Abstract
In structural studies of large proteins by NMR, global fold determination plays an increasingly important role in providing a first look at a target’s topology and reducing assignment ambiguity in NOESY spectra of fully-protonated samples. In this work, we demonstrate the use of ultrasparse sampling, a new data processing algorithm, and a 4-D time-shared NOESY experiment (1) to collect all NOEs in 2H/13C/15N-labeled protein samples with selectively-protonated amide and ILV methyl groups at high resolution in only four days, and (2) to calculate global folds from this data using fully automated resonance assignment. The new algorithm, SCRUB, incorporates the CLEAN method for iterative artifact removal, but applies an additional level of iteration, permitting real signals to be distinguished from noise and allowing nearly all artifacts generated by real signals to be eliminated. In simulations with 1.2% of the data required by Nyquist sampling, SCRUB achieves a dynamic range over 10000:1 (250× better artifact suppression than CLEAN) and completely quantitative reproduction of signal intensities, volumes, and lineshapes. Applied to 4-D time-shared NOESY data, SCRUB processing dramatically reduces aliasing noise from strong diagonal signals, enabling the identification of weak NOE crosspeaks with intensities 100× less than diagonal signals. Nearly all of the expected peaks for interproton distances under 5 Å were observed. The practical benefit of this method is demonstrated with structure calculations for 23 kDa and 29 kDa test proteins using the automated assignment protocol of CYANA, in which unassigned 4-D time-shared NOESY peak lists produce accurate and well-converged global fold ensembles, whereas 3-D peak lists either fail to converge or produce significantly less accurate folds. The approach presented here succeeds with an order of magnitude less sampling than required by alternative methods for processing sparse 4-D data.
INTRODUCTION
The size limit for protein structure determination by NMR has been steadily expanding to encompass medium and large targets—but with these have come challenges, including severe resonance overlap in NOESY spectra due to the large numbers of proton signals. The standard strategy for overcoming this is to label the sample using selective protonation, typically of amide as well as Ile δ1, Leu, and Val (ILV) methyl positions in an otherwise deuterated background, in order to simplify NOESY spectra while maintaining sufficient long-range NOE information for an initial structure determination1,2. The global folds derived from these sparse distance constraints constitute a critical first step in the calculation of high-resolution structures, by greatly reducing the number of possible assignments for the observed crosspeaks in subsequent NOESY spectra collected with fully protonated samples. However, the low density and limited redundancy of observable NOE information in sparsely-protonated samples makes it critical to identify and assign unambiguously a high percentage of the NOE crosspeaks. Unfortunately, this is a challenging task using conventional 3-D NOESY experiments, as either the donor or acceptor group of each NOE crosspeak is encoded with only a single frequency. With large proteins, even sparsely protonated samples produce 3-D NOESY spectra with significant degeneracy, particularly in the poorly dispersed methyl region, preventing complete and unambiguous crosspeak assignment.
A simple solution to this problem would be to collect high-resolution 4-D NOESY spectra, but the prolonged measurement times (many weeks) required by conventional Nyquist sampling of 4-D spectra have been a major obstacle precluding their use. The development of sparse sampling methods for biomolecular NMR spectroscopy has the potential to remove this barrier3–15. Instead of recording a complete multidimensional grid of time domain samples, spaced in each dimension at the Nyquist interval, a sparse dataset includes only a small subset of these data points, typically spaced irregularly and at a density far below that required by the Nyquist criterion. When data are processed by the Fourier transform (FT), such a reduction in the sampling rate leads to aliasing artifacts8,11,16,17, but by arranging the sampling points with either partial or total randomness, these artifacts can be made to take the form of low-level random noise9,11. Alternatively, one can employ maximum entropy reconstruction5,7,12 (MaxEnt) or multidimensional decomposition6,18,19 (MDD) with sparse data in place of the FT. Sparse sampling makes it possible to obtain very high resolution spectral information with only a small fraction of the data required conventionally.
The processing of sparse datasets to produce useful spectra becomes increasingly challenging as the amount of data is diminished, however—especially if the dynamic range is high, and if quantitative accuracy is important, as is often the case with NOESY. For 4-D NOESY experiments, MaxEnt and MDD require the sparse setto contain at least 10% of the conventional data12,19. The FT can be used with any dataset, including ultrasparse datasets incorporating less than 2% of the conventional data, but reducing the number of samples increases the aliasing artifact level. Since each signal generates aliasing artifacts proportional to its intensity, the artifacts from the strongest peaks are frequently so large as to obscure the weakest peaks20. Thus FT processing of sparse data is, by itself, generally inadequate for the recovery of the weak signals that are essential in the determination of protein structures.
To facilitate the rapid determination of global folds, we introduce here a new data processing algorithm, SCRUB, which makes it possible to apply ultrasparse sampling to 4-D NOESY. SCRUB is derived from the CLEAN artifact-suppression method developed in the radioastronomy community in the 1970s21, but it achieves a much more thorough result, producing spectra with a very high dynamic range (>10000:1 in the absence of noise, or otherwise at the level of the thermal noise) and quantitative reproduction of signal intensities, volumes, and lineshapes. Using this new method, we demonstrate the application of sparse sampling to structure determination by solving the global folds of two proteins, one 23 kDa in size and the other 29 kDa, in each case using only a single ultrasparse time-shared NOESY experiment and fully automated crosspeak assignment.
RESULTS AND DISCUSSION
Design of SCRUB
We and other groups have previously addressed artifact problems in FT-processed sparsely sampled data using variations of the CLEAN algorithm20–28. The principle behind CLEAN is that each peak S in a sparsely sampled spectrum will be surrounded by a pattern of artifacts A that can be predicted a priori, through knowledge of the sampling pattern and its associated point response P. Thus one could calculate the artifacts associated with each peak and subtract them to produce an artifact-free spectrum. The actual calculation is the convolution of S with P and it yields the sum S + A.
Three challenging problems arise in applying this principle. First, the determination of S + A for a peak S depends on S’s shape and intensity. Second, the artifacts from any one signal distort all the other signals; thus one cannot know the true intensities of the stronger signals at first, and one frequently cannot see the weaker signals at all. Third, one must distinguish signals from noise and artifacts.
The first problem was traditionally solved by fitting the shape of each peak to an analytical function and then calculating the convolution. Fitting requires a priori knowledge of the proper shape of the signal, and is difficult to do in the presence of artifacts from other signals, thermal noise, and signal overlap. We proposed an alternative solution, decomposing the peak into individual volume elements (“voxels”) and processing these as if they were independent signals25. This decomposition is valid because of the linearity of the FT, and it eliminates any need for fitting or calculating convolutions. Determining S + A for a particular voxel requires only that P be aligned on that voxel and scaled according to the voxel’s intensity.
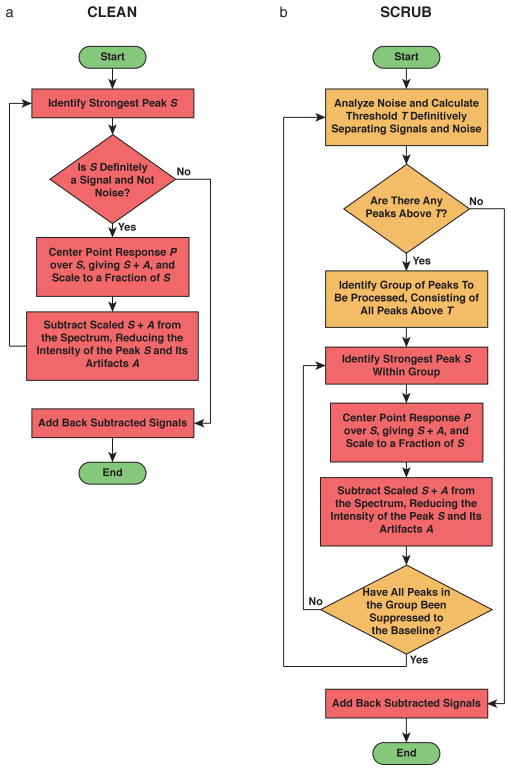
Because of the second and third problems, the original inventor of CLEAN, the radioastronomer J.A. Högbom, proposed an iterative procedure21 (Figure 1a). One begins with the strongest peak, which is the most likely to be a real signal out of all peaks present. S + A is calculated for this signal, scaled to some fraction of the peak height, and then subtracted from the original spectrum to remove a portion of the signal and its artifacts. This is repeated many times. As artifacts are removed, the true intensities of the other strong peaks become apparent, and the weaker peaks are uncovered. At some point, the remaining intensity or residual of the first peak becomes weaker than one of the other peaks, and the subtraction shifts to this second peak. Eventually, a point is reached when it is no longer possible to distinguish the residuals of the strong signals and/or unprocessed weak signals from remaining artifacts and thermal noise. At this point, the signals that were removed are restored by adding back an artifact-free S, appropriately scaled, at every position where S + A was subtracted. This iterative procedure produces a spectrum with significantly reduced artifacts.
