the EETs formation in the heart. In agreement with our conclusion, it has been previously reported that CYP2C11 and especially CYP2J3 are highly expressed in the heart; however, CYP2C11 activity is many fold higher than CYP2J3 (41). On the other hand, 20-HETE formation could be attributed to the constitutive expression of CYP4Fs and/or CYP4A1/2.
With respect to the lungs, CYP2B1/2, CYP2C11, and CYP2J3 proteins were each found to be significantly expressed. Lung produces a mixture of 14,15-, 11,12-, and 8,9-EET in a ratio of 0.8:1:0.5. Therefore, CYP2J3 and CYP2B2 could be excluded due to their different pattern of regioselectivity (39,42). As aforementioned, CYP2C11 has a characteristic feature of producing equal amounts of 14,15- and 11,12-EET. For CYP2B1, it produces a mixture of 14,15-, 11,12-, and 8,9-EET in a ratio of 0.8:1:0.7, as previously reported (42). Regioselectivity suggests that CYP2B1 dominates EETs formation in the lung, although CYP2C11 may also play a role. This is consistent with earlier study showed high CYP2B1 expression in the lung (19). Concerning HETEs formation, it can be catalyzed by CYP4A1/2 and/or CYP4F isoforms.
Kidney expresses several epoxygenases, CYP2C11, CYP2C23, and CYP2J3, in addition to CYP4A1/2/3 and CYP4Fs as hydroxylases. Kidney produced EETs mixture of 0.3:1:0.6 corresponding to 14,15-, 11,12-, and 8,9-EET, respectively. There is a close similarity between the regioselectivity of kidney microsomal fraction and CYP2C23, which produces 14,15-, 11,12-, and 8,9-EET in a ratio of 0.2:1:0.4 (40). CYP2C24 which is believed to be an important epoxygenase in the kidney has a substantially different ratio of 1:1:0.4 (40). This suggests that CYP2C23 is the most dominant epoxygenase in the kidney, which is consistent with previously published studies (41,43). Although it was suggested that CYP4A2 is the major ω-hydroxylases in the kidney (44), it was reported elsewhere that CYP4A2 catalyzes the formation of 19- and 20-HETE in a ratio of 1:3.6 (37,42,45). This ratio does not conform with our results. The kidney microsomal fraction produced 19- and 20-HETE in a ratio of 1:12.9 which is close to the reported ratio of CYP4A1 which is 1:12.6 (37). Hence, CYP4A1 but not CYP4A2 seems to be the major enzyme implicated in renal 19/20-HETE formation. This conclusion is consistent with earlier study demonstrating no inhibitory effect of specific CYP4A2 antisense oligonucleotides on renal 20-HETE production, whereas, specific CYP4A1 antisense oligonucleotides were able to inhibit 20-HETE production in vivo (46).
In the liver, all epoxygenases, CYP2B1/2, CYP2C11, CYP2C23, and CYP2J3, and hydroxylases, CYP4A1/2/3 and CYP4Fs were highly expressed. Therefore, deriving a conclusion regarding the dominant CYP epoxygenases and CYP hydroxylases is difficult. However, regioselectivity of liver microsomal incubate was found to be 0.4:1:0.5 for 14,15-, 11,12-, and 8,9-EET, respectively. From the aforementioned regioselectivity of CYP2B1, CYP2C11, and CYP2C23, probably CYP2C23 together with CYP2C11 are the main epoxygenases in the liver. In this regard, it was previously reported that CYP2C11 is a major epoxygenase in the male rat liver (47).
The statistical analysis that was performed on Km values was in agreement with the activity and protein expression of P450. Our results suggested that the dominant epoxygenases were CYP2C11, CYP2B1, CYP2C23, and CYP2C23/CYP2C11 for the heart, lung, kidney, and liver, respectively. This conclusion was substantiated by the fact that there are statistical differences in epoxygenation kinetics between the four organs. Moreover, these differences were less apparent between kidney and liver, especially for 8,9-EET which did not significantly differ. This could be attributed to CYP2C23 which is shared between the kidney and the liver. On the other hand, 19-HETE formation kinetics significantly differed between lung, kidney, and liver, suggesting the involvement of different P450s. 20-HETE formation kinetics did not differ between heart and kidney or between lung and liver. These results suggest that CYP4A1 is the main ω-hydroxylase in the heart and kidney, whereas CYP4A2 and/or CYP4F isoforms are the main ω-hydroxylase enzymes in the lung and liver.
