Abstract
Transcellular calcium transport is an essential activity in mineralized tissue formation, including dental hard tissues. In many organ systems, this activity is regulated by membrane-bound sodium/calcium (Na+/Ca2+) exchangers, which include the NCX and NCKX [sodium/calcium-potassium (Na+/Ca2+-K+ ) exchanger] proteins. During enamel maturation, when crystals expand in thickness, Ca2+ requirements vastly increase but exactly how Ca2+ traffics through ameloblasts remains uncertain. Previous studies have shown that several NCX proteins are expressed in ameloblasts, although no significant shifts in expression were observed during maturation which pointed to the possible identification of other Ca2+ membrane transporters. NCKX proteins are encoded by members of the solute carrier gene family, Slc24a, which include 6 different proteins (NCKX1–6). NCKX are bidirectional electrogenic transporters regulating Ca2+ transport in and out of cells dependent on the transmembrane ion gradient. In this study we show that all NCKX mRNAs are expressed in dental tissues. Real-time PCR indicates that of all the members of the NCKX group, NCKX4 is the most highly expressed gene transcript during the late stages of amelogenesis. In situ hybridization and immunolocalization analyses clearly establish that in the enamel organ, NCKX4 is expressed primarily by ameloblasts during the maturation stage. Further, during the mid-late maturation stages of amelogenesis, the expression of NCKX4 in ameloblasts is most prominent at the apical poles and at the lateral membranes proximal to the apical ends. These data suggest that NCKX4 might be an important regulator of Ca2+ transport during amelogenesis.
Keywords: Amelogenesis, Enamel, Solute carrier gene, Sodium/calcium exchanger, Sodium/calcium/potassium exchanger, Mouse
Introduction
Enamel and dentin formation both begin with an extracellular proteinaceous matrix that directs the subsequent events of hydroxyapatite (Hap) formation and biomineralization. Calcium (Ca2+) and phosphate (PO3−4) are core molecular building blocks of Hap, yet their transport from the circulation to the developing dentin and enamel matrix is poorly understood. The importance of this ionic movement is, nevertheless, well appreciated [Smith, 1998; Hubbard, 2000]. During dentin formation, Ca2+ is transported from the pulpal vascular network through the odontoblast layer, by what is believed to be a transcellular pathway, to be incorporated in the mineral phase at the interface between the nonmineralized pre-dentin and the mineralized dentin [Linde, 1995; Tsumura et al., 2010]. Mature enamel is composed of densely packed Hap crystallites arranged into enamel prisms [Paine et al., 2001]. Despite the high mineral content of enamel, we are only starting to understand which ion transporters, including Ca2+, HCO3− and Cl− transporters, are critical to amelogenesis and thus allow for ion transport between the papillary layer and the forming enamel crystals [Hubbard, 1995, 1996, 2000; Franklin et al., 2001; Turnbull et al., 2004; Josephsen et al., 2010; Lacruz et al., 2010b; Lacruz et al., 2010c; Okumura et al., 2010; Lacruz et al., 2012b].
One of the mechanisms used by cells for Ca2+ import and export are the sodium/calcium (Na+/Ca2+) exchangers, where the main function is to control Ca2+ flux across the plasma membranes and/or intracellular compartments [Quednau et al., 2004; Schnetkamp, 2004]. Members of the Slc8a and Slc24a gene families encode for Na+/Ca2+ exchangers. NCX proteins (NCX1, NCX2 and NCX3) are coded by 3 Slc8a genes (Slc8a1, Slc8a2 and Slc8a3, respectively) and are bidirectional transporters capable of exchanging Na+ and Ca2+ in either a Ca2+ efflux or influx depending on the transmembrane ion gradient [Uehara et al., 2004]. There are 6 sodium/calcium-potassium exchangers (NCKX) coded by the Slc24a gene family (NCKX1–6 coded by Slc24a1–6, respectively) [Schnetkamp, 2004; Uehara et al., 2004]. NCKX1 was discovered in the outer segments of retinal rod photoreceptors, which extrude Ca2+ based on the Na+ gradient, and at the same time requires K+ cotransport [Schnetkamp et al., 1989]. The additional 5 NCKX proteins were shown to have a broader tissue distribution. NCKX2 is expressed in cone photoreceptors [Winkfein et al., 2003] and in neurons throughout the brain [Tsoi et al., 1998]. NCKX3 and NCKX4 proteins are abundant in the brain, but have a wide expression distribution [Tsoi et al., 1998; Li et al., 2002]. The expression of NCKX5 varied among different tissues, with a predominant level in the skin and eyes [Lamason et al., 2005]. NCKX6 (also known as NCLX; Na+/Ca2+-Li+ exchanger) displays a broad pattern of tissue expression including the brain, lung and heart, but its biological importance is unclear [Cai and Lytton, 2004]. Like the products of the Slc8a gene family, the products of the Slc24a gene family are also bidirectional electrogenic transporters capable of exchanging Na+ for Ca2+, K+ in either a Ca2+ efflux or influx with a stochiometry of 4Na+:1Ca2+, 1K+ [Li et al., 2002; Schnetkamp, 2004; Uehara et al., 2004].