Figure 1.
Flow charts illustrating (a) CLEAN and (b) SCRUB.
We have successfully applied a voxel-by-voxel CLEAN to several ultrasparse datasets20,25,28,29, including 4-D NOESY spectra without diagonals20,28, and others have also applied versions of CLEAN in NMR22–24,26,27. However, the dynamic range achieved by CLEAN was insufficient for the most challenging cases, such as 4-D NOESY spectra containing strong diagonals and weak crosspeaks20,28—as would be needed for structure determination.
We reasoned that CLEAN was not removing all of the artifacts, and that the dynamic range could be improved by additional suppression. Conventional implementations of CLEAN can only carry out suppression until the residuals of signals can no longer be distinguished from the apparent noise; the remaining residuals wouldbe expected to contribute significant artifacts. If one could suppress these residual signal components to the baseline, removing the rest of their artifacts, the dynamic range would be expected to improve substantially.
To achieve this, we designed a new algorithm, SCRUB (Scrupulous CLEANing to Remove Unwanted Baseline Artifacts). SCRUB follows the same logic as CLEAN, but organizes the suppression in a different way (Figure 1b). Instead of processing all signals and artifacts in one pass, SCRUB processes signals in groups over multiple passes. For each pass, a threshold is defined based on an analysis of the spectrum’s apparent noise, such that there is a very strong probability that all peaks above the threshold are signals rather than thermal noise or artifacts. This group of peaks, and only this group of peaks, is subjected to the iterative subtraction procedure extending all the way to the baseline. The noise statistics are then reevaluated, a new threshold is calculated, and the next pass begins.
The essential difference between traditional CLEAN and SCRUB is that each iteration of CLEAN approaches the spectrum anew, with no knowledge of which peaks in the spectrum might be residuals leftover from previous subtractions. By comparison, SCRUB identifies peaks from noise at the start of each pass and remembers this identification throughout the run, allowing it to apply the subtractive process with confidence all the way to the baseline.
For a full description of SCRUB, please see the methods section. Two movies demonstrating the operation of the algorithm on simulated datasets are included in the Supplementary Information.
Tests of SCRUB on Simulated Data
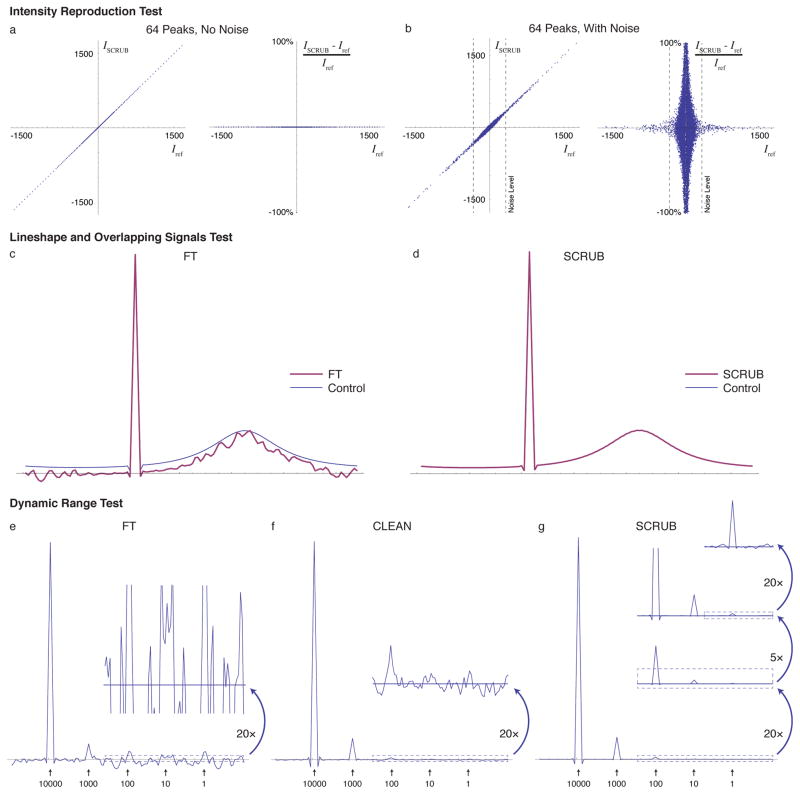
We have evaluated the capabilities of SCRUB using both simulated and experimental datasets. Imitating the 4-D experimental data described below, the simulations were conducted with three sparse dimensions, sampled at 1.2% of conventional Nyquist sampling using a cosine-weighted pattern. To measure the ability of SCRUB to remove artifacts from a spectrum without disturbing the accuracy of the signal intensities, we carried out simulations with a large number of signals of varied strengths and compared the intensity of every data point before and after SCRUB. Figures 2a–b contains correlation plots demonstrating the quantitative accuracy of SCRUB. With 64 signals concentrated on one plane in the 3-D matrix, in the absence of thermal noise the voxels representing signals are unchanged after SCRUB processing, and in the presence of noise they pick up only a very small error, comparable in magnitude to the noise.
Figure 2.
Tests of SCRUB Accuracy and Performance with Simulated Data. (a) Correlation and relative error plots comparing the intensity values in a simulated 3-D sparsely sampled (3189 time domain points, or 1.2% of conventional, distributed with cosine weighting, transformed into a 128 × 128 × 128 frequency domain, as is used for the 4-D experimental data later in the paper) spectrum of 64 peaks and no noise after SCRUB processing with that of a control spectrum without artifacts. SCRUB introduces no measureable error. (b) The same for a dataset also including simulated noise at a level of ~20% of the strongest signal. In the presence of noise, only a very slight error (comparable to the noise) is introduced by SCRUB. (c) 1-D trace of a 3-D simulated sparsely sampled (1.2%) spectrum containing two signals, one nondecaying of relative height 1.0 and the other Lorentzian with a full width at half height of ~25% of the spectral width and a relative height of 0.2, after FT processing. The control spectrum calculated using Nyquist sampling is shown as a thin trace. With sparse sampling, the lineshape of the broad peak is severely corrupted by artifacts. Note that the apparent vertical downshift is due to the convolution of the sparse sampling point response with the broad peak. (d) The same after artifact suppression with SCRUB, showing recovery of the correct lineshape and no problems due to the overlap of the two peaks (in this plot, the SCRUB trace exactly overlays the control trace). (e) 1-D trace of a simulated sparsely sampled spectrum (1.2%) containing five signals with relative intensities varying by five orders of magnitude, with white noise at ~10% of the weakest signal’s intensity; when processed using the FT alone, only the strongest two peaks are visible due to the sampling artifacts. (f) The same after processing with CLEAN. The third peak is now visible. (g) The same after processing with SCRUB. All peaks are visible. The residual noise and artifacts are only visible with 2000× magnification of the baseline.
The simulations in Figures 2c–d demonstrate that SCRUB is able to reproduce lineshapes correctly, including overlapped lineshapes. The simulation consists of two overlapping peaks, one nondecaying and the other a Lorentzian with a full width at half height of ~25% of the spectral width. Once again, simulated 3-D sparse time domain data were generated for 1.2% of the Nyquist sampling points. After FT, both peaks are visible, but the broad peak’s line-shape is severely corrupted by artifacts. SCRUB processing removes the artifacts, revealing the true lineshape.
Figures 2e–g contain simulation results demonstrating the dynamic range achievable with SCRUB, in comparison to CLEAN and to an FT without artifact suppression. Once again, only 1.2% of the conventional data are used. Five signals are present, each an order of magnitude weaker than the next, arranged on a 1-D trace within a 3-D space. The artifacts after FT present a noise floor that reaches 6.64% of the tallest signal’s height, obscuring those peaks that are less than 1/10 the strongest peak’s intensity. CLEAN reduces the artifact level to 0.3% of the tallest peak, which allows the detection of the 1/100 peak but nothing weaker. By comparison, SCRUB reduces the artifact level another two orders of magnitude, to 0.00115% of the tallest peak, allowing five orders of magnitude of signals to be detected. The dynamic range is improved from 15:1 with FT alone to ~87000:1 (whereas CLEAN achieves ~300:1).
