Abstract
Protein aggregation is a major problem for biopharmaceuticals. While the control of aggregation is critically important for the future of protein pharmaceuticals, mechanisms of aggregate assembly, particularly the role that structure plays, are still poorly understood. Increasing evidence indicates that partially folded intermediates critically influence the aggregation pathway. We have previously reported the use of the basic leucine zipper (bZIP) domain of Activating Transcription Factor 5 (ATF5) as a partially folded model system to investigate protein aggregation. This domain contains three regions with differing structural propensity: a N-terminal polybasic region, a central helical leucine zipper region, and a C-terminal extended valine zipper region. Additionally, a centrally positioned cysteine residue readily forms an intermolecular disulfide bond that reduces aggregation. Computational analysis of ATF5 predicts that the valine zipper region facilitates self-association. Here we test this hypothesis using a truncated mutant lacking the C-terminal valine zipper region. We compare the structure and aggregation of this mutant to the wild-type (WT) form under both reducing and non-reducing conditions. Our data indicate that removal of this region results in a loss of alpha-helical structure in the leucine zipper and a change in the mechanism of self-association. The mutant form displays increased association at low temperature but improved resistance to thermally induced aggregation.
Keywords: ATF5, ATFx, bZIP, zipper, helix, aggregation, stability
Introduction
Protein aggregation is a major concern of the biopharmaceutical industry.1 The aggregation of protein products may not only lead to a decrease in drug potency, but aggregated species can also display enhanced immunogenicity.2 Consequently, controlling the formation of protein aggregates in a pharmaceutical formulation is of great importance. The difficulty in preventing aggregation partially stems from our lack of understanding of the mechanism by which aggregates form. In particular, the role that protein structure plays in this process is poorly understood. Increasing evidence suggests that partially folded protein intermediates critically influence the aggregation pathway; however, this relationship remains ill-defined.3
Two general categories of protein aggregates have been reported in the literature: amyloid fibrils and amorphous aggregates.4 Fibrillar aggregates have received much attention due to their association with a growing number of serious disease states for which limited treatment options exist.4c, 5 They are characterized by a cross-beta structure, in which the beta strands align in a highly organized format along the long-axis of the fibril.4c, 5a, 6 Amorphous aggregates are often associated with recombinant protein production and purification in vitro; however increasing evidence indicates that a number of proteins can develop into both fibrillar and amorphous aggregates in vitro.7 The nature of the aggregated species is highly dependent upon protein sequence and solution conditions that control protein conformation.7 Amorphous aggregates are so named because unlike fibrillar aggregates, they do not display long-range order.3h, 4a, 4d However, these aggregates do possess structure and, typically, an increase in intermolecular beta-sheet content is observed compared to the native state.3h, 4a, 8 This is not true in all cases, and the development of alternative structures, including alpha helix, has been reported in aggregates.3g, 9 It is currently unclear how the amino acid sequence and higher order structure directs aggregate assembly and organization.
We have previously reported the production of the basic leucine zipper (bZIP) domain of activating transcription factor 5 (ATF5), a notable protein target for treatment of glioblastoma.10 Because of its propensity to aggregate in vitro, we have used ATF5 as a model system to investigate protein aggregation.11 Our previous data indicate that disulfide bond formation prevents the formation of large amorphous protein aggregates through improved retention of alpha-helical structure.11 Computational analysis predicted that the C-terminal region, which contains a series of three valine residues (V257, V264 and V271) in lieu of the conserved leucine residues typically observed in these positions, most likely facilitates aggregation.11 We suggested that disulfide bond formation minimized solution exposure of this C-terminal region through improved structural retention, resulting in diminished protein aggregation.11 The work presented here further characterizes the contribution of this C-terminal region to the aggregation of ATF5.
A mutant form of the ATF5 protein that lacks the C-terminal region (residues 257–282) was created using site-directed mutagenesis. This variant (V257STOP) was created via the insertion of a stop codon immediately preceding the first valine residue (V257) in this region. The structure of the purified mutant form of ATF5 was evaluated using circular dichroism spectroscopy (CD), Fourier transform infrared (FTIR) spectroscopy and two-dimensional nuclear magnetic resonance (NMR) spectroscopy. The results were compared with that of the wild-type (WT) form of ATF5 to determine the effect of C-terminal truncation on protein structure. The extent of thermal aggregation of both proteins was monitored using static light scattering (SLS) analyses and dynamic light scattering (DLS) analyses. Structural changes that occurred during the aggregation process were evaluated using FTIR analysis. These data were collectively used to compare the formation and growth of protein aggregates of both the WT and V257STOP forms of ATF5 and identify correlations between protein structure and aggregate development. Our data indicate that removal of the C-terminal region of ATF5 changes the structure of the protein and the mechanism of thermally induced protein self-association.
Materials and Methods
Protein Expression and Purification
The cDNA of ATF5 was obtained through ATCC (MGC-842), and the bZIP domain was PCR amplified and inserted into a Novagen (San Diego, CA) pET-42b vector as described previously.10a The V257STOP truncated mutant was generated using site-directed mutagenesis to insert a stop codon at the valine in position 257. Mutagenesis was performed by GenScript USA (Piscataway, NJ). Plasmids were transformed into Novagen BL21(DE3) E. coli for expression. Cell growth, lysis and purification were performed using the procedure described previously with the following modification.10a Unlike wild type, the truncated mutant was present largely in the soluble fraction following cell lysis and centrifugation. This fraction was heated at 60 °C for 1 hour to precipitate the majority of cellular proteins. The mutant remained soluble under these conditions. To ensure that any aggregates formed during this purification step were eliminated the protein was denatured and subsequently subjected to the same refolding procedure as previously described for the WT.10a Protein concentrations were determined using a standard Bradford assay. The purity of ATF5 was determined using densitometry of Coomassie stained SDS-PAGE gels.10a This method was also used to confirm intermolecular disulfide bond formation using non-reducing conditions. Isotopic labeling for two-dimensional NMR was accomplished during cellular expression with the use of 15N-ammonium chloride (>99% N-15; Isotec, St. Louis, MO).
Circular Dichroism Spectroscopy
Circular dichroism (CD) spectra were acquired with an Applied Photophysics Chirascan (Leatherhead, UK) equipped with a Peltier-type temperature controller and a four-position sample holder. ATF5 WT and V257STOP samples were prepared at a final protein concentration of 10 μM in 20 mM MES buffer at pH 6.0. Scans were performed at 10 °C in a 0.1-cm path length cell from 260 to 190 nm. The selected bandwidth was 4 nm, the data pitch was 1 nm and the sampling time was 0.5 sec. Samples were prepared and analyzed in duplicate. Raw data were converted into units of molar ellipticity using the standard formula, [θ] = θ*M*10/C*l, where [θ] is the molar ellipticity, θ is the ellipticy in mdeg, M is the molecular weight in mg/mmol, C is the concentration in mg/mL and l is the path length in cm. Multiplying by 10 facilitates the conversion from millimoles to decimoles, resulting in the final units of deg*cm2*dmol−1. The resulting average and standard deviations were plotted.