Based on studies in a variety of tissues, Na+/Ca2+ exchangers demonstrated a tissue-specific expressed pattern. Recently the expression of NCX1 and NCX3 was demonstrated in ameloblasts [Okumura et al., 2010] and odontoblasts [Tsumura et al., 2010]. In this study, we present evidence for mRNA expression for all the 6 members of the Slc24a gene family in the developing mouse molar organ, with the highest expression by Slc24a4/NCKX4. These data complement recently presented DNA array data indicating that in rat incisors NCKX4 is one of the most highly upregulated gene transcripts when maturation-stage enamel organ cells are compared to secretory-stage enamel organ cells [Lacruz et al., 2012a]. We further characterize the sublocalization of NCKX4 mRNA and protein in ameloblasts. Increased expression of NCKX4 mRNA in the late stages of amelogenesis suggests that it may have an important function in regulating Ca2+ transport during enamel maturation.
Materials and Methods
Animals and Tissue Preparation
Swiss Webster mice were treated in accordance with Institutional and Federal guidelines. For PCR and Western blot analysis, the mandibular first molars from mice on postnatal days 3, 6 and 9 were extracted. Using expression profiling of enamelin described elsewhere [Simmer et al., 2009], a specific secretory-stage enamel gene, it is inferred that in our sample, a maturation-stage ameloblast may only be identified on day 9 postnatal whereas ameloblasts at various secretory stages are readily identified on postnatal days 3 and 6. It is estimated that on days 7–8 the mouse first molars are transitioning from a primarily secretory to a maturation function [Simmer et al., 2009]. The mandibles of 7-day-old mice were used for in situ hybridization. Brain and eye tissue were collected from adult mice and used as control tissues for mRNA expression analyses.
RT-PCR and Real-Time PCR
Total RNA was extracted from whole mandibular first molars of day-3, day-6 and day-9 mice using RNeasy Mini Kit (Qiagen, Gaithersburg, Md., USA) as described previously [Lacruz et al., 2010a]. Total RNA was isolated from brain and eye tissue of adult mice to act as control tissues. Complementary DNA (cDNA) was generated from molars and control tissues using RETROscript® Kit (Ambion, Austin, Tex., USA). Gene-specific primers were synthesized for RT-PCR and real-time PCR (table 1). RT-PCR conditions were 95°C denaturation, 55°C annealing and 65°C extension times over 40 cycles and PCR-generated DNA products were resolved by electrophoresis on a 1.5% agarose gel.
Table 1.
PCR primer design
| Gene product (symbol) | Primer sequence | Product size |
|---|---|---|
| RT-PCR | ||
| NCKX1 (Slc24a1) | F: 5′-CACAGGAGAGGCGGTTACTC-3′ R: 5′-CCACTGCCATTTCATTGTTG-3′ |
198 |
| NCKX2 (Slc24a2) | F: 5′-ATCTCCACCAAAGCCCCACAG-3′ R: 5′-CCCAATGACATGCAGAATGATGGC-3′ |
422 |
| NCKX3 (Slc24a3) | F: 5′-CTATGGATCGCAGCCTTCTC-3′ R: 5′-CGGAGTAGATGAGCCCTCTG-3′ |
301 |
| NCKX4 (Slc24a4) | F: 5′-CTGGCTAAGAACTGCACAGATCCTG-3′ R: 5′-CCACAATCGTTCCTACACCGACA-3′ |
307 |
| NCKX5 (Slc24a5) | F: 5′-CTCTGACAACACCAGACTGCAGGA-3′ R: 5′-ACCAGGGAAGACCTAGGCATAGCA-3′ |
300 |
| NCKX6 (Slc24a6) | F: 5′-TTCACTGCACTCTATCTCGGC-3′ R: 5′-ACAGGAAGGAGACCGCCGAT-3′ |
521 |
|
| ||
| Real-time PCR (qPCR) | ||
| NCKX1 | F: 5′-GCCTACGCTCTCTATGTGTTC-3′ R: 5′-ACTGGGCTTGCTGAGGTC-3′ |
123 |
| NCKX2 | F: 5′-CCAACTTTACCGTCGAAGC-3′ R: 5′-AGTGGGTCAAGGGTGTGTATC-3′ |
113 |
| NCKX3 | F: 5′-GTCTTTGTCACGGTCTTTGG-3′ R: 5′-GATGGAGAAACAGAGGAACAC-3′ |
105 |
| NCKX4 | F: 5′-GTCGCTCTCACTGTCCTTG-3′ R: 5′-AAGCACAGGAAGACAGCATAG-3′ |
98 |
| NCKX5 | F: 5′-GCCTTCTTGGGTGTATTTATC-3′ R: 5′-GAGAGTGTTGAAACCATGTTG-3′ |
128 |
| NCKX6 | F: 5′-CTAGACTGCACTGGCTCTTTG-3′ R: 5′-CCGTAATATGTTCACCACCTC-3′ |
92 |
| β-actin | F: 5′-GGGAAATCGTGCGTGACATC-3′ R: 5′-GCGGCAGTGGCCATCTC-3′ |
76 |
|
| ||
| In situ probe synthesis | ||
| NCKX4 | F: 5′-GGTCGACTTGATCAACTCAGCCAATGGTGTTA-3′ R: 5′-GGGATCCTGATGAAGGTGACCATGAAGAA-3′ |
328 |
Primers used the detection of NCKX family members in RT-PCR and real-time PCR. The primers of NCKX1, NCKX2 and NCKX3 used for RT-PCR followed the primer sequence reported before [Pyrski et al., 2007]. Other RT-PCR primer sequences were identified using MacVector software. Real-time primers were identified from the AutoPrimer Bank (http://www.autoprime.de/AutoPrimeWeb). For the generation of the NCKX4 in situ probe, the primers included a SalI and BamHI site at their 5′ end (underlined) to aid in the subsequent cloning into plasmid pSP73.