Application of SCRUB to 4-D Time-Shared NOESY
In order to collect all possible NOE information for ILV methyl- and amide-protonated proteins, we designed a 4-D time-shared (TS) NOESY experiment that simultaneously records amide-amide, methyl-methyl, amide-methyl, and methyl-amide NOEs in 15N-HSQC-NOESY-15N-TROSY, 13C-HSQC-NOESY-13C-HSQC, 15N-HSQC-NOESY-13C-HSQC, and 13C-HSQC-NOESY-15N-TROSY spectra, respectively (Figure S1). This is made possible by implementing concatenated 13C and 15N coherence transfer elements and joint 13C/15N evolution periods that provide optimal resolution for both nuclei30. Our experiment is conceptually similar to a previous implementation31, but features sensitivity-enhanced coherence transfers both before and after the NOE transfer period.
We tested the 4-D TS NOESY experiment on two ILV methyl- and amide-protonated proteins, the 23 kDa phosphatase Ssu72 and 29 kDa human carbonic anhydrase II (HCAII), using a cold probe-equipped 800 MHz spectrometer. For each dataset, 3189 complex points were sampled, representing just 1.2% of the measurements required by conventional Nyquist sampling at equivalent resolution. This ultrasparse sampling allowed each 4-D dataset to be collected in only 4 days. The four simultaneously-recorded NOESY pathways were separated and processed with either the Fourier transform (FT) alone or the FT followed by artifact removal with CLEAN or SCRUB. This produced four 4-D spectra for each protein with dimensions (N1, HN1, N2, HN2), (C1, HC1, C2, HC2), (N1, HN1, C2, HC2) and (C1, HC1, N2, HN2), where label numbers 1 and 2 denote NOE donor and acceptor groups, respectively, and the directly detected dimension is HN2/HC2. The final resolution of each indirect dimension is 128 points.
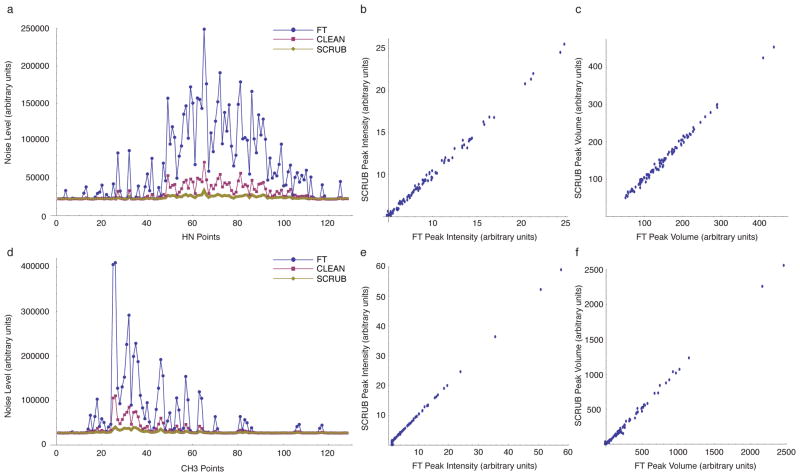
As expected, certain regions of the FT-processed methyl-methyl and amide-amide spectra have severely elevated noise levels due to the aliasing artifacts generated by strong diagonal signals. With SCRUB processing, however, there was a dramatic reduction in the aliasing artifacts, with noise levels decreased to, or close to, the baseline experimental noise (Figures 3 and 4). Maximum noise reductions of 7.37-fold and 11.39-fold were observed for the HCAII amide-amide and methyl-methyl spectra, respectively. The FT-processed amide-methyl and methyl-amide spectra, which do not contain diagonal peaks, have significantly lower levels of aliasing artifacts, and SCRUB processing effectively suppressed artifacts in these spectra as well (data not shown). By comparison, CLEAN reduces but does not completely eliminate the artifacts. As the correlation plots in Figure 3 show, the intensities and volumes of the stronger peaks are not substantially affected by SCRUB processing (the comparison does not include weaker peaks, as they cannot be observed without the help of artifact suppression).
Figure 3.
Noise Reduction with SCRUB in 4-D NOESY Experiments for HCAII. (a) Comparison of the apparent noise level (thermal noise + artifacts) measured for each F1/F2/F3 cube of the sparsely sampled (1.2%) amide-amide 4-D NOESY of HCAII after processing with the FT alone, CLEAN, or SCRUB. (b) Comparison of peak intensities with and without SCRUB for the 100 strongest peaks in the amide-amide 4-D NOESY of HCAII. The peak list was picked automatically by NMRView and was not edited. (c) Comparison of peak volumes with and without SCRUB for the 100 strongest peaks in the amide-amide 4-D NOESY of HCAII. The same peak list was used as in (b), and the volumes were calculated by NMR-View. (d)–(f) The same for the methyl-methyl 4-D NOESY of HCAII.
Figure 4.
Artifact removal with SCRUB in 4-D TS NOESY spectra. Representative F1/F2 planes from the amide-amide and methyl-methyl NOESY spectra of HCAII are shown after processing with either the FT (left panel), CLEAN (middle panel), or SCRUB (right panel). Panels are plotted at identical contour levels, and 1-D slices along the green dashed lines are shown above each panel to highlight long-range crosspeaks that are lost to aliasing noise in the FT- and CLEAN- processed spectra. Planes in (a) and (b) are taken from F1/F2/F3 cubes #75 and #31, respectively, which have 5.52-fold and 7.73-fold reductions in artifact noise with SCRUB processing, respectively. The marked crosspeak in (a) is from T207 HN to G139 HN, and the marked crosspeak in (b) is from V68 γ2 to V159 γ2.
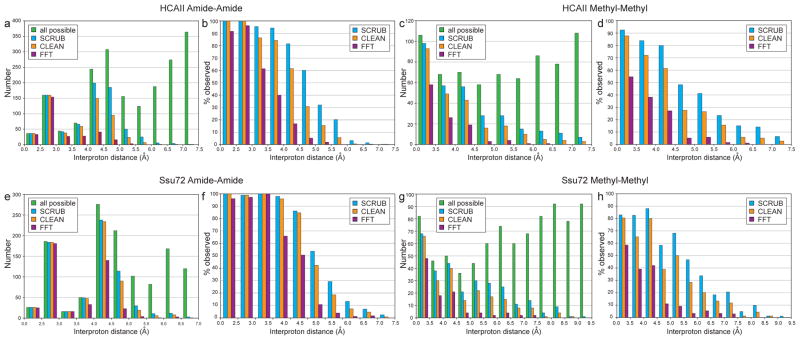
The success of SCRUB processing in removing artifact noise from the diagonal-containing TS spectra should aid significantly in the identification of NOE crosspeaks. To measure this benefit quantitatively, we assigned automatically picked and manually edited peak lists from the full SCRUB-processed TS datasets for both test proteins as described in the Methods section, and filtered them based on the noise levels of the FT-, CLEAN- or SCRUB- processed datasets. Peaks with intensities greater than four times the noise level at their resident HC2/HN2 points were matched to their assigned interproton distances based on a reference crystal structure. Figure 5 shows the resulting histograms for the methyl-methyl and amide-amide crosspeaks.
Figure 5.
Removal of aliasing artifacts promotes peak identification in the 4-D TS NOESY spectra for HCAII (top) and Ssu72 (bottom). NOE crosspeaks with intensities over four times the noise level in spectra processed with either the FT, CLEAN, or SCRUB were matched to their corresponding interproton distances, and unique distances are plotted as histograms in (a, c, e, g). Each interproton distance was counted as observed if either corresponding NOE (Hi → Hj or Hj → Hi) surpassed the noise threshold. “All expected” refers to the total number of interproton distances calculated from a reference crystal structure (PDB code 2ILI for HCAII and PDB code 3P9Y for Ssu72). The histograms in (b, d, f, h) show the percent of observed distances relative to all expected. Short interproton distances that were not identified in the SCRUB-processed methyl-methyl spectra are the result of signal degeneracy. For example, the four unidentified distances under 3.5 Å (out of 53 total) for HCAII correspond to intraresidue methyl-methyl NOE crosspeaks for residues V109, V121, L143, and L223 that overlap with diagonal signals. The small number of very long distances (>8.0 Å) observed in the methyl-methyl spectrum of Ssu72 likely result from incorrect rotamer assignments in the reference crystal structure due to the insufficient resolution of the model (2.1 Å) for precisely defining ILV sidechain orientations. For example, the two longest distances—9.31 Å between I173 δ1 and L176 δ2 and 8.55 Å between V43 γ2 and L45 δ2—are substantially shorter in a second crystal structure (PDB code 3FDF, 3.2 Å resolution)—6.99 Å and 7.87 Å, respectively—as the result of different rotamer selections for residues L45 and I173.
