Abstract
We report here the transcriptional responses in Saccharomyces cerevisiae to deletion of the RNH201 gene encoding the catalytic subunit of RNase H2. Deleting RNH201 alters RNA expression of 349 genes by ≥1.5-fold (q-value <0.01), of which 123 are upregulated and 226 are downregulated. Differentially expressed genes (DEGs) include those involved in stress responses and genome maintenance, consistent with a role for RNase H2 in removing ribonucleotides incorporated into DNA during replication. Upregulated genes include several that encode subunits of RNA polymerases I and III, and genes involved in ribosomal RNA processing, ribosomal biogenesis and tRNA modification and processing, supporting a role for RNase H2 in resolving R-loops formed during transcription of rRNA and tRNA genes. A role in R-loop resolution is further suggested by a higher average GC-content proximal to the transcription start site of downregulated as compared to upregulated genes. Several DEGs are involved in telomere maintenance, supporting a role for RNase H2 in resolving RNA-DNA hybrids formed at telomeres. A large number of DEGs encode nucleases, helicases and genes involved in response to dsRNA viruses, observations that could be relevant to the nucleic acid species that elicit an innate immune response in RNase H2-defective humans.
1. Introduction
Ribonucleases H (RNase H) cleave the RNA strand of RNA-DNA hybrids (1, 2). Most organisms encode two types of RNases H, Types 1 and 2, that have different substrate preferences (2). The subject of this study is RNase H2, a 3-subunit enzyme complex (3) that cleaves substrates with one to multiple consecutive ribonucleotides (2–6). RNase H2 has been implicated in degrading R-loops formed during transcription (7–10) and at telomeres (11–13), digesting RNA primers that initiate Okazaki fragment synthesis during DNA replication (14–17) and removing ribonucleotides incorporated by DNA polymerases during replication (18–22). The importance of RNase H2 function is illustrated by the fact that loss of RNase H2 is associated with cellular stress and genome instability (21, 23, 24) (for review see (2, 25, 26)) and by the fact that mutations in the RNH201, RNH202 and RNH203 genes encoding the three subunits of human RNase H2 are causative for Aicardi-Goutières syndrome (AGS) (6, 27–31), an autosomal recessive autoinflammatory disorder mimicking viral infection of the brain and exhibiting similarities to Systemic Lupus Erythematosis. AGS has been suggested to result from the accumulation of nucleic acid species that activate the innate immune response (32–34). The goal of this study is to further probe RNase H2 functions and the consequences of loss of RNase H2 activity by investigating for the first time (to our knowledge) the global changes in mRNA expression associated with deleting Saccharomyces cerevisiae RNH201, which encodes the RNase H2 catalytic subunit. The results reveal an extensive transcriptional response indicative of cellular stress and consistent with roles for RNase H2 in removing rNMPs from DNA and in resolving RNA-DNA hybrids formed at telomeres and during transcription by RNA polymerases I and III. Also observed are changes in expression of several nuclease and helicase genes involved in processing DNA and RNA, which could be relevant to the pathogenesis of autoimmune disease in humans defective in RNase H2.
2. Materials and Methods
2.1 Materials and Reagents
S. cerevisiae strains were previously described (21). Yeast Gene Expression Microarrays, Design ID 015072, were from Agilent Technologies, Inc. (San Clara, CA).
2.2 Isolation of Total RNA
Strains were streaked onto yeast peptone dextrose adenine (YPDA) plates and grown at 30°C for 3 days. Seven individual colonies per strain were grown in 20 mL YPDA at 30°C for 6–8 hours. Cells were counted and a calculated volume from the day cultures were diluted into 25 ml of YPDA and grown at 30°C overnight with constant shaking to achieve a cell density of 1–3×107 cells/ml. Cells (3×108) were harvested by centrifugation and washed with cold, deionized H2O (Figure 1A). Total RNA was isolated using a RiboPure yeast kit (Ambion), as per manufacturer’s instructions. RNA quality was evaluated using a denaturing 0.8% agarose gel and a Bioanalyzer Nano chip. RNA was quantified using a NanoDrop spectrophotometer. RNA was stored at −80°C until analyzed using microarray.
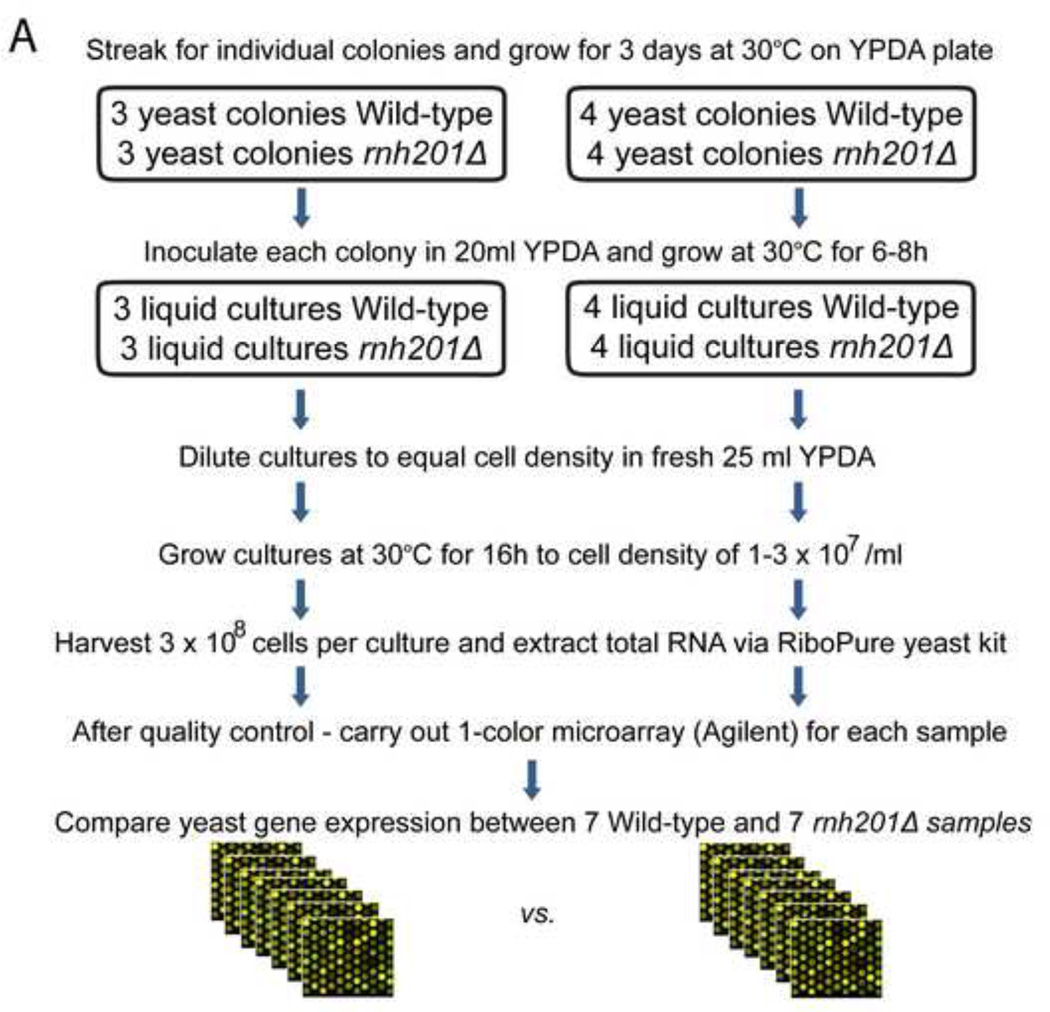
Figure 1. Experimental design and workflow of data analysis.
Processing of samples and data are depicted in Panels A and B, respectively. As shown in Panel A, WT and rnh201Δ strains were grown at 30C and total RNA isolated. Total RNA was used for 1-color microarray analysis. Yeast gene expression was compared between WT and rnh201Δ samples and data processed as indicated in Panel B. As shown in Panel A, comparisons between WT and rnh201Δ strains were performed in two separate batches: the first experiment involved three samples per strain and the second involved four samples per strain.
2.3. Microarray processing
Gene expression analysis was conducted using Agilent Whole Yeast Genome 4×44 multiplex format oligo arrays (015072) (Agilent Technologies) following the Agilent 1-color microarray-based gene expression analysis protocol. Starting with 500 ng of total RNA, Cy3 labeled cRNA was produced according to manufacturer’s protocol. For each sample, 1.65 µg of Cy3 labeled cRNAs were fragmented and hybridized for 17 hours in a rotating hybridization oven. Slides were washed and then scanned with an Agilent scanner. Pixel intensity data was acquired from the array images using Agilent Feature Extraction software (v9.5), using the 1-color defaults for all parameters. The raw data can be found in the Gene Expression Omnibus (GEO) database (GEO accession number GSE34668).