For real-time PCR, molar cDNA was generated from 1 μg RNA in a 20-μl reaction system using the QuantiTect Rev Transcription Kit (Qiagen Inc., Valencia, Calif., USA). Real-time PCR was performed with a C1000™ Thermal (Bio-Rad Life Sciences, Hercules, Calif., USA). IQ SYBR Green Supermix Kit (Bio-Rad Life Sciences) was used in real-time PCR assays according to the manufacturer’s protocol. Real-time PCR data was normalized to β-actin (Actb) mRNA using methods described in Schmittgen and Livak [2008]. Statistical analysis included multivariate analysis of variance (MANOVA) to assess whether the 6 dependent variables (gene variants) could distinguish the 3 experimental groups (days 3, 6 and 9). In the presence of a significant MANOVA, individual ANOVAs were run using the individual dependent variables. In the presence of a significant ANOVA post hoc Tukey’s tests were performed to determine which group differences were significant.
In situ Hybridization
The mandibles of 7-day-old postnatal mice were fixed with 4% paraformaldehyde, decalcified in 10% EDTA solution for 14 days, embedded in Tissue-Tek® optimal cutting temperature compound, sectioned at 6 μm thickness and mounted. A NCKX4-specific, 328 bp probe suitable for in situ hybridization was prepared from the unique central cytoplasmic loop of the encoded protein [Li et al., 2002] using primers listed in table 1. This amplified region was sub-cloned into vector pSP73 (Promega Corporation, Madison, Wisc., USA). This vector was then digested with SalI and transcribed with SP6 RNA polymerase (Roche, Mannheim, Germany) to prepare the antisense probe;BamHI and T7 polymerase (Roche) were likewise used to get the NCKX4 sense (negative control) probe using digoxigenin-labeled RNA mixture (Roche). In situ hybridizations were performed following standard procedures [Wilkinson, 1992].
Western Blot
Mandibular first molars were isolated on days 3, 6 and 9 and lysed in ice-cold RIPA buffer (Santa Cruz Biotechnology, Inc., Santa Cruz, Calif., USA) and sonicated on ice. The lysate was then centrifuged at 14,000 rpm for 10 min at 4°C, the protein-containing supernatant collected and the protein concentration determined. Total protein obtained from molars (100 μg) were separated on 4–12% SDS-PAGE gels (Invitrogen, Carlsbad, Calif., USA) and then transferred to Immobilon-P membrane (Millipore, Bedford, Mass., USA). The membrane was incubated with polyclonal NCKX4-specific antibody (1: 1,000 dilution; Protein-Tech Group, Inc., catalogue # 18992–1-AP), followed by incubation with HRP-conjugated goat anti-rabbit IgG (Santa Cruz Biotechnology). Proteins were visualized by SuperSignal West Chemiluminescent Substrate kit (Thermo Scientific, Rockford. Ill., USA). The optical densities of bands were determined using ImagineJ Software, and all data was normalized to the signal for β-actin (Sigma, St. Louis, Mo., USA; catalogue # A1978).
Immunohistochemistry
Four 3-week-old Swiss Webster mice were anesthetized and perfused as follows. Lactated Ringer solution (6–8 ml) was injected directly into the heart followed by injection of 15 ml of fixative containing 4% paraformaldehyde, 0.1% glutaraldehyde in 0.08 M sodium cacodylate buffer and 0.05% calcium chloride, pH 7.2–7.4. The mandibles were dissected and soft tissues were removed and kept in the same fixative overnight. After washing the mandibles, they were decalcified in 4.13% disodium EDTA (pH 7.3) at 4°C for 3 weeks, embedded in paraffin and sectioned (approx. 5 μm thick). Tissue sections were dewaxed and rehydrated and endogenous peroxidase was blocked with 0.3% H2O2 in water. Sections were blocked with 1% bovine serum albumin and incubated overnight with anti-NCKX4 antibody produced in rabbit (1: 50 dilution; Sigma-Aldrich, catalogue # SAB2105562). This antibody was generated against a NCKX4 peptide region that has 100% identity between human and mouse. After washing, sections were incubated with a biotinylated anti-rabbit IgG secondary antibody (Vector Laboratories, Burlingame, Calif., USA). Sections were then incubated with the ready-to-use Vectastain Elite ABC reagent (Cat # PK 7100) (Vector Laboratories) prior to incubation with AEC peroxidase substrate kit (Vector Laboratories, Cat# SK-4200) as described in the protocol provided by the manufacturer. Sections were counter-stained with Mayer’s hematoxylin, dehydrated through ethanol series, dewaxed and coverslipped.
