Abstract
Mutations of the KRAS oncogene are predictive for resistance to treatment with antibodies against the epithelial growth factor receptor in patients with colorectal cancer. Overcoming this therapeutic dilemma could potentially be achieved by the introduction of drugs that inhibit signaling pathways that are activated by KRAS mutations. To identify comprehensively such signaling pathways we profiled pretreatment biopsies and normal mucosa from 65 patients with locally advanced rectal cancer - 30 of which carried mutated KRAS - using global gene expression microarrays. By comparing all tumor tissues exclusively to matched normal mucosa, we could improve assay sensitivity, and identified a total of 22,297 features that were differentially expressed (adjusted P-value <0.05) between normal mucosa and cancer, including several novel potential rectal cancer genes. We then used this comprehensive description of the rectal cancer transcriptome as the baseline for identifying KRAS-dependent alterations. The presence of activating KRAS mutations is significantly correlated to an upregulation of 13 genes (adjusted P-value <0.05), among them DUSP4, a MAP-kinase phosphatase, and SMYD3, a histone methyltransferase. Inhibition of the expression of both genes has previously been shown using the MEK1-inhibitor PD98059 and the antibacterial compound Novobiocin, respectively. These findings suggest a potential approach to overcome resistance to treatment with antibodies against the epithelial growth factor receptor in patients with KRAS-mutant rectal carcinomas.
INTRODUCTION
The introduction of therapeutic antibodies for cancer treatment was a first step towards the implementation of targeted therapies, and, consequently, an important milestone towards the realization of individualized treatment. The most heralded target for a rational therapy of patients with colorectal cancer was an antibody against the EGF receptor, Cetuximab. The gene that encodes this protein maps to 7p, which is subject to recurrent genomic amplification in CRC (Platzer et al., 2002). Treatment with Cetuximab leads to higher response rates and to a significant prolongation of the progression-free interval in metastatic colorectal cancer. However, recent evidence strongly suggests that treatment failure in patients receiving chemotherapy in combination with anti-EGFR antibodies is caused by activating mutations of the KRAS proto-oncogene (Lievre et al., 2006, 2008; Di Fiore et al., 2007; Karnoub and Weinberg, 2008). Mutations of this gene occur in 35-45% of all colorectal cancers (Brink et al., 2003; Baldus et al., 2010), and result in the continuous activation of the KRAS signaling pathway, now independent of EGFR-dependent stimulation. Therefore, targets other than EGFR are currently pursued for the treatment of patients with KRAS mutated colorectal cancer. Alternatively, one could envision that drugs that counteract the effect of mutant KRAS or its downstream targets and would thus overcome the resistance of KRAS mutant tumors to EGFR inhibitors, could evolve as valuable treatment options.
We therefore aimed to analyze systematically and comprehensively the influence of KRAS mutations on the rectal cancer transcriptome. Towards this goal, we performed whole genome expression profiling of locally advanced rectal cancers, for which the respective KRAS mutation status had recently been analyzed (Gaedcke et al., 2010). We focused exclusively on rectal carcinomas and normalized gene expression levels for all carcinomas to matched normal mucosa biopsies. We defined these two criteria in an attempt to reduce the noise induced by the idiosyncrasies of individual patient samples and by differences as a consequence of the anatomical location. We hypothesized that the delineation of a “KRAS signature”, and with it a comprehensive and definitive description of the rectal cancer transcriptome will lead to the identification of novel critical pathways and potential target genes, and hence unexplored potential alternative therapeutic strategies.
MATERIALS AND METHODS
Selection of Patients, Sample Ascertainment and RNA Isolation
Sixty-five patients with rectal adenocarcinomas were included in this study (Supplementary Table 1). All tumors were located within 12 cm from the anocutaneous verge, and diagnosed as locally advanced stages of the disease (UICC II/III). From each patient we collected pretreatment tumor biopsies adhering to the guidelines set by the local ethical review board. Biopsies were immediately stored in RNAlater (Qiagen, Hilden, Germany). Using a second forceps normal rectal mucosa biopsies were obtained from all 65 patients at a minimum distance of 3 cm from the tumor site.
Subsequently, RNA was isolated using TRIzol (Invitrogen, Carlsbad, CA) following standard procedures as previously described (Grade et al., 2006, 2007). Nucleic acid quantity, quality and purity were determined using a spectrophotometer (Nanodrop, Rockland, DE) and a 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). RNA samples with an RNA Integrity Number (RIN) of > 5 were included.
Gene Expression Profiling
Expression profiling was performed as previously described (Grade et al., 2010). Briefly, 1 μg of total RNA was labeled with Cy3 using the Low RNA Input Fluorescent Linear Amplification Kit according to the manufacturer’s recommendations (Agilent Technologies, Santa Clara, CA). Quantity and efficiency of the labeled amplified cRNA were determined using the NanoDrop ND-1000 UV-VIS Spectrophotometer version 3.2.1. Subsequently, 1.5 μg of Cy3-labeled cRNA was hybridized to an oligonucleotide-based Whole Human Genome Microarray (4×44K, Agilent Technologies) and incubated at 65°C for 17 h. Slides were washed and scanned using an Agilent G2565BA scanner. Raw data were extracted using the Feature Extraction software version 9.1 (Agilent Technologies).
Data Normalization and Processing
Statistical analyses were performed with the free software R (version 2.8, www-r-project.org). The R-package ‘limma’ (www.bioconductor.org) was used for data normalization and identification of differentially expressed genes. Raw expression data from all 130 microarrays were log2-transformed and quantile normalized (Bolstad et al., 2003). Features that showed in 90% of all arrays an expression that was lower than the average “Dark Corner” values were removed.
Statistical Analysis and Pathway Information
Genes with significantly different expression level ratios between tumor and mucosa samples were identified using the Limma method (Smyth, 2004). To control for multiple testing, raw p-values were adjusted using the method of Benjamini and Hochberg (Benjamini and Hochberg, 1995). Genes were regarded as differentially expressed when the adjusted P-value was smaller than 0.05. For a more stringent assessment we applied additional filter criteria: a 2-fold change in expression and a “tumor marker” criterion (the lowest expression of a given feature in the tumor samples always had to be higher than the highest expression in all the matched normal mucosae, or vice versa (in the following referred to as Min/Max criterion)).