2.4.1 Gene expression data analysis
Probes on the array were annotated for gene representation according to the University of California Santa Cruz Saccharomyces cerevisiae genome June 2008 genome assembly (SGD/sacCer2 version). The array data were quantile normalized and log-2 transformed. Analyses were performed using the Partek Genomics Suite software (version 6.6) (Partek Incorporated, St. Louis, MO). Analysis of data is shown in a flow chart in Figure 1B.
2.4.2 Batch correction and contrast between genotypes
For each probe, a two-way analysis of variance (ANOVA) mixed effects linear model, Yijk = µ + αi + βj + εijk, where µ is the grand mean expression and Yijk represents the measurement of the ith genotype, jth scan date (random effect) and kth sample, was applied to batch correct the data. εijk represents the random error. The errors are assumed to be normally and independently distributed, with a mean of 0 and a standard deviation of δ for all measurements. For each probe, a student t-test was then applied to statistically compare the difference between the means of the rnh201Δ and wild type strains expression measurements. The Benjamini-Hochberg false discovery rate (FDR) method was used to adjust the p-values (p) for multiple testing (35). An FDR q-value ≤ 0.01 was used as a threshold for significance.
2.4.3 GO-ANOVA analysis
To find differentially expressed functional groups of genes, the genes were annotated to functional categories according to the GO database. For each GO functional category, a two-way ANOVA model, Yijkl = µ + αi + βj + γ(α)ik + (αβ)ij + εijkl, where Yijkl represents the lth functional group expression measurement for the ith genotype, jth gene and kth sample, was used for analysis of the log base 2 transformation of the batch corrected expression data. µ is the grand mean expression of the functional group and εijkl represents the random error. Errors were assumed to be normally and independently distributed, with a mean of 0 and a standard deviation of δ for all measurements. (αβ) represents the gene-by-genotype effect and γ(α) is a sample-to sample effect. Sample is a random effect in the model. The gene expression of the functional group was evaluated as a weighted average (least mean square) of the corresponding factor levels according to the sum of the model components representing the grand mean, the individual effects (genotype and gene) and the interaction effects (genotype by gene and genotype by sample). The analysis was performed with GO categories of maximum size = 200. Only significant categories of size > 5 were considered. The Benjamini-Hochberg FDR method was used to adjust the p-values (p) for multiple testing.
2.5. Functional enrichment analysis
DAVID v6.7 (36, 37) was used to find gene ontologies and classes enriched in lists of DEGs. Functional annotation cluster analysis was performed using the default settings except that Gene Ontology (GO) term Cellular Component was removed. DEGs were submitted as Ensemble Gene IDs and Agilent YeastV2 was used as a background. The list of enriched gene classes was filtered based on the following criteria: 1) Benjamini-Hochberg corrected p-value < 0.05, 2) lowest p-value of an individual DAVID Annotation Cluster and, 3) not a member of another Annotation Cluster where a class exists with a lower p-value.
2.6. GC content of flanking sequences
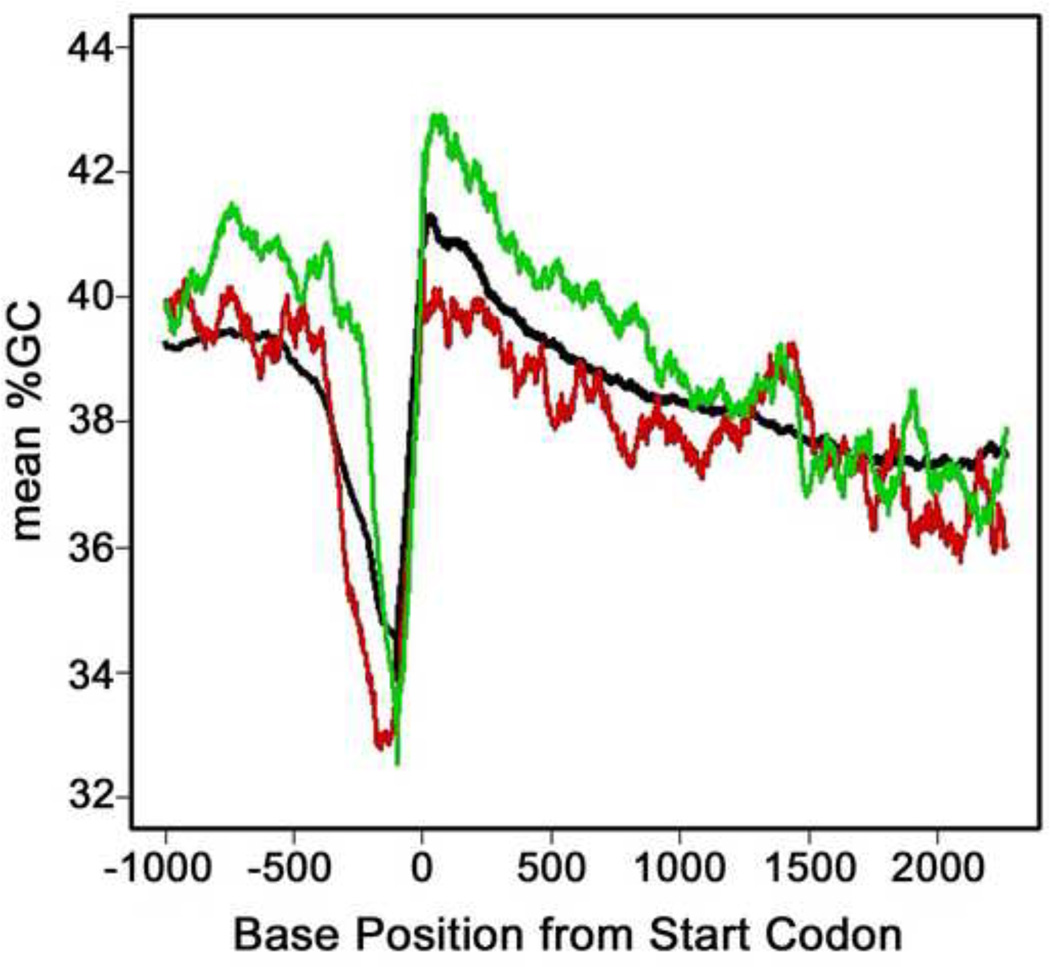
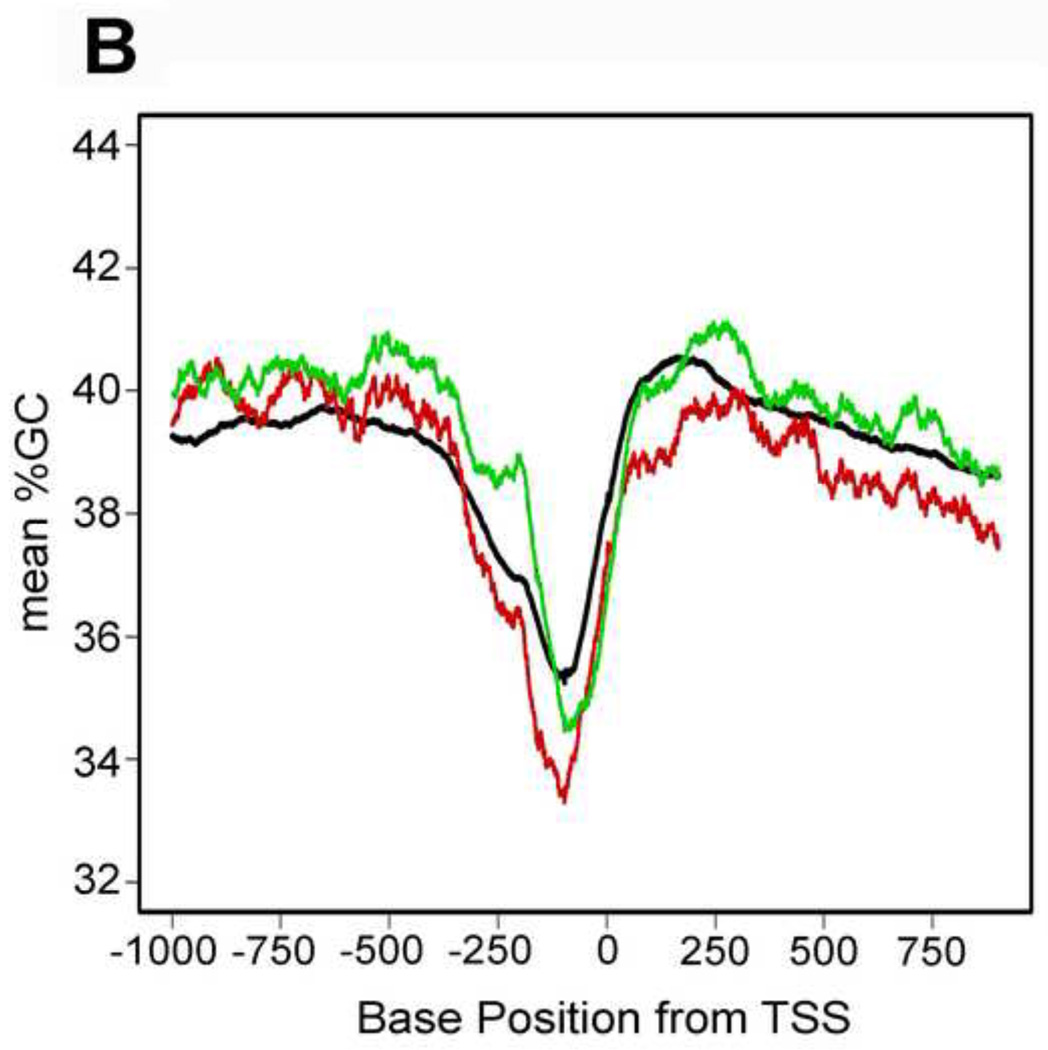
Genomic sequences of the ORFs with introns and untranslated regions (UTRs) were downloaded from the University of California Santa Cruz (UCSC) genome database. The data are based on DNA sequence dated June 2008 (sacCer2) in the S. cerevisiae Genome Database (38, 39). We used either the genomic location for the AUG start codon (Figure 4A) or experimentally determined transcription start sites (TSSs) coordinates (Figure 4B) as the reference points for the evaluating the GC content in the vicinity of the TSS. The locations of TSSs were obtained from supplemental Table S4 in an article that reports the transcribed regions of S. cerevisiae by RNA sequencing (40). For use of TSSs as a reference point, any ORF without a detected 5´ UTR had the genome coordinate of the start codon (AUG) as the reference point. For each ORF, the number of G and C bases was counted from the flanking sequence in a 100-base pair (bp) sliding window from −1000 to +2500. GC content was calculated separately for all of the ORFs, as well as the downregulated and upregulated DEGs. YNL072W (the gene knocked out) was omitted from the analysis and the DNA sequences for six DEGs (Q0275, YGR272C, YCL027C-A, YDR474C, YGL046W and YOL153C) were not available.
