Abstract
Post-translational modifications (PTMs) of histones are an essential feature in the dynamic regulation of chromatin. One of these modifications, ubiquitylation, has been speculated to directly influence the stability of the nucleosome, which represents the basic building block of chromatin. Here we report a strategy for the semisynthesis of site-specifically ubiquitylated histone H2A (uH2A). This branched protein was generated through a three-piece expressed protein ligation (EPL) approach including a traceless ligation at valine. uH2A could be efficiently incorporated into nucleosomes, thereby opening the way to detailed biochemical and biophysical studies on the function of this PTM. Accordingly, we used uH2A, as well as a previously generated ubiquitylated H2B (uH2B), in chaperone-coupled nucleosome stability assays to demonstrate that the direct effect of ubiquitylated histones on nucleosomal stability is in fact modest.
The genome in eukaryotic cells is packaged in the form of chromatin - a dynamic nucleoprotein complex between DNA and the basic histone proteins H2A, H2B, H3 and H4, with the nucleosome as the fundamental repeating unit1. The dynamic nature of chromatin manifests itself on multiple levels. In the cell, the structure of chromatin is highly dynamic, thereby regulating transactions such as replication, transcription and repair2. Dynamics at the nucleosomal level include transient DNA unwrapping (‘breathing’), which allows access to the nucleosomal DNA and facilitates nucleosome disassembly3. Further, histones can be transiently removed either to facilitate access to the DNA template4,5 or to be exchanged for a variety of specialized histone variants6.
The first steps in nucleosome disassembly involve partial unwrapping of DNA and opening up the interface between the H2A-H2B dimers and the (H3-H4)2 tetramer, followed by the removal of either one or both of the H2A-H2B dimers3,7. This leaves behind the DNA-(H3-H4)2 tetramer complex referred to as the tetrasome, which can further dissociate into free DNA and (H3-H4)2 tetramers8,9. Conversely, the assembly of nucleosomes is thought to occur through a reversal of these processes10. The highly basic nature of the histones predisposes them to interact non-specifically with DNA. This is prevented by a heterogenous family of histone chaperones which bind free histones11. Nucleosome assembly protein 1 (Nap1) is among the best characterized of these histone chaperones12. Nap1 binds to linker histone H1, H2A-H2B dimers and (H3-H4)2 tetramers by recognizing their common histone-fold13. The chaperone activity of Nap1 has been widely used in in vitro reconstitution of chromatin14 and has recently found utility in determining nucleosome stability under equilibrium conditions10,15.
Nature strategically employs histone PTMs to alter the steric and electrostatic properties of histones, thereby influencing the stability and dynamics of chromatin through both cisand trans-acting mechanisms16–20. Of the many known PTMs, perhaps the most dramatic is ubiquitylation, in which the ~8 kDa protein ubiquitin is attached through its C-terminus to the ε-amino group of a target lysine. The best-known ubiquitylation sites on chromatin are lysine 119 of H2A (uH2A) and lysine 120 of H2B (uH2B). Both are tightly associated with transcription regulation and DNA damage repair21, but their mechanistic functions are less well understood. Several studies have suggested a stabilizing effect of ubiquitylation22,23 on nucleosomal structure, while others arrive at the opposite conclusion24,25. The lack of access to chemically defined uH2A and uH2B has hindered detailed biophysical analysis of ubiquitylated nucleosomes, and thus progress on this problem. Previously, we developed a robust synthesis of uH2B using an EPL approach26,27. In this study, we report on the successful synthesis of uH2A. Access to both these ubiquitylated histones has allowed us to interrogate the effect of the modification on nucleosome stability by employing Nap1-mediated nucleosome assembly assays.
For the synthesis of uH2A, we developed a strategy based on sequential, regioselective EPL reactions. EPL allows the connection of a synthetic peptide containing an N-terminal cysteine (or functional equivalent) with a recombinant protein carrying a C-terminal thioester, forming an amide bond. Building on the established synthesis of uH2B26,27, we devised a synthetic route to uH2A in which ubiquitin is first ligated to a peptide corresponding to the C-terminal part of H2A, via the ε-amine of the canonical lysine 119 (Fig. 1a). In a second step, the peptide-ubiquitin conjugate is ligated to the remainder of H2A. Critical to this strategy was the choice of the ligation junctions, which must be compatible with efficient reactions, based on well-established criteria 28,29, whilst minimizing sequence changes to the native ubiquitylated protein.