Fourier Transform Infrared Spectroscopy
Absorption spectra (256 scans) were collected at a 5 mg/mL concentration from 900 to 4000 cm−1 at a resolution of 4 cm−1 on a Bruker (Billerica, MA) Tensor 27 Fourier transform infrared spectrophotometer instrument using a Harrick Scientific Products (Pleasentville, NY) BioATR Cell II sample apparatus and a Thermo Scientific (Waltham, MA) Haake DC30-K20 refrigerated circulator bath under constant nitrogen purge. The spectrophotometer was equipped with a KBr beam splitter and a liquid-nitrogen-cooled mercury cadmium telluride (MCT) detector. Reference spectra consisting of 20 mM MES buffer pH 6.0 with or without 10 mM dithiothreitol (DTT) were collected prior to each sample solution and subtracted as background. Resultant spectra were first processed using OPUS 6.5 software (Bruker Optics). A correction was performed to minimize absorption interference from atmospheric water and carbon dioxide. The minimum and maximum values within the amide I region (1600–1700 cm−1) were normalized. Data were then further processed using GRAMS/AI 8.0 spectroscopy software (Thermo Fisher Scientific). A baseline correction was performed, and the data were smoothed using a five-point Savitsky-Golay function. Fourier self-deconvolution was performed and the data were fitted by inserting individual Gausssian bands to match the second derivative peak position. Secondary structure assignments were made based upon reference spectra reported in the literature.12
The temperature was increased from 10 °C to 90 °C to facilitate thermal analysis of stability. Scans (256) were collected every 2.5 °C at a ramping rate of 2.5 °C per minute. A reference melt consisting of 20 mM MES buffer pH 6.0 with or without 10 mM dithiothreitol (DTT) was collected immediately prior to sample analysis and subtracted as background. The resultant spectra were processed using OPUS 6.5 software. A correction was performed to minimize absorption interference from atmospheric water and carbon dioxide. The minimum and maximum values within the amide I region (1600–1700 cm−1) were normalized. The absorbance spectra in the Amide I region were plotted as a function of temperature.
Nuclear Magnetic Resonance Spectroscopy
Two-dimensional 1H-15N heteronuclear single quantum coherence (HSQC) spectra were recorded at 25 °C using a Bruker AVANCE 800 MHz spectrometer equipped with a triple-resonance CRYO-probe with pulse field gradients. Samples were prepared at a concentration of 1 mM in 20 mM MES buffer pH 6.0, containing 5% D2O. Water suppression was accomplished using flip-back pulses. Data were acquired in 8 scans with 1024 points in 1H and 128* increments in 15N. 1H chemical shifts were referenced with respect to an external DSS standard in D2O.13 Indirect referencing relative to 1H was determined for 15N, assuming a ratio of 15N/1H = 0.101329118. Data were processed using NMRPipe and Sparky software.14
Static Light Scattering
Static light scattering (SLS) data were acquired with a Photon Technology International (PTI) spectrofluorometer (Lawrenceville, NJ) equipped with a Peltier-type temperature controller and a four-position sample holder. Samples were prepared at a final protein concentration of 100 μM in 20 mM MES buffer at pH 6.0 in the presence or absence of 10 mM DTT. 10 μM and 1 μM samples were analyzed to examine the effect of protein concentration on aggregation. The intensity of scattered light was measured as a function of temperature and detected at an angle of 90° to the light source by a photomultiplier tube. An arc lamp white light source was used in all cases. The excitation wavelength was set at 350 nm. The emission wavelength range was set from 300 nm to 400 nm. Spectra were obtained following a 5-min equilibration period at each temperature. Data were collected every 2.5 °C between the temperature range of 4 to 81.5 °C. The background was subtracted from each data point based on a blank containing the corresponding buffer solution. Two samples were prepared and analyzed for each set of solution conditions. The resulting average and standard deviations were plotted.
Dynamic Light Scattering
The average hydrodynamic radius was monitored as a function of temperature using a Wyatt Technology Corporation (Santa Barbara, CA) DynaPro Plate Reader Plus instrument. ATF5 WT and V257STOP samples were prepared in 20 mM MES buffer pH 6.0 at a final concentration of 100 μM in the presence or absence of 10 mM DTT. Samples containing the isolated V257STOP aggregate or monomer species were prepared at a 40 μM concentration in phosphate buffered saline (PBS) at pH 7.4. All samples were filtered before analysis using a 0.2 μm Anotop 10 Whatman (Piscataway, NJ) 10 mm inorganic syringe filter. A 384-well Corning (Corning, NY) polystyrene plate was used for these experiments. Data were obtained in three consecutive acquisition periods, each 15 seconds in length, and averaged. Acquisitions were obtained every 2.5 °C over a temperature range of 10 °C to 67.5 °C with a ramping rate of 1 °C/min. A constant nitrogen purge at 60 cc/min and 60 psi was performed to prevent condensation. Average hydrodynamic radii and polydispersity indices were calculated using cumulant analysis.15 Two samples were prepared and analyzed for each set of solution conditions. The resulting average and standard deviations were plotted. Particle distribution was evaluated using a nonnegative least squares regularization method of analysis.16 A representative data set for each form of ATF5 was plotted.
Thioflavin T Fluorescence
Fluorescence data were acquired with a Photon Technology International (PTI) spectrofluorometer (Lawrenceville, NJ) equipped with a Peltier-type temperature controller and a four-position sample holder. Samples were prepared at a final protein concentration of 100 μM in 20 mM MES buffer at pH 6.0 in the presence or absence of 10 mM DTT. Thioflavin T dye (Sigma-Aldrich; St. Loius, MO) was added at a final concentration of 10 μM. Fluorescence intensity was measured as a function of temperature and detected at an angle of 90° to the light source by a photomultiplier tube. An arc lamp white light source was used in all cases. The excitation wavelength was set at 450 nm. The emission wavelength range was set from 400 nm to 500 nm. Spectra were obtained following a 5-min equilibration period at each temperature. Data were collected every 2.5 °C between the temperature range of 10 to 80 °C. The background was subtracted from each data point based on a blank containing the corresponding buffer solution. Two samples were prepared and analyzed for each set of solution conditions. The resulting average and standard deviations were plotted.
Results
Effects of C-terminal Truncation on ATF5 Structure
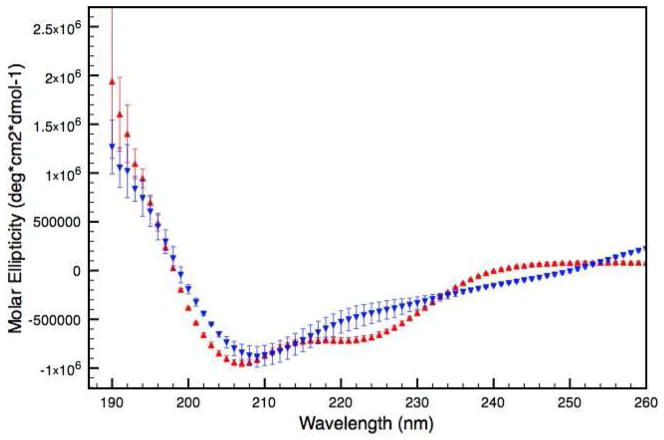
Circular dichroism (CD) spectroscopy was performed to evaluate differences in secondary structure between the wild type (WT) and the truncated mutant (V257STOP) forms of ATF5. The mutant protein displays a CD absorption spectrum that is distinct from the WT form (Figure 1). The WT possesses a double-minima absorption pattern with negative peaks at 222 nm and 208 nm. This pattern is indicative of α-helical protein structure.17 Given that the extended C-terminal portion of the protein was removed to make the V257STOP protein, one would expect that the remaining leucine zipper region would retain helicity. The V257STOP mutant displays a single absorption minimum at 209 nm and a broad shoulder between 260 nm and 230 nm, which is not consistent with alpha-helical structure and indicates a substantial structural change has occurred in the absence of the valine zipper region. Negative absorption at 209 nm has been observed for proteins possessing β-structure.18 Additionally, it has been reported that certain types of β-bend can give negative absorption at 208 nm.19 The absorption observed between 260 nm and 230 nm could be attributed to β-turn structure, which has been demonstrated to display CD signal in this region.20
Figure 1.
CD absorption spectra of the WT (red) and the V256STOP mutant (blue) forms of ATF5 in 20 mM MES pH 6.0 at 4 °C.