Figure 4. Genomic GC content of rnh201Δ DEGs.
A. G+C (in %) was calculated for a 100-base sliding window of genomic DNA starting 1000 bases up stream of start codons and extending 2500 downstream. B. G+C content (in %) was calculated for a 100-base sliding window of genomic DNA starting 1000 bases up stream of TSSs and extending 1000 downstream. Upregulated genes are represented by a red line, downregulated genes are represented by a green line, and the black line represents the mean %G+C content for all yeast ORFs.
2.7. Network analysis
Transcriptional networks were identified by overlaying DEGs onto the YeastNet v. 2 probabilistic functional gene network (41) and visualized in Cytoscape (42). Discreet subnetworks were identified by filtering for log likelihood score (LLS) ≥ 2.303, thus increasing the confidence in while decreasing the number of observed interactions. The significance (p < 10−4) of each sub-network was confirmed by a bootstrap approach to test if the number of sub-network interactions was greater than the number of edges of 10,000 randomly generated networks with equal numbers of nodes.
3. Results
3.1 Differentially expressed genes (DEGs) in the rnh201Δ yeast strain
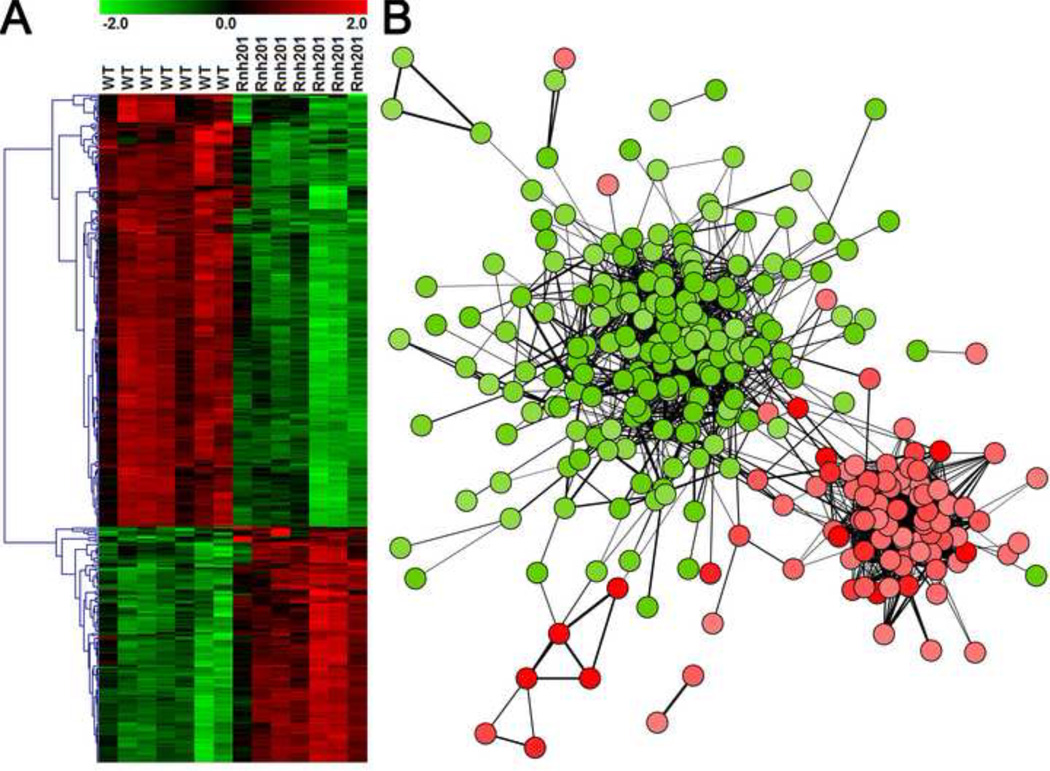
We measured mRNA levels for seven independent cultures of asynchronously growing wild type and rnh201Δ yeast strains in the exponential phase of growth (Figure 1A), and analyzed the data as shown in Figure 1B. A comparison of the expression profiles of the rnh201Δ strain to that of its wild type parent strain identified a large number of differentially expressed genes (DEGs), (Figures 2A and 2B). Supplementary Table 1 lists all the genes analyzed. A total of 349 probes corresponding to 349 unique genes (Supplementary Table 1) changed expression by 1.5-fold or greater (q-value < 0.01). Among these, 123 were up regulated and 226 were down regulated. When the expression patterns for these genes were compared by hierarchical clustering analysis, clear differences between the wild type and rnh201Δ strains were observed for the up and downregulated genes (Figure 2A).
Figure 2. Expression and Clustering.
Visualization of Differential Expression. A. Row/gene normalized hierarchical clustering of DEGs (n=349) in rnh201Δ vs. wild type. Quantile-normalized, log2 intensity values for rnh201Δ DEGs (n=349) were loaded into TMEV. Intensity values were transformed by the Normalize Gene/Rows function which subtracts the gene/row mean and divides by the gene/row standard deviation. Hierarchical clustering of the gene tree was performed using Pearson correlation as the distance metric and complete linkage clustering. B. Transcriptional network of rnh201Δ vs. wild type DEGs from YeastNet v. 2. Edge width is proportional to LLS (2.3 to 4.3). Visualization of normalized data is centered on zero (black) with 2.0 (red) and −2.0 (green) as the upper and lower bounds, respectively.
3.2 Functional enrichment and GO-ANOVA analysis of DEGs
Analysis of the 349 DEGs by DAVID (see Materials and Methods) revealed that deleting the RNH201 gene alters expression of genes involved in several cellular processes (Table 1). A GOANOVA analysis (GO category by GO-category basis [see Materials and Methods] (35)) further revealed statistically significant differences in gene expression for several biological process terms related to DNA replication and transcription (Table 2). These biological categories, and others not shown, partially overlap with the functional enrichment analysis in Table 2.
Table 1.