Figure 1.
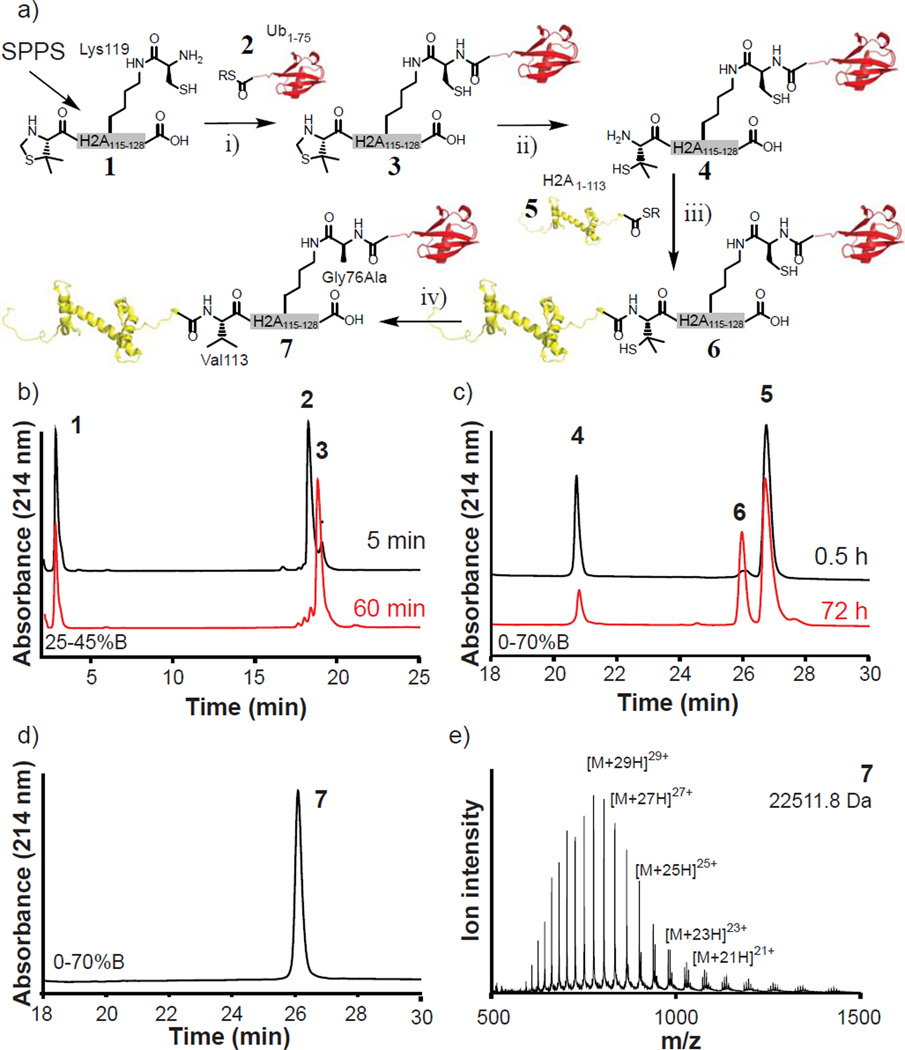
Synthesis of uH2A: a) Synthesis scheme: i) Ligation of peptide 1 to ubiquitin(1-75) α-thioester 2, forming the branched protein 3. ii) Deprotection of 3 by methoxylamine treatment. iii) Formation of 6 by ligation of H2A(1-113) α-thioester 5 to 4. iv) Final desulfurization to form uH2A, 7. R: C6H4CH2COOH. b) RP-HPLC data for the ligation between 1 and 2. c) RP-HPLC data for the ligation between 4 and 5. d-e) RP-HPLC and ESI-MS data for uH2A, 7, (M+H)+ = 22511.8 Da, [(M+H)+expected = 22512.0 Da]
For the ligation junction between ubiquitin and the C-terminal peptide of H2A, we replaced glycine 76 of ubiquitin by a cysteine residue, attached via an iso-peptide linkage to the ε-amine of K119 within the synthetic peptide. This cysteine serves as a reactive handle for the ligation to an ubiquitin (1–75)-α-thioester. Upon chemical desulfurization, the cysteine is converted to alanine. This glycine to alanine substitution at position 76 in ubiquitin is known to be functionally silent in the case of uH2B27. For the second EPL step, inspection of the H2A sequence (Figure S1 in the Supporting Information) indicated that the optimal ligation junction would be between serine 113 and valine 114. This allows for use of the non-natural amino acid penicillamine as a valine surrogate, which upon desulfurization affords the canonical amino acid and hence a traceless process30,31.