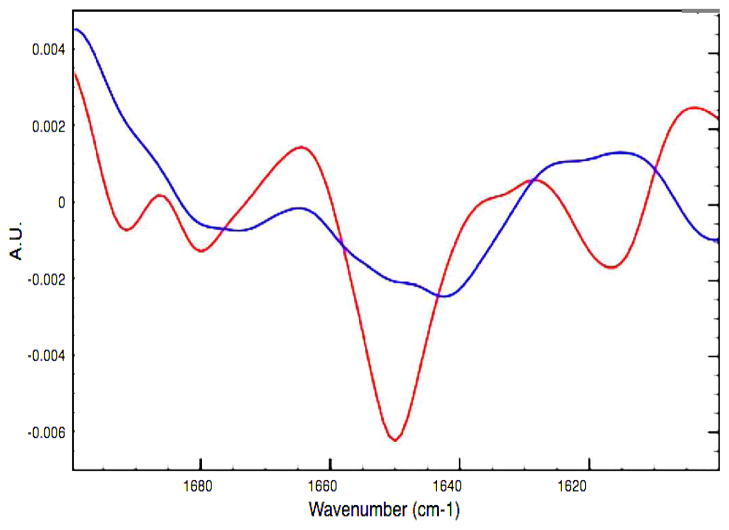
Further experimental analysis of secondary structure was performed using Fourier transform infrared spectroscopy (FTIR) to better understand these structural differences. Deconvolution of FTIR absorption in the Amide I region for the WT and V257STOP forms of ATF5 also indicates that the two proteins possess distinctly different secondary structure (Figures 2 and 3). The absorption peaks observed here are broader than those typically observed for a well-structured system.21 The data for both forms of ATF5 are suggestive of a mixed state or partially folded system. The WT form displays absorption peaks at 1621 cm−1, 1650 cm−1, 1676 cm−1 and 1688 cm−1. The absorption band at 1621 cm−1 corresponds to intermolecular β-sheet structure.12a, 22 The dominant absorption peak observed at 1650 cm−1 corresponds to α-helical structure.12a, 22 The absorption peak observed at 1676 cm−1 could represent β-sheet or β-turn structure.12a, 22 Strong β-sheet absorption at low wavenumbers in the Amide I region (1620 cm−1 to 1639 cm−1) is often accompanied by weaker absorption at higher wavenumbers (1670 cm−1 to 1680 cm−1).21c, 23 It is likely, based upon the absorption observed at 1621 cm−1, that the peak at 1676 cm−1 primarily represents β-sheet structure. Lastly, the absorption peak at 1688 cm−1 is reflective of β-turn structure.12a, 22
Figure 2.
Second derivative of FTIR spectra for the WT (red) and V257STOP mutant (blue) forms of ATF5 at 10 °C.
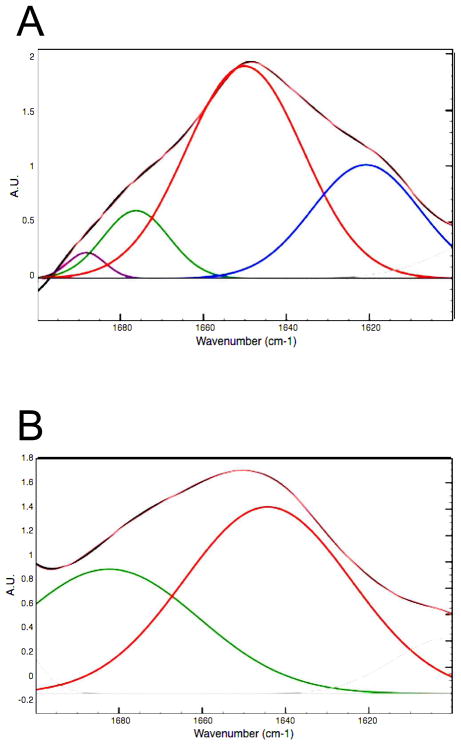
Figure 3.
Deconvolution of the FTIR absorption spectra for the WT (A) and V257STOP mutant (B) forms of ATF5 at 10 °C. Absorption in the Amide I (1600–1700 cm−1) and Amide II (1500–1600 cm−1) regions is plotted. The black outline denotes the parent absorption spectrum. The pink overlay indicates the fitted spectrum. Individual absorption peaks in the Amide I region are color-coded according the structure they represent (blue = extended structure; red = alpha helix/unordered; green = extended structure/beta turn; purple = beta turn).
The V257STOP mutant form of ATF5 displays broad absorption peaks at 1644 cm−1 and 1682 cm−1. While the fitting procedure resulted in the placement of two broad peaks for the mutant form, the second derivative data suggest the presence of two states, one of which may be α-helical (1650 cm−1) and the other unordered structure (1641 cm−1).12a, 22, 24 The broad absorption peak at 1682 cm−1 could represent β-sheet and/or β-turn structure.12a, 22 The absorption band observed at 1601 cm−1 likely corresponds to side chain vibrations and is not predictive of secondary structure.25
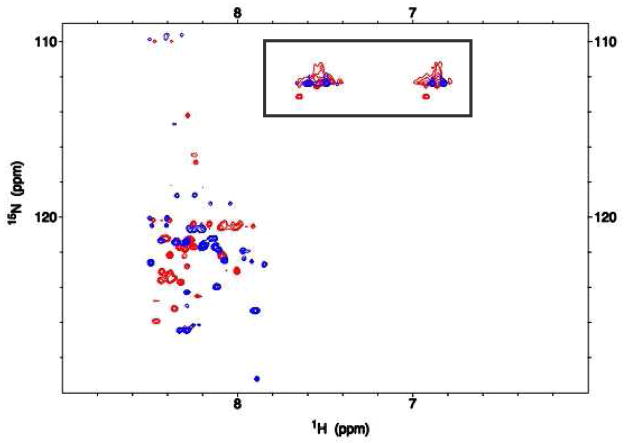
Additional studies were performed using two-dimensional NMR to compare chemical shift signal dispersion for the two forms of ATF5 (Figure 4). The 1H-15N heteronuclear single quantum coherence (HSQC) experiment used here gives an indication of overall protein fold and structure based upon signal dispersion. This experiment can also provide general insight into protein dynamics, which are reflected in and often dictate peak shape and intensity. Both forms of ATF5 display NH signals largely between 8.0 and 8.5 ppm on the 1H-axis (Figure 5). Much of the signal observed between 6.5 ppm and 7.5 ppm arises from NH2 groups in the Asn and Gln side chains, which typically have chemical shifts distinct from those of the NH groups in the amide backbone. While some overlap in backbone signal is observed between the two forms of ATF5, their chemical shifts are largely distinct, which reaffirms the substantial differences in their overall structure and/or conformational dynamics. A qualitative comparison of peak shape between the two spectra suggests that both forms of ATF5 possess well-defined structural regions as well as more conformationally labile regions. This is illustrated by the presence of both sharp, circular cross-peaks that correspond to well-defined structure and broad, misshapen cross-peaks, which are indicative of conformational exchange. The NMR signal intensity is proportional to the concentration of protein and approximately reflects the amount of protein that is present in the monomeric/dimeric state. Large molecular weight species are not observed in the standard HSQC experiment, in this case self-associated species composed of more than approximately 8–10 protomers. At the same molar concentration the signal to noise ratio observed in these experiments was 1.8-fold greater for the WT form than for the truncated mutant. This suggests that roughly 45% of the mutant protein’s NMR signal was not observed due to protein aggregation. No visible precipitation was observed in the NMR sample tube, which indicates that the aggregate species must be soluble in nature.
Figure 4.
2D 1H-15N HSQC spectrum of 1 mM 15N-labeled ATF5 WT (red peaks) and V257STOP mutant (blue peaks) in 20 mM MES buffer, pH 6.0. Peaks corresponding to NH2 side chains rather than NH backbone groups are boxed.