Results
NCKX4 mRNA Expression Increases in Late Developmental Stages of Dental Formation
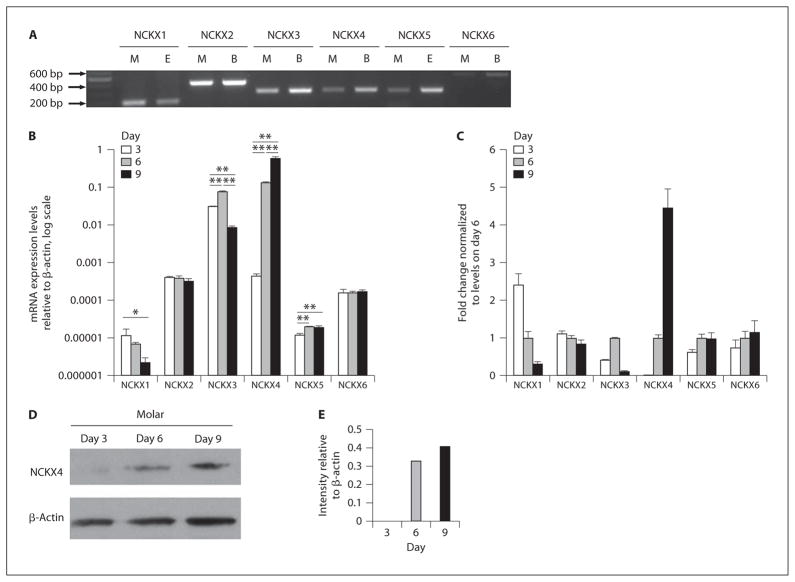
Gene-specific primers demonstrated expression of all 6 members of the Slc24a gene family (NCKX1–6) in the molars of 3-day-old mice (fig. 1A). Mouse eye cDNA was used as a positive control tissue for NCKX1 and NCKX5, while whole brain cDNA served as a control tissue for NCKX2, NCKX3, NCKX4 and NCKX6 [Cai and Lytton, 2004; Schnetkamp, 2004]. The sizes of all amplified DNA were as expected, with only a single product in each case. mRNA expression levels in mandibular first molars, relative to β-actin, were determined for each of the Slc24a genes on days 3, 6 and 9 using real-time PCR (fig. 1B). MANOVA indicated significant group differences when all dependent variables (gene variant expressions) were considered together (Wilks Lambda = 0.000; d.f. = 6,1; p = 0.019). When comparing the expression levels of individual genes on days 3, 6 and 9, ANOVA showed significant group differences for NCXK1, NCKX3, NCKX4 and NCKX5, but not for NCKX2 or NCKX6 (online suppl. table S1; www.karger.com/doi/10.1159/000337493). Comparing transcript levels for each gene at each time point revealed that NCKX1 expression was numerically lower on day 9 relative to days 3 and 6, but a significant difference was only detected when comparing day 3 to day 9 (p ≤ 0.05). NKCX3 significantly decreased on day 9 relative to day 3 (p ≤ 0.05) but increased on day 6 relative to day 3 (p ≤ 0.05). NCKX4 and NCKX5 expression increased sequentially throughout the developmental series sampled (from day 3 to day 9). For NCKX4, each increase in expression at each time point was significant (p ≤ 0.005). For NCKX5, differences were significant when comparing day 3 to days 6 or 9 (p ≤ 0.005), but no significant difference was found between days 6 and 9 (p = 0.931). Transcript fold changes are presented relative to the expression seen on day 6 (fig. 1C).
Fig. 1.
Identification of Slc24a family members in mouse mandibular first molars isolated on days 3, 6 and 9. A All 6 Slc24a gene transcripts could be amplified by standard RT-PCR from 3-day-old mandibular first molars (M). Amplified fragments were of the predicted sizes (NCKX1, 198 bp; NCKX2, 422 bp; NCKX3, 301 bp; NCKX4, 307 bp; NCKX5, 300 bp; NCKX6, 521 bp). Control tissues included were brain (B) and eye (E). A DNA marker was included on the left of A. B The mRNA expression level for Slc24a family members in the 3-, 6- and 9-day-old molars were examined using real-time PCR. Messenger RNA levels were normalized to β-actin using the delta CT method. Statistical significance was assessed using ANOVA, and the post hoc Tukey’s significant-difference test was used to assess which group differences were significant (*p < 0.05 and ** p < 0.01). Normalized mRNA levels were log-transformed for graphical representation (B). C Fold changes of the mRNA expression level for Slc24a genes calculated using the 2(−Delta Delta C(T)) method normalizing transcript levels relative to values on day 6. D Western blot analysis for NCKX4 expression in mouse mandibular first molars isolated on days 3, 6 and 9. The band size seen for NCKX4 was of predicted size, being approximately 75 kDa. Levels of β-actin in each sample serve as a control. E Protein levels noted in D were quantitated by laser densitometry, and normalized against β-actin expression.