For HCAII, only 58% and 60% of the interproton distances 4.5 Å or shorter exceed the noise threshold in the FT-processed methyl-methyl and amide-amide spectra. CLEAN processing improves the spectral quality, with 83% and 88% of interproton distances under 4.5 Å exceeding the noise threshold in the methyl-methyl and amide-amide spectra. In the SCRUB-processed spectra, however, the data completeness improves even more dramatically, to 90% in the methyl-methyl spectrum and 96% in the amide-amide spectrum, again for distances under 4.5 Å. Over the full range of interproton distances, there are 137 distance constraints (22% of the total number) that exceed the noise threshold in the two SCRUB-processed spectra but not in the CLEAN-processed spectra, underscoring the practical advantage provided by this new method. Similar results were obtained for Ssu72. As shown in Figure 5, longer interproton distances—which correspond to weaker NOE cross-peaks—are preferentially lost in the CLEAN- and FT-processed datasets. Many of these lower-intensity peaks provide long-range contacts that are crucial for successfully defining a target’s topology (see examples in Figure 4). It should also be noted that in the SCRUB-processed spectra it was almost always possible to observe crosspeaks for both magnetization transfer directions (Hi → Hj as well as Hj → Hi), but the CLEAN- and FT-processed spectra were significantly less redundant. For example, for HCAII, 92% and 89% of interproton distances under 4.5 Å were derived from two cross-peaks in the methyl-methyl and amide-amide spectra, respectively, versus 83% and 82% in the CLEAN-processed spectra and 45% and 72% in the FT-processed spectra. In the amide-methyl and methyl-amide datasets for both test proteins, nearly all peaks in the SCRUB-processed spectra were also identifiable in the CLEAN- and FT-processed spectra, a result that is consistent with the lower artifact levels observed in these diagonal-free datasets.
Calculation of Global Folds
To illustrate the impact of the high-resolution, high-dimensionality TS NOESY data on structure determination, we used the automated assignment protocol of CYANA32 to calculate the global folds of Ssu72 and HCAII. The only input constraints were dihedral restraints derived from the backbone chemical shifts33 and peak lists from all four TS NOESY spectra. Calculations were performed with either (1) automatically picked peak lists, filtered by computer to eliminate peaks with no possible assignment, but unedited by human examination, or (2) peak lists that were automatically picked and filtered and then manually edited to eliminate artifact signals. As a positive control illustrating the best possible structures for this data, we also calculated global folds using automatically picked and manually edited peak lists that were assigned manually in reference to the crystal structures of the two test proteins. For each input, five independent calculations were performed. Out of the five calculations conducted for each protein, the ensemble with the median RMSD of backbone atoms in the core domain (RMSDbb,core) is shown for HCAII in Figure 6 and Ssu72 in Figure 7, and evaluations of automated peak assignment correctness and ensemble convergence and accuracy are presented in Tables S1–S4. As described in the Methods section, it was necessary to make modifications to the automated assignment protocol of CYANA to account for the sparseness of NOE data involving only methyl and amide groups.
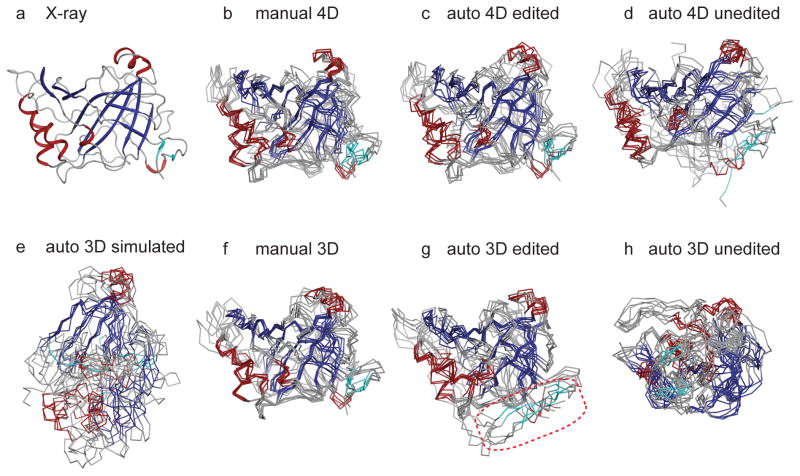
Figure 6.
CYANA structure calculations for HCAII using manually assigned and manually edited peaks (b, f), auto-assigned and manually edited peaks (c, g), and auto-assigned and unedited peaks (d, h). In (b–d), peak lists are from SCRUB-processed 4-D TS spectra. Cyan coloring highlights two small parallel strands that are part of the central β-sheet that are misplaced in the structures calculated with manually edited 3-D peak lists (boxed in red in (g)). In (f–h), peak lists are from conventional 3-D TS spectra. In (e), calculations are based on simulated 3-D peak lists derived from 4-D spectra. The reference crystal structure (PDB code 2ILI) is shown in (a). The ensemble in (e) is aligned over the largest segment of converged structure. The unconverged N-terminus of HCAII (21 residues) is omitted from (b)–(h) for clarity.
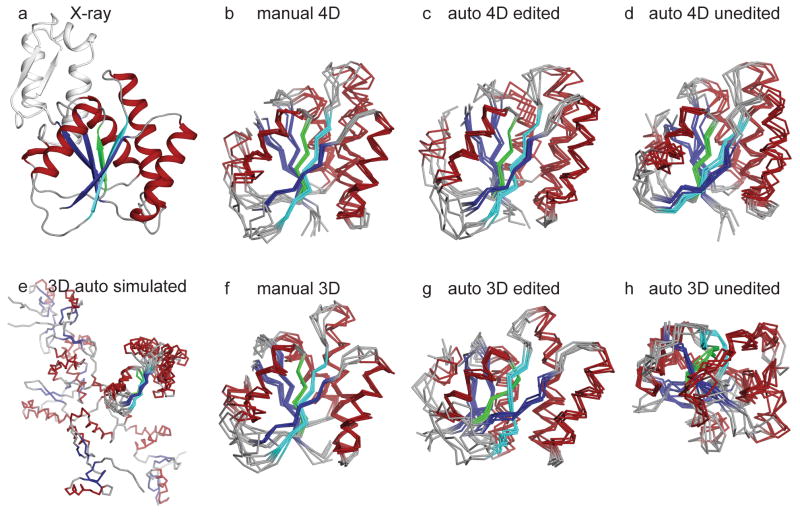
Figure 7.
CYANA structure calculations for Ssu72 using manually assigned and manually edited peaks (b, f), auto-assigned and manually edited peaks (c, g), and auto-assigned and unedited peaks (d, h). In (b–d), peak lists are from SCRUB-processed 4-D TS spectra. Green and cyan coloring highlights the characteristic β-propeller twist of the central β-sheet that is lost in the ensemble produced by auto-assigned and manually edited 3-D peak lists (compare (c) and (g)) In (f–h), peak lists are from conventional 3-D TS spectra. In (e), calculations are based on simulated 3-D peak lists derived from 4-D spectra. The reference crystal structure (PDB code 3FDF) is shown in (a). The substrate-binding subdomain of Ssu72, which is not constrained to the main phosphatase domain by the TS NOESY data, is omitted from (b–h), and is aligned separately in Figure S2. The ensemble in (e) is aligned over the largest segment of converged structure.