Functional enrichment analysis of DEGS
| Term | Count | List Total | Pop Hits | Pop Total | Benjamini |
|---|---|---|---|---|---|
| All, n=349 | |||||
| BP:0009266~response to temperature stimulus | 54 | 259 | 216 | 4716 | 3.59E- 19 |
| BP:0007039~vacuolar protein catabolic process | 40 | 259 | 117 | 4716 | 6.49E- 19 |
| PIR~ribosome biogenesis | 33 | 343 | 148 | 6104 | 3.94E- 09 |
| BP:0006112~energy reserve metabolic process | 17 | 259 | 42 | 4716 | 2.98E- 08 |
| KEGG~sce00500:Starch and sucrose metabolism | 14 | 67 | 39 | 1437 | 1.84E- 07 |
| COG~Carbohydrate transport and metabolism | 10 | 51 | 31 | 975 | 5.53E- 05 |
| BP:0042775~mitochondrial ATP synthesis coupled electron transport | 11 | 259 | 28 | 4716 | 7.72E- 05 |
| BP:0000462~maturation of SSU- rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) |
18 | 259 | 83 | 4716 | 8.78E- 05 |
| BP:0016052~carbohydrate catabolic process | 18 | 259 | 84 | 4716 | 1.59E- 04 |
| BP:0042273~ribosomal large subunit biogenesis | 16 | 259 | 71 | 4716 | 1.82E- 04 |
| BP:0022900~electron transport chain | 13 | 259 | 68 | 4716 | 5.86E- 03 |
| BP:0043462~regulation of ATPase activity | 4 | 259 | 4 | 4716 | 1.15E- 02 |
| BP:0016310~phosphorylation | 24 | 259 | 211 | 4716 | 1.79E- 02 |
| BP:0033753~establishment of ribosome localization | 9 | 259 | 43 | 4716 | 3.17E- 02 |
| KEGG~sce00020:Citrate cycle (TCA cycle) | 7 | 67 | 33 | 1437 | 4.75E- 02 |
| Upregulated, n=123* | |||||
| BP:0042254~ribosome biogenesis | 45 | 88 | 351 | 4716 | 4.37E- 25 |
| BP:0000462~maturation of SSU- rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) |
18 | 88 | 83 | 4716 | 2.14E- 12 |
| BP:0042273~ribosomal large subunit biogenesis | 16 | 88 | 71 | 4716 | 3.85E- 11 |
| BP:0033750~ribosome localization | 9 | 88 | 43 | 4716 | 1.56E- 05 |
| MF:0008173~RNA methyltransferase activity | 7 | 63 | 30 | 4047 | 3.61E- 04 |
| PIR~rna- binding | 15 | 121 | 229 | 6104 | 3.81E- 03 |
| BP:0017183~peptidyl-diphthamide biosynthetic process from peptidyl-histidine |
3 | 88 | 5 | 4716 | 3.88E- 02 |
| Downregulated, n=226* | |||||
| BP:0009266~response to temperature stimulus | 53 | 171 | 216 | 4716 | 8.11E- 28 |
| BP:0007039~vacuolar protein catabolic process | 40 | 171 | 117 | 4716 | 2.65E- 26 |
| BP:0015980~energy derivation by oxidation of organic compounds |
33 | 171 | 178 | 4716 | 1.20E- 12 |
| KEGG~sce00500:Starch and sucrose metabolism | 14 | 60 | 39 | 1437 | 3.94E- 08 |
| KEGG~sce00500:Starch and sucrose metabolism | 14 | 60 | 39 | 1437 | 3.94E- 08 |
| BP:0044275~cellular carbohydrate catabolic process | 17 | 171 | 76 | 4716 | 3.79E- 07 |
| BP:0055114~oxidation reduction | 34 | 171 | 351 | 4716 | 7.08E- 06 |
| BP:0042773~ATP synthesis coupled electron transport | 10 | 171 | 28 | 4716 | 1.16E- 05 |
| BP:0006793~phosphorus metabolic process | 27 | 171 | 289 | 4716 | 2.81E- 04 |
| BP:0043462~regulation of ATPase activity | 4 | 171 | 4 | 4716 | 3.41E- 03 |
| BP:0008643~carbohydrate transport | 8 | 171 | 40 | 4716 | 8.08E- 03 |
| BP:0006096~glycolysis | 7 | 171 | 30 | 4716 | 9.62E- 03 |
| KEGG~sce00020:Citrate cycle (TCA cycle) | 7 | 60 | 33 | 1437 | 2.56E- 02 |
| BP:0006914~autophagy | 14 | 171 | 143 | 4716 | 2.68E- 02 |
| BP:0006732~coenzyme metabolic process | 14 | 171 | 149 | 4716 | 3.78E- 02 |
Benjamini Benjamini-Hochberg multiple-test corrected p; BP Gene Ontology Consortium Biological Process; COG Clusters of Orthologous Groups; Count number of genes in both list and gene class; Kegg Kyoto Encyclopedia of Genes and Genomes; List Total number of genes in list; MF Gene Ontology Consortium Molecular Function; PIR Protein Information Resource SuperFamily; Pop Hits number of genes in both gene class and array; Pop Total number of genes in array.
The p-values for the n=123 up- and n=226 downregulated DEGs do not account for these lists as a subset of the n=349 dataset; therefore, these should be viewed as less-confident approximations than would be yielded by a multivariate approach.
Table 2.
GO-ANOVA analysis
| Term | n | p | Benjamini |
|---|---|---|---|
| BP:0042273~ribosomal large subunit biogenesis | 23 | 2.65E-06 | 4.53E-05 |
| BP:0042274~ribosomal small subunit biogenesis | 11 | 6.90E-06 | 7.48E-05 |
| BP:0009117~nucleotide metabolic process | 89 | 7.55E-06 | 7.83E-05 |
| BP:0006974~response to DNA damage stimulus | 179 | 8.85E-06 | 8.56E-05 |
| BP:0000075~cell cycle checkpoint | 13 | 1.57E-05 | 1.18E-04 |
| BP:0006338~chromatin remodeling | 58 | 2.63E-05 | 1.64E-04 |
| BP:0006302~double-strand break repair | 54 | 3.07E-05 | 1.80E-04 |
| BP:0006310~DNA recombination | 76 | 1.33E-04 | 5.25E-04 |
| BP:0030001~metal ion transport | 51 | 7.32E-02 | 1.08E-01 |
Benjamini Benjamini-Hochberg multiple-test corrected p; BP Gene Ontology Consortium Biological Process; n: number of genes; p: Nominal p-value.
3.3 Deletion of RNH201 generates a specific transcriptional response
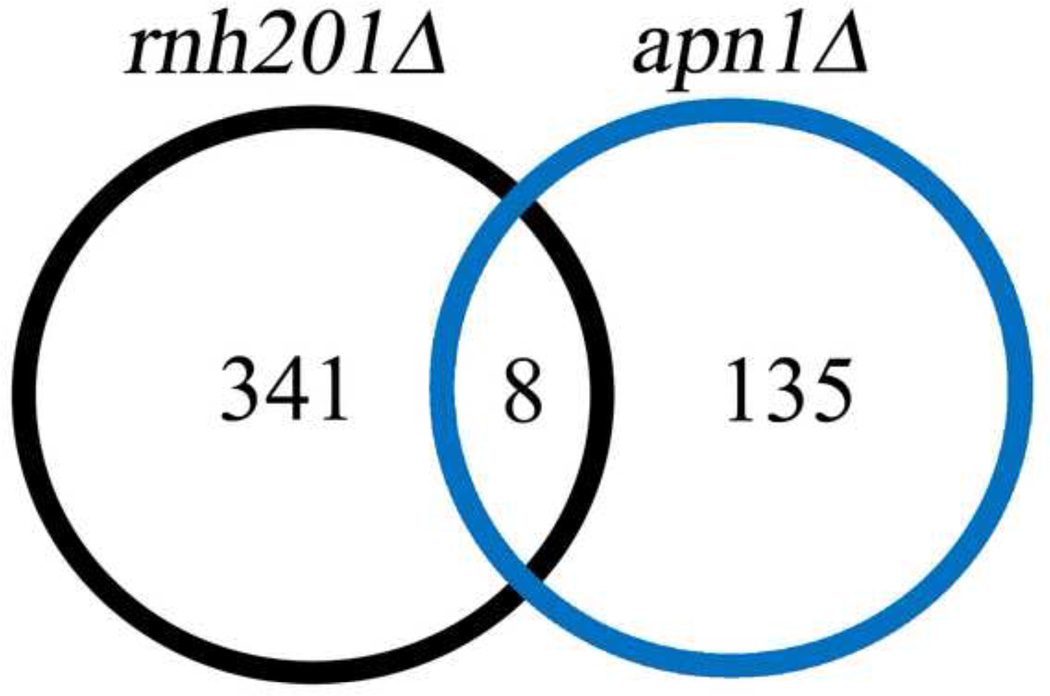
RNase H2 is involved in repairing ribonucleotides in DNA (20, 21, 24). As one in silico test of the specificity of the transcriptional response to loss of RNase H2, we compared the 349 DEGs in the rnh201Δ strain to the 143 DEGs observed (43) upon deletion of a different DNA repair gene, APN1. APN1 encodes the major endonuclease involved in repair of a different lesion, an abasic site. Only eight DEGs (YAR066W, YRO2, HSP30, TDH1, CDA1, FET3, CMK2 and GPH1) were common to the rnh201Δ and apn1Δ strains (Figure 3).