These results were also supported by immunoinhibition studies that CYP2C11 is the predominant epoxygenase in the heart; whereas CYP2B1/2 is the predominant in lung. In addition, CYP2C23 and CYP2C23/CYP2C11 are the dominant epoxygenases in the kidney and liver, respectively. On the other hand, CYP4A1/2/3 plays a significant role in AA hydroxylation in the kidney and heart.
In this study, the formation of each EET was determined as EET plus the corresponding DHET, because the degradation of EETs through hydrolysis by sEH to DHET is the major metabolic pathway (41). Pooled microsomal fractions for each organ were used because investigating inter-animal variability was not the scope of the current study. Nevertheless, it is recognized that individual animals may differ in their expression levels and/or enzymatic activity of P450 enzymes. The study of 5,6-EET is associated with an additional problem of instability in physiological buffer and consequent spontaneous degradation to lactone, which may be associated with an underestimation of enzyme activity. Therefore, 5,6-EET data shown should be interpreted with some degree of caution.
CONCLUSIONS
In conclusion, the current study provides a unique explicit comparison of the total P450 epoxygenase and P450 hydroxylase activity in the heart, lung, kidney, and liver. We concluded that AA is metabolized to EETs and HETEs in all organs tested but with varying metabolic activities. P450 epoxygenase activity in the heart and lung is higher than the P450 hydroxylase activity of the same organ. P450 hydroxylase activity in the liver is significantly higher than its P450 epoxygenase activity, while similar activities were found in the kidney. In light of the data presented, CYP2C11 is the predominant epoxygenase in the heart, whereas CYP2B1 is the predominant in lung. In addition, CYP2C23 and CYP2C23/CYP2C11 are the dominant epoxygenases in the kidney and liver, respectively. CYP4A1 is the major hydroxylase in the kidney and heart. On the other hand, CYP4A2 and/or CYP4F1/4 are probably the dominant ω-hydroxylases in the liver and lung. The data derived from this work may assist in the identification of targets for the treatment, or prevention, of diseases that are associated with disturbances in AA metabolism.
ACKNOWLEDGMENTS
The authors wish to thank Dr. Dion Brocks for his valuable comments on this manuscript. This work was supported by a grant from the Canadian Institutes of Health Research (CIHR) MOP 106665 to AOSE. AAE is the recipients of Egyptian Government Scholarship. BNMZ is the recipient of Alberta Innovates-Health Solutions Studentship. AA-M is the recipient of Alberta Ingenuity Graduate Scholarship and Izaak Walton Killam Memorial Graduate Scholarship.
Conflict of Interest
The authors declare no conflict of interest.
REFERENCES
- 1.Panigrahy D, Kaipainen A, Greene ER, Huang S. Cytochrome P450-derived eicosanoids: the neglected pathway in cancer. Cancer Metastasis Rev. 2010;29(4):723–735. doi: 10.1007/s10555-010-9264-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roman RJ. P-450 metabolites of arachidonic acid in the control of cardiovascular function. Physiol Rev. 2002;82(1):131–185. doi: 10.1152/physrev.00021.2001. [DOI] [PubMed] [Google Scholar]