Western Blot Analysis Shows Increased NCKX4 Expression in 9-Day-Old Mice
Western blot analysis was carried out with total protein isolated from mouse mandibular first molars on days 3, 6 and 9. NCKX4 protein was not detectable on day 3; however, NCKX4 was clearly evident on day 6 and this expression increases further on day 9 (fig. 1D). This procedure was repeated many times and the results were similar each time. Results from Western blot analysis were quantitated and normalized to the signal for β-actin (fig. 1E). For NCKX4, the fold changes seen at the protein level reflect a similar trend to that observed at the mRNA level.
In situ Hybridization Identifies NCKX4 in Ameloblasts during the Late Stages of Amelogenesis
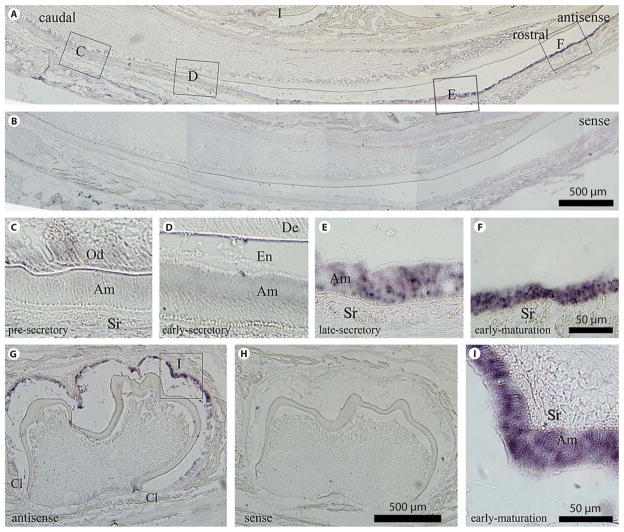
To characterize the spatial mRNA expression of NCKX4 in the developing tooth organ, in situ hybridization was performed on a 7-day-old mouse mandibular incisor (fig. 2A, C–F) and mandibular first molar (fig. 2G, I). Cytoplasmic expression of NCKX4 mRNA in ameloblasts was absent in the presecretory and early secretory stage (fig. 2C, D), becoming evident during the late secretory stage (fig. 2E) and being most highly expressed during the maturation stage (fig. 2F). This expression profile is also seen in molar ameloblasts where no expression is noted in presecretory ameloblasts located at, or in close proximity to the cervical loop (fig. 2G), with highest expression at the cusp tip (fig. 2G, I). Data also show some expression of NCKX4 in cell populations other than late-secretory-stage and maturation-stage ameloblasts; being weakly expressed in the cells of the papillary layer (e.g. cells of the stellate reticulum) and in odontoblasts. The sense probe (negative control) gave no signal (fig. 2B, H). The in situ data presented here is consistent with our real-time PCR results that show NCKX4 mRNA levels more highly expressed on day 9 when maturation-stage ameloblasts are found in relatively large quantities in the tooth organ.
Fig. 2.
In situ hybridization analysis of NCKX4 in the developing mandibular dentition of a 7-day-old mouse. Mouse incisors (A–F) and molars (G–I) in sagittal section using the antisense riboprobe (A, C–F, G, I) versus the sense (negative control) riboprobe (B, H). No hybridization signal is evident in the negative control sections. C–F, I, are enlarged regions identified in A and G. Ameloblast cell populations are presecretory (C), early-secretory (D), late-secretory (E) and early-maturation (F, I). The highest level of NCKX4 mRNA is seen during late-secretory and maturation-stage amelogenesis as evidenced in the incisors (E and F compared to C and D) and in the molar (I). Am = Ameloblasts; Cl = cervical loop; De = dentin; En = enamel; Od = odontoblasts; Sr = stellate reticulum. A, B, G, H Scale bars: 500 μm. C–F, I Scale bars: 50 μm.
Immunolocalization Identifies NCKX4 in Ameloblasts and the Papillary Layer Cells during Maturation-Stage Amelogenesis
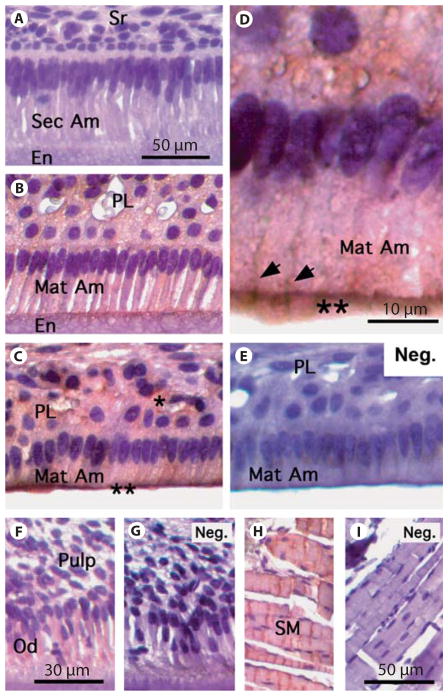
Using the polyclonal antibody generated against an N-terminal peptide region of NCKX4 (Sigma-Aldrich) on the incisors of 3-week-old mice, the expression of NCKX4 was largely restricted to maturation-stage enamel organ cells, and most prominently in mid-late maturation-stage ameloblasts (fig. 3B–D). Lighter staining was noted in the overlying papillary layer cells (fig. 3B, C). Observing the same section prepared and processed in an identical manner, no NCKX4 expression was noted in the presecretory-stage (data not shown) or secretory-stage enamel organ cells (fig. 3A). Using secondary antibody only, no background staining was observed in the control tissue of maturation ameloblasts (fig. 3E). Spatially, within maturation ameloblasts, greater NCKX4 expression was noted at the apical poles (fig. 3B–D) and at the lateral membranes proximal to the apical ends (fig. 3D). In addition, NCKX4 is expressed in odontoblast cells (fig. 3F). An additional control included skeletal muscle known to express NCKX4 [Li et al., 2002] (fig. 3H). In figure 3, G and I are included as additional controls (no primary antibody) for odontoblast and pulp tissues and skeletal muscle, respectively.