For both test proteins, the control global fold calculations based on manually-assigned 4-D TS NOESY crosspeaks yielded accurate and well-converged structural ensembles (Figures 6b and 7b), a result in line with the excellent completeness of observed interproton distances. For HCAII, the ensembles from five independent calculations have a mean RMSDbb,core of 0.904 ± 0.084 Å and a bias to the reference crystal structure over the same set of atoms of 1.634 ± 0.075 Å. For Ssu72, the mean RMSDbb,core is 0.923 ± 0.128 Å and the bias is 2.076 ± 0.126 Å. Both structure calculations also demonstrate the limitations of the ILV methyl- and amide-protonated labeling strategy. For example, the first 21 residues of HCAII lack an ILV residue and do not feature any long-range amide-mediated contacts to the main body of the enzyme, and hence are not converged in the structure calculations. In the case of Ssu72, the enzyme contains a substrate-binding subdomain which lacks long-range ILV methyl- and amide-mediated contacts to the core domain and is omitted from Figure 7. While the subdomain structure converges (see Figure S2), its orientation relative to the core of the enzyme is poorly defined.
The results from structure calculations with automated assignment of the 4-D TS NOESY peak lists highlight the advantage of the high dimensionality datasets. For both targets, automated assignment of the manually edited 4-D peak lists by CYANA was remarkably accurate, with low levels of assignment ambiguity. For HCAII, 98% of all crosspeaks were assigned correctly when compared to the manual assignments, including 97% of the critical long-range NOEs, and only 13% of crosspeaks were assigned ambiguously. A similar level of success was observed in the Ssu72 calculations, with 97% of crosspeaks assigned correctly, including 94% of the long-range NOEs, and only 11% assigned ambiguously. These calculations produced high-quality structural ensembles, with mean RMSDsbb,core of 0.958 ± 0.081 Å and 1.009 ± 0.158 Å for HCAII and Ssu72, respectively (Figures 6c and 7c). The biases of the ensembles were also close to those observed in ensembles calculated with manually assigned peak lists, indicating that the automated assignment did not introduce any systematic structural distortions. For both target proteins, using the automatically picked and unedited 4-D peak lists as input to CYANA resulted in ensembles that are well-converged but with slightly more deviation from the crystal structure (Figures 6d and 7d).
To assess the benefits conferred by the high dimensionality of the 4-D TS data, we performed control global fold calculations with two types of 3-D data. For the first, we used simulated 3-D peak lists, which were generated by removing the first indirect heteronuclear dimension (C1/N1) of the 4-D peak lists, and which therefore have the same resolution and sensitivity as the 4-D data. The resulting calculations produced structures that either failed to converge or converged poorly to inaccurate conformations. For HCAII, small portions of the HCAII structures converge accurately along the central β-sheet, however, the remainder of the protein is pulled into a vastly non-native conformation (Figure 6e). Similarly, the C-terminal portion of the central β-sheet in Ssu72 converges, but the N-terminus is largely unrestrained (Figure 7e).
In a second approach, we collected an experimental 3-D TS dataset at high resolution and picked peaks in the same fashion described above for the 4-D dataset, to give two analogous sets of peak lists: (1) automatically picked/filtered, without any human intervention and (2) automatically picked/filtered and then manually edited. As with the 4-D data, a positive control was calculated to gauge the accuracy of automated assignment by CYANA, using the 3-D manually edited peak lists after assignment by an experienced spectroscopist, with reference to the crystal structure. Compared to their 4-D counterparts, the 3-D control calculations based on manually assigned peak lists produced marginally better ensembles due to the higher sensitivity of the 3-D spectra and therefore increased numbers of crosspeaks (Figures 6f and 7f).
In contrast, the ensembles generated with the automated assignment protocol using manually edited 3-D peak lists were much worse than those generated using the equivalent 4-D peak lists, particularly in terms of the bias from the reference crystal structure. In the case of HCAII, two small parallel strands (colored cyan in Figure 6) that are part of the central β-sheet are misplaced in the structures calculated with manually edited 3-D peak lists, disrupting the integrity of the central β-sheet (compare Figures 6c and 6g). In the case of Ssu72, the characteristic β-propeller twist of the central β-sheet is lost in the ensembles produced by automated assignment of manually edited 3-D peak lists (compare Figures 7c and 7g). On the other hand, there is no obvious structural distortion in the ensembles generated by the automated assignment protocol of CYANA when using manually edited 4-D peak lists: the structures maintain the correct protein fold, and they have statistics similar to the ensembles derived from manually assigned peak lists.
With 3-D peak lists that were automatically picked and filtered but unedited by a spectroscopist, CYANA produced ensembles which converged tightly, but to protein folds that are wildly inaccurate (Figures 6h and 7h). This is in sharp contrast with the ensembles based on the unedited 4-D peak lists, which converged to the correct fold, with only a slight increase in the ensemble RMSD and structural bias.
CONCLUSIONS
We have demonstrated that the SCRUB algorithm’s much more aggressive treatment of residual signal intensities during artifact suppression makes it possible to rescue weak NOE crosspeaks from aliasing artifact noise, finally permitting the application of ultrasparse sampling to NOESY experiments with large dynamic range. In our analysis, we were routinely able to identify NOE crosspeaks with intensities 100-fold less than diagonal signals. For example, the peaks marked in Figures 4a and 4b are 112-fold and 203-fold weaker, respectively, than the strongest diagonal signals at their resident HC2 points. Coupling ultrasparse sampling with the TS NOESY technique maximizes the efficiency of data collection, allowing us to collect all the 4-D data necessary for global fold calculations—at a resolution unachievable with conventional methods—in only four days. As shown here, this high-resolution, high-dimensionality data has an enormous practical benefit in automated NOE assignment procedures. For proteins like HCAII and Ssu72, where there is sufficient long-range distance constraint information available in sparsely protonated samples, it is possible to go from NMR sample to global fold in a matter of days.
Several other methods have been developed for processing sparsely-sampled NMR data, including three—coupled-multidimensional decomposition, forward maximum entropy reconstruction, and CLEAN—that have been applied to 4-D NOESY experiments 18,20,25,27,34,35. The first two methods reconstruct the full time domain dataset by iterative fitting against the frequency domain spectrum. It should be noted that these procedures require sampling rates of at least 10% of the Nyquist grid, or nearly an order of magnitude more than the ultrasparse sampling rate used in this study (1.2%). As we show in this study, much lower sampling rates are sufficient for observing nearly complete interproton distances through 4.5 Å in selectively-protonated proteins of up to 29 kDa. It should also be noted that SCRUB is relatively fast, as it involves only additions and subtractions in the frequency domain. Typical runtimes for a full 4-D spectrum on a 2.4 GHz CPU vary from twenty minutes to two hours.
CLEAN, from which SCRUB is derived, also works on extremely sparse data, and is also a computationally fast method20,25–28. Conventional CLEAN is not able to achieve sufficient artifact suppression to be applied to most NOESY spectra with diagonals, a major impediment to its use in structure determination20. A recent implementation from Stanek and Kozminski26,27 comes closer, but it relies on fitting decaying sinusoids to time domain data, which is always challenging, especially for complicated spectra containing overlapping, broad, or distorted signals and in the presence of noise. SCRUB achieves much more thorough artifact suppression without any reliance on fitting. It does this by processing individual voxels rather than signals. A voxel-by-voxel procedure makes it possible to deconvolute the artifacts from any spectrum, regardless of complexity, without any impact on the intensities, volumes, or lineshapes of the signals.
Due to the advantages afforded by 4-D NOESY data, we expect that the methods presented here will be particularly useful as NMR structure determination continues to push toward higher molecular weight targets.
COMPUTATIONAL METHODS
SCRUB
Prior to the start of processing, the point response is calculated from the sampling pattern, and an artifact-free point response is prepared by extracting the point response’s central peak. At all stages, noise levels are estimated by computing histograms of the intensities of data points and fitting the sum of two Gaussians, one to fit the noise distribution and the other to fit the signal distribution. We derive as follows the highest intensity value Inmax that a noise voxel is likely to assume in a spectrum of N voxels given that the noise follows a normal distribution with a mean of zero and a standard deviation σ. The probability P(−I < x < I) that a noise voxel’s intensity x will be within the range −I to I is:
where erf is the error function. We are seeking the intensity Inmax for which P(x < −Inmax ∨ x > Inmax) is less than or equal to 1/N:
Solving for Inmax, we obtain:
SCRUB processes a 4-D spectrum in 3-D cubes, and each cube in batches of voxels that undergo artifact suppression simultaneously. All steps are carried out in the frequency domain. The voxels to be suppressed in each batch are selected based on a threshold Tmain that begins as the noise floor plus a margin τ which changes after each batch:
where i is the batch number. For the first batch i = 0, τ0 is set to Inmax, meaning that the initial threshold is 2Inmax. At the end of each batch, τ is decreased by half the current standard deviation of the noise:
To reduce the likelihood of an artifact peak being treated as a signal, the selection of voxels for a SCRUB batch is interwoven with the first cycles of artifact suppression on that batch. Processing begins with the voxel that has the strongest intensity, and others are added to the batch later using several criteria. The first step of processing a batch is to check the intensity of the strongest voxel in the spectrum against Tmain; if it qualifies, the batch processing begins. If it does not exceed Tmain in intensity, Tmain is reduced by successive σ/2 increments until the strongest voxel qualifies or the stopping criterion is met (see below). A portion of the artifacts generated by the strongest voxel is then removed by translating the artifact-corrupted point response so that its central peak is aligned with the strongest voxel, scaling it to a height of g times the strongest voxel’s intensity, where g is the user-specified “loop gain” parameter, and then subtracting the scaled and translated point response. The loop gain is a number between 0 and 1 specifying the fraction of a signal and its artifacts to be removed during each suppression operation; unless otherwise stated, we use a value of 10%. Note that g is always multiplied by the current intensity of a voxel rather than the original intensity, so that the intensity changes by the same relative value but progressively smaller absolute values in subsequent cycles.