Both gene lists were screened for known interactions and involvement in biological networks using the software package Ingenuity Pathway Analysis (IPA; Ingenuity, Mountain View, CA). The genes showing a 2-fold change in expression were queried as to their enrichment at certain chromosomal locations.
From statistical theory it is anticipated that the analysis of paired tumor and mucosa samples from the same patients is more powerful than a similar analysis with unpaired tumor and mucosa samples from different patients (Fisher, 1925). To demonstrate further the superiority of a paired tumor and mucosa samples in our data we performed some random sampling experiments. In each run, 30 patients were randomly chosen from our studied collective and their tumor samples were compared to their related mucosa samples. In the same run the tumor samples from the 30 selected patients were also compared to 30 randomly chosen unrelated mucosa samples (Supplementary Fig. 1).
The potential of differentially expressed genes detected between tumors with and without a KRAS mutation to distinguish between those two groups was evaluated using discriminant analysis within a Leave-One-Out-Cross-Validation (LOOCV).
Semi-Quantitative Real-time PCR
The mRNA expression levels of distinct genes were validated by semi-quantitative real-time PCR (qPCR) using iQ™ SYBR® Green Supermix (BIO-RAD Laboratories, Hercules, CA). Gene-specific primers were designed using Primer3 (http://frodo.wi.mit.edu/) and obtained from MWG Biotech AG (Ebersberg, Germany). All nucleotides were optimized according to standard protocols and shown to produce single amplicons and no primer-dimer artifacts. The efficiency of amplification was validated using LinRegPCR (http://www.gene-quantification.de/download.html#linregpcr). Corresponding primer sequences are listed in Supplementary Table 2.
Briefly, total RNA was reverse-transcribed into cDNA using Superscript III Reverse Transcriptase (Invitrogen, Carlsbad, CA) and subsequently diluted 1:5. Triplicate quantifications were performed for each gene in an iCycler (Bio-Rad Laboratories GmbH, Munich, Germany), and each data point was calculated as the median of the three measured CT values. Relative mRNA levels were calculated and normalized to the expression levels of OTUB1, FBXL12 and RAB35 using the ΔΔCt technique. These genes were specifically chosen because their expression levels were stable among all samples. A detailed protocol can be found at www.riedlab.nci.nih.gov/protocols.asp.
Analysis of KRAS Status
KRAS mutation status was assessed by Sanger sequencing of DNA extracted from tumor biopsies. Analysis included exons 1, 2 and 3 as reported previously (Gaedcke et al., 2010). Gene expression profiles and KRAS mutation status were analyzed from identical biopsies (Supplementary Table 1).
RESULTS
The signaling pathway governed by the oncogene KRAS is crucially involved in tumorigenesis. In addition, there is sound evidence that mutations in the KRAS oncogene determine response to treatments that target the MAP kinase pathway, a promising molecular target for individualized therapy.
In order to identify downstream pathways of mutated KRAS that could explain resistance to MAP-kinase pathway inhibition, we first created a baseline for the systematic exploration of the consequences of activated ras signaling by comprehensively cataloguing transcriptional alterations in 65 rectal carcinomas for which we had previously established KRAS mutation status. In order to account for potential differences between rectal and colonic carcinomas, including different therapeutic regimen, we concentrated in this study exclusively on rectal cancer, and we compensated for inter-patient transcriptional differences by normalizing changes in the tumor transcriptome to patient-matched normal mucosa.
The Rectal Cancer Transcriptome: Differentially Expressed Genes
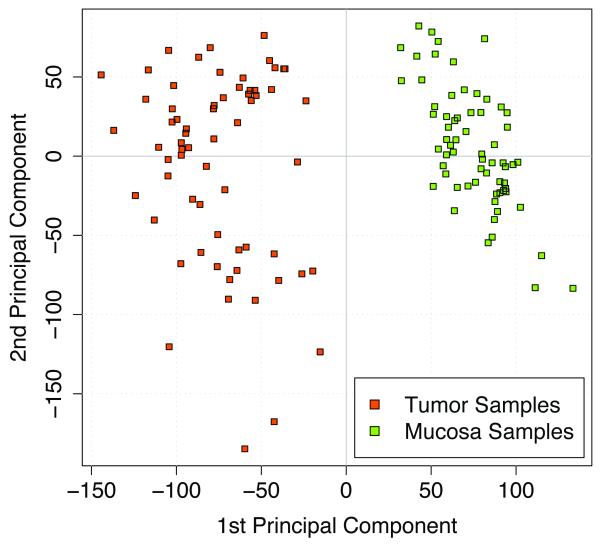
We profiled a series of 65 locally advanced rectal cancers. In contrast to previously performed microarray analyses of rectal carcinomas (Alon et al., 1999; Zou et al., 2002; Friederichs et al., 2005; Bianchini et al., 2006) matched normal rectal mucosa samples were used for comparison, to increase the power of the tests due to likely smaller variances. As expected using paired samples yielded in significantly more differentially expressed genes than using unpaired samples (P<0.01, Supplementary Fig. 1). Data were normalized and filtered as described in Materials and Methods. Of the 29,149 remaining features 22,297 were differentially expressed according to the FDR-adjusted P-values; they allowed a clear separation between tumors and matched normal mucosa samples (Fig. 1). To increase further the biological relevance of differentially expressed genes we applied the additional filter criteria of 2-fold difference in expression which reduced the number to 3,174 genes.
Figure 1.
Principal component plot representing tumor and mucosa samples from 65 patients.