Figure 5.
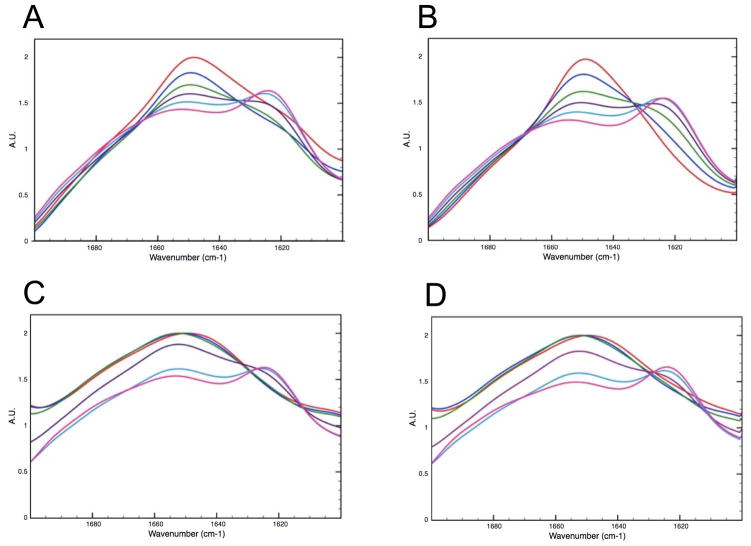
FTIR absorption in the Amide I region is shown for the WT and V257STOP forms both with and without 10 mM DTT at varying temperature. Panels A and B show data collected on the WT form in the absence (A) and presence (B) of DTT. Panels C and D show data collected on the V257STOP mutant form in the absence (C) and presence (D) of DTT. The data shown was collected at 10 °C (red), 25°C (blue), 40 °C (green), 55 °C (purple), 70 °C (light blue) and 90 °C (pink).
Analysis of Change in Structure Observed During ATF5 Aggregation
In order to investigate the structural changes that accompany aggregate growth and development, FTIR analysis was employed. This technique permits the analysis of sample both in solution and in the solid state, thereby allowing us to monitor structural changes that occur as the protein precipitates. In these experiments, FTIR absorption was monitored for both forms of ATF5, each under reducing and non-reducing conditions, as a function of temperature (Figure 5). The WT form begins to change structure between 10 °C and 25 °C. This transition is characterized by a loss in α-helical structure at 1650 cm−1 and an increase in β-sheet structure at 1620 cm−1 (See SI, Figures S1 and S2). Increased absorption at ~1620 cm−1 is typically observed during protein aggregation and is indicative of intermolecular β-sheet structure.21a, 21c, 23 In the presence of DTT, two isobestic points are observed for the WT melt (~1630 cm−1 and 1670 cm−1). This suggests that the loss of helical structure is directly linked to a gain in structure at lower and higher wavenumber. In contrast, a more complex structural transition is observed for the WT form in the absence of DTT. In this case, an initial loss of signal is observed between 10 °C and 25 °C at lower wavenumber, followed by a subsequent increase in β-sheet signal at 1620 cm−1.
Interestingly, the V257STOP mutant form of ATF5 does not change structure until the temperature rises above 40 °C. A decrease in unordered absorption at 1644 cm−1 and an increase in intermolecular β-sheet absorption at 1620 cm−1 are then observed. A loss in absorption at higher wavenumber (between 1670 cm−1 to 1700 cm−1) is also observed as a function of temperature. This suggests a reduction in β-sheet or β-turn structure. This loss continues until the temperature exceeds 70 °C, where a slight increase in absorption in this region is then observed. The structural transitions observed for the V257STOP mutant are largely unaffected by DTT addition.
Effects of C-terminal Truncation on ATF5 Aggregation
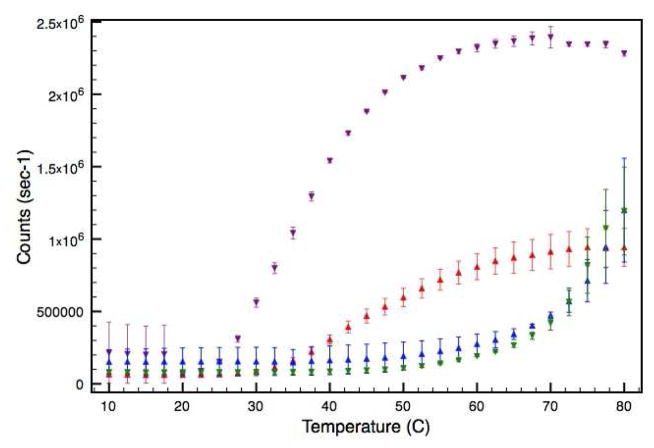
Static light scattering (SLS) analysis was performed to compare the growth of thermally induced protein aggregates between the WT and V257STOP forms of ATF5. In these experiments the average intensity of scattered light was measured in counts per second as a function of temperature under both reducing and non-reducing conditions (Figure 6). The onset of aggregation occurs at a much lower temperature for the WT form of ATF5 than for the V257STOP mutant. In the presence of DTT, WT aggregation is characterized by a sharp increase in scattering intensity between 25 °C and 30 °C. The scattering intensity eventually levels off and starts to decline at high temperature (between 70°C and 80 °C) due to the settling of large particles out of solution. This was confirmed by visual inspection of samples following the melt. In the absence of DTT, the WT onset of aggregation is shifted to higher temperature. An increase in scattering intensity is not observed until almost 40 °C. Additionally, particulate growth appears to be more gradual under these conditions.
Figure 6.
SLS analysis of the 100 μM WT and V257STOP truncated forms of ATF5 in both the absence and presence of 10 mM DTT. The intensity of scattered light is plotted as a function of temperature. Data for the WT form in the absence and presence of DTT is illustrated in red and purple, respectively. Data for the V257STOP mutant in the absence and presence DTT is illustrated in blue and green, respectively.
Aggregation onset for the V257STOP mutant form of ATF5 is not observed until the temperature exceeds 50 °C. Aggregation of this species is depicted by a gradual increase in scattering intensity observed between 60 °C and 70 °C, followed by a sharper increase in scattering intensity between 70 °C and 80 °C. Unlike WT, the aggregation of this form of ATF5 is unaffected by the addition of DTT. Additional SLS studies were performed at lower concentration (10 and 1 μM; data not shown). Notably, the WT form aggregated less readily at these lower concentrations under both reducing and non-reducing solutions conditions, whereas the particle size and onset temperature of V257STOP mutant remained unaffected by altering the concentration. Further investigation was performed using dynamic light scattering (DLS) analysis to evaluate particle size distribution as a function of temperature using the 100-μM samples.
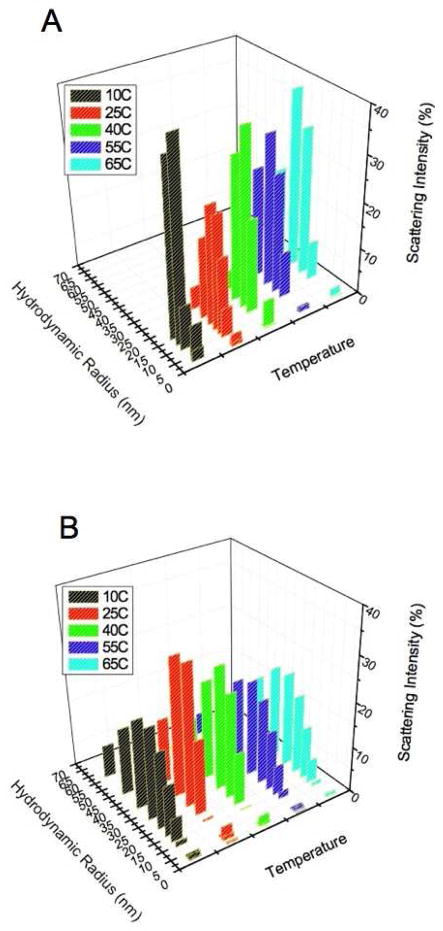
DLS uses autocorrelation analysis to measure the time-dependent fluctuations in light scattering intensity. The autocorrelation function reflects particle diffusion in solution, which is directly related to particle size. In these experiments the autocorrelation function was acquired at increasing temperatures for the two forms of ATF5 under both reducing and non-reducing solution conditions. Cumulant analysis provided an average hydrodynamic radius (RH) value and polydispersity index for the particles in solution (Figure 7).15
Figure 7.