Figure 3. Overlap comparison of rnh201Δ DEGs with DEGs from another published study.
Genes differentially expressed in rnh201Δ vs. wild type show little commonality with DEGs from an apn1Δ study (42).
3.4. Up and downregulated genes have different GC content
RNase H2 degrades R-loops that form during transcription (44, 45) and R-loops are more stable in GC-rich sequences (46, 47). Therefore, we evaluated whether the average GC content around the AUG or transcription start site differed between upregulated and downregulated DEGs. We plotted the average GC content of a sliding 100 base pair (bp) window for all yeast genes and compared it to the matched regions of the genes that are either upregulated or downregulated in response to deleting RNH201. When compared to the average of all genes up to about 1000 base pairs downstream of the start codon, the GC content of upregulated genes was slightly lower, whereas the GC content of downregulated genes was slightly higher (Figure 4A). This difference in GC content extends into the ORF following the AUG. Analysis using transcriptional start sites (TSSs) as the reference point showed a similar inverse relationship between GC content and transcriptional response upon deletion of RNH201 (Figure 4B).
3.5. Transcriptional networks affected by deletion of RNH201
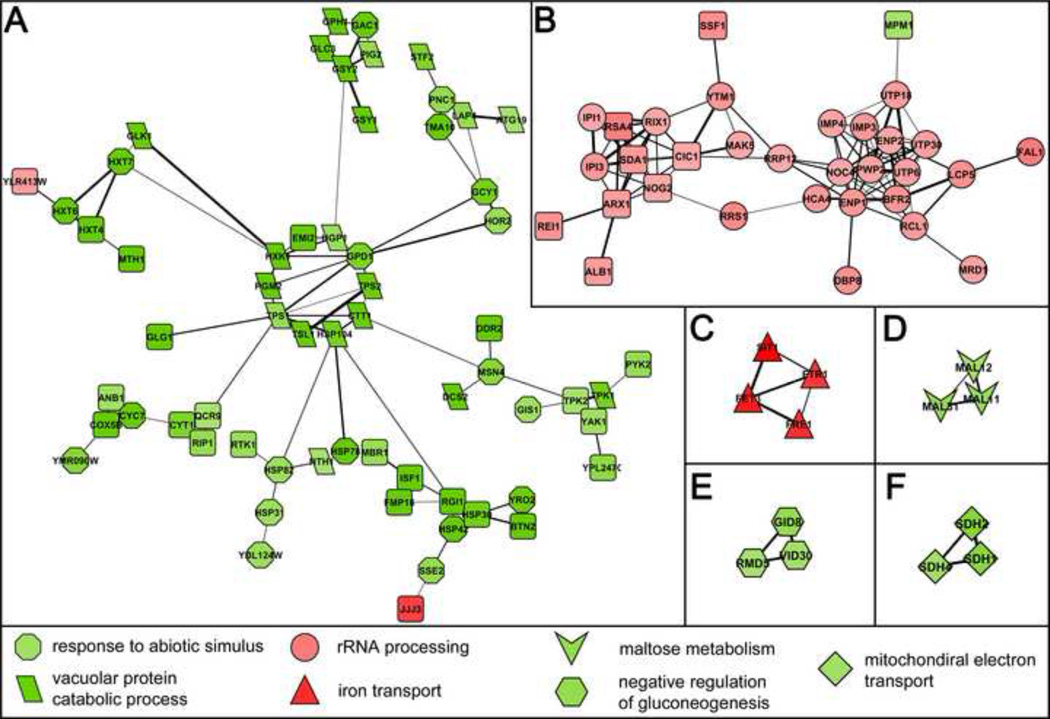
We overlaid the 349 DEGs onto YeastNet v. 2, a probabilistic functional gene network composed of 102,803 linkages among 5,483 proteins (41, 48), and visualized the resulting networks in Cytoscape (42). This analysis revealed distinct networks for the up and downregulated genes (Figure 2B). Filtering the network for higher confidence interactions revealed six distinct sub-networks (Figure 5A-F) that were largely or entirely up or downregulated. Each sub-network was enriched for functional ontologies and pathways as follows: downregulated sub-network A, GO9628~response to abiotic stimulus (p=6.30x10−19) and GO7039~vacuolar protein catabolic process (p=1.22x10−15); upregulated sub-network B, GO6364~rRNA processing (p=1.39x10−21); upregulated sub-network C, enriched for PIR~iron transport (p=1.3x10−6); downregulated sub-network D, enriched for PIR~maltose metabolism (p=4.25x10−5); downregulated sub-network E, enriched for GO45721 ~negative regulation of gluconeogenesis (p=1.52x10−4); and, downregulated sub-network F, enriched for GO:0006121 ~mitochondrial electron transport, succinate to ubiquinone (p=2.25x10−5).
Figure 5. Transcriptional Networks.
High resolution yeast transcriptional networks differentially regulated in rnh201Δ vs. wild type. Discreet rnh201Δ subnetworks were identified by filtering (LLS >= 2.303) the YeastNet v. 2 probabilistic functional gene network prior to overlay with DEGs (n=349). A. Downregulated subnetwork enriched for GO9628~response to abiotic stimulus (p=6.30x10−19) and GO7039~vacuolar protein catabolic process (p=6.09x10−15). B. Upregulated subnetwork enriched for GO6364~rRNA processing (p=4.18x10−21). C. Upregulated subnetwork enriched for PIR~iron transport (p=1.30x10−6). D. Downregulated subnetwork enriched for PIR~maltose metabolism (p=4.25x10−5). E. Downregulated subnetwork enriched for GO45721~negative regulation of gluconeogenesis (p=1.52x10−4). F. Downregulated subnetwork enriched for GO:0006121~mitochondrial electron transport, succinate to ubiquinone (p=2.25x10−5). Edge width is proportional to LLS (2.3 to 4.3). Up and downregulated genes are shaded red and green, respectively. Membership in enriched gene ontologies is denoted by node shape as indicated in the figure key. Squares with rounded corners represent enriched gene ontologies that do not have a nomenclature and therefore default to this shape. This figure only includes genes whose fold change values were ≥1.5.
4. Discussion
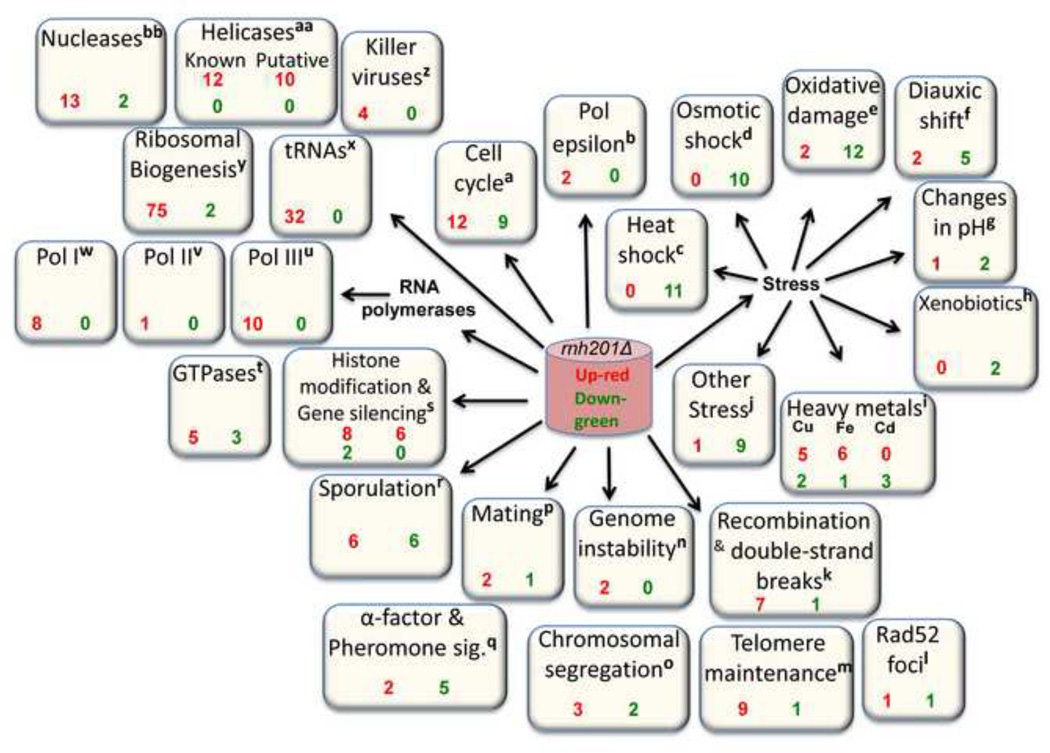
The DEGs observed here can be considered in light of the phenotypic consequences of loss of yeast RNase H2 and its known and proposed cellular functions (Figure 6). For discussion purposes, in addition to those genes whose expression changed at least 1.5-fold and were used for pathway analyses (Supplementary Table 1, Figure 6) we also consider genes whose expression changed by ≥ 1.25-fold with q values ≤ 0.01 (Supplementary Table 2, Figure 6).