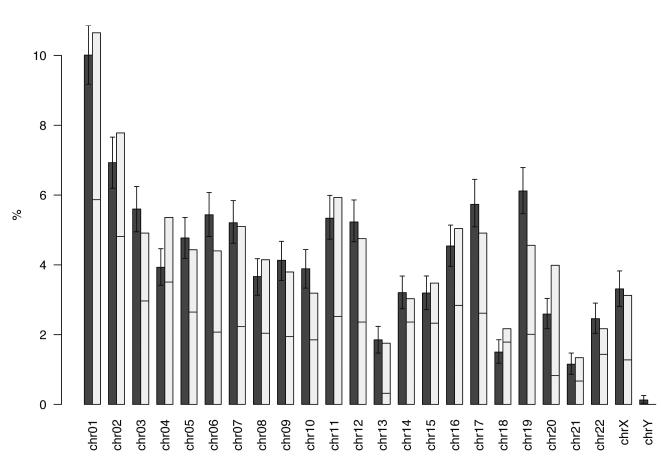
Since we have previously shown that a disproportional number of deregulated genes maps to specific chromosomes (Grade et al., 2007), we now explored whether this observation holds true when individual tumors were compared to matched normal mucosa. Of the 3,174 differentially expressed genes (false discovery rate adjusted P value <0.05 and a >2-fold difference in expression), 3,136 had a defined chromosomal location. One thousand six hundred seventy one genes were down-regulated and 1,503 were up-regulated in the tumors. In order to compare the observed percentage of differentially expressed genes per chromosome with the percentage of genes expected to be differentially expressed by chance, we calculated 1,000 random distributions of these 3,174 genes. In fact, chromosomes 4, 18 and 20 showed significantly more differentially expressed genes than expected by chance, and, interestingly, chromosome 19 showed fewer genes (Fig. 2). Most genes on chromosomes 13 and 20 were upregulated; in contrast, most genes on chromosomes 14, 15 and 18 were underexpressed in the tumors, which is consistent with the frequent gain of chromosomes 13 and 20, and losses that include chromosomes 4 and 18.
Figure 2.
The dark bars display the expected percentage including the 95% confidence interval, the light bars display the observed percentages of DEGs per chromosome. Chromosomes 4, 18 and 20 show significantly more DEGs than expected; chromosome 19 shows less DEGs. The horizontal line in the light bar indicates the proportion of up- and downregulated genes displaying upregulation of most of the genes on chromosome 13 and 20 and downregulation on chromosomes 14, 15 and 18. (DEG, differentially expressed genes)
Of the 3,174 features, 1,481 were up-regulated and 1,693 were down-regulated in the tumors (Supplementary Table 3). To identify potential novel cancer genes, differentially expressed genes were filtered to be either always higher expressed in the tumors compared to the mucosa, or vice versa, our so called Min/Max criteria. Nineteen features fulfilled this criterion, representing 17 different genes (two of the genes were represented with two features). Eleven of these features (ten genes) were highly expressed in the tumors, while eight features (seven genes) were highly expressed in the normal mucosa (Table 1).
TABLE 1.
Expression Ratio for Genes of Interest
| a) Features fulfilling the Min >< Max criteria | |||||
|---|---|---|---|---|---|
| Systematic name |
Gene name | Description | log2 FC tumor vs. mucosa |
P | Map |
| NM_013227 | ACAN | Aggrecan | 3.07 | 6.60E-36 | 15q26.1 |
| NM_004673 | ANGPTL1 | Angiopoietin-like 1 | −2.66 | 1.22E-37 | 1q25.2 |
| NM_018270 | ANO5 | Anoctamin 5 | −2.9 | 3.78E-32 | 11p14.3 |
| NM_018270 | C20orf20 | Chromosome 20 open reading frame 20 | 1.67 | 1.05E-33 | 20q13.33 |
| NM_018140 | CEP72 | Centrosomal protein 72kDa | 1.95 | 5.92E-35 | 5p15.33 |
| NM_021101 | CLDN1 | Claudin 1 | 4.29 | 5.01E-48 | 3q28-q29 |
| NM_015036 | ENDOD1 | Endonuclease domain containing 1 | −1.91 | 6.00E-37 | 11q21 |
| NM_001079675 | ETV4 | Ets variant 4 | 3.25 | 1.26E-41 | 17q21 |
| NM_001079675 | ETV4 | Ets variant 4 | 2.97 | 9.72E-39 | 17q21 |
| NM_001445 | FABP6 | Fatty acid binding protein 6, ileal | 4.82 | 4.94E-43 | 5q33.3-q34 |
| NM_000148 | FUT1 | Fucosyltransferase 1 | 2.49 | 2.00E-35 | 19q13.3 |
| NM_003641 | IFITM1 | Interferon induced transmembrane protein 1 (9-27) |
2.82 | 1.36E-38 | 11p15.5 |
| NM_021034 | IFITM3 | Interferon induced transmembrane protein 3 (1-8U) |
2.68 | 2.72E-38 | 11p15.5 |
| NM_006790 | MYOT | Myotilin | −1.91 | 3.97E-41 | 5q31 |
| NM_005012 | ROR1 | Receptor tyrosine kinase-like orphan receptor 1 |
−1.67 | 9.35E-38 | 1p32-p31 |
| NM_005012 | ROR1 | Receptor tyrosine kinase-like orphan receptor 1 |
−1.81 | 1.32E-35 | 1p32-p31 |
| NM_003759 | SLC4A4 | Solute carrier family 4, sodium bicarbonate cotransporter, member 4 |
−4.81 | 8.58E-30 | 4q21 |
| NM_006714 | SMPDL3A | Sphingomyelin phosphodiesterase, acid- like 3A |
−2.53 | 3.09E-34 | 6q22.31 |
| NM_021158 | TRIB3 | Tribbles homolog 3 (Drosophila) | 3.51 | 3.07E-41 | 20p13-p12.2 |
| b) Genes being differentially expressed between KRAS WT and KRAS mutant. | |||||
|---|---|---|---|---|---|
| Systematic name |
Gene name | Description | log2 FC WT vs. mutant |