We proceeded with the solid phase synthesis of branched peptide 1 using the fluorenylmethoxycarbonyl (Fmoc) Nα protection logic. Orthogonal protection of lysine 119 using the Mtt group allowed selective attachment of cysteine to the ε-amino group of this residue. The penicillamine moiety was introduced protected as 5,5’-dimethyl-1,3-thiazolidine-4-carboxylic acid, which set the desired regioselectivity in the first ligation reaction. Accordingly, excess purified peptide 1 was combined with ubiquitin(1-75)-α-thioester 2 (produced by thiolysis of a fusion protein of ubiquitin with the Mxe GyrA in-tein, see Figure S1) affording branched protein 3 with essentially peak-to-peak conversion after 1 h reaction (Figure 1b, Figure S1). Treatment of the ligation product in situ with 0.5 M methoxylamine under mildly acidic conditions gave conjugate 4 in which the N-terminal penicillamine moiety is revealed (Figure S2). The isolated yield of conjugate 4 was 65% relative to 2 (i.e. over two steps). In the second ligation reaction, 4 was combined with a 2.7- fold excess of recombinantly generated H2A(1-113)-α-thioester (5, produced similarly to 2, Figure S1) in the presence of 50 mM 4-mercaptophenyl acetic acid (MPAA) under inert atmosphere. The reaction was allowed to proceed for 72 h at which point the ligation product 6 was isolated by RP-HPLC in 42% yield relative to 4 (Figure 1c and Figure S2). Of note, the use of MPAA as a thiol catalyst crucially reduced the buildup of unreactive internal disulfides of 4, thus increasing the ligation efficiency.
The last step in the process, radical-based desulfurization of 6 to give uH2A 7, gave unexpected results. We initially employed the standard protocol which involved dissolving 6 in a buffer containing 250 mM TCEP, 2 mM radical starter VA-041 and t-butylmercaptan/ethanethiol for radical propagation32. This procedure led to significant accumulation of two side products, in addition to the desired product 7 (Figure S2). Interestingly, the HPLC retention times of these side-products were similar to the retention times of the starting materials of the second ligation reaction. Mass spectrometric analysis revealed that the side products corresponded to 4,lacking the penicillamine residue, as well as to 5,with a C-terminal carboxylic acid. Analogous cleavage products were not observed at the cysteine desulfurization site (i.e. ligation site 1). Thus, the observed backbone cleavage events seem to be related to specific steric and/or electronic properties of the thiyl radical generated on the penicillamine moiety. Fortunately, these side-reactions could be suppressed by using an optimized protocol based on ref. 30, involving more radical starter (16 mM VA-041) and glutathione (40 mM) for radical propagation. Under these conditions the final product 7 was obtained in 53% isolated yield (Figure 1d-e).