Figure 6. Expression changes observed upon deleting RNH201.
This schematic depicts numerous changes in gene expression when RNH201 is deleted. Each white box represents a category based on descriptions in the Saccharomyces Gene Database (SGD) and enumerates genes whose expression was either up- (red) or downregulated (green). The superscript for each category is used in Supplementary Table 1 to highlight the genes listed in that category. Some genes are in more than one category, which therefore may contain more than one superscript. In this figure, all genes are considered whose expression was changed by ≥ 1.25 with q values of ≤ 0.01 (See Supplementary Table 1). The total number of DEGs in the different functional groups exceeds the total number DEGs because some genes have multiple functional assignments.
4.1. DEGs reflecting altered DNA metabolism
The ability of yeast RNase H2 to incise the backbone in DNA duplexes containing a single ribonucleotide led to the proposal that RNase H2 might initiate removal of ribonucleotides incorporated by DNA polymerases (4, 19–21). In support of this idea, yeast replicative Pols α, δ and ε incorporate large numbers of ribonucleotides into DNA in vitro (49), a large number of unrepaired ribonucleotides are observed in the nuclear genome of yeast rnh201Δ strains, and this number increases in rnh201Δ strains encoding a pol2-M644G variant of the leading strand replicase Pol ε that is promiscuous for ribonucleotide incorporation (21). The pol2-M644G rnh201Δ strain progresses slowly through S phase, has elevated dNTP pools (21), and is sensitive to treatment with the replication inhibitor, hydroxyurea (HU) (24). These phenotypes are indicators of replicative stress, which could result from difficulty in replicating DNA templates containing rNMPs (50), and/or from impaired replication due to unresolved R-loops formed during transcription (see below). Supporting the former possibility is a recent study (24) describing the consequences of concomitantly deleting RNH201 and RNH1, the latter encoding the second known yeast RNase H. When grown in the presence of HU, rnh1Δ rnh201Δ double mutant cells exhibit a strong cell cycle arrest accompanied by phosphorylation of the Rad53 checkpoint kinase, both indicative of replicative stress. Survival of these cells depends on two lesion tolerance pathways, MMS2-dependent template switching and translesion DNA synthesis (TLS), the latter requiring both Rev1 and the catalytic (Rev3) subunit of DNA polymerase ζ (24).
These cellular phenotypes are consistent with significant (q ≤ 0.01) changes in expression of numerous genes observed here (Figure 6 and Supplementary Table 1). REV1, which is required for TLS by Pol ζ, is upregulated. So too is OGG1, which encodes the glycoslyase that removes 8-oxo-guanine from DNA. Activation of DNA repair is accompanied by cell cycle arrest and here we see twelve genes associated with progression through the cell cycle are upregulated. Increased expression is observed for two genes (PES4 and SLD5) associated with the function of Pol ε, the leading strand replicase. SLD5 encodes one of the subunits of the GINS complex that interacts and stimulates the polymerase activity but not the primase activity of DNA polymeases/primase α (51). Loss of RNase H2 leads to changes in expression of least 74 genes associated with some type of cellular stress, including heat, osmotic shock, oxidative damage, diauxic shift, changes in pH, and exposure to xenobiotics or heavy metals (Figure 6). Stress due to heavy metal exposure may be related to the observation that many DEGs observed here are involved in metal homeostasis, especially iron and zinc metabolism (Figure 5C, Table 2). Because stalled replication forks can yield double strand breaks in DNA, and because the 2´- OH of a ribose renders the DNA backbone sensitive to hydrolysis to create nicks, another potential outcome of loss of RNase H2 activity is recombination, a possibility supported by increased expression of several genes associated with recombination and formation of Rad52 foci.
Another possible outcome of unrepaired ribonucleotides in DNA is genome instability initiated by spontaneous or enzymatic cleavage of the DNA backbone. We previously reported genome instability in rnh201Δ strains, primarily in the form of short deletions in tandem-repeat sequences (21, 52). In the absence of RNase H2, ribonucleotides are incorporated in DNA and these are targeted by topoisomerase 1 generating deletions (23). These deletions are initiated when topoisomerase 1, an enzyme that relieves DNA super-coiling generated during transcription and which also has RNase activity (53), nicks the backbone of duplex DNA containing ribonucleotides (23). These nicks contain ends that must be processed to create 3´-O and 5´-P DNA ends to allow ligation to proceed. This processing is thought to provide the opportunity for strand misalignment in repetitive sequences that gives rise to the short deletions. Short deletion mutagenesis is exacerbated in the pol2-M644G rnh201Δ strains that contain a large number of unrepaired ribonucleotides in the genome (21, 52). In light of this ribonucleotide-dependent mutagenesis, it is notable that seven DEGs are associated with genome instability and chromosomal segregation (Figure 6).
In our initial study of rNTP incorporation by yeast replicases (54), we speculated that because rNMPs distort DNA helix geometry, their transient presence in DNA might have cellular signaling functions. Ribonucleotides are already implicated as signals for mating type switching in fission yeast (55). Other theoretical possibilities include signaling for mismatch repair, reloading nucleosomes after replication, for chromatin remodeling and for gene silencing. In this regard, we note that several DEGs observed here (Table 2Figure 6) are associated with mating, α-factor and pheromone signaling, sporulation, and histone modification and gene silencing. Also, eight DEGs encode proteins that are known or suggested to be GTPases, which are often associated with signaling functions.
4.2. DEGs reflecting altered R-loop resolution
Several studies (7, 45) indicate that yeast RNase H2 participates in resolving R-loops formed during transcription. Because R-loop stability increases with increasing GC content (46, 47), we wondered if there was a relationship between the GC content and changes in gene expression upon loss of RNase H2. The data in Figure 4 show that GC content near the start codons and TSSs of downregulated genes is slightly higher than the average. This observation is consistent with the possibility that, in the absence of RNH201, stable R-loops preferentially hinder expression of genes with higher GC content. The proposed role of RNase H2 in resolving R-loops formed during transcription is further supported by increased expression of eight genes encoding subunits of RNA polymerase I (Figure 6), which transcribes 35S rDNA. Production of 35S ribosomal RNA accounts for more than half of all transcription in yeast and therefore provides great potential for R-loop formation. The present study extends this possibility to transcription of 5S rDNA and tRNA genes by RNA polymerase III, because expression is also increased for genes encoding ten subunits of RNA polymerase III (See Supplementary Table 3). Also striking is the increased expression of many genes encoding proteins that modify rRNAs and tRNAs (Figure 6). These include 32 genes that modify tRNAs to enhance expression of genes that are needed for stress responses, and are thought to regulate the rate and fidelity of translation (see (56) and references therein). Thus, the simplest hypothesis is that a defect in RNase H2 results in a stress response that could be due to unresolved R-loops, unrepaired ribonucleotides in DNA, or both, and that the cell responds to this stress by up-regulation expression of TRM genes to increase translation of genes needed to respond to this stress Equally striking is the increased expression of a large number of genes involved in ribosomal biogenesis and translation (Table 1Figure 6 and Supplementary Table 1). These data strongly support the idea that loss of RNase H2 reduces the ability to resolve R-loops that form as RNA polymerases I and III try to produce large quantities of rRNA and tRNA. Collision of unresolved R-loops with replication forks can also promote transcription-associated recombination (8, 57), consistent with several DEG’s implicated in recombination and formation of Rad52 foci (Figure 6). Nine genes associated with telomere function are upregulated (Figure 6), consistent with a proposed role for RNase H2 in resolving DNA-RNA hybrids formed at telomeres (12, 13, 58).