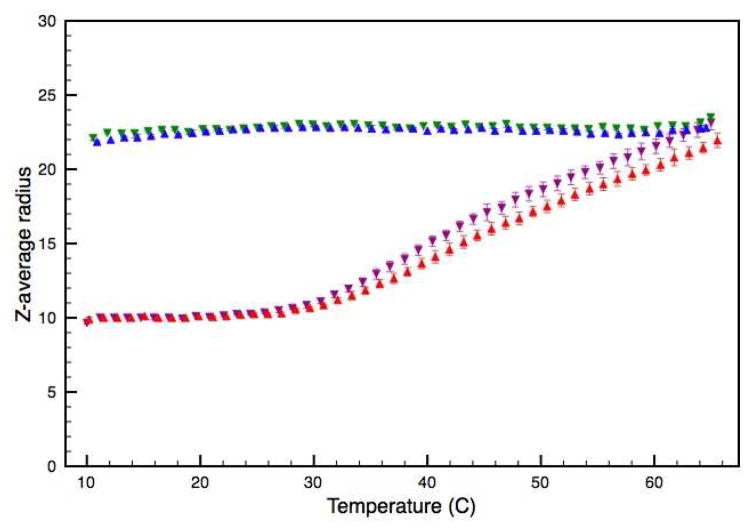
DLS analysis of the WT and V257STOP truncated forms of ATF5 in both the absence and presence of 10 mM DTT. The average hydrodynamic radii are plotted as a function of temperature. Data for the WT form in the absence and presence of DTT is illustrated in red and purple, respectively. Data for the V257STOP mutant in the absence and presence DTT is illustrated in blue and green, respectively.
The WT form of ATF5 displays an average RH of 10 nm at 10 °C under both reducing and non-reducing solution conditions. This average value is unchanged at low temperatures but begins to gradually increase in size between 25 °C and 30 °C. While this trend is the same for the WT protein under both reducing and non-reducing conditions, the average value becomes slightly larger for the reduced sample as a function of temperature. In contrast, the V257STOP mutant form of ATF5 maintains an average RH of approximately 22 nm at nearly all temperatures measured. A slight increase in average RH occurs for the V257STOP form as the temperature exceeds 60 °C. (Analysis above 65 °C was not performed, because the polystyrene plates used in these experiments cannot be used above this temperature.) Again, there is no distinction between data collected under reducing and non-reducing solution conditions for the V257STOP mutant for ATF5. When these experiments were performed at 10 and 1-μM protein concentrations, the WT again displayed concentration dependence, whereas the V257STOP mutant remained unaffected.
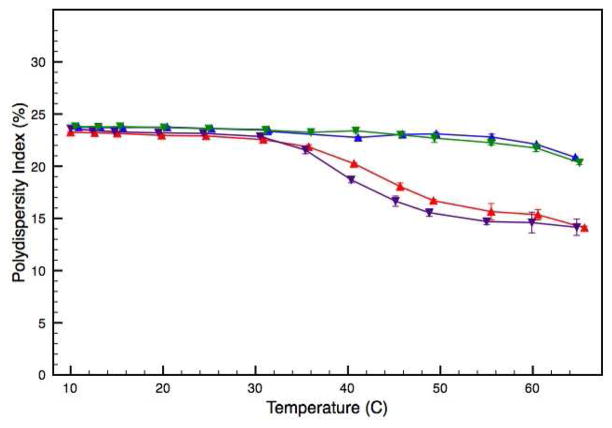
While this data provides useful information about trends associated with increasing temperature, the cumulant analysis provides an average RH value. The polydispersity index reported with the average value provides an estimation of distribution about the mean. The polydispersity values reported for both the WT and V257STOP systems suggest multimodal sample populations (Figure 8). Additionally, the average values reported here are much larger than what would be expected for ATF5 monomer or dimer. For example an IgG mononclonal antibody with an estimated molecular weight of 150 kDa typically displays an average hydrodynamic radius of approximately 12 nm. 26 This suggests that self-associated protein species are present in both the WT and V257STOP samples. Solution polydispersity decreases for both forms of ATF5 as particle size increases (Figure 8). This could be related to the development of micron-size protein aggregates that exceed the 1 μm size limit of DLS detection. As protein precipitates from solution, the polydispersity of the remaining soluble species may be lowered.
Figure 8.
DLS analysis of the WT and V257STOP truncated forms of ATF5 in both the absence and presence of 10 mM DTT. The polydispersity index is plotted as a function of temperature. Data for the WT form in the absence and presence of DTT is illustrated in red and purple, respectively. Data for the V257STOP mutant in the absence and presence DTT is illustrated in blue and green, respectively.
Regularization analysis is an alternative method for analyzing DLS data that makes no assumptions about sample homogeneity and permits characterization of multimodal sample populations.16 This analysis was performed on the dynamic light scattering data collected for both forms of ATF5 between 10 °C and 65 °C. The data is plotted as a distribution illustrating the relative intensity of light scattered as a function of particle size in solution (Figure 9). The data for both forms of ATF5 indicate heterogeneous, multimodal sample populations. The WT sample contains two sample populations, a smaller species ranging from approximately 2 nm to 4 nm in radii that likely represents monomeric or dimeric protein and a larger population that ranges from 8 nm to 35 nm in size and consists of aggregated protein species. The V257STOP sample also contains two sample populations. A smaller species ranging again from approximately 2 nm to 4 nm in radii that reflects monomeric or dimeric protein and a larger population ranging from 10 nm to 70 nm in size that is consistent with aggregated species.
Figure 9.
DLS distribution data for the (A) WT and (B) V257STOP truncated form of ATF5. The percent intensity of scattered light is displayed as a function of hydrodynamic radius.
The range in size observed for protein monomer/dimer by DLS is consistent with the NMR data, which indicates a partially folded and highly flexible monomer/dimer. DLS analysis of the aggregated species indicates the existence of particles of varying sizes. Interestingly, the V257STOP mutant form consistently displays a much broader size range of aggregated protein than the WT form. Moreover, this distribution remains relatively constant across the temperatures and concentrations measured. The lack of concentration dependence of V257STOP as determined by SLS further indicates the 22-nm, soluble aggregates are stable. This result coupled with the consistently elevated thermal transition temperature of the mutant implies the protein molecules are associated in an organized structure. This is in contrast to the WT, where the aggregated population steadily increases in size as a function of temperature. This suggests that aggregation of the WT form occurs through growth to increase the size of soluble protein aggregates to 10–35 nm.
To further probe the structure of the soluble aggregates, Thioflavin T fluorescence was measured in the presence of both forms of ATF5 as a function of temperature (See SI, Figure S3). Thioflavin T is a fluorescent dye commonly used for the detection of cross-beta structure typically found in amyloid fibrils.27 Soluble aggregates of the truncated mutant form of ATF5 were able to induce Thioflavin T fluorescence, whereas those composed of the WT protein did not induce fluorescence. The emission intensity at 480 nm for the mutant was 710,813 counts/second at 10 °C and the emission intensity for the WT at the same temperature was 875 counts/second. Despite the presence of aggregates in both samples a 1000-fold increase in emission intensity was observed for V257STOP, indicating the structures of the aggregates differ. At lower temperatures aggregates of the truncated variant exhibit amyloid-like behavior in this assay, suggesting the helical leucine zipper region in ATF5 adopts an alternative conformation to form these assemblies. In the WT protein, no fluorescence is detected, suggesting this region is protected from adopting the same intermolecular structure as V257STOP by the adjacent valine zipper.