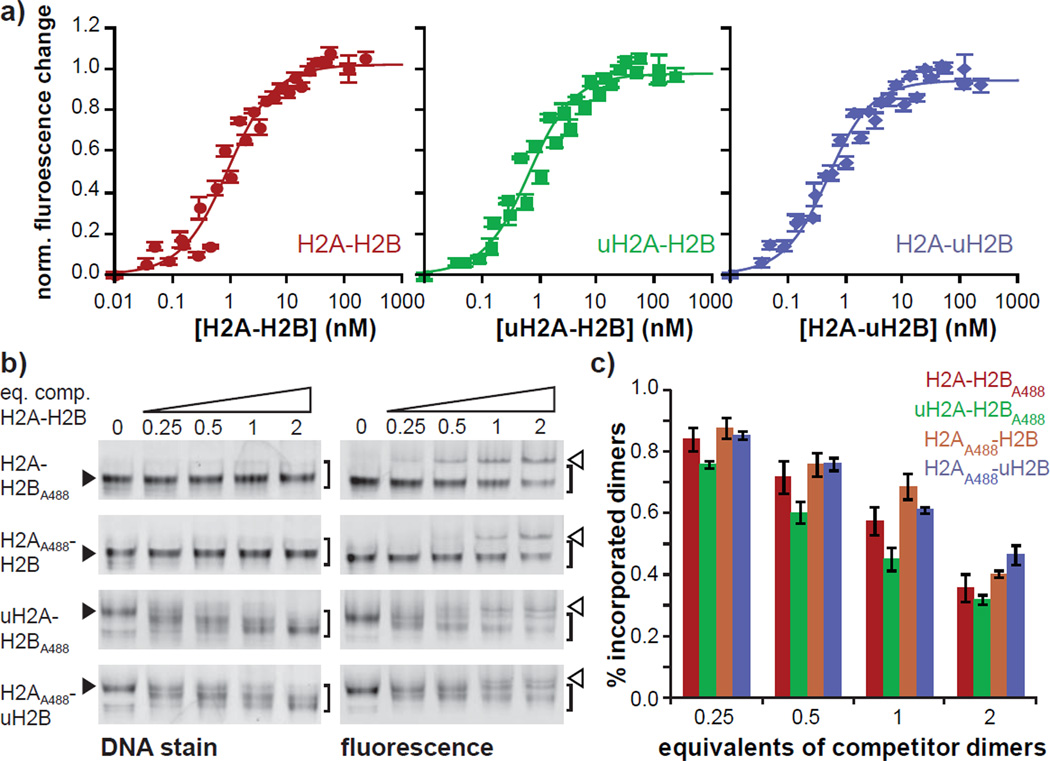
Together with uH2B, which we produced according to ref. 27 (see Figure S3), we moved forward to investigate the effects of histone ubiquitylation on nucleosome assembly and stability. The stability of a nucleosome is determined by the dissociation constants (KD) between all the components of the nucleoprotein complex. However, the high energy barriers encountered in the process, as well as the high number of possible non-native interactions between histones and DNA result in dominant kinetic effects and direct measurements of equilibrium constants are difficult33. Histone chaperones act by preventing such non-native interactions and therefore render the assembly and disassembly reactions reversible15. Therefore, histone chaperones can be used as molecular tools to determine nucleosome stability. We decided to employ mouse Nap1 (mNap1) to investigate the propensity of ubiquitylated histones to form nucleosomes (Fig. S4). Nap1 is implicated in nucleosome disassembly at promoters and coding regions34, sites where histone ubiquitylation plays an important regulatory role21. Thus, we were also directly interested to measure possible effects of histone ubiquitylation on Nap1 binding. We first determined the KD of the mNap1 complexes with unmodified, uH2A or uH2B containing dimers, using a florescence based binding assay13. A mutant of mNap1 was generated with all but one (Cys 388) of its cysteine residues mutated to serine. The unique cysteine residue was site-specifically labeled with the fluorescent dye Alexa 546-C5-maleimide (mNap1A546, Figure S4). We then refolded H2A-H2B dimers or (H3-H4)2 tetramers using xenopus laevis histones as well as the synthetic uH2A and uH2B molecules (Figure S5). Unmodified, uH2A and uH2B containing dimers were subsequently titrated into a solution containing 0.25 nM of labeled mNap1. Dimer binding to mNap1 resulted in a 5–10% reduction of A546 fluorescence, which allowed the generation of binding isotherms (Figure 2a). For analysis, a reversible binding model (one-site, non-cooperative) was fit to the data, resulting in KD values of 0.8 ± 0.1 nM for unmodified, and 0.5 ± 0.1 nM and 0.4 ± 0.1 nM for uH2A and uH2B containing dimers, respectively (see Table S1). Thus, we conclude that histone ubiquitylation results in an increase in mNap1 binding affinity, but the effects are small.