P | Map |
| NM_000719 | CACNA1C | Calcium channel, voltage dependent, L type, alpha 1C subunit |
1.2 | 2.16E-05 | 12p13.3 |
| NM_003217 | TEGT | Testis enhanced gene transcript (BAX- inhibitor-1) |
0.54 | 4.75E-06 | 12q12-q13 |
| NM_016057 | COPZ1 | Coatomer protein complex, subunit zeta 1 |
0.52 | 7.25E-07 | 12q13.2- q13.3 |
| NM_172240 | WDR51B | WD repeat domain 51B | 0.6 | 2.21E-06 | 12q21.33 |
| NM_206819 | MYBPC1 | Myosin binding protein C, slow type | 0.68 | 3.86E-06 | 12q23.2 |
| NM_020672 | S100A14 | S100 calcium binding protein A14 | 0.94 | 1.02E-06 | 1q21.3 |
| NM_018178 | GOLPH3L | Golgi phosphoprotein 3 like | 0.57 | 1.73E-05 | 1q21.3 |
| NM_001001552 | LEMD1 | LEM domain containing 1 | 2.02 | 7.58E-07 | 1q32.1 |
| NM_022743 | SMYD3 | SET and MYND domain containing 3 | 0.49 | 3.40E-06 | 1q44 |
| NM_144659 | TCP10L | T-complex 10 (mouse)-like | 0.52 | 1.57E-05 | 21q22.11 |
| NM_030666 | SERPINB1 | Serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
0.56 | 1.32E-05 | 6p25 |
| NM_001394 | DUSP4 | Dual specificity phosphatase 4 | 0.9 | 1.10E-05 | 8p12-p11 |
| NM_138969 | RDHE2 | Epidermal retinal dehydrogenase 2 | 1.26 | 1.22E-06 | 8q12.1 |
FC, Fold Change
Validation of Gene Expression Levels Using Semi-quantitative Real-time PCR
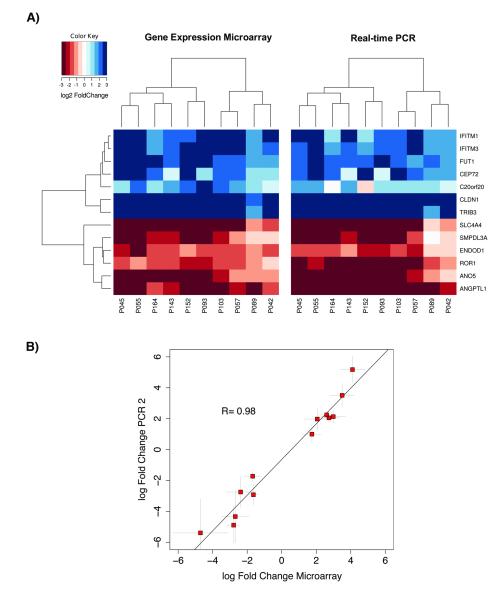
To validate independently the gene expression measurements derived from the microarray experiments, semi-quantitative RT-PCR (qPCR) was performed for 13 out of the 17 different genes in 10 tumors and 10 matched mucosa samples. As shown in Figure 3, the differential expression levels were confirmed for all genes analyzed with both methods. To assess the correlation of microarray data and qPCR results Pearson’s correlation coefficient R was calculated using the fold changes between tumor and matched mucosa. We found a highly significant correlation between both techniques (Pearson‘s R=0.98, P<0.01; Fig. 3).
Figure 3.
(A) Comparison of log2 FC between qPCR and gene expression array (B) Correlation between log fold changes of qPCR measurements versus those of microarray features. Plot represents 13 of the detected tumor markers. Microarray fold changes deviate from those in Tab. 1 because they were derived from only 9 patients.
The Rectal Cancer Transcriptome: Biological Networks
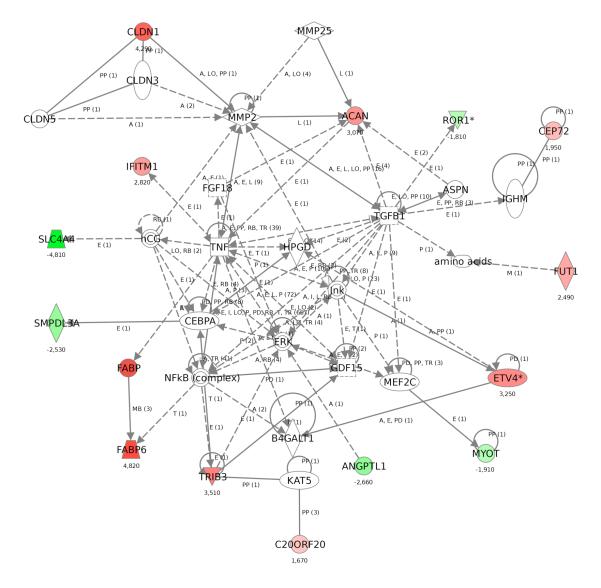
We then interrogated in which networks or pathways these 17 genes operate using the software package IPA. Strikingly, 14 of the genes clustered together in one particular network, which was connected through CEBPA and NFkB complex, which are transcription factors, GDF 15 and TNF, both of which are BMP superfamily members, and ERK and JNK (also known as MAPK1 and MAPK8), two components of the MAP Kinase pathway. Furthermore, TGFB1 was a central gene within the network (Fig. 4).
Figure 4.
Ingenuity pathway analysis reveals the close relationship between 14 of 17 of most relevant genes in rectal cancer
For a more global analysis of the differentially expressed genes we expanded the IPA analysis using genes that passed the filter criteria based on a fold change larger than two (n=3,174). As expected a large number of networks emerged. The top five, based on P-value, were functionally associated to cell cycle, cell mediated immune response, cell-cell signaling, tumor and organ morphology, and, most prominently, cancer. The most outstanding intersections were centered on IL-6 (P=10−41), MMP3 and KRAS (P=10−38), NR3C1 (P=10−36), BRCA1 and CDKN2A (P=10−36), as well as ERK and TRIB3 (P=10−34). The most relevant functions described for the differentially expressed genes included tumorigenesis (P=10−61), cancer (P=10−58), neoplasia (P=10−57), genetic disorders (P=10−42) and colorectal cancer (P=10−39) (Supplementary Fig. 2).
Effect of KRAS Mutation Status on the Rectal Cancer Transcriptome
After we had now carefully annotated the transcriptional changes associated with rectal cancer we aimed to identify the consequences of KRAS mutations on the rectal cancer transcriptome. This analysis is relevant because (i) activating mutations are known to play a fundamental role in carcinogenesis, (ii) KRAS status is used for stratification of anti-EGFR therapy with Cetuximab and, (iii) it is of clinical importance to identify strategies to overcome the resistance against such antibody-based treatment.