- 3.Sudhahar V, Shaw S, Imig JD. Epoxyeicosatrienoic acid analogs and vascular function. Curr Med Chem. 2010;17(12):1181–1190. doi: 10.2174/092986710790827843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keseru B, Barbosa-Sicard E, Popp R, Fisslthaler B, Dietrich A, Gudermann T, et al. Epoxyeicosatrienoic acids and the soluble epoxide hydrolase are determinants of pulmonary artery pressure and the acute hypoxic pulmonary vasoconstrictor response. FASEB J. 2008;22(12):4306–4315. doi: 10.1096/fj.08-112821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pokreisz P, Fleming I, Kiss L, Barbosa-Sicard E, Fisslthaler B, Falck JR, et al. Cytochrome P450 epoxygenase gene function in hypoxic pulmonary vasoconstriction and pulmonary vascular remodeling. Hypertension. 2006;47(4):762–770. doi: 10.1161/01.HYP.0000208299.62535.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salvail D, Cloutier M, Rousseau E. Functional reconstitution of an eicosanoid-modulated Cl− channel from bovine tracheal smooth muscle. Am J Physiol Cell Physiol. 2002;282(3):C567–C577. doi: 10.1152/ajpcell.00029.2001. [DOI] [PubMed] [Google Scholar]
- 7.Roman RJ, Maier KG, Sun CW, Harder DR, Alonso-Galicia M. Renal and cardiovascular actions of 20-hydroxyeicosatetraenoic acid and epoxyeicosatrienoic acids. Clin Exp Pharmacol Physiol. 2000;27(11):855–865. doi: 10.1046/j.1440-1681.2000.03349.x. [DOI] [PubMed] [Google Scholar]
- 8.Sacerdoti D, Gatta A, McGiff JC. Role of cytochrome P450-dependent arachidonic acid metabolites in liver physiology and pathophysiology. Prostaglandins Other Lipid Mediat. 2003;72(1–2):51–71. doi: 10.1016/S1098-8823(03)00077-7. [DOI] [PubMed] [Google Scholar]
- 9.Yoshida S, Hirai A, Tamura Y. Possible involvement of arachidonic acid metabolites of cytochrome P450 monooxygenase pathway in vasopressin-stimulated glycogenolysis in isolated rat hepatocytes. Arch Biochem Biophys. 1990;280(2):346–351. doi: 10.1016/0003-9861(90)90340-5. [DOI] [PubMed] [Google Scholar]
- 10.Zou AP, Fleming JT, Falck JR, Jacobs ER, Gebremedhin D, Harder DR, et al. 20-HETE is an endogenous inhibitor of the large-conductance Ca(2+)-activated K+ channel in renal arterioles. Am J Physiol. 1996;270(1 Pt 2):R228–R237. doi: 10.1152/ajpregu.1996.270.1.R228. [DOI] [PubMed] [Google Scholar]
- 11.Aboutabl ME, Zordoky BN, El-Kadi AO. 3-methylcholanthrene and benzo(a)pyrene modulate cardiac cytochrome P450 gene expression and arachidonic acid metabolism in male Sprague Dawley rats. Br J Pharmacol. 2009;158(7):1808–1819. doi: 10.1111/j.1476-5381.2009.00461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yousif MH, Benter IF, Roman RJ. Cytochrome P450 metabolites of arachidonic acid play a role in the enhanced cardiac dysfunction in diabetic rats following ischaemic reperfusion injury. Auton Autacoid Pharmacol. 2009;29(1–2):33–41. doi: 10.1111/j.1474-8673.2009.00429.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lv X, Wan J, Yang J, Cheng H, Li Y, Ao Y, et al. Cytochrome P450 omega-hydroxylase inhibition reduces cardiomyocyte apoptosis via activation of ERK1/2 signaling in rat myocardial ischemia-reperfusion. Eur J Pharmacol. 2008;596(1–3):118–126. doi: 10.1016/j.ejphar.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 14.Kroetz DL, Huse LM, Thuresson A, Grillo MP. Developmentally regulated expression of the CYP4A genes in the spontaneously hypertensive rat kidney. Mol Pharmacol. 1997;52(3):362–372. doi: 10.1124/mol.52.3.362. [DOI] [PubMed] [Google Scholar]
- 15.Certikova Chabova V, Kramer HJ, Vaneckova I, Thumova M, Skaroupkova P, Tesar V, et al. The roles of intrarenal 20-hydroxyeicosatetraenoic and epoxyeicosatrienoic acids in the regulation of renal function in hypertensive Ren-2 transgenic rats. Kidney Blood Press Res. 2007;30(5):335–346. doi: 10.1159/000107710. [DOI] [PubMed] [Google Scholar]
- 16.Jacobs ER, Effros RM, Falck JR, Reddy KM, Campbell WB, Zhu D. Airway synthesis of 20-hydroxyeicosatetraenoic acid: metabolism by cyclooxygenase to a bronchodilator. Am J Physiol. 1999;276(2 Pt 1):L280–L288. doi: 10.1152/ajplung.1999.276.2.L280. [DOI] [PubMed] [Google Scholar]