Discussion
In this report we compare the structure and aggregation of the wild-type (WT) form of ATF5 with a truncated mutant form that lacks the valine zipper region (V257STOP). The truncated ATF5 displays a distinctly different structure than the WT form. Removal of the valine zipper region results in a loss of α-helical structure. This is interesting because the leucine zipper region of ATF5 is the region with the highest propensity to adopt α-helical structure, whereas the valine zipper region is predicted to be disordered but have beta propensity.11 Based on this information, one would expect that removal of the valine zipper region would result in an increased overall helical structure for the mutant, but the opposite effect is observed. This indicates that the valine zipper region is important for maintaining the α-helical structure of the adjacent helical region. If as predicted, the central leucine zipper region is the most structured region of ATF5, then the valine zipper region must help stabilize this helical structure. It is not clear whether structural stabilization is accomplished through backbone or side-chain interactions or both.
The dynamic light scattering data indicate that both forms of ATF5 tend to self-associate at low temperature (10 °C). The conformation of the monomer/dimer appears to be highly dynamic by NMR analysis, but it is not clear to what extent exchange occurs between these two species and the soluble aggregate at lower temperatures. Based on the NMR data acquired using low millimolar concentrations of protein, a large percentage of these samples must be present in their low molecular weight monomeric/dimeric form in order to be observed on the NMR timescale.28 The structural data collected using CD and FTIR reflect the structure of both free and self-associated protein species. Stable beta structure would result in NH chemical shifts appearing further downfield, above 8.5 ppm, and no such signal is observed in the NMR spectra. As such, it is likely that the helical signal observed for the WT form of ATF5 is largely reflective of the base protomer structure and the intermolecular β-structure observed reflects the presence of self-associated protein species.
The structural data collected on the mutant form of ATF5 indicate that the protein possesses β-sheet and β-turn structure. However, the data also suggest that the system contains non-standard structural features. Again, because ATF5 self-association occurs at low temperature, the CD and FTIR data may reflect signal from both monomeric or dimeric species and larger soluble aggregates. The complexity of this system precludes high-resolution delineation of the structure of the V257STOP mutant.
To further probe the nature of the soluble aggregates, the samples were assayed for binding to Thioflavin T. These data indicate that the initial aggregated or self-associated state of the V257STOP mutant possesses a structure or conformation that is able to bind and induce Thioflavin T fluorescence. Given that Thioflavin T fluorescence is typically observed in the presence of fibrillar or amyloid aggregates, this suggests that the initial aggregated state of the mutant protein possesses structural features that are similar to the cross-beta structure present in amyloid fibrils.27 The fact that the WT form of ATF5 does not induce Thioflavin T fluorescence further indicates a structural difference exists between the soluble aggregated forms of the two proteins.
Despite a difference in the structure of the soluble aggregates, the FTIR data indicate that the final aggregated states of the two forms of ATF5 are structurally similar. The increased FTIR absorption signal at 1620 cm−1 is reflective of intermolecular β-structure and has been observed for numerous aggregated protein species.21a, 21c, 23, 25 This suggests that while the pathway or mechanism leading to the formation of the final state may differ between proteins, the structure of the final state is sequence independent.21a, 21c, 23, 25
It appears that the WT form of ATF5 protein possesses a conformation that readily permits intermolecular β-sheet addition or elongation. Our initial hypothesis was that solution exposure of the valine zipper region facilitated protein self-association through the initiation of intermolecular β-structure. Therefore, under reducing conditions, where the valine zipper region is more exposed, protein aggregation occurs more readily. Our data show that upon removal of the valine zipper region, the bZIP domain still self-associates, but these soluble, structured aggregates remain thermally stable and resist further aggregate growth up to high temperature. This data suggests the mutant protein forms a structured, stable aggregate at low temperature that involves the portion of the protein that is helical in the native state. Moreover, the data suggest the adjacent sequence, although lacking in definable regular secondary structure, supports retention of helical structure in this region of the native protein, preventing formation of stable aggregates at low temperature. In addition, no difference in aggregation is observed between the reduced and non-reduced forms of the ATF5 mutant. Removal of the valine zipper region changes the conformation of the alpha-helical region such that intermolecular disulfide bond formation is no longer a determining factor in aggregate formation, structure or thermal stability.
Conclusions
We have shown that removal of the valine zipper region that lacks definable secondary structure from ATF5 affects the structure of the adjacent, primarily α-helical leucine zipper region, leading to a more disordered state. While both the WT and mutant forms of ATF5 readily self-associate, the soluble associated species differ in size and structure. The differences between these two forms of ATF5 are also reflected in their thermal stability. A much higher temperature is required to induce rapid aggregate growth for the mutant form than for the WT. Additionally, aggregate growth for the V257STOP mutant is unaffected by the presence of DTT. This study illustrates the critical relationship between protein structure and aggregate assembly. Furthermore, this work lays the foundation for future high-resolution studies that can be performed on these systems using multi-dimensional NMR.
Supplementary Material
Acknowledgments
We thank Drs. C. Olsen and Q. Ye for technical assistance. This publication was made possible by NIH Grant Number P20 RR-17708 from the National Center for Research Resources and the Kansas University Center for Research. Support for N. Ciaccio was provided by the Madison and Lila Self Graduate Fellowship. T.S. Reynolds was supported by a KU Honors Undergraduate Research Award.