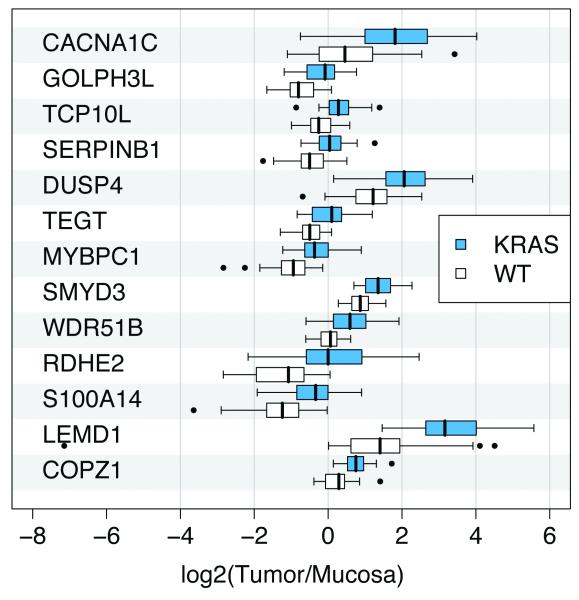
Unsupervised clustering did not result in separation according to the KRAS mutation status, however, we identified a set of 13 genes that were differentially expressed between the two groups, based on an adjusted P-value smaller than 0.05. These genes are: COPZ1, LEMD1, S100A14, RDHE2, WDR51B, SMYD3, MYBPC1, TEGT, DUSP4, SERPINB1, TCP10L, GOLPH3L, and CACNA1C. Interestingly, KRAS mutation caused upregulation of all of these genes (Fig. 5 and Table 1). The potential of these differentially expressed genes to distinguish tumors with and without mutations was evaluated using a Leave-One-Out-Cross-Validation (LOOCV). With that estimate we achieved a test accuracy of 96.9% (sensitivity 93.3%; specificity 97.1%). Of those 13 genes, only one, DUSP4, had been previously reported to be linked to KRAS and the MAPK pathway.
Figure 5.
Differentially expressed genes between KRAS wild type and mutant rectal carcinomas
DISCUSSION
Activating mutations of the KRAS oncogene play an important role in colorectal carcinogenesis. Mutations of this gene result in the GTP-dependent activation of the MAPK pathway, which, in turn impairs cell differentiation and apoptosis, and increases cell proliferation.
Furthermore, KRAS mutations have implications above and beyond basic tumor biology because successful targeting of the EGFR axis using Cetuximab depends on the maintenance of wild type KRAS (Lievre et al., 2006, 2008; Di Fiore et al., 2007; De Roock et al., 2008; Karapetis et al., 2008). Nevertheless, nothing is known about the transcriptional differences between KRAS mutant and wild-type tumors in rectal carcinomas and their impact on the whole transcriptome. In an attempt to identify such differences, we assessed KRAS mutation status and its consequences on the cancer transcriptome by analyzing 65 locally advanced rectal cancers and their corresponding normal mucosa. With this considerably large dataset, we were in a position to screen for KRAS mutation dependent transcriptional consequences on downstream targets.
Impact of KRAS on the Rectal Cancer Transcriptome
Forty-seven percent of rectal carcinomas in our dataset revealed activating KRAS mutations (Gaedcke et al., 2010). Our data are therefore congruent with published data on the prevalence of the mutations. Comparison of KRAS mutant and wild-type rectal cancers revealed thirteen differentially expressed genes which were always, and with high-fold and high-significance differentially expressed between tumors with and without mutations. All genes were upregulated in the mutant tumors. Relatively little is known about most of these genes. Only for one of the upregulated genes an association to the MAPK pathway had been reported previously: MAP-2 kinase phosphatase (DUSP4) has previously been reported to be upregulated in various cancer types (Yip-Schneider et al., 2001; Wang et al., 2003). Our own data confirm the significant upregulation of DUSP4 in rectal cancer (P=10−21).
Khambata-Ford and colleagues (2007) investigated the impact of DUSP4 expression levels on outcome of patients with metastatic colorectal cancer. Gene expression profiling from 80 patients with metastatic colorectal carcinomas enrolled in a Cetuximab monotherapy trial revealed DUSP4 as one of the top resistance markers. Since KRAS mutations are currently considered as some of the most relevant resistance markers for treatment failure, overexpression of DUSP4 within the same group confirms the finding of a mutation dependent regulation. Lung cancers with EGFR mutations respond well to Cetuximab, and it was recently shown that DUSP4 is downregulated in those tumors. The overexpression of DUSP4 in rectal cancer in the presence of KRAS mutations which are resistant to Cetuximab is therefore a possible explanation for the mode of action. DUSP4 expression levels could therefore serve as biomarkers for treatment stratification therapies with Cetuximab.
In cDNA microarray analysis, the gene LEMD1 (LEM domain-containing 1) has previously been found to be upregulated in colorectal cancer and was shown to be a member of the cancer-testis antigens (Yuki et al., 2004). TEGT is a regulator of apoptosis (Grzmil et al., 2006), SERPINB1 was reported to be upregulated in oral cancer (Tseng et al., 2009) and SMYD3, a histone methyltransferase, is involved in the proliferation of cancer cells (Hamamoto et al., 2004, 2006; Zou et al., 2009). Nine of the thirteen genes showed connections when analyzed with IPA which suggests a functional relationship between these genes and could explain why they are jointly deregulated as a consequence of KRAS mutation (Supplementary Fig. 3).
We queried the relevance of identifying KRAS-related genes for clinical considerations. For instance, if resistance to Cetuximab as a consequence of KRAS mutation depends on KRAS regulated genes one could hypothesize that transcriptional modification of these genes would restore the sensitivity of colorectal carcinomas to Cetuximab. DUSP4 is a good example because low levels of DUSP4 sensitize tumors to Cetuximab and decreasing DUSP4 levels using the agent PD98059 could therefore be used in treatment of KRAS mutated tumors in combination with Cetuximab (Yip-Schneider et al., 2001). Another potential target for such an intervention would be SMYD3, another one of the differentially expressed genes in our dataset, because the drug Novobiocin lowers the expression level of this gene (Luo et al., 2009).