Fig. 3.
NCKX4 immunolocalization in the developing mandibular incisor of a 3-week-old mouse. A No NCKX4 staining was visible in secretory-stage ameloblasts. B Staining is evident at the apical pole of early maturation-stage ameloblasts. Weak staining is also visible in the papillary layer (PL). C Mid-late maturation-stage ameloblasts and PL stain for NCKX4 with stronger signals detected at the apical pole of ameloblasts identified by double asterisks (**). D Higher magnification (× 100) image of mid-late maturation-stage ameloblasts showing strong reactivity at apical pole as marked by double asterisks, and extending to the lateral membranes (arrowheads). E Negative control: mid-late maturation-stage ameloblasts were incubated with no primary NCKX4 antibody but all other steps were performed. No positive staining was visible in this tissue section. F, G Odontoblasts showed positive staining when incubated with NCKX4 (F), whereas the negative control (no primary antibody) showed no staining (G). H, I Skeletal muscle (tissue control): H shows positive staining, but the negative control (no primary antibody) did not stain (I ). En = enamel; Mat Am = maturation ameloblasts; Neg. = no primary antibody; Od = odontoblasts; PL = papillary layer; Sec Am = secretory ameloblasts; SM = skeletal muscle; Sr = stellate reticulum. A–C, E, H, I Scale bars: 50 μm. D Scale bar: 10 μm. F, G Scale bars: 30 μm.
Discussion
Na+/Ca2+ exchangers contribute to calcium transport in many tissues, including the tooth organ as discussed recently for NCX1 and NCX3 [Okumura et al., 2010; Tsumura et al., 2010]. In this study, we demonstrate that NCKX4 is expressed in late secretory-stage and maturation-stage ameloblasts, with significantly increased mRNA and protein expression during the late stages of amelogenesis. Calcium handling is a critical activity of ameloblasts in order to meet the needs of enamel Hap biomineralization [Smith, 1998; Hubbard, 2000]. Relatively high Ca2+ deposition was found during maturation-stage amelogenesis, as demonstrated by 45Ca tracer [Takano, 1994, 1995], radiographic [Suga, 1982] and chemical analysis [Robinson et al., 1978]. Therefore, ameloblasts must adjust their function to allow for Ca2+ transport to the enamel matrix at a rate that meets requirements for crystal growth.
In this study, we identified that the expression of NCKX4 in the enamel organ is primarily restricted to the ameloblasts, being most highly expressed in maturation-stage ameloblasts. Some expression is also noted in the papillary layer cells during enamel maturation. Given the increase requirements for Ca2+ transport during maturation [Smith, 1998; Hubbard, 2000] and the spatiotemporal expression pattern of NCKX4 described here, it is likely that NCKX4 participates in Ca2+ transport during this critical period. NCKX4 is a bidirectional transporter capable of exchanging Na+/Ca2+-K+ in either a Ca2+ efflux or influx depending on the transmembrane ion gradient [Li et al., 2002; Schnetkamp, 2004; Uehara et al., 2004]. Thus, the function(s) of NCKX4 during amelogenesis can be delineated into at least 3 possible scenarios. First, NCKX4 may be involved in Ca2+ export from the ameloblasts into the forming enamel, i.e. supporting and/or duplicating the activities of NCX1 and NCX3 [Okumura et al., 2010]. Based on the immunohistochemical data presented here, where greater expression of NCKX4 is observed at the apical poles and the lateral membranes proximal to the apical ends of maturation-stage ameloblasts, this is a preferred scenario. Secondly, NCKX4 may facilitate Ca2+ import from the papillary layer. This scenario is also possible, based on the absolute need for Ca2+ in the formation of enamel crystals and the fact that other known NCX (NCX1 and NCX3) are localized to the apical pole of maturation-stage ameloblasts [Okumura et al., 2010]; however, the immunohistochemical data does not support such activity. A third alternative, but possibly the least likely, is that NCKX4 participates in both import and export activities during amelogenesis.