Footnotes
The alpha-helical and intermolecular beta-sheet content of the WT and V257STOP forms of ATF5, represented by the FTIR absorption at 1650 cm−1 and 1620 cm−1 respectively, was evaluated as a function of temperature. Differences in the structure of these aggregates were probed using Thioflavin T. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.(a) Carpenter JF, Randolph TW, Jiskoot W, Crommelin DJ, Middaugh CR, Winter G, Fan YX, Kirshner S, Verthelyi D, Kozlowski S, Clouse KA, Swann PG, Rosenberg A, Cherney B. Overlooking subvisible particles in therapeutic protein products: gaps that may compromise product quality. J Pharm Sci. 2009;98(4):1201–5. doi: 10.1002/jps.21530. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Bee JS, Davis M, Freund E, Carpenter JF, Randolph TW. Aggregation of a monoclonal antibody induced by adsorption to stainless steel. Biotechnol Bioeng. 2010;105(1):121–9. doi: 10.1002/bit.22525. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Ludwig DB, Carpenter JF, Hamel JB, Randolph TW. Protein adsorption and excipient effects on kinetic stability of silicone oil emulsions. J Pharm Sci. 2010;99(4):1721–33. doi: 10.1002/jps.21982. [DOI] [PubMed] [Google Scholar]; (d) Wang W. Protein aggregation and its inhibition in biopharmaceutics. Int J Pharm. 2005;289(1–2):1–30. doi: 10.1016/j.ijpharm.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 2.(a) Hermeling S, Crommelin DJ, Schellekens H, Jiskoot W. Structure-immunogenicity relationships of therapeutic proteins. Pharm Res. 2004;21(6):897–903. doi: 10.1023/b:pham.0000029275.41323.a6. [DOI] [PubMed] [Google Scholar]; (b) Hermeling S, Schellekens H, Maas C, Gebbink MF, Crommelin DJ, Jiskoot W. Antibody response to aggregated human interferon alpha2b in wild-type and transgenic immune tolerant mice depends on type and level of aggregation. J Pharm Sci. 2006;95(5):1084–96. doi: 10.1002/jps.20599. [DOI] [PubMed] [Google Scholar]; (c) Rosenberg AS. Effects of protein aggregates: an immunologic perspective. AAPS J. 2006;8(3):E501–7. doi: 10.1208/aapsj080359. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Fradkin AH, Carpenter JF, Randolph TW. Immunogenicity of aggregates of recombinant human growth hormone in mouse models. J Pharm Sci. 2009;98(9):3247–64. doi: 10.1002/jps.21834. [DOI] [PubMed] [Google Scholar]; (e) Purohit VS, Middaugh CR, Balasubramanian SV. Influence of aggregation on immunogenicity of recombinant human Factor VIII in hemophilia A mice. J Pharm Sci. 2006;95(2):358–71. doi: 10.1002/jps.20529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(a) Munishkina LA, Fink AL, Uversky VN. Accelerated fibrillation of alpha-synuclein induced by the combined action of macromolecular crowding and factors inducing partial folding. Curr Alzheimer Res. 2009;6(3):252–60. doi: 10.2174/156720509788486491. [DOI] [PubMed] [Google Scholar]; (b) Bauer R, Carrotta R, Rischel C, Ogendal L. Characterization and isolation of intermediates in beta-lactoglobulin heat aggregation at high pH. Biophys J. 2000;79(2):1030–8. doi: 10.1016/S0006-3495(00)76357-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Gomez-Orellana I, Variano B, Miura-Fraboni J, Milstein S, Paton DR. Thermodynamic characterization of an intermediate state of human growth hormone. Protein Sci. 1998;7(6):1352–8. doi: 10.1002/pro.5560070611. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Speed MA, Morshead T, Wang DI, King J. Conformation of P22 tailspike folding and aggregation intermediates probed by monoclonal antibodies. Protein Sci. 1997;6(1):99–108. doi: 10.1002/pro.5560060111. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Calloni G, Lendel C, Campioni S, Giannini S, Gliozzi A, Relini A, Vendruscolo M, Dobson CM, Salvatella X, Chiti F. Structure and dynamics of a partially folded protein are decoupled from its mechanism of aggregation. J Am Chem Soc. 2008;130(39):13040–50. doi: 10.1021/ja8029224. [DOI] [PubMed] [Google Scholar]; (f) Khare SD, Dokholyan NV. Molecular mechanisms of polypeptide aggregation in human diseases. Curr Protein Pept Sci. 2007;8(6):573–9. doi: 10.2174/138920307783018703. [DOI] [PubMed] [Google Scholar]; (g) Watzlawik J, Skora L, Frense D, Griesinger C, Zweckstetter M, Schulz-Schaeffer WJ, Kramer ML. Prion protein helix1 promotes aggregation but is not converted into beta-sheet. J Biol Chem. 2006;281(40):30242–50. doi: 10.1074/jbc.m605141200. [DOI] [PubMed] [Google Scholar]; (h) Chi EY, Krishnan S, Randolph TW, Carpenter JF. Physical stability of proteins in aqueous solution: mechanism and driving forces in nonnative protein aggregation. Pharm Res. 2003;20(9):1325–36. doi: 10.1023/a:1025771421906. [DOI] [PubMed] [Google Scholar]
- 4.(a) Weiss WFt, Young TM, Roberts CJ. Principles, approaches, and challenges for predicting protein aggregation rates and shelf life. J Pharm Sci. 2009;98(4):1246–77. doi: 10.1002/jps.21521. [DOI] [PubMed] [Google Scholar]; (b) Lee CF. Self-assembly of protein amyloids: a competition between amorphous and ordered aggregation. Phys Rev E Stat Nonlin Soft Matter Phys. 2009;80(3 Pt 1):031922. doi: 10.1103/PhysRevE.80.031922. [DOI] [PubMed] [Google Scholar]; (c) Maji SK, Wang L, Greenwald J, Riek R. Structure-activity relationship of amyloid fibrils. FEBS Lett. 2009;583(16):2610–7. doi: 10.1016/j.febslet.2009.07.003. [DOI] [PubMed] [Google Scholar]; (d) Fawzi NL, Yap EH, Okabe Y, Kohlstedt KL, Brown SP, Head-Gordon T. Contrasting disease and nondisease protein aggregation by molecular simulation. Acc Chem Res. 2008;41(8):1037–47. doi: 10.1021/ar800062k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.(a) Eisenberg D, Nelson R, Sawaya MR, Balbirnie M, Sambashivan S, Ivanova MI, Madsen AO, Riekel C. The structural biology of protein aggregation diseases: Fundamental questions and some answers. Acc Chem Res. 2006;39(9):568–75. doi: 10.1021/ar0500618. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Stefani M, Dobson CM. Protein aggregation and aggregate toxicity: new insights into protein folding, misfolding diseases and biological evolution. J Mol Med. 2003;81(11):678–99. doi: 10.1007/s00109-003-0464-5. [DOI] [PubMed] [Google Scholar]; (c) Fandrich M, Meinhardt J, Grigorieff N. Structural polymorphism of Alzheimer Abeta and other amyloid fibrils. Prion. 2009;3(2):89–93. doi: 10.4161/pri.3.2.8859. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Bellotti V, Nuvolone M, Giorgetti S, Obici L, Palladini G, Russo P, Lavatelli F, Perfetti V, Merlini G. The workings of the amyloid diseases. Ann Med. 2007;39(3):200–7. doi: 10.1080/07853890701206887. [DOI] [PubMed] [Google Scholar]
- 6.(a) Fernandez-Busquets X, de Groot NS, Fernandez D, Ventura S. Recent structural and computational insights into conformational diseases. Curr Med Chem. 2008;15(13):1336–49. doi: 10.2174/092986708784534938. [DOI] [PubMed] [Google Scholar]; (b) Squires AM, Devlin GL, Gras SL, Tickler AK, MacPhee CE, Dobson CM. X-ray scattering study of the effect of hydration on the cross-beta structure of amyloid fibrils. J Am Chem Soc. 2006;128(36):11738–9. doi: 10.1021/ja063751v. [DOI] [PubMed] [Google Scholar]
- 7.(a) Bellesia G, Shea JE. Effect of beta-sheet propensity on peptide aggregation. J Chem Phys. 2009;130(14):145103. doi: 10.1063/1.3108461. [DOI] [PubMed] [Google Scholar]; (b) Rezaei-Ghaleh N, Zweckstetter M, Morshedi D, Ebrahim-Habibi A, Nemat-Gorgani M. Amyloidogenic potential of alpha-chymotrypsin in different conformational states. Biopolymers. 2009;91(1):28–36. doi: 10.1002/bip.21079. [DOI] [PubMed] [Google Scholar]; (c) Qin Z, Hu D, Zhu M, Fink AL. Structural characterization of the partially folded intermediates of an immunoglobulin light chain leading to amyloid fibrillation and amorphous aggregation. Biochemistry. 2007;46(11):3521–31. doi: 10.1021/bi061716v. [DOI] [PubMed] [Google Scholar]