Identification of Novel Rectal Cancer Tumor markers
The most stringent criteria to select differentially expressed genes was introduced to reveal new tumor markers (Min >< Max rule). This rule filtered genes that are always higher expressed in any of the tumors compared to all mucosa samples, or vice versa. Of the 19 features identified eleven were higher and eight were lower expressed in the tumor. The expression levels of 13 of these genes were validated using qPCR. As in previous validation experiments, the results between arrays and qPCR were extremely reproducible (R=0.98) attesting to the robustness of either methodology. Within the validated genes ETV4 (Liu et al., 2007), ROR1 (Katoh, 2005) or CLDN1 (Kinugasa et al., 2007; Huo et al., 2009), C20orf20 (Cai et al., 2003; Carvalho et al., 2009) and FUT1 (Hallouin et al., 1999) have already been linked to colorectal cancer. Others are known to play a role in carcinogenesis in general, such as TRIB3 (Du et al., 2003), ACAN (Skandalis et al., 2006; Stylianou et al., 2008) and CEP72 (Kang et al., 2008) but have not been directly associated with colorectal cancer whereas an involvement of MYOT, ENDOD1 and ANO5 in epithelial tumorigenesis is a novel finding. All genes that we previously found differentially expressed or overexpressed in a more limited dataset of colorectal cancer were confirmed to be deregulated in the same direction (Grade et al., 2006, 2007).
Interestingly, when we analyzed these 17 genes using IPA we found 14 of them operating in one network (Fig. 4). This network was connected through TNF, TGFB1, ERK, and the NFkB complex which highlights the central role that these signaling pathways assume in CRC (Glick, 2004; Fang and Richardson, 2005; Zhang et al., 2007; Balkwill, 2009). Expanding the numbers of genes for pathway analysis we used the differentially expressed genes based on a FC >2. The main interceptions within the networks like MMP3, KRAS, p16 or ERK again confirm the relevance of the retrieved genes.
In summary, this is the most comprehensive and systematic gene expression study of rectal carcinomas and normal mucosa. Using matched samples rather than a normal reference pool was important to retrieve more differentially expressed genes. In addition, this is the first systematic exploration of gene expression changes that are a consequence of activating KRAS mutations in rectal cancer. We identified DUSP4 and SMYD3 as attractive targets for a potential combination therapy of patients with Cetuximab resistant tumors.
Supplementary Material
Supplementary figure 2. Top five networks (from an IPA analysis of genes differentially expressed between tumor and mucosa based on a fold change larger than two (n=3,174).
ACKNOWLEDGEMENTS
The authors are indebted to Ms. Jessica Eggert and Mr. Chan Rong Lai for excellent technical support. We also thank Dr. Gabriela Salinas-Riester and Mr. Lennart Opitz for help with microarray hybridizations, Drs. Laszlo Füzesi and Hilka Rothe for pathology reports, and Dr. Reinhard Ebner for critically reading the manuscript. The authors are indebted to Drs. Nikolas Stoecklein and Achim Weber for their review of this manuscript.
Supported by the intramural research program of the National Institutes of Health, National Cancer Institute, and by the Deutsche Forschungsgemeinschaft (KFO 179).
Footnotes
Accession Number The gene expression data have been deposited in the NCBI Gene Expression Omnibus (GSE20842).
REFERENCES
- Alon U, Barkai N, Notterman DA, Gish K, Ybarra S, Mack D, Levine AJ. Broad patterns of gene expression revealed by clustering analysis of tumor and normal colon tissues probed by oligonucleotide arrays. Proc Natl Acad Sci U S A. 1999;96:6745–6750. doi: 10.1073/pnas.96.12.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldus SE, Schaefer KL, Engers R, Hartleb D, Stoecklein NH, Gabbert HE. Prevalence and heterogeneity of KRAS, BRAF, and PIK3CA mutations in primary colorectal adenocarcinomas and their corresponding metastases. Clin Cancer Res. 2010;16:790–799. doi: 10.1158/1078-0432.CCR-09-2446. [DOI] [PubMed] [Google Scholar]
- Balkwill F. Tumour necrosis factor and cancer. Nat Rev Cancer. 2009;9:361–371. doi: 10.1038/nrc2628. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statist. Soc. B. 1995;57:289–300. [Google Scholar]
- Bianchini M, Levy E, Zucchini C, Pinski V, Macagno C, De Sanctis P, Valvassori L, Carinci P, Mordoh J. Comparative study of gene expression by cDNA microarray in human colorectal cancer tissues and normal mucosa. Int J Oncol. 2006;29:83–94. [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Brink M, de Goeij AF, Weijenberg MP, Roemen GM, Lentjes MH, Pachen MM, Smits KM, de Bruine AP, Goldbohm RA, van den Brandt PA. K-ras oncogene mutations in sporadic colorectal cancer in The Netherlands Cohort Study. Carcinogenesis. 2003;24:703–710. doi: 10.1093/carcin/bgg009. [DOI] [PubMed] [Google Scholar]
- Cai Y, Jin J, Tomomori-Sato C, Sato S, Sorokina I, Parmely TJ, Conaway RC, Conaway JW. Identification of new subunits of the multiprotein mammalian TRRAP/TIP60-containing histone acetyltransferase complex. J Biol Chem. 2003;278:42733–42736. doi: 10.1074/jbc.C300389200. [DOI] [PubMed] [Google Scholar]