- 17.Quigley R, Baum M, Reddy KM, Griener JC, Falck JR. Effects of 20-HETE and 19(S)-HETE on rabbit proximal straight tubule volume transport. Am J Physiol Ren Physiol. 2000;278(6):F949–F953. doi: 10.1152/ajprenal.2000.278.6.F949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zordoky BN, Aboutabl ME, El-Kadi AO. Modulation of cytochrome P450 gene expression and arachidonic acid metabolism during isoproterenol-induced cardiac hypertrophy in rats. Drug Metab Dispos. 2008;36(11):2277–2286. doi: 10.1124/dmd.108.023077. [DOI] [PubMed] [Google Scholar]
- 19.Imaoka S, Hashizume T, Funae Y. Localization of rat cytochrome P450 in various tissues and comparison of arachidonic acid metabolism by rat P450 with that by human P450 orthologs. Drug Metab Pharmacokinet. 2005;20(6):478–484. doi: 10.2133/dmpk.20.478. [DOI] [PubMed] [Google Scholar]
- 20.Thum T, Borlak J. Gene expression in distinct regions of the heart. Lancet. 2000;355(9208):979–983. doi: 10.1016/S0140-6736(00)99016-0. [DOI] [PubMed] [Google Scholar]
- 21.Minamiyama Y, Takemura S, Akiyama T, Imaoka S, Inoue M, Funae Y, et al. Isoforms of cytochrome P450 on organic nitrate-derived nitric oxide release in human heart vessels. FEBS Lett. 1999;452(3):165–169. doi: 10.1016/S0014-5793(99)00612-2. [DOI] [PubMed] [Google Scholar]
- 22.Delozier TC, Kissling GE, Coulter SJ, Dai D, Foley JF, Bradbury JA, et al. Detection of human CYP2C8, CYP2C9, and CYP2J2 in cardiovascular tissues. Drug Metab Dispos. 2007;35(4):682–688. doi: 10.1124/dmd.106.012823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marji JS, Wang MH, Laniado-Schwartzman M. Cytochrome P-450 4A isoform expression and 20-HETE synthesis in renal preglomerular arteries. Am J Physiol Ren Physiol. 2002;283(1):F60–F67. doi: 10.1152/ajprenal.00265.2001. [DOI] [PubMed] [Google Scholar]
- 24.Barakat MM, El-Kadi AO, du Souich P. L-NAME prevents in vivo the inactivation but not the down-regulation of hepatic cytochrome P450 caused by an acute inflammatory reaction. Life Sci. 2001;69(13):1559–1571. doi: 10.1016/S0024-3205(01)01241-3. [DOI] [PubMed] [Google Scholar]
- 25.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193(1):265–275. [PubMed] [Google Scholar]
- 26.Gharavi N, El-Kadi AO. tert-Butylhydroquinone is a novel aryl hydrocarbon receptor ligand. Drug Metab Dispos. 2005;33(3):365–372. doi: 10.1124/dmd.104.002253. [DOI] [PubMed] [Google Scholar]
- 27.Nithipatikom K, Grall AJ, Holmes BB, Harder DR, Falck JR, Campbell WB. Liquid chromatographic-electrospray ionization-mass spectrometric analysis of cytochrome P450 metabolites of arachidonic acid. Anal Biochem. 2001;298(2):327–336. doi: 10.1006/abio.2001.5395. [DOI] [PubMed] [Google Scholar]
- 28.Powell PK, Wolf I, Jin R, Lasker JM. Metabolism of arachidonic acid to 20-hydroxy-5,8,11,14-eicosatetraenoic acid by P450 enzymes in human liver: involvement of CYP4F2 and CYP4A11. J Pharmacol Exp Ther. 1998;285(3):1327–1336. [PubMed] [Google Scholar]
- 29.Xu F, Falck JR, Ortiz de Montellano PR, Kroetz DL. Catalytic activity and isoform-specific inhibition of rat cytochrome p450 4F enzymes. J Pharmacol Exp Ther. 2004;308(3):887–895. doi: 10.1124/jpet.103.059626. [DOI] [PubMed] [Google Scholar]
- 30.Brash AR. Arachidonic acid as a bioactive molecule. J Clin Invest. 2001;107(11):1339–1345. doi: 10.1172/JCI13210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu S, Moomaw CR, Tomer KB, Falck JR, Zeldin DC. Molecular cloning and expression of CYP2J2, a human cytochrome P450 arachidonic acid epoxygenase highly expressed in heart. J Biol Chem. 1996;271(7):3460–3468. doi: 10.1074/jbc.271.7.3460. [DOI] [PubMed] [Google Scholar]
- 32.Capdevila JH, Falck JR, Harris RC. Cytochrome P450 and arachidonic acid bioactivation. Molecular and functional properties of the arachidonate monooxygenase. J Lipid Res. 2000;41(2):163–181. [PubMed] [Google Scholar]