After the first subtraction operation on the strongest voxel, it will have an intensity which we shall call the suppression level Isupp. SCRUB then surveys all other voxels in the spectrum, looking for any with an intensity greater than Tmain and also greater than Isupp + τi to include in the batch. If none are found, further subtractions are carried out on the strongest voxel, with an additional test after each subtraction, until either a voxel is found to add to the batch or the stopping criterion (described below) is reached with respect to the strongest voxel. The Isupp + τi test ensures that voxels that are added to a batch belong to real signals by requiring them to remain strong (above Tmain) through enough suppression operations to put the originally strongest voxel at least τi below them. This is based on the idea that an artifact peak with enough intensity to exceed Tmain would almost always be formed with a substantial contribution from the artifact pattern of the strongest peak, so the suppression of the strongest peak would be expected to weaken any such artifact peaks.
A different standard is used for voxels that are directly adjacent to voxels already belonging to the batch. Instead of being required to exceed Tmain and Isupp + τi in intensity, they instead must exceed Tadj = Inmax + τi/2 and Isupp + τi/2. This weaker standard is based on the idea that signals usually occupy more than one voxel. Once the strongest voxel of the signal has been identified and validated as a real signal, the likelihood that adjacent voxels contain signals rather than noise or artifacts is also much higher. A relaxed test makes it easier for SCRUB to identify and validate all of the voxels belonging to a signal, ensuring that all of them undergo artifact suppression.
When a voxel is found that meets either the Tmain and Isupp + τi standard (if it is not adjacent to a batch member) or the Tadj = Inmax + τi/2 and Isupp + τi/2 standard (if it is adjacent), that voxel is added to the batch. It is then subjected to cycles of artifact suppression until its intensity drops to Isupp, and it has the same intensity as the other members of the batch.
The suppression level Isupp is then lowered through iterative artifact suppression. In each cycle, the point response is translated, scaled, and subtracted from each member of the group in turn, reducing all of them by the same increment of gIsupp. The spectrum is then checked for additional voxels which may qualify to be added to the batch before the next cycle begins. Note that Tmain and Tadj are recalculated after each cycle of applying suppression to all voxels in the batch, taking into account reductions in Inmax due to artifact suppression. This may make it possible to add voxels to a batch that were just below the threshold initially.
Suppression of a batch continues until the following condition is met. Since the level of artifacts still remaining in the spectrum from a given suppressed voxel is proportional to Isupp, and since the artifact contributions from multiple such voxels are additive, the upper limit for the level of artifacts Iart remaining from a batch of voxels that have been suppressed to Isupp should be roughly proportional to nbatchIsupp, where nbatch is the number of voxels in the batch. Our goal is to suppress the batch until the remaining artifacts Iart can be deemed to be making a negligible contribution, at most, to the observed noise level Inmax, i.e. Iart ≫ Inmax. Since Iart ∝ nbatchIsupp, we know that Iart ≫ Inmax if nbatchIsupp ≫ Inmax. The stopping criterion is therefore to require:
where b is a user-specified small number, here set to 1%.
Batches of voxels are processed until it becomes difficult to be certain that any remaining peaks are signals rather than noise. We consider this condition to be reached when Tmain has been decreased to within s standard deviations of the noise floor, where s is a user-specified constant, here set to two.
SCRUB maintains a table of all suppression operations that are carried out and uses it, together with the artifact-free point response, to restore the signals at the end of the run. The artifact-free point response is translated to each voxel where signal intensity was suppressed, scaled to match the original signal height at that position (less any residual height), and added.
The software is written in C++ and available from the authors by request. It has been compiled and tested on Mac OS X, Windows, and Linux systems.
Sampling Pattern
A sampling pattern was constructed using the randomized concentric shell sampling method11 containing 3189 points arranged with cosine weighting on 64 shells, adapted to a 64 × 64 × 64 point grid. Each sampling point was assigned a weight based on its Voronoi volume. The sampling pattern contained 1.217% of the data that would be collected conventionally for a 64 × 64 × 64 point time domain. All spectra were calculated at 128 × 128 × 128 point digital resolution in the indirect dimensions.
Simulations
In all simulations, 3-D sparse time domains (imitating one cube out of a 4-D spectrum) were populated with sinusoids corresponding to the desired signals, the FT was computed, and SCRUB processing was carried out, using default settings unless otherwise noted. Frequencies were chosen to place all signals on the same plane, and for small numbers of signals, on the same line.
For the simulation assessing the accuracy of intensity reproduction, 64 non-decaying signals were used. Because the sampling points were distributed according to a cosine weighting pattern, these signals automatically acquired a finite, cosine-apodized line-shape upon FT. Half of the signals were given positive intensities and half negative; within each group, intensities ranged from 0.03125c to 1.0c in multiples of 0.03125, where c is an arbitrary constant. The control spectrum was constructed in the frequency domain by placing a copy of the artifact-free point response (generated from the sampling pattern) at each expected signal position, scaled according to the expected signal height; this method ensures that the control signals have the same lineshape as the sparse signals, including any subtle variations from the cosine shape arising from randomization and/or imperfections in the sparse pattern’s statistical approximation of a cosine apodization function. In a second test, white noise with a mean of zero and a standard deviation set so that the noise level would be at ~20% of the tallest peak’s intensity after FT was added to the sparse dataset. For comparison, the same white noise values were transformed and added to the frequency domain control; by preparing the noise for the control in this manner, any effects of sparse sampling on the behavior of the noise would be accounted for.
For the lineshape and overlapping signals test, two signals were used, the first nondecaying with a relative height of 1.0 and the second decaying with an amplitude and relaxation rate set to produce a peak with a relative height of 0.2 and a full width at half height of ~25% of the spectral width (33 data points in a spectrum of 128 points). Because of the very wide second signal, SCRUB required many more subtractions than for normal signals, and a gain of 75% was used to speed up the calculation. For the control, a 64 × 64 × 64 conventional time domain was populated with values corresponding to the same two signals, multiplied by the inverse Fourier transform of the artifact-free point response (to apply the same window function that the cosine-weighted sampling pattern imparts implicitly to the sparsely sampled data), and then Fourier transformed.
To assess the dynamic range achievable with SCRUB, non-decaying signals at relative intensities of 10000, 1000, 100, 10, and 1 were used. White noise was added such that it would have a level of ~10% of the weakest peak after FT.
EXPERIMENTAL METHODS
Sample Preparation
Full-length human carbonic anhydrase II (HCAII) and Ssu72 were expressed and purified as described previously36,37. The Ssu72 construct contains an active site mutation (C13S) that abolishes its phosphatase activity38. Perdeuterated proteins were expressed in D2O M9 minimal media with 15N-NH4Cl and 2H/13C-glucose, with the addition of 85 mg [3-2H] 13C-α-ketoisovalerate and 50 mg [3,3-2H2] 13C-α-ketobutyrate ~1 hour prior to induction for selective protonation of ILV methyl groups39. Samples prepared from 10% 13C-glucose M9 minimal media were used to stereospecifically assign valine and leucine methyl groups40. All isotopes were purchased from Cambridge Isotope Laboratories, Inc. Final sample conditions were 25 mM Tris-HCl pH 8.0, 25 mM KCl, 2 mM dithiothreitol, and 5% D2O for Ssu72, and 100 mM potassium phosphate pH 6.8, 2 mM dithiothreitol, and 10% D2O for HCAII. All samples contained ~1 mM protein.