4.3. DEGs involved in nucleic acid processing - possible implications for autoimmunity?
Failure to resolve R-loops has been associated with neurodegenerative diseases including spinocerebellar atatxia type 1, myotonic dystrophy and fragile X type A (46, 47, 59, 60). Moreover, mutations in the genes encoding human RNase H2 that result in Aicardi-Goutières syndrome are thought to allow DNA/RNA species to accumulate that elicit an innate immune-mediated inflammation that mimics viral infection. The nucleic acid species responsible for immune-mediated inflammation remain uncertain, but could be byproducts of viral infection, transcription, replication, repair or transposition (29, 61), and for a review see (62) and references therein). With these possibilities in mind, we note increased expression of several genes implicated in yeast response to killer viruses, and increased expression of several genes implicated in nucleic acid transactions, including 22 genes encoding known or putative helicases and 13 genes encoding nucleases. The increased expression of these genes could be relevant to the pathogenesis of autoimmune disease in humans defective in RNase H2.
Supplementary Material
Highlights.
Deleting RNH201 affects expression of many genes in budding yeast
Deleting RNH201 elicits a strong cellular stress response
Deleting RNH201 influences transcription by RNA Polymerases I and III
The transcriptional response to deleting RNH201 suggests multiple roles for RNase H2
Acknowledgements
We thank Keith Shockley and Jessica Williams for helpful comments on the manuscript. This work was supported by Projects Z01 ES065070 (to T.A.K.) and Z01 ES102345-04 (to P. R. B.) from the Division of Intramural Research of the NIH, NIEHS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
None declared
References
- 1.Stein H, Hausen P. Enzyme from calf thymus degrading the RNA moiety of DNA-RNA Hybrids: effect on DNA-dependent RNA polymerase. Science. 1969;166:393–395. doi: 10.1126/science.166.3903.393. [DOI] [PubMed] [Google Scholar]
- 2.Cerritelli SM, Crouch RJ. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 2009;276:1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jeong HS, Backlund PS, Chen HC, Karavanov AA, Crouch RJ. RNase H2 of Saccharomyces cerevisiae is a complex of three proteins. Nucleic Acids Res. 2004;32:407–414. doi: 10.1093/nar/gkh209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eder PS, Walder RY, Walder JA. Substrate specificity of human RNase H1 and its role in excision repair of ribose residues misincorporated in DNA. Biochimie. 1993;75:123–126. doi: 10.1016/0300-9084(93)90033-o. [DOI] [PubMed] [Google Scholar]
- 5.Frank P, Braunshofer-Reiter C, Wintersberger U, Grimm R, Busen W. Cloning of the cDNA encoding the large subunit of human RNase HI, a homologue of the prokaryotic RNase HII. Proc Natl Acad Sci U S A. 1998;95:12872–12877. doi: 10.1073/pnas.95.22.12872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chon H, Vassilev A, DePamphilis ML, Zhao Y, Zhang J, Burgers PM, Crouch RJ, Cerritelli SM. Contributions of the two accessory subunits, RNASEH2B and RNASEH2C, to the activity and properties of the human RNase H2 complex. Nucleic Acids Res. 2009;37:96–110. doi: 10.1093/nar/gkn913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drolet M, Phoenix P, Menzel R, Masse E, Liu LF, Crouch RJ. Overexpression of RNase H partially complements the growth defect of an Escherichia coli delta topA mutant: R-loop formation is a major problem in the absence of DNA topoisomerase I. Proc Natl Acad Sci U S A. 1995;92:3526–3530. doi: 10.1073/pnas.92.8.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huertas P, Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell. 2003;12:711–721. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 9.Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 10.Drolet M. Growth inhibition mediated by excess negative supercoiling: the interplay between transcription elongation, R-loop formation and DNA topology. Mol Microbiol. 2006;59:723–730. doi: 10.1111/j.1365-2958.2005.05006.x. [DOI] [PubMed] [Google Scholar]
- 11.Forstemann K, Lingner J. Telomerase limits the extent of base pairing between template RNA and telomeric DNA. EMBO reports. 2005;6:361–366. doi: 10.1038/sj.embor.7400374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 13.Luke B, Panza A, Redon S, Iglesias N, Li Z, Lingner J. The Rat1p 5' to 3' exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol Cell. 2008;32:465–477. doi: 10.1016/j.molcel.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 14.Qiu J, Qian Y, Frank P, Wintersberger U, Shen B. Saccharomyces cerevisiae RNase H(35) functions in RNA primer removal during lagging-strand DNA synthesis, most efficiently in cooperation with Rad27 nuclease. Mol Cell Biol. 1999;19:8361–8371. doi: 10.1128/mcb.19.12.8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ayyagari R, Gomes XV, Gordenin DA, Burgers PM. Okazaki fragment maturation in yeast. I. Distribution of functions between FEN1 AND DNA2. J Biol Chem. 2003;278:1618–1625. doi: 10.1074/jbc.M209801200. [DOI] [PubMed] [Google Scholar]
- 16.Jin YH, Ayyagari R, Resnick MA, Gordenin DA, Burgers PM. Okazaki fragment maturation in yeast. II. Cooperation between the polymerase and 3'-5'-exonuclease activities of Pol delta in the creation of a ligatable nick. J Biol Chem. 2003;278:1626–1633. doi: 10.1074/jbc.M209803200. [DOI] [PubMed] [Google Scholar]
- 17.Balakrishnan L, Bambara RA. Eukaryotic lagging strand DNA replication employs a multi-pathway mechanism that protects genome integrity. J Biol Chem. 2011;286:6865–6870. doi: 10.1074/jbc.R110.209502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eder PS, Walder JA. Ribonuclease H from K562 human erythroleukemia cells. Purification, characterization, and substrate specificity. J Biol Chem. 1991;266:6472–6479. [PubMed] [Google Scholar]
- 19.Ide H, Yagi R, Yamaoka T, Kimura Y. Misincorporation of ribonucleotides by DNA polymerase during in vitro DNA replication. Nucleic Acids Symp Ser. 1993:133–134. [PubMed] [Google Scholar]
- 20.Rydberg B, Game J. Excision of misincorporated ribonucleotides in DNA by RNase H (type 2) and FEN-1 in cell-free extracts. Proc Natl Acad Sci U S A. 2002;99:16654–16659. doi: 10.1073/pnas.262591699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nick McElhinny SA, Kumar D, Clark AB, Watt DL, Watts BE, Lundstrom EB, Johansson E, Chabes A, Kunkel TA. Genome instability due to ribonucleotide incorporation into DNA. Nat Chem Biol. 2010;6:774–781. doi: 10.1038/nchembio.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rychlik MP, Chon H, Cerritelli SM, Klimek P, Crouch RJ, Nowotny M. Crystal structures of RNase H2 in complex with nucleic acid reveal the mechanism of RNA-DNA junction recognition and cleavage. Mol Cell. 2010;40:658–670. doi: 10.1016/j.molcel.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim N, Huang SN, Williams JS, Li YC, Clark AB, Cho JE, Kunkel TA, Pommier Y, Jinks-Robertson S. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science. 2011;332:1561–1564. doi: 10.1126/science.1205016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lazzaro F, Novarina D, Amara F, Watt DL, Stone JE, Costanzo V, Burgers PM, Kunkel TA, Plevani P, Muzi-Falconi M. RNase H and Postreplication Repair Protect Cells from Ribonucleotides Incorporated in DNA. Mol Cell. 2012;45:99–110. doi: 10.1016/j.molcel.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clark AB, Kunkel TA. The importance of being DNA. Cell Cycle. 2010;9:4422–4424. doi: 10.4161/cc.9.22.14052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poveda AM, Le Clech M, Pasero P. Transcription and replication: Breaking the rules of the road causes genomic instability. Transcription. 2010;1:99–102. doi: 10.4161/trns.1.2.12665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Figiel M, Chon H, Cerritelli SM, Cybulska M, Crouch RJ, Nowotny M. The structural and biochemical characterization of human RNase H2 complex reveals the molecular basis for substrate recognition and Aicardi-Goutières syndrome defects. J Biol Chem. 2011;286:10540–10550. doi: 10.1074/jbc.M110.181974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goutières F. Aicardi-Goutières syndrome. Brain Dev. 2005;27:201–206. doi: 10.1016/j.braindev.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 29.Crow YJ, Leitch A, Hayward BE, Garner A, Parmar R, Griffith E, Ali M, Semple C, Aicardi J, Babul-Hirji R, Baumann C, Baxter P, Bertini E, Chandler KE, Chitayat D, Cau D, Dery C, Fazzi E, Goizet C, King MD, Klepper J, Lacombe D, Lanzi G, Lyall H, Martinez-Frias ML, Mathieu M, McKeown C, Monier A, Oade Y, Quarrell OW, Rittey CD, Rogers RC, Sanchis A, Stephenson JB, Tacke U, Till M, Tolmie JL, Tomlin P, Voit T, Weschke B, Woods CG, Lebon P, Bonthron DT, Ponting CP, Jackson AP. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutières syndrome and mimic congenital viral brain infection. Nat Genet. 2006;38:910–916. doi: 10.1038/ng1842. [DOI] [PubMed] [Google Scholar]