Figure 2.
mNap1 mediated nucleosome formation. a) mNap1A546 binding to histone dimers (data from 3 independent experiments are included in the plots, error bars: instrumental error, solid lines: binding isotherm, for parameters see Table S1). b) Native PAGE analysis of nucleosome formation experiment with fluorescent histone dimers under competition with unmodified histone dimers (filled triangles: nucleosome in the absence of competitor, open triangles: mNap1:dimer complexes). c) Quantification of the relative fluorescence intensity of the nucleosomal bands (averaged data from 3 independent experiments, error bars denote S.E.M). The brackets in b) indicated the integrated area for quantification. For full gels, see Fig. S12.
H2A or H2B ubiquitylation may influence H2A-H2B:(H3- H4)2 or H2A-H2B:DNA interactions, thereby modulating nucleosome stability. We therefore turned our attention to the deposition of H2A-H2B dimers onto tetrasomes. Using fluorescently labeled histones, the formation of nucleosomes can be followed using gel electrophoresis followed by fluorescence scanning. We labeled previously characterized single-cysteine mutants of H2A (at N110C, ref. 18) and H2B (at T115C, ref. 13) with Alexa 488-C5-maleimide (H2AA488 and H2BA488, Figures S6). To follow uH2A incorporation, we used H2BA488 to prepare the dimer pair H2A-H2BA488 and uH2AH2BA488 (Figure S7). Similarly, we used H2AA488 to refold the dimer pair H2AA488-H2B and H2AA488-uH2B. All histone dimers formed defined nucleosomes as judged from native PAGE (Figure S8). To determine the relative nucleosome formation propensity of the ubiquitylated histone dimers, we implemented a competition assay. The fluorescent and differently ubiquitylated histone dimers were mixed with increasing amounts of non-fluorescent competitor H2A-H2B, and subsequently combined with pre-formed tetrasomes in the presence of mNap1. The reaction mixtures