- Carvalho B, Postma C, Mongera S, Hopmans E, Diskin S, van de Wiel MA, van Criekinge W, Thas O, Matthai A, Cuesta MA, Droste JSTS, Craanen M, Schrock E, Ylstra B, Meijer GA. Multiple putative oncogenes at the chromosome 20q amplicon contribute to colorectal adenoma to carcinoma progression. Gut. 2009;58:79–89. doi: 10.1136/gut.2007.143065. [DOI] [PubMed] [Google Scholar]
- De Roock W, Piessevaux H, De Schutter J, Janssens M, De Hertogh G, Personeni N, Biesmans B, Van Laethem JL, Peeters M, Humblet Y, Van Cutsem E, Tejpar S. KRAS wild-type state predicts survival and is associated to early radiological response in metastatic colorectal cancer treated with cetuximab. Ann Oncol. 2008;19:508–515. doi: 10.1093/annonc/mdm496. [DOI] [PubMed] [Google Scholar]
- Di Fiore F, Blanchard F, Charbonnier F, Le Pessot F, Lamy A, Galais MP, Bastit L, Killian A, Sesboue R, Tuech JJ, Queuniet AM, Paillot B, Sabourin JC, Michot F, Michel P, Frebourg T. Clinical relevance of KRAS mutation detection in metastatic colorectal cancer treated by Cetuximab plus chemotherapy. Br J Cancer. 2007;96(8):1166–1169. doi: 10.1038/sj.bjc.6603685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du KY, Herzig S, Kulkarni RN, Montminy M. TRB3: A tribbles homolog that inhibits Akt/PKB activation by insulin in liver. Science. 2003;300:1574–1577. doi: 10.1126/science.1079817. [DOI] [PubMed] [Google Scholar]
- Fang JY, Richardson BC. The MAPK signalling pathways and colorectal cancer. Lancet Oncol. 2005;6:322–327. doi: 10.1016/S1470-2045(05)70168-6. [DOI] [PubMed] [Google Scholar]
- Fisher RA. Applications of ‘Student‘s’ Distribution. Metron. 1925;5:90–104. [Google Scholar]
- Friederichs J, Rosenberg R, Mages J, Janssen KP, Maeckl C, Nekarda H, Holzmann B, Siewert JR. Gene expression profiles of different clinical stages of colorectal carcinoma: toward a molecular genetic understanding of tumor progression. Int J Colorectal Dis. 2005;20:391–402. doi: 10.1007/s00384-004-0722-1. [DOI] [PubMed] [Google Scholar]
- Gaedcke J, Grade M, Jung K, Schirmer M, Jo P, Obermeyer C, Wolff HA, Herrmann MK, Beissbarth T, Becker H, Ried T, Ghadimi M. KRAS and BRAF mutations in patients with rectal cancer treated with preoperative chemoradiotherapy. Radiother Oncol. 2010;94:76–81. doi: 10.1016/j.radonc.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick AB. TGFbeta1, back to the future: revisiting its role as a transforming growth factor. Cancer Biol Ther. 2004;3:276–283. doi: 10.4161/cbt.3.3.849. [DOI] [PubMed] [Google Scholar]
- Grade M, Ghadimi BM, Varma S, Simon R, Wangsa D, Barenboim-Stapleton L, Liersch T, Becker H, Ried T, Difilippantonio MJ. Aneuploidy-dependent massive deregulation of the cellular transcriptome and apparent divergence of the Wnt/beta-catenin signaling pathway in human rectal carcinomas. Cancer Res. 2006;66(1):267–282. doi: 10.1158/0008-5472.CAN-05-2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grade M, Hormann P, Becker S, Hummon AB, Wangsa D, Varma S, Simon R, Liersch T, Becker H, Difilippantonio MJ, Ghadimi BM, Ried T. Gene expression profiling reveals a massive, aneuploidy-dependent transcriptional deregulation and distinct differences between lymph node-negative and lymph node-positive colon carcinomas. Cancer Res. 2007;67:41–56. doi: 10.1158/0008-5472.CAN-06-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grade M, Hummon AB, Camps J, Emons G, Spitzner M, Gaedcke J, Hoermann P, Ebner R, Becker H, Difilippantonio MJ, Ghadimi BM, Beissbarth T, Caplan NJ, Ried T. A genomic strategy for the functional validation of colorectal cancer genes identifies potential therapeutic targets. Int J Cancer. 2010 May 12; doi: 10.1002/ijc.25453. 2010. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzmil M, Kaulfuss S, Thelen P, Hemmerlein B, Schweyer S, Obenauer S, Kang TW, Burfeind P. Expression and functional analysis of Bax inhibitor-1 in human breast cancer cells. J Pathol. 2006;208:340–349. doi: 10.1002/path.1902. [DOI] [PubMed] [Google Scholar]
- Hallouin F, Goupille C, Rocher J, Le Pendu J. A rat experimental model for the design of vaccines against tumor associated antigens Tn and Sialyl-Tn. Glycoconjugate Journal. 1999;16:681–684. doi: 10.1023/a:1007199124074. [DOI] [PubMed] [Google Scholar]
- Hamamoto R, Furukawa Y, Morita M, Iimura Y, Silva FP, Li M, Yagyu R, Nakamura Y. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat Cell Biol. 2004;6:731–740. doi: 10.1038/ncb1151. [DOI] [PubMed] [Google Scholar]
- Hamamoto R, Silva FP, Tsuge M, Nishidate T, Katagiri T, Nakamura Y, Furukawa Y. Enhanced SMYD3 expression is essential for the growth of breast cancer cells. Cancer Sci. 2006;97:113–118. doi: 10.1111/j.1349-7006.2006.00146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo Q, Kinugasa T, Wang L, Huang J, Zhao J, Shibaguchi H, Kuroki M, Tanaka T, Yamashita Y, Nabeshima K, Iwasaki H, Kuroki M. Claudin-1 protein is a major factor involved in the tumorigenesis of colorectal cancer. Anticancer Research. 2009;29:851–857. [PubMed] [Google Scholar]