- 8.Wang L, Maji SK, Sawaya MR, Eisenberg D, Riek R. Bacterial inclusion bodies contain amyloid-like structure. PLoS Biol. 2008;6(8):e195. doi: 10.1371/journal.pbio.0060195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.(a) Kunjithapatham R, Oliva FY, Doshi U, Perez M, Avila J, Munoz V. Role for the alpha-helix in aberrant protein aggregation. Biochemistry. 2005;44(1):149–56. doi: 10.1021/bi048564t. [DOI] [PubMed] [Google Scholar]; (b) Thompson AJ, Barnham KJ, Norton RS, Barrow CJ. The Val-210-Ile pathogenic Creutzfeldt-Jakob disease mutation increases both the helical and aggregation propensities of a sequence corresponding to helix-3 of PrP(C) Biochim Biophys Acta. 2001;1544(1–2):242–54. doi: 10.1016/s0167-4838(00)00225-9. [DOI] [PubMed] [Google Scholar]
- 10.(a) Ciaccio NA, Moreno ML, Bauer RL, Laurence JS. High-yield expression in E. coli and refolding of the bZIP domain of activating transcription factor 5. Protein Expr Purif. 2008;62(2):235–43. doi: 10.1016/j.pep.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Angelastro JM, Canoll PD, Kuo J, Weicker M, Costa A, Bruce JN, Greene LA. Selective destruction of glioblastoma cells by interference with the activity or expression of ATF5. Oncogene. 2006;25(6):907–16. doi: 10.1038/sj.onc.1209116. [DOI] [PubMed] [Google Scholar]; (c) Monaco SE, Angelastro JM, Szabolcs M, Greene LA. The transcription factor ATF5 is widely expressed in carcinomas, and interference with its function selectively kills neoplastic, but not nontransformed, breast cell lines. Int J Cancer. 2007;120(9):1883–90. doi: 10.1002/ijc.22469. [DOI] [PubMed] [Google Scholar]
- 11.Ciaccio NA, Laurence JS. Effects of disulfide bond formation and protein helicity on the aggregation of activating transcription factor 5. Mol Pharm. 2009;6(4):1205–15. doi: 10.1021/mp900058t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.(a) Byler DM, Susi H. Examination of the secondary structure of proteins by deconvolved FTIR spectra. Biopolymers. 1986;25(3):469–87. doi: 10.1002/bip.360250307. [DOI] [PubMed] [Google Scholar]; (b) Krimm S, Bandekar J. Vibrational spectroscopy and conformation of peptides, polypeptides, and proteins. Adv Protein Chem. 1986;38:181–364. doi: 10.1016/s0065-3233(08)60528-8. [DOI] [PubMed] [Google Scholar]
- 13.Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley JL, Sykes BD. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J Biomol NMR. 1995;6(2):135–40. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- 14.(a) Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6(3):277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]; (b) Goddard T, Kneller D. Sparky. Vol. 3. University of California; San Francisco: [Google Scholar]
- 15.Koppel DE. Analysis of Macromolecular Polydispersity in Intensity Correlation Spectroscopy: The Method of Cumulants. J Chem Phys. 1972;57(11):4814–20. [Google Scholar]
- 16.Provencher SW. Inverse problems in polymer characterization: Direct analysis of polydispersity with photon correlation spectroscopy. Die Makromolekulare Chemie. 1979;180(1):201–209. [Google Scholar]
- 17.Chen YH, Yang JT, Martinez HM. Determination of the secondary structures of proteins by circular dichroism and optical rotatory dispersion. Biochemistry. 1972;11(22):4120–31. doi: 10.1021/bi00772a015. [DOI] [PubMed] [Google Scholar]
- 18.(a) Fan H, Vitharana SN, Chen T, O’Keefe D, Middaugh CR. Effects of pH and polyanions on the thermal stability of fibroblast growth factor 20. Mol Pharm. 2007;4(2):232–40. doi: 10.1021/mp060097h. [DOI] [PubMed] [Google Scholar]; (b) Liu C, Chu D, Wideman RD, Houliston RS, Wong HJ, Meiering EM. Thermodynamics of denaturation of hisactophilin, a beta-trefoil protein. Biochemistry. 2001;40(13):3817–27. doi: 10.1021/bi002609i. [DOI] [PubMed] [Google Scholar]; (c) Green NM, Wrigley NG, Russell WC, Martin SR, McLachlan AD. Evidence for a repeating cross-beta sheet structure in the adenovirus fibre. EMBO J. 1983;2(8):1357–65. doi: 10.1002/j.1460-2075.1983.tb01592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woody R. Study of Theoretical Circular Dichrosim of Polypeptides: Contribution of Beta-turns. In: Blout E, Bovey F, Goodman M, Lotan N, editors. Peptides, Polypeptides and Proteins. John Wiley; NY: 1974. [Google Scholar]
- 20.(a) Chang CT, Wu CS, Yang JT. Circular dichroic analysis of protein conformation: inclusion of the beta-turns. Anal Biochem. 1978;91(1):13–31. doi: 10.1016/0003-2697(78)90812-6. [DOI] [PubMed] [Google Scholar]; (b) Yang JT, Wu CS, Martinez HM. Calculation of protein conformation from circular dichroism. Methods Enzymol. 1986;130:208–69. doi: 10.1016/0076-6879(86)30013-2. [DOI] [PubMed] [Google Scholar]
- 21.(a) Dong A, Prestrelski SJ, Allison SD, Carpenter JF. Infrared spectroscopic studies of lyophilization- and temperature-induced protein aggregation. J Pharm Sci. 1995;84(4):415–24. doi: 10.1002/jps.2600840407. [DOI] [PubMed] [Google Scholar]; (b) Prestrelski SJ, Tedeschi N, Arakawa T, Carpenter JF. Dehydration-induced conformational transitions in proteins and their inhibition by stabilizers. Biophys J. 1993;65(2):661–71. doi: 10.1016/S0006-3495(93)81120-2. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Clark AH, Saunderson DH, Suggett A. Infrared and laser-Raman spectroscopic studies of thermally-induced globular protein gels. Int J Pept Protein Res. 1981;17(3):353–64. doi: 10.1111/j.1399-3011.1981.tb02002.x. [DOI] [PubMed] [Google Scholar]
- 22.Susi H, Byler DM. Protein structure by Fourier transform infrared spectroscopy: second derivative spectra. Biochem Biophys Res Commun. 1983;115(1):391–7. doi: 10.1016/0006-291x(83)91016-1. [DOI] [PubMed] [Google Scholar]
- 23.Casal HL, Kohler U, Mantsch HH. Structural and conformational changes of beta-lactoglobulin B: an infrared spectroscopic study of the effect of pH and temperature. Biochim Biophys Acta. 1988;957(1):11–20. doi: 10.1016/0167-4838(88)90152-5. [DOI] [PubMed] [Google Scholar]
- 24.Priddy TS, Middaugh CR, Carlson GM. Electrostatic changes in phosphorylase kinase induced by its obligatory allosteric activator Ca2+ Protein Sci. 2007;16(3):517–27. doi: 10.1110/ps.062577507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwegman JJ, Carpenter JF, Nail SL. Evidence of partial unfolding of proteins at the ice/freeze-concentrate interface by infrared microscopy. J Pharm Sci. 2009;98(9):3239–46. doi: 10.1002/jps.21843. [DOI] [PubMed] [Google Scholar]
- 26.Hawe A, Kasper JC, Friess W, Jiskoot W. Structural properties of monoclonal antibody aggregates induced by freeze-thawing and thermal stress. Eur J Pharm Sci. 2009;38(2):79–87. doi: 10.1016/j.ejps.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 27.(a) Hawe A, Sutter M, Jiskoot W. Extrinsic fluorescent dyes as tools for protein characterization. Pharm Res. 2008;25(7):1487–99. doi: 10.1007/s11095-007-9516-9. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) LeVine H., 3rd Thioflavine T interaction with synthetic Alzheimer’s disease beta-amyloid peptides: detection of amyloid aggregation in solution. Protein Sci. 1993;2(3):404–10. doi: 10.1002/pro.5560020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.(a) Sapienza PJ, Lee AL. Using NMR to study fast dynamics in proteins: methods and applications. Curr Opin Pharmacol. 2010;10(6):723–30. doi: 10.1016/j.coph.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Spyracopoulos L, Sykes BD. Thermodynamic insights into proteins from NMR spin relaxation studies. Curr Opin Struct Biol. 2001;11(5):555–9. doi: 10.1016/s0959-440x(00)00261-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.