were resolved by native PAGE and visualized by fluorescence scanning and DNA staining (Figure 2b). Quantification of the fluorescent bands from nucleosomal species indicated that uH2A-H2B dimers are more easily competed away compared to the other species (Figure 2c). We therefore conclude that H2A ubiquitylation reduces nucleosome stability – although the effect is subtle.
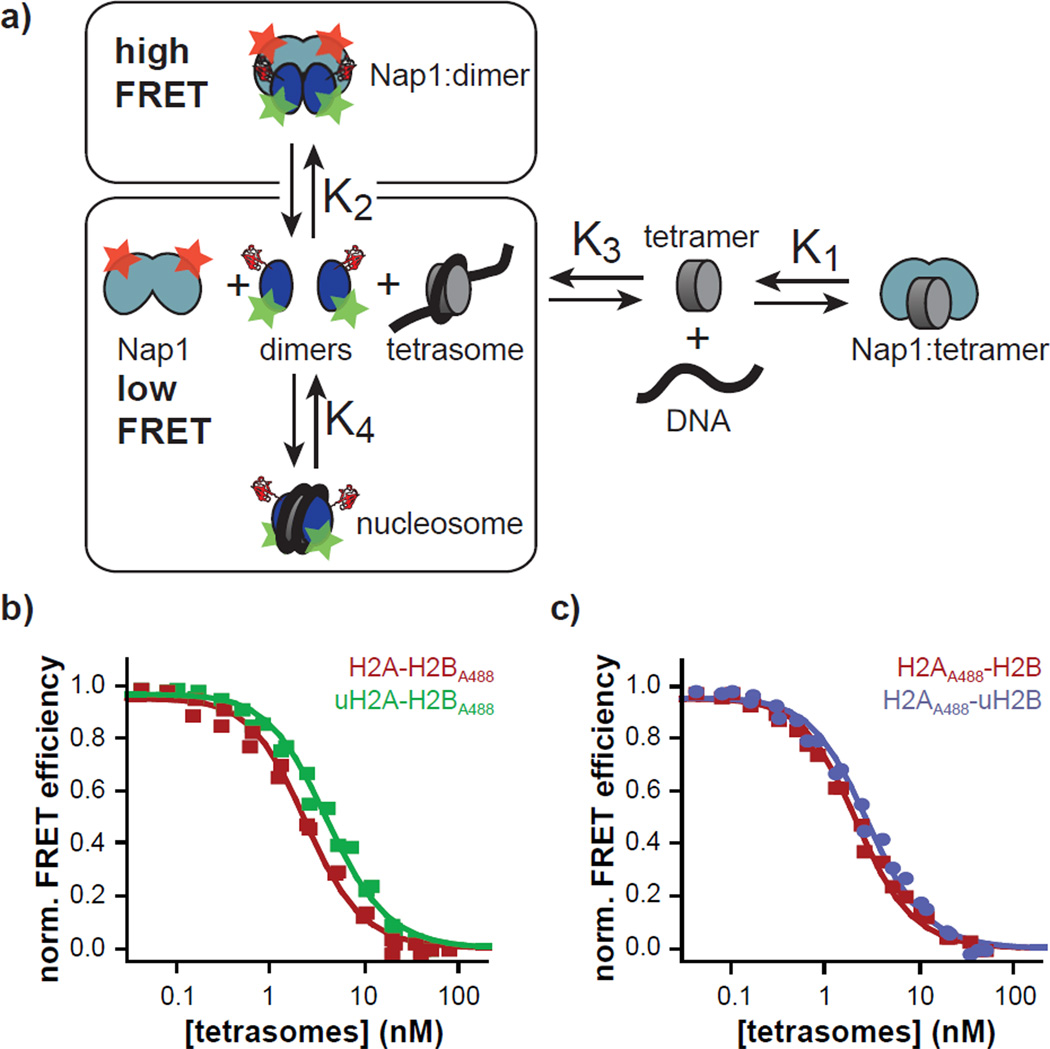
To more closely investigate the thermodynamic aspect of nucleosome stability, we further employed a coupled equilibrium stability assay, using the well-characterized S. cerevisiae Nap1 homolog, yNap1, to ensure full reversibility of all the reactions15. This assay is based on monitoring fluorescence resonance energy transfer (FRET) between H2A-H2B dimers carrying a donor chromophore (Figure 3a) and yNap1, carrying an acceptor chromophore. Nucleosome formation is initiated by titrating in pre-formed tetrasomes, which shifts the binding equilibrium between dimers and yNap1 towards free yNap1 and nucleosomes (Figure 3a). If the KD’s of the complexes between yNap1 and (H3-H4)2 (K1), between yNap1 and H2A-H2B (K2) and between (H3-H4)2 and DNA (K3) are known, the KD between H2A-H2B and tetrasomes (nucleo-some assembly, K4) can be calculated from the titration experiment. We stress that K4 is directly related to nucleosome stability as it measures the association – dissociation equilibrium of dimers from nucleosomes.
Figure 3.
Stability of ubiquitylated nucleosomes. a) Scheme of the chaperone assisted, FRET based assay to measure nucleosome stability, following ref. 15. The boxes indicate equilibria measured in this work. b, c) Nucleosome stability measured by FRET for the indicated histone dimers. (data from 2 independent experiments are included in the plot, error bars: instrumental error, they are smaller than the symbols; solid lines: Model in shown in a), for parameters, see Table s1).