- Kang GH, Lee S, Cho NY, Gandamihardja T, Long TI, Weisenberger DJ, Campan M, Laird PW. DNA methylation profiles of gastric carcinoma characterized by quantitative DNA methylation analysis. Lab Invest. 2008;88:161–170. doi: 10.1038/labinvest.3700707. [DOI] [PubMed] [Google Scholar]
- Karapetis CS, Khambata-Ford S, Jonker DJ, O’Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S, Price TJ, Shepherd L, Au HJ, Langer C, Moore MJ, Zalcberg JR. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol. 2008;9:517–531. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M. Comparative genomics on ROR1 and ROR2 orthologs. Oncol Rep. 2005;14:1381–1384. doi: 10.3892/or.14.5.1381. [DOI] [PubMed] [Google Scholar]
- Khambata-Ford S, Garrett CR, Meropol NJ, Basik M, Harbison CT, Wu S, Wong TW, Huang X, Takimoto CH, Godwin AK, Tan BR, Krishnamurthi SS, Burris HA, 3rd, Poplin EA, Hidalgo M, Baselga J, Clark EA, Mauro DJ. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J Clin Oncol. 2007;25:3230–3237. doi: 10.1200/JCO.2006.10.5437. [DOI] [PubMed] [Google Scholar]
- Kinugasa T, Huo Q, Higash D, Shibaguchi H, Kuroki M, Tanaka T, Futami K, Yamashita Y, Hachimine K, Maekawa S, Nabeshima K, Iwasaki H, Kuroki M. Selective up-regulation of claudin-1 and claudin-2 in colorectal cancer. Anticancer Research. 2007;27:3729–3734. [PubMed] [Google Scholar]
- Lievre A, Bachet JB, Boige V, Cayre A, Le Corre D, Buc E, Ychou M, Bouche O, Landi B, Louvet C, Andre T, Bibeau F, Diebold MD, Rougier P, Ducreux M, Tomasic G, Emile JF, Penault-Llorca F, Laurent-Puig P. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol. 2008;26:374–379. doi: 10.1200/JCO.2007.12.5906. [DOI] [PubMed] [Google Scholar]
- Lievre A, Bachet JB, Le Corre D, Boige V, Landi B, Emile JF, Cote JF, Tomasic G, Penna C, Ducreux M, Rougier P, Penault-Llorca F, Laurent-Puig P. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992–3995. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- Liu HY, Zhou B, Wang L, Li Y, Zhou ZG, Sun XF, Xu B, Zeng YJ, Song JM, Luo HZ, Yang L. Association of E1AF mRNA expression with tumor progression and matrilysin in human rectal cancer. Oncology. 2007;73:384–388. doi: 10.1159/000136158. [DOI] [PubMed] [Google Scholar]
- Luo XG, Zou JN, Wang SZ, Zhang TC, Xi T. Novobiocin decreases SMYD3 expression and inhibits the migration of MDA-MB-231 human breast cancer cells. IUBMB Life. 2009;62:194–199. doi: 10.1002/iub.288. [DOI] [PubMed] [Google Scholar]
- Platzer P, Upender MB, Wilson K, Willis J, Lutterbaugh J, Nosrati A, Willson JK, Mack D, Ried T, Markowitz S. Silence of chromosomal amplifications in colon cancer. Cancer Res. 2002;62:1134–1138. [PubMed] [Google Scholar]
- Skandalis SS, Theocharis AD, Vynios DH, Papageorgakopoulou N, Hjerpe A, Karamanos NK, Theocharis DA. Cartilage aggrecan undergoes significant compositional and structural alterations during laryngeal cancer. Biochim Biophys Acta. 2006;1760:1046–1053. doi: 10.1016/j.bbagen.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- Stylianou M, Skandalis SS, Papadas TA, Mastronikolis NS, Theocharis DA, Papageorgakopoulou N, Vynios DH. Stage-related decorin and versican expression in human laryngeal cancer. Anticancer Res. 2008;28:245–251. [PubMed] [Google Scholar]
- Tseng MY, Liu SY, Chen HR, Wu YJ, Chiu CC, Chan PT, Chiang WF, Liu YC, Lu CY, Jou YS, Chen JY. Serine protease inhibitor (SERPIN) B1 promotes oral cancer cell motility and is over-expressed in invasive oral squamous cell carcinoma. Oral Oncol. 2009;45:771–776. doi: 10.1016/j.oraloncology.2008.11.013. [DOI] [PubMed] [Google Scholar]
- Wang HY, Cheng Z, Malbon CC. Overexpression of mitogen-activated protein kinase phosphatases MKP1, MKP2 in human breast cancer. Cancer Lett. 2003;191:229–237. doi: 10.1016/s0304-3835(02)00612-2. [DOI] [PubMed] [Google Scholar]
- Yip-Schneider MT, Lin A, Marshall MS. Pancreatic tumor cells with mutant K-ras suppress ERK activity by MEK-dependent induction of MAP kinase phosphatase-2. Biochem Biophys Res Commun. 2001;280:992–997. doi: 10.1006/bbrc.2001.4243. [DOI] [PubMed] [Google Scholar]
- Yuki D, Lin YM, Fujii Y, Nakamura Y, Furukawa Y. Isolation of LEM domain-containing 1, a novel testis-specific gene expressed in colorectal cancers. Oncol Rep. 2004;12:275–280. [PubMed] [Google Scholar]
- Zhang X, Jin B, Huang C. The PI3K/Akt pathway and its downstream transcriptional factors as targets for chemoprevention. Curr Cancer Drug Targets. 2007;7:305–316. doi: 10.2174/156800907780809741. [DOI] [PubMed] [Google Scholar]
- Zou JN, Wang SZ, Yang JS, Luo XG, Xie JH, Xi T. Knockdown of SMYD3 by RNA interference down-regulates c-Met expression and inhibits cells migration and invasion induced by HGF. Cancer Lett. 2009;280:78–85. doi: 10.1016/j.canlet.2009.02.015. [DOI] [PubMed] [Google Scholar]
- Zou TT, Selaru FM, Xu Y, Shustova V, Yin J, Mori Y, Shibata D, Sato F, Wang S, Olaru A, Deacu E, Liu TC, Abraham JM, Meltzer SJ. Application of cDNA microarrays to generate a molecular taxonomy capable of distinguishing between colon cancer and normal colon. Oncogene. 2002;21:4855–4862. doi: 10.1038/sj.onc.1205613. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure 2. Top five networks (from an IPA analysis of genes differentially expressed between tumor and mucosa based on a fold change larger than two (n=3,174).