This study contributes to the knowledge of the roles of NCXs in enamel formation. In particular, NCKX4 expression was negligible or absent during early amelogenesis and is highly upregulated during the maturation stage. The expression of NCKX4 in enamel epithelial cells suggests a function in transcellular Ca2+ movement during enamel Hap formation.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Joseph J. Miller for his assistance with the statistical analyses and Drs. Malcolm Snead and Michael Hubbard for their continued discussions related to this and other ongoing projects of mutual interest. This work was supported by grant DE019629 from the National Institutes of Health.
Abbreviations used in this paper
- Hap
hydroxyapatite
- NCKX
sodium/calcium-potassium exchanger
- NCX
sodium/calcium exchanger
- Slc
solute carrier gene family
References
- Cai X, Lytton J. Molecular cloning of a sixth member of the K+-dependent Na+/Ca2+ exchanger gene family, NCKX6. J Biol Chem. 2004;279:5867–5876. doi: 10.1074/jbc.M310908200. [DOI] [PubMed] [Google Scholar]
- Franklin IK, Winz RA, Hubbard MJ. Endoplasmic reticulum Ca2+-ATPase pump is up-regulated in calcium-transporting dental enamel cells: a non-housekeeping role for SERCA2b. Biochem J. 2001;358:217–224. doi: 10.1042/0264-6021:3580217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard MJ. Calbindin28kDa and calmodulin are hyperabundant in rat dental enamel cells. Identification of the protein phosphatase calcineurin as a principal calmodulin target and of a secretion-related role for calbindin28kDa. Eur J Biochem. 1995;230:68–79. doi: 10.1111/j.1432-1033.1995.tb20535.x. [DOI] [PubMed] [Google Scholar]
- Hubbard MJ. Abundant calcium homeostasis machinery in rat dental enamel cells. Up-regulation of calcium store proteins during enamel mineralization implicates the endoplasmic reticulum in calcium transcytosis. Eur J Biochem. 1996;239:611–623. doi: 10.1111/j.1432-1033.1996.0611u.x. [DOI] [PubMed] [Google Scholar]
- Hubbard MJ. Calcium transport across the dental enamel epithelium. Crit Rev Oral Biol Med. 2000;11:437–466. doi: 10.1177/10454411000110040401. [DOI] [PubMed] [Google Scholar]
- Josephsen K, Takano Y, Frische S, Praetorius J, Nielsen S, Aoba T, Fejerskov O. Ion transporters in secretory and cyclically modulating ameloblasts: a new hypothesis for cellular control of preeruptive enamel maturation. Am J Physiol Cell Physiol. 2010;299:C1299–1307. doi: 10.1152/ajpcell.00218.2010. [DOI] [PubMed] [Google Scholar]
- Lacruz RS, Hilvo M, Kurtz I, Paine ML. A survey of carbonic anhydrase mRNA expression in enamel cells. Biochem Biophys Res Commun. 2010a;393:883–887. doi: 10.1016/j.bbrc.2010.02.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacruz RS, Nanci A, Kurtz I, Wright JT, Paine ML. Regulation of pH during amelogenesis. Calcif Tissue Int. 2010b;86:91–103. doi: 10.1007/s00223-009-9326-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacruz RS, Nanci A, White SN, Wen X, Wang H, Zalzal SF, Luong VQ, Schuetter VL, Conti PS, Kurtz I, Paine ML. The sodium bicarbonate cotransporter (NBCe1) is essential for normal development of mouse dentition. J Biol Chem. 2010c;285:24432–24438. doi: 10.1074/jbc.M110.115188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacruz RS, Smith CE, Bringasjr P, Chen YB, Smith SM, Snead ML, Kurtz I, Hacia JG, Hubbard MJ, Paine ML. Identification of novel candidate genes involved in mineralization of dental enamel by genome-wide transcript profiling. J Cell Physiol. 2012a;227:2264–2275. doi: 10.1002/jcp.22965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacruz RS, Smith CE, Moffatt P, Chang EH, Bromage TG, Bringas P, Jr, Nanci A, Baniwal SK, Zabner J, Welsh MJ, Kurtz I, Paine ML. Requirements for ion and solute transport, and pH regulation, during enamel maturation. J Cell Physiol. 2012b;227:1776–1785. doi: 10.1002/jcp.22911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamason RL, Mohideen MA, Mest JR, Wong AC, Norton HL, Aros MC, Jurynec MJ, Mao X, Humphreville VR, Humbert JE, Sinha S, Moore JL, Jagadeeswaran P, Zhao W, Ning G, Makalowska I, McKeigue PM, O’Donnell D, Kittles R, Parra EJ, Mangini NJ, Grunwald DJ, Shriver MD, Canfield VA, Cheng KC. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science. 2005;310:1782–1786. doi: 10.1126/science.1116238. [DOI] [PubMed] [Google Scholar]