K1 and K3 have been measured previously15, but K2, the affinity between yNap1 and H2A-H2B with or without ubiquitin, had to be determined. This was done in a manner analogous to the titration carried out for mNap1. We labeled a single cysteine mutant (D201C) of yNap1 with A546-maleimide (yNap1A546, Figure S9) and measured binding curves of unmodified, uH2A or uH2B containing histone dimers (Table 1 and Figure S10). A direct comparison between the histone binding ability of mNap1 and yNap1 revealed a 10-fold higher affinity of mNap1 for histone dimers. H2A and H2B are highly conserved between higher eukaryotes but diverge in sequence from the yeast homologs, which might explain the lower affinity of the yeast chaperone for the Xenopus histones. With values for K1, K2 and K3 in hand, we proceeded to elucidate possible ubiquitylation-dependent differences in K4. Using the same set of A488 labeled histone dimers as previously employed for the competition assays, we observed a strong FRET signal upon histone dimer binding to yNap1A546 (Figure S11). Titration of pre-formed tetrasomes led to nucleosome formation (Figure S8), which resulted in a FRET loss and, at 50 nM of tetrasomes, in the complete recovery of the donoronly spectrum (Figure S11). The experimental data of the complete titrations for all 4 histone dimer types (Figure 3b,c) were then analyzed using a numerical model of four coupled equilibria (Figure 3a) using the measured parameters for K1-K3 and K4 as a fitting parameter (Table 1). The analysis revealed that both uH2A and uH2B lead to slightly less than a ~2 fold increase in K4 relative to the appropriate unmodified controls, from 0.8 ± 0.2 nM (H2A-H2BA488) and 0.7 ± 0.1 nM (H2AA488-H2B) to 1.5 ± 0.2 nm (uH2A-H2BA488) and 1.2 ± 0.2 nM (H2AA488-uH2B). Thus, we find that the thermodynamic impact of uH2A and uH2B on nucleosome stability is in fact modest, although both do lead to a net destabilization of the particle. We further find a stronger binding affinity of canonical H2A-H2B dimer to tetrasome than published previously17. This is likely due to experimental differences, as we used tetrasomes that were preformed by salt dialysis, whereas the previous study combined H3-H4 and DNA together under physiological conditions. Therefore, the current experiments may contain a higher fraction of DNA that is properly occupied by (H3-H4)2.
In this study, we tackled a long-standing question in the chromatin field, namely if histone ubiquitylation alters the stability of nucleosomes. Using a competition assay and semisynthetic ubiquitylated histones, we found that uH2A is less effectively incorporated into nucleosomes by mNap1. Further, employing yNap1 in a chaperone-assisted coupled equilibrium assay, we demonstrated a mild nucleosome destabilizing effect of both uH2A and uH2B. We conclude that the direct impact of histone ubiquitylation on nucleosome stability is marginal. The availability of a full set of ubiquitylated histones, including the new reagent presented here, sets the stage for additional biochemical studies on the function of histone ubiquitylation, in particular in the presence of more specialized histone chaperones (e.g. facilitates chromatin transcription, FACT25) and/or ATP-dependent remodelers.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank members of the Muir laboratory for valuable discussions. Supported from grants by the US National Institutes of Health (GM086868 and GM095880), by the Swiss National Science Foundation (BF) and by the Novo Scholarship Programme (SK). KL and AH are supported by the Howard Hughes Medical Institute and by P01 GM088409.
Footnotes
ASSOCIATED CONTENT
Supporting Information
Biochemical, synthetic and analysis methods, analytical characterization of all purified reagents and raw experimental data. This information is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interests.
REFERENCES
- 1.van Holde K. Chromatin. New York: Springer; 1989. [Google Scholar]
- 2.Bell O, Tiwari VK, Thoma NH, Schubeler D. Nature Reviews Genetics. 2011;12:554. doi: 10.1038/nrg3017. [DOI] [PubMed] [Google Scholar]
- 3.Li G, Levitus M, Bustamante C, Widom J. Nat Struct Mol Biol. 2005;12:46. doi: 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