Data Acquisition and Processing
Time-shared (TS) NOESY datasets were collected at 30°C for Ssu72 and 25°C for HCAII on an 800 MHz Agilent Inova spectrometer equipped with a triple-resonance, cryogenically cooled probe. 4-D data collection used the sparse sampling pattern described above and the pulse sequence given in Figure S1. Modification of the TS NOESY pulse sequence code allowed sampling from an explicit schedule of evolution times. Maximum evolution times were 0.0119 s for HN1/HC1 (5400 Hz spectral width), 0.0237 s for N1 and N2 (2700 Hz spectral width), and 0.0162 s for C1 and C2 (3950 Hz spectral width). Signals in the first proton dimension are frequency-shifted in order to center the amide region while leaving the transmitter frequency on water. Therefore, the indirect proton spectral width covers from water to the edge of the amide proton spectrum, and methyl protons are aliased. With a recycle delay of 1.4 s and 4 scans per FID, the total acquisition time for each 4-D dataset was 98 hours.
The first step in processing the 4-D TS NOESY datasets was to separate overlapping NOESY pathways31. Magnetization pathways detected on different proton types (e.g. amide-methyl and amide-amide NOESY) are readily separated by setting the spectral width of the directly detected dimension to cover both amide and methyl proton signals and then extracting the appropriate region when computing the Fourier transformation. Pathways detected on the same proton type (e.g. amide-methyl and methyl-methyl) overlap due to the aliasing of methyl protons in the second indirect dimension, and are instead separated by collecting two interleaved datasets for each set of frequency labeling delays, with magnetization pathways originating on methyl groups selectively inverted in the second, which are then added and subtracted to select pathways originating solely from amide and methyl groups, respectively. The second processing step is to resolve the sensitivity-enhanced signals recorded in the first two dimensions into quadrature components as described elsewhere41. Finally, the indirect dimension data were converted from hypercomplex encoding in one octant to complex encoding in four octants using formulas described previously39,40.
Time domain datasets were then processed with either the FT alone, or the FT followed by artifact removal with CLEAN or SCRUB. All spectra were calculated at 128 point resolution in each indirect dimension42,43, and direct dimensions were zero-filled to give a final resolution of ~128 points after extracting the appropriate region of proton signals. SCRUB processing times for the amide-amide, amide-methyl, methyl-amide, and methyl-methyl spectra were 35 minutes, 4 minutes, 2 minutes, and 65 minutes, respectively, for the HCAII dataset, and 22 minutes, 5 minutes, 3 minutes, and 115 minutes, respectively, for the Ssu72 dataset.
Control 3-D datasets were collected with conventional sampling and a TS pulse sequence that lacks a 13C/15N coherence transfer and chemical shift evolution before NOE transfer. Evolution times and spectral widths were identical to the 4-D experiment with the exception of the indirect proton dimension HN1/HC1, which was collected at twice the spectral width (10800 Hz) to avoid the overlap of amide and methyl signals. With a recycle delay of 1.4 s; 8 scans per FID; and 128 and 48 complex points for the indirect 1H and 15N/13C dimensions, respectively, the total acquisition time for each 3-D dataset was 92.5 hours. The indirect dimensions were linear predicted and zero filled to generate final matrices of 512 point for the indirect 1H dimension after extracting the appropriate region and 128 points for the indirect 13C/15N dimension.
Data Analysis
The 3-D and 4-D TS NOESY spectra were analyzed with NMRView44. Peaks were picked at three times the noise level using NMRView’s automated peak picking function, and filtered by computer against the target protein’s methyl and amide chemical shifts to ensure that each peak had at least one assignment possibility within the chemical shift tolerances used by CYANA (see below). Next, peaks were examined manually by an experienced spectroscopist to remove obvious noise or artifacts. Finally, the automatically picked and manually edited peak lists were assigned manually with reference to the crystal structure, for use in control structure calculations and as benchmarks to measure the success of automated assignments by CYANA. We also tested CYANA with 3-D and 4-D peak lists generated without any human intervention. These peak lists were picked automatically with NMRView at four times the noise level and then filtered against the methyl and amide chemical shifts. No further editing was performed on these automatically picked and unedited peak lists. Simulated 3-D peak lists were generated by removing the first heteronuclear dimension (C1/N1) of the manually edited 4-D peak lists.
For the histogram analysis in Figure 5, the automatically picked and manually edited 4-D peaks were matched to their corresponding interproton distance using reference crystal structures 2ILI45 (1.05 Å resolution) and 3P9Y37 (2.1 Å resolution) for HCAII and Ssu72, respectively. Stereospecific assignments of leucine and valine methyls were used to improve the accuracy of distance matching, but were not used as restraints in structure calculations. Each distance was counted as observed if either corresponding NOE crosspeak (Hi → Hj or Hj → Hi) was above the noise threshold. Interproton distances involving methyl protons were calculated based on pseudoatom positions. Interproton distances involving amide groups without observable peaks in the 15N-HSQC-TROSY spectra of Ssu72 and HCAII were not included in the “all expected” numbers. Although the Ssu72 sample used in this study is in the apo form, we chose to use the substrate-bound crystal structure (3P9Y) to calculate interproton distances rather than the available apo form crystal structure (3FDF), which is at much lower resolution (3.2 Å). While the backbone conformations of the two states are nearly identical37, substantial discrepancies were observed for a number of methyl-methyl interproton distances due to different rotamer fitting for certain ILV sidechains. The higher-resolution substrate-bound model should allow for better rotamer fitting for these sidechains and therefore more accurate interproton distances.
Structure Calculations
All structure calculations were performed with CYANA 3.032 with five independent runs for each setup. Dihedral angle restraints were derived from backbone carbon chemical shifts using TALOS+33; 292 and 390 dihedral restraints were calculated for Ssu72 and HCA2, respectively. Upper distance limits were calculated by CYANA based on peak volumes with the average distance limit set to 4.5 Å and lower and upper cutoffs set to 2.0 Å and 7.0 Å, respectively. For calculations with manually-assigned peak lists, CYANA was run with default parameters and 20,000 torsion angle dynamics steps. One hundred structures were calculated for each run with the five lowest energy structures represented in the final ensemble.
For calculations with automated assignment of peak lists, significant modifications to the default CYANA protocol were required to account for the sparse distance constraint information inherent to global fold calculations. CYANA uses seven cycles of combined automated NOE assignment and structure calculation, and in each cycle potential peak assignments are scored on several criteria, including how well they are fulfilled in the ensemble from the previous cycle. This structure-based probability is controlled by the “violation cutoff” variable, which decreases from cycle to cycle. Due to the limited amount of distance constraint information available in selectively-protonated samples, global fold calculations converge relatively slowly over the seven cycles of the CYANA calculation, causing large numbers of potential assignments to be prematurely discarded based on the default violation cutoff values. To overcome this issue, the structure-based probabilities were relaxed (by setting the violation cutoffs to ten times their default values in the noeassign.cya macro). A second criterion used by CYANA for judging peak assignment possibilities is network anchoring, which measures each potential peak assignment’s fit into the network formed by the assignment possibilities of all other peaks. In general, network anchoring probabilities were unexpectedly low in the global fold calculations, presumably due to the low redundancy and limited distance constraint information, which led to many peaks left unassigned, particularly in the amide-amide datasets. Therefore, the weight of the network anchoring probability was decreased relative to the other assignment criteria (by setting the variable alignfactor to 10 in noeassign.cya), and the overall probability threshold for assignment acceptance was lowered (by setting the quality variable to 0.30 in noeassign.cya). Importantly, these modifications were critical for the success of global fold calculations based on both 3-D and 4-D peak lists.
For calculations with 4-D peak lists, the chemical shift tolerance for determining peak assignment possibilities was set to 0.02 ppm in the direct methyl dimension (HC2), and the tolerances in all other dimensions were scaled to this value based on digital resolutions, giving tolerances of 0.06 ppm for HN2, 0.08 ppm for HC1/HN1, 0.24 ppm for C1/C2, and 0.41 ppm for N1/N2. The same strategy was used for the calculations with experimental 3-D peak lists, giving tolerances of 0.02 ppm for all proton dimensions, 0.24 ppm for C2, and 0.41 ppm for N2. Simulated 3-D peaks in the amide-amide and methyl-methyl NOESY spectra that overlap with the diagonal (and therefore have the diagonal assignment within the chemical shift tolerances) were discarded by CYANA. One hundred structures were annealed per cycle using 20,000 torsion angle dynamics steps, and the ten lowest energy structures were chosen for structure-based probability scoring of assignment possibilities in the subsequent cycle. The final cycle calculated 100 structures with the five lowest energy structures represented in the final ensemble. Assignment accuracy was judged by comparing peak lists outputted after the last cycle of NOE assignment by CYANA (name-cycle7-ref.peaks) with the corresponding manually-assigned lists.