- Li XF, Kraev AS, Lytton J. Molecular cloning of a fourth member of the potassium-dependent sodium-calcium exchanger gene family, NCKX4. J Biol Chem. 2002;277:48410–48417. doi: 10.1074/jbc.M210011200. [DOI] [PubMed] [Google Scholar]
- Linde A. Dentin mineralization and the role of odontoblasts in calcium transport. Connect Tissue Res. 1995;33:163–170. doi: 10.3109/03008209509016997. [DOI] [PubMed] [Google Scholar]
- Okumura R, Shibukawa Y, Muramatsu T, Hashimoto S, Nakagawa K, Tazaki M, Shimono M. Sodium-calcium exchangers in rat ameloblasts. J Pharmacol Sci. 2010;112:223–230. doi: 10.1254/jphs.09267fp. [DOI] [PubMed] [Google Scholar]
- Paine ML, White SN, Luo W, Fong H, Sarikaya M, Snead ML. Regulated gene expression dictates enamel structure and tooth function. Matrix Biol. 2001;20:273–292. doi: 10.1016/s0945-053x(01)00153-6. [DOI] [PubMed] [Google Scholar]
- Pyrski M, Koo JH, Polumuri SK, Ruknudin AM, Margolis JW, Schulze DH, Margolis FL. Sodium/calcium exchanger expression in the mouse and rat olfactory systems. J Comp Neurol. 2007;501:944–958. doi: 10.1002/cne.21290. [DOI] [PubMed] [Google Scholar]
- Quednau BD, Nicoll DA, Philipson KD. The sodium/calcium exchanger family-SLC8. Pflugers Arch. 2004;447:543–548. doi: 10.1007/s00424-003-1065-4. [DOI] [PubMed] [Google Scholar]
- Robinson C, Fuchs P, Deutsch D, Weatherell JA. Four chemically distinct stages in developing enamel from bovine incisor teeth. Caries Res. 1978;12:1–11. doi: 10.1159/000260309. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Schnetkamp PP, Basu DK, Szerencsei RT. Na+-Ca2+ exchange in bovine rod outer segments requires and transports K+ Am J Physiol. 1989;257:C153–157. doi: 10.1152/ajpcell.1989.257.1.C153. [DOI] [PubMed] [Google Scholar]
- Schnetkamp PP. The SLC24 Na+/Ca2+-K+ exchanger family: vision and beyond. Pflugers Arch. 2004;447:683–688. doi: 10.1007/s00424-003-1069-0. [DOI] [PubMed] [Google Scholar]
- Simmer JP, Hu Y, Lertlam R, Yamakoshi Y, Hu JC. Hypomaturation enamel defects in Klk4 knockout/LacZ knockin mice. J Biol Chem. 2009;284:19110–19121. doi: 10.1074/jbc.M109.013623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CE. Cellular and chemical events during enamel maturation. Crit Rev Oral Biol Med. 1998;9:128–161. doi: 10.1177/10454411980090020101. [DOI] [PubMed] [Google Scholar]
- Suga S. Progressive mineralization pattern of developing enamel during the maturation stage. J Dent Res. 1982;(Spec No):1532–1542. [PubMed] [Google Scholar]
- Takano Y. Histochemical aspects of calcium regulation by the enamel forming cells during matrix formation and maturation. Kaibogaku Zasshi. 1994;69:106–122. [PubMed] [Google Scholar]
- Takano Y. Enamel mineralization and the role of ameloblasts in calcium transport. Connect Tissue Res. 1995;33:127–137. doi: 10.3109/03008209509016992. [DOI] [PubMed] [Google Scholar]
- Tsoi M, Rhee KH, Bungard D, Li XF, Lee SL, Auer RN, Lytton J. Molecular cloning of a novel potassium-dependent sodium-calcium exchanger from rat brain. J Biol Chem. 1998;273:4155–4162. doi: 10.1074/jbc.273.7.4155. [DOI] [PubMed] [Google Scholar]
- Tsumura M, Okumura R, Tatsuyama S, Ichikawa H, Muramatsu T, Matsuda T, Baba A, Suzuki K, Kajiya H, Sahara Y, Tokuda M, Momose Y, Tazaki M, Shimono M, Shibukawa Y. Ca2+ extrusion via Na+-Ca2+ exchangers in rat odontoblasts. J Endod. 2010;36:668–674. doi: 10.1016/j.joen.2010.01.006. [DOI] [PubMed] [Google Scholar]
- Turnbull CI, Looi K, Mangum JE, Meyer M, Sayer RJ, Hubbard MJ. Calbindin independence of calcium transport in developing teeth contradicts the calcium ferry dogma. J Biol Chem. 2004;279:55850–55854. doi: 10.1074/jbc.M409299200. [DOI] [PubMed] [Google Scholar]
- Uehara A, Iwamoto T, Nakamura Y, Imanaga I. Forefront of Na+/Ca2+ exchanger studies: physiology and molecular biology of monovalent cation sensitivities in Na+/Ca2+ exchangers. J Pharmacol Sci. 2004;96:19–22. doi: 10.1254/jphs.fmj04002x4. [DOI] [PubMed] [Google Scholar]
- Wilkinson DG. In Situ Hybridization: A Practical Approach. (IRL Press) Oxford University Press; Oxford: 1992. [Google Scholar]
- Winkfein RJ, Szerencsei RT, Kinjo TG, Kang K, Perizzolo M, Eisner L, Schnetkamp PP. Scanning mutagenesis of the alpha repeats and of the transmembrane acidic residues of the human retinal cone Na/Ca-K exchanger. Biochemistry. 2003;42:543–552. doi: 10.1021/bi026982x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.