- 33.Choudhary D, Jansson I, Stoilov I, Sarfarazi M, Schenkman JB. Metabolism of retinoids and arachidonic acid by human and mouse cytochrome P450 1b1. Drug Metab Dispos. 2004;32(8):840–847. doi: 10.1124/dmd.32.8.840. [DOI] [PubMed] [Google Scholar]
- 34.Poloyac SM, Tortorici MA, Przychodzin DI, Reynolds RB, Xie W, Frye RF, et al. The effect of isoniazid on CYP2E1- and CYP4A-mediated hydroxylation of arachidonic acid in the rat liver and kidney. Drug Metab Dispos. 2004;32(7):727–733. doi: 10.1124/dmd.32.7.727. [DOI] [PubMed] [Google Scholar]
- 35.Laethem RM, Balazy M, Falck JR, Laethem CL, Koop DR. Formation of 19(S)-, 19(R)-, and 18(R)-hydroxyeicosatetraenoic acids by alcohol-inducible cytochrome P450 2E1. J Biol Chem. 1993;268(17):12912–12918. [PubMed] [Google Scholar]
- 36.Theken KN, Deng Y, Kannon MA, Miller TM, Poloyac SM, Lee CR. Activation of the acute inflammatory response alters cytochrome P450 expression and eicosanoid metabolism. Drug Metab Dispos. 2011;39(1):22–29. doi: 10.1124/dmd.110.035287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguyen X, Wang MH, Reddy KM, Falck JR, Schwartzman ML. Kinetic profile of the rat CYP4A isoforms: arachidonic acid metabolism and isoform-specific inhibitors. Am J Physiol. 1999;276(6 Pt 2):R1691–R1700. doi: 10.1152/ajpregu.1999.276.6.R1691. [DOI] [PubMed] [Google Scholar]
- 38.Carroll MA, Balazy M, Huang DD, Rybalova S, Falck JR, McGiff JC. Cytochrome P450-derived renal HETEs: storage and release. Kidney Int. 1997;51(6):1696–1702. doi: 10.1038/ki.1997.234. [DOI] [PubMed] [Google Scholar]
- 39.Wu S, Chen W, Murphy E, Gabel S, Tomer KB, Foley J, et al. Molecular cloning, expression, and functional significance of a cytochrome P450 highly expressed in rat heart myocytes. J Biol Chem. 1997;272(19):12551–12559. doi: 10.1074/jbc.272.19.12551. [DOI] [PubMed] [Google Scholar]
- 40.Holla VR, Makita K, Zaphiropoulos PG, Capdevila JH. The kidney cytochrome P-450 2C23 arachidonic acid epoxygenase is upregulated during dietary salt loading. J Clin Invest. 1999;104(6):751–760. doi: 10.1172/JCI7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Imig JD. Epoxides and soluble epoxide hydrolase in cardiovascular physiology. Physiol Rev. 2012;92(1):101–130. doi: 10.1152/physrev.00021.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Capdevila JH, Karara A, Waxman DJ, Martin MV, Falck JR, Guenguerich FP. Cytochrome P-450 enzyme-specific control of the regio- and enantiofacial selectivity of the microsomal arachidonic acid epoxygenase. J Biol Chem. 1990;265(19):10865–10871. [PubMed] [Google Scholar]
- 43.Imig JD, Navar LG, Roman RJ, Reddy KK, Falck JR. Actions of epoxygenase metabolites on the preglomerular vasculature. J Am Soc Nephrol. 1996;7(11):2364–2370. doi: 10.1681/ASN.V7112364. [DOI] [PubMed] [Google Scholar]
- 44.Ito O, Nakamura Y, Tan L, Ishizuka T, Sasaki Y, Minami N, et al. Expression of cytochrome P-450 4 enzymes in the kidney and liver: regulation by PPAR and species-difference between rat and human. Mol Cell Biochem. 2006;284(1–2):141–148. doi: 10.1007/s11010-005-9038-x. [DOI] [PubMed] [Google Scholar]
- 45.Helvig C, Dishman E, Capdevila JH. Molecular, enzymatic, and regulatory characterization of rat kidney cytochromes P450 4A2 and 4A3. Biochemistry. 1998;37(36):12546–12558. doi: 10.1021/bi981048g. [DOI] [PubMed] [Google Scholar]
- 46.Wang MH, Guan H, Nguyen X, Zand BA, Nasjletti A, Laniado-Schwartzman M. Contribution of cytochrome P-450 4A1 and 4A2 to vascular 20-hydroxyeicosatetraenoic acid synthesis in rat kidneys. Am J Physiol. 1999;276(2 Pt 2):F246–F253. doi: 10.1152/ajprenal.1999.276.2.F246. [DOI] [PubMed] [Google Scholar]
- 47.Imig JD. Epoxygenase metabolites. Epithelial and vascular actions. Mol Biotechnol. 2000;16(3):233–251. doi: 10.1385/MB:16:3:233. [DOI] [PubMed] [Google Scholar]