Supplementary Material
Acknowledgments
We thank Prof. Peter Güntert of the Goethe-Universität, Frankfurt am Main, and Prof. Douglas Kelly of the University of North Carolina at Chapel Hill for helpful discussions. This work was supported by the NIH (grant GM079376 to P.Z.) and Duke University institutional funds (to B.E.C.). J.W.W. is the recipient of a Kamin Fellowship.
Footnotes
Supporting Information. Figure of 4-D TS pulse sequence; figure of Ssu72 subdomain alignment; tables reporting CYANA assignment percentages and structural statistics; movies of SCRUB operation. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Goto NK, Gardner KH, Mueller GA, Willis RC, Kay LE. J Biomol NMR. 1999;13:369. doi: 10.1023/a:1008393201236. [DOI] [PubMed] [Google Scholar]
- 2.Gardner KH, Rosen MK, Kay LE. Biochemistry. 1997;36:1389. doi: 10.1021/bi9624806. [DOI] [PubMed] [Google Scholar]
- 3.Barna JCJ, Laue ED. J Magn Reson. 1987;75:384. [Google Scholar]
- 4.Barna JCJ, Laue ED, Mayger MR, Skilling J, Worrall SJP. J Magn Reson. 1987;73:69. [Google Scholar]
- 5.Schmieder P, Stern AS, Wagner G, Hoch JC. J Biomol NMR. 1994;4:483. doi: 10.1007/BF00156615. [DOI] [PubMed] [Google Scholar]
- 6.Orekhov VY, Ibraghimov IV, Billeter M. J Biomol NMR. 2001;20:49. doi: 10.1023/a:1011234126930. [DOI] [PubMed] [Google Scholar]
- 7.Rovnyak D, Frueh DP, Sastry M, Sun ZY, Stern AS, Hoch JC, Wagner G. J Magn Reson. 2004;170:15. doi: 10.1016/j.jmr.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 8.Kazimierczuk K, Kozminski W, Zhukov I. J Magn Reson. 2006;179:323. doi: 10.1016/j.jmr.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 9.Kazimierczuk K, Zawadzka A, Kozminski W, Zhukov I. J Biomol NMR. 2006;36:157. doi: 10.1007/s10858-006-9077-y. [DOI] [PubMed] [Google Scholar]
- 10.Pannetier N, Houben K, Blanchard L, Marion D. J Magn Reson. 2007;186:142. doi: 10.1016/j.jmr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 11.Coggins BE, Zhou P. J Magn Reson. 2007;184:207. doi: 10.1016/j.jmr.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hyberts SG, Frueh DP, Arthanari H, Wagner G. J Biomol NMR. 2009;45:283. doi: 10.1007/s10858-009-9368-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coggins BE, Venters RA, Zhou P. Progress in NMR Spectroscopy. 2010;57:381. doi: 10.1016/j.pnmrs.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kazimierczuk K, Stanek J, Zawadzka-Kazimierczuk A, Kozminski W. Prog Nucl Magn Reson Spectrosc. 2010;57:420. doi: 10.1016/j.pnmrs.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 15.Kazimierczuk K, Orekhov VY. Angew Chem, Int Ed Engl. 2011;50:5556. doi: 10.1002/anie.201100370. [DOI] [PubMed] [Google Scholar]
- 16.Coggins BE, Zhou P. J Magn Reson. 2006;182:84. doi: 10.1016/j.jmr.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 17.Marion D. J Biomol NMR. 2006;36:45. doi: 10.1007/s10858-006-9066-1. [DOI] [PubMed] [Google Scholar]
- 18.Tugarinov V, Kay LE, Ibraghimov IV, Orekhov VY. J Am Chem Soc. 2005;127:2767. doi: 10.1021/ja044032o. [DOI] [PubMed] [Google Scholar]
- 19.Hiller S, Ibraghimov I, Wagner G, Orekhov VY. J Am Chem Soc. 2009;131:12970. doi: 10.1021/ja902012x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Werner-Allen JW, Coggins BE, Zhou P. J Magn Reson. 2010;204:173. doi: 10.1016/j.jmr.2010.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Högbom JA. Astron Astrophys, Suppl Ser. 1974;15:417. [Google Scholar]
- 22.Barna JCJ, Tan SM, Laue ED. J Magn Reson. 1988;78:327. [Google Scholar]
- 23.Kupče E, Freeman R. J Magn Reson. 2005;173:317. doi: 10.1016/j.jmr.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 24.Kazimierczuk K, Zawadzka A, Kozminski W, Zhukov I. J Magn Reson. 2007;188:344. doi: 10.1016/j.jmr.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 25.Coggins BE, Zhou P. J Biomol NMR. 2008;42:225. doi: 10.1007/s10858-008-9275-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stanek J, Kozminski W. J Biomol NMR. 2010;47:65. doi: 10.1007/s10858-010-9411-2. [DOI] [PubMed] [Google Scholar]
- 27.Stanek J, Augustyniak R, Kozminski W. J Magn Reson. 2011;214:91. doi: 10.1016/j.jmr.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 28.Wen J, Zhou P, Wu J. J Magn Reson. 2012;218:128. doi: 10.1016/j.jmr.2012.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wen J, Wu J, Zhou P. J Magn Reson. 2011;209:94. doi: 10.1016/j.jmr.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parella T, Nolis P. Concept Magn Reson A. 2010;36A:1. [Google Scholar]
- 31.Frueh DP, Vosburg DA, Walsh CT, Wagner G. J Biomol NMR. 2006;34:31. doi: 10.1007/s10858-005-5338-4. [DOI] [PubMed] [Google Scholar]
- 32.Herrmann T, Guntert P, Wuthrich K. J Mol Biol. 2002;319:209. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 33.Shen Y, Delaglio F, Cornilescu G, Bax A. J Biomol NMR. 2009;44:213. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hyberts SG, Frueh DP, Arthanari H, Wagner G. J Biomol NMR. 2009;45:283. doi: 10.1007/s10858-009-9368-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hiller S, Ibraghimov I, Wagner G, Orekhov VY. J Am Chem Soc. 2009;131:12970. doi: 10.1021/ja902012x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Venters RA, Huang CC, Farmer BT, 2nd, Trolard R, Spicer LD, Fierke CA. J Biomol NMR. 1995;5:339. doi: 10.1007/BF00182275. [DOI] [PubMed] [Google Scholar]
- 37.Werner-Allen JW, Lee CJ, Liu P, Nicely NI, Wang S, Greenleaf AL, Zhou P. J Biol Chem. 2011;286:5717. doi: 10.1074/jbc.M110.197129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Werner-Allen JW, Zhou P. Biomol NMR Assign. 2011 doi: 10.1007/s12104-011-9325-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goto NK, Gardner KH, Mueller GA, Willis RC, Kay LE. J Biomol NMR. 1999;13:369. doi: 10.1023/a:1008393201236. [DOI] [PubMed] [Google Scholar]
- 40.Neri D, Szyperski T, Otting G, Senn H, Wuthrich K. Biochemistry. 1989;28:7510. doi: 10.1021/bi00445a003. [DOI] [PubMed] [Google Scholar]
- 41.Xia Y, Sze K, Zhu G. J Biomol NMR. 2000;18:261. doi: 10.1023/a:1026590201757. [DOI] [PubMed] [Google Scholar]
- 42.Coggins BE, Venters RA, Zhou P. Prog Nucl Magn Reson Spectrosc. 2010;57:381. doi: 10.1016/j.pnmrs.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coggins BE, Zhou P. J Magn Reson. 2006;182:84. doi: 10.1016/j.jmr.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 44.Johnson BA. Methods Mol Biol. 2004;278:313. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 45.Fisher SZ, Maupin CM, Budayova-Spano M, Govindasamy L, Tu C, Agbandje-McKenna M, Silverman DN, Voth GA, McKenna R. Biochemistry. 2007;46:2930. doi: 10.1021/bi062066y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.