- 4.Groth A, Rocha W, Verreault A, Almouzni G. Cell. 2007;128:721. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 5.Li B, Carey M, Workman JL. Cell. 2007;128:707. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 6.Talbert PB, Henikoff S. Nat Rev Mol Cell Bio. 2010;11:264. doi: 10.1038/nrm2861. [DOI] [PubMed] [Google Scholar]
- 7.Gansen A, Hauger F, Toth K, Langowski J. Anal Biochem. 2007;368:193. doi: 10.1016/j.ab.2007.04.047. [DOI] [PubMed] [Google Scholar]
- 8.Bohm V, Hieb AR, Andrews AJ, Gansen A, Rocker A, Toth K, Luger K, Langowski J. Nucleic Acids Research. 2011;39:3093. doi: 10.1093/nar/gkq1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Cell. 2004;116:51. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 10.Mazurkiewicz J, Kepert JF, Rippe K. Journal of Biological Chemistry. 2006;281:16462. doi: 10.1074/jbc.M511619200. [DOI] [PubMed] [Google Scholar]
- 11.De Koning L, Corpet A, Haber JE, Almouzni G. Nature Structural & Molecular Biology. 2007;14:997. doi: 10.1038/nsmb1318. [DOI] [PubMed] [Google Scholar]
- 12.Park YJ, Luger K. Curr Opin Struct Biol. 2008;18:282. doi: 10.1016/j.sbi.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andrews AJ, Downing G, Brown K, Park YJ, Luger K. J Biol Chem. 2008 doi: 10.1074/jbc.M805918200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lusser A, Kadonaga JT. Nature Methods. 2004;1:19. doi: 10.1038/nmeth709. [DOI] [PubMed] [Google Scholar]
- 15.Andrews AJ, Chen X, Zevin A, Stargell LA, Luger K. Mol Cell. 2010;37:834. doi: 10.1016/j.molcel.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Science. 2006;311:844. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 17.Neumann H, Hancock SM, Buning R, Routh A, Chapman L, Somers J, Owen-Hughes T, van Noort J, Rhodes D, Chin JW. Molecular Cell. 2009;36:153. doi: 10.1016/j.molcel.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fierz B, Chatterjee C, McGinty RK, Bar-Dagan M, Raleigh DP, Muir TW. Nat Chem Biol. 2011;7:113. doi: 10.1038/nchembio.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simon M, North JA, Shimko JC, Forties RA, Ferdinand MB, Manohar M, Zhang M, Fishel R, Ottesen JJ, Poirier MG. P Natl Acad Sci USA. 2011;108:12711. doi: 10.1073/pnas.1106264108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith E, Shilatifard A. Mol Cell. 2010;40:689. doi: 10.1016/j.molcel.2010.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weake VM, Workman JL. Mol Cell. 2008;29:653. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 22.Davies N, Lindsey GG. Biochim Biophys Acta. 1994;1218:187. doi: 10.1016/0167-4781(94)90009-4. [DOI] [PubMed] [Google Scholar]
- 23.Chandrasekharan MB, Huang F, Sun ZW. Proc Natl Acad Sci U S A. 2009;106:16686. doi: 10.1073/pnas.0907862106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li W, Nagaraja S, Delcuve GP, Hendzel MJ, Davie JR. Biochemical Journal. 1993;296:737. doi: 10.1042/bj2960737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Cell. 2006;125:703. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 26.McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. Nature. 2008;453:812. doi: 10.1038/nature06906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGinty RK, Koehn M, Chatterjee C, Chiang KP, Pratt MR, Muir TW. ACS Chem Biol. 2009 doi: 10.1021/cb9002255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hackeng TM, Griffin JH, Dawson PE. Proc Natl Acad Sci U S A. 1999;96:10068. doi: 10.1073/pnas.96.18.10068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muralidharan V, Muir TW. Nat Methods. 2006;3:429. doi: 10.1038/nmeth886. [DOI] [PubMed] [Google Scholar]
- 30.Haase C, Rohde H, Seitz O. Angew Chem Int Ed Engl. 2008;47:6807. doi: 10.1002/anie.200801590. [DOI] [PubMed] [Google Scholar]
- 31.Chen J, Wan Q, Yuan Y, Zhu JL, Danishefsky SJ. Angew Chem Int Edit. 2008;47:8521. doi: 10.1002/anie.200803523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wan Q, Danishefsky SJ. Angew Chem Int Ed Engl. 2007;46:9248. doi: 10.1002/anie.200704195. [DOI] [PubMed] [Google Scholar]
- 33.Thastrom A, Gottesfeld JM, Luger K, Widom J. Biochemistry. 2004;43:736. doi: 10.1021/bi0302043. [DOI] [PubMed] [Google Scholar]
- 34.Walfridsson J, Khorosjutina O, Matikainen P, Gustafsson CM, Ekwall K. Embo J. 2007;26:2868. doi: 10.1038/sj.emboj.7601728. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.