- 30.Rohman MS, Koga Y, Takano K, Chon H, Crouch RJ, Kanaya S. Effect of the disease-causing mutations identified in human ribonuclease (RNase) H2 on the activities and stabilities of yeast RNase H2 and archaeal RNase HII. FEBS J. 2008;275:4836–4849. doi: 10.1111/j.1742-4658.2008.06622.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reijns MA, Bubeck D, Gibson LC, Graham SC, Baillie GS, Jones EY, Jackson AP. The structure of the human RNase H2 complex defines key interaction interfaces relevant to enzyme function and human disease. J Biol Chem. 2011;286:10530–10539. doi: 10.1074/jbc.M110.177394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goutières F, Aicardi J, Barth PG, Lebon P. Aicardi-Goutières syndrome: an update and results of interferon-alpha studies. Ann Neurol. 1998;44:900–907. doi: 10.1002/ana.410440608. [DOI] [PubMed] [Google Scholar]
- 33.Rigby RE, Leitch A, Jackson AP. Nucleic acid-mediated inflammatory diseases. Bioessays. 2008;30:833–842. doi: 10.1002/bies.20808. [DOI] [PubMed] [Google Scholar]
- 34.Rice GI, Bond J, Asipu A, Brunette RL, Manfield IW, Carr IM, Fuller JC, Jackson RM, Lamb T, Briggs TA, Ali M, Gornall H, Couthard LR, Aeby A, Attard-Montalto SP, Bertini E, Bodemer C, Brockmann K, Brueton LA, Corry PC, Desguerre I, Fazzi E, Cazorla AG, Gener B, Hamel BC, Heiberg A, Hunter M, van der Knaap MS, Kumar R, Lagae L, Landrieu PG, Lourenco CM, Marom D, McDermott MF, van der Merwe W, Orcesi S, Prendiville JS, Rasmussen M, Shalev SA, Soler DM, Shinawi M, Spiegel R, Tan TY, Vanderver A, Wakeling EL, Wassmer E, Whittaker E, Lebon P, Stetson DB, Bonthron DT, Crow YJ. Mutations involved in Aicardi-Goutieres syndrome implicate SAMHD1 as regulator of the innate immune response. Nat Genet. 2009;41:829–832. doi: 10.1038/ng.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 36.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 37.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 38. [November 2010];SGD project. http://downloads.yeastgenome.org/
- 39.Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, Fisk DG, Hirschman JE, Hitz BC, Karra K, Krieger CJ, Miyasato SR, Nash RS, Park J, Skrzypek MS, Simison M, Weng S, Wong ED. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 2012;40:D700–D705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee I, Li Z, Marcotte EM. An improved, bias-reduced probabilistic functional gene network of baker's yeast, Saccharomyces cerevisiae. PLoS One. 2007;2:e988. doi: 10.1371/journal.pone.0000988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rusyn I, Fry RC, Begley TJ, Klapacz J, Svensson JP, Ambrose M, Samson LD. Transcriptional networks in S. cerevisiae linked to an accumulation of base excision repair intermediates. PLoS One. 2007;2:e1252. doi: 10.1371/journal.pone.0001252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tous C, Aguilera A. Impairment of transcription elongation by R-loops in vitro. Biochem Biophys Res Commun. 2007;360:428–432. doi: 10.1016/j.bbrc.2007.06.098. [DOI] [PubMed] [Google Scholar]
- 45.El Hage A, French SL, Beyer AL, Tollervey D. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 2010;24:1546–1558. doi: 10.1101/gad.573310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin Y, Dent SY, Wilson JH, Wells RD, Napierala M. R loops stimulate genetic instability of CTG.CAG repeats. Proc Natl Acad Sci U S A. 2010;107:692–697. doi: 10.1073/pnas.0909740107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McIvor EI, Polak U, Napierala M. New insights into repeat instability: role of RNA*DNA hybrids. RNA biology. 2010;7:551–558. doi: 10.4161/rna.7.5.12745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee I, Date SV, Adai AT, Marcotte EM. A probabilistic functional network of yeast genes. Science. 2004;306:1555–1558. doi: 10.1126/science.1099511. [DOI] [PubMed] [Google Scholar]
- 49.Nick McElhinny SA, Watts BE, Kumar D, Watt DL, Lundstrom EB, Burgers PM, Johansson E, Chabes A, Kunkel TA. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc Natl Acad Sci U S A. 2010;107:4949–4954. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Watt DL, Johansson E, Burgers PM, Kunkel TA. Replication of ribonucleotide-containing DNA templates by yeast replicative polymerases. DNA Repair (Amst) 2011;10:897–902. doi: 10.1016/j.dnarep.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Falco M, Ferrari E, De Felice M, Rossi M, Hubscher U, Pisani FM. The human GINS complex binds to and specifically stimulates human DNA polymerase alpha-primase. EMBO Rep. 2007;8:99–103. doi: 10.1038/sj.embor.7400870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clark AB, Lujan SA, Kissling GE, Kunkel TA. Mismatch repair-independent tandem repeat sequence instability resulting from ribonucleotide incorporation by DNA polymerase epsilon. DNA Repair (Amst) 2011;10:476–482. doi: 10.1016/j.dnarep.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sekiguchi J, Shuman S. Site-specific ribonuclease activity of eukaryotic DNA topoisomerase I. Mol Cell. 1997;1:89–97. doi: 10.1016/s1097-2765(00)80010-6. [DOI] [PubMed] [Google Scholar]
- 54.Nick McElhinny SA, Kissling GE, Kunkel TA. Differential correction of lagging-strand replication errors made by DNA polymerases {alpha} and {delta} Proc Natl Acad Sci U S A. 2010;107:21070–21075. doi: 10.1073/pnas.1013048107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vengrova S, Dalgaard JZ. The wild-type Schizosaccharomyces pombe mat1 imprint consists of two ribonucleotides. EMBO Rep. 2006;7:59–65. doi: 10.1038/sj.embor.7400576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chan CT, Dyavaiah M, DeMott MS, Taghizadeh K, Dedon PC, Begley TJ. A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet. 2010;6:e1001247. doi: 10.1371/journal.pgen.1001247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mischo HE, Gomez-Gonzalez B, Grzechnik P, Rondon AG, Wei W, Steinmetz L, Aguilera A, Proudfoot NJ. Yeast Sen1 helicase protects the genome from transcription-associated instability. Molecular cell. 2011;41:21–32. doi: 10.1016/j.molcel.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luke B, Lingner J. TERRA: telomeric repeat-containing RNA. Embo J. 2009;28:2503–2510. doi: 10.1038/emboj.2009.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reddy K, Tam M, Bowater RP, Barber M, Tomlinson M, Nichol Edamura K, Wang YH, Pearson CE. Determinants of R-loop formation at convergent bidirectionally transcribed trinucleotide repeats. Nucleic acids research. 2011;39:1749–1762. doi: 10.1093/nar/gkq935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wongsurawat T, Jenjaroenpun P, Kwoh CK, Kuznetsov V. Quantitative model of R-loop forming structures reveals a novel level of RNA-DNA interactome complexity. Nucleic Acids Res. 2012;40:e16. doi: 10.1093/nar/gkr1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Coffin SR, Hollis T, Perrino FW. Functional consequences of the RNase H2A subunit mutations that cause Aicardi-Goutieres syndrome. J Biol Chem. 2011;286:16984–16991. doi: 10.1074/jbc.M111.228833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Crow YJ, Rehwinkel J. Aicardi-Goutieres syndrome and related phenotypes: linking nucleic acid metabolism with autoimmunity. Hum Mol Genet. 2009;18:R130–R136. doi: 10.1093/hmg/ddp293. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.