Abstract
Introduction
Mdm2 inhibits p53 transactivation by forming a p53-Mdm2 complex on chromatin. Upon DNA damage-induced complex disruption, such latent p53 can be activated, but in cells overexpressing Mdm2 because of a homozygous single nucleotide polymorphism at position 309 (T→G) of mdm2, the complex is highly stable and cannot be disrupted by DNA damage, rendering p53 inactive.
Methods
To determine whether the p53 response phenotype is influenced differentially in cells with variable mdm2 genotypes, we compared responses to DNA damage and targeted p53-Mdm2 complex disruption by Nutlin-3 in the following wild-type p53 human cancer cell lines: A875 and CCF-STTG-1 (G/G for mdm2 SNP309), SJSA-1 (mdm2 genomic amplification and T/T for mdm2 SNP309), MCF-7 (estrogen-induced Mdm2 overexpression and T/G for mdm2 SNP309), ML-1 and H460 (T/T for mdm2 SNP309), and K562 (p53-null and T/G for mdm2 SNP309). We also examined mdm2 gene-splicing patterns in these lines by cloning and sequencing analyses.
Results
While Mdm2-overexpressing G/G cells were resistant to p53 activation by DNA damage, they were sensitive to Nutlin-3. Strikingly, the p53 G1 checkpoint in G/G cells was activated by Nutlin-3 but not by etoposide, whereas in other Mdm2-overexpressing cells, both drugs activated p53 and subsequent G1 arrest or apoptosis. cDNA clones lacking exons 5–9 were generated at a high frequency in cells overexpressing Mdm2.
Conclusion
Nutlin-3 and DNA damage distinguish a differential phenotype in human cancer cells with G/G mdm2 SNP309 from other Mdm2 overexpressers. Categorization of the Mdm2 isoforms produced and their influence on p53 activity will help in characterization and treatment development for different cancers.
Keywords: p53, Mdm2, SNP309, Phenotype, Nutlin-3
Introduction
The p53 tumor suppressor protein plays an important role in preventing cancer development by arresting or killing potentially tumorigenic cells. The protein has a short half-life in normally proliferating cells due to rapid proteosomal degradation but is stabilized after stress.1 The p53 protein must be restrained in healthy cells to allow normal cell growth but can be rapidly stimulated in response to cellular stress.2 The p53 protein is a transcription factor that activates several downstream targets of different cellular functions, including apoptosis, growth arrest, and DNA repair.2 Consequently, malignant progression is dependent upon the loss of p53 function, either through p53 mutationsordefectsinp53 signaling pathways.
p53 transactivation can be inhibited by Mouse Double Minute 2 (Mdm2) overexpression through gene duplication, enhanced transcription, or translation.3 Wild-type p53 human tumor cells often show Mdm2 overexpression. The Mdm2 protein is a key inhibitor of p53 stability; p53 protein promotes mdm2 transcription via a p53-dependent promoter in mdm2, and the Mdm2 protein inhibits p53 protein function.3
Mdm2 inactivates p53 by tagging the protein for proteosomal degradation. However, hypomorphic mdm2 mice expressing reduced Mdm2 levels demonstrate enhanced p53 activity without increased p53 protein levels, indicating ubiquitination is not the only way that Mdm2 inactivates p53.4 Mdm2 can complex with p53 on chromatin to keep the protein latent until activation is required, then the complex can be released to facilitate critical gene transcription.5 However, we have demonstrated that in cells with Mdm2 overexpression from a homozygous single nucleotide polymorphism at position 309 in mdm2 (SNP309 T→G), Mdm2 protein localizes with stabilized p53 at p53 binding sites on chromatin and blocks p53 transactivation.6 SNP309 causes increased SP1 affinity, directly leading to Mdm2 overexpression from its P2 promoter.7 The mdm2 SNP309 also accelerates tumor formation in a sex-specific and hormone-dependent manner.8 Additionally, Mdm2 protein has been found at p53 binding sites on chromatin in other cell lines, including the wild-type Mdm2-overexpressing cell line SJSA-1.9
Disruption of the p53-Mdm2 complex represents the conceptual basis for targeted chemotherapy to activate p53 transcription.10 Cytotoxic agents used in cancer treatment act mainly by inducing genotoxic stress that leads to p53 post-translational modifications and disruption of the p53-Mdm2 complex.11 Recently, there have been efforts to identify potent and selective small-molecule inhibitors of the p53-Mdm2 interaction with in vitro and in vivo antitumor activity, without causing DNA damage. One such class is a series of cis-imidazoline analogs named Nutlins.12 Nutlins are designed to fit into the p53 binding pocket of Mdm2, thus preventing an interaction between the two proteins. By doing so, Nutlin treatment can induce p53 transcriptional activity. Nutlins are ineffective in cells lacking either p53 or Mdm2 and are nontoxic to mice.
We compared the effects of Nutlin-3 and etoposide on cell cycle transition and p53 transactivation in human cells with Mdm2 overexpression resulting from several different genotypes including SNP309. Etoposide is a clinically used chemotherapeutic agent known to activate p53. The 50% inhibitory concentration (IC50) for etoposide varies widely across cell lines but has been reported to range from 0.1 µM to 38 µM. Etoposide induces cellular stress by inhibiting the action of topoisomerase II, thus causing double strand breaks; etoposide is cytotoxic to cells regardless of p53 status.
Methods
Cell Culture
A875 and CCF-STTG-1 cells were a gift from Arnold Levine; SJSA-1, MCF-7, H460, and K562 cells were purchased from American Type Culture Collection; and ML-1 cells were a gift from Michael Kastan. MANCA cells were a gift from Andrew Koff. Cells were grown in RPMI 1640 (Mediatech, Herndon, Va) containing 10% fetal bovine serum (Gemini, West Sacramento, Calif) and 2500 units of penicillin-streptomycin (Mediatech). MCF-7 cells were supplemented with 10 nM estrogen (Sigma, Saint Louis, Mo), charcoal-stripped serum, and grown without phenol red. All cells were incubated with 5% CO2 at 37°C (98.6°F). Etoposide (Sigma) and Nutlin-3 (Calbiochem, San Diego, Calif) were added where indicated.
Flow Cytometry
Fluorescence-activated cell sorting (FACS) was performed on a FACScan (BD Biosciences, San Jose, Calif). Cells were centrifuged at 2000 rpm for seven minutes at 4°C (39.2°F), washed with phosphate-buffered saline (PBS), resuspended in PBS containing 2% bovine serum albumin, 0.1% sodium azide, and 1% pluronic F-68, fixed in 30% ethanol, and stored overnight at 4°C (39.2°F). Before sorting, propidium iodide staining and RNase treatment were performed for 30 minutes at 37°C (98.6°F).
cDNA
RNA was isolated by using QIA-shredder columns and the RNeasy Mini Kit (Qiagen, Valencia, Calif). Five micrograms of RNA were used for cDNA synthesis using the High Capacity cDNA Archive Kit (Applied Biosystems, Foster City, Calif).
Quantitative Reverse Transcription-PCR (qRT-PCR)
The primer probes for gapdh (Applied Biosystems Pre-developed Assay Reagents) mdm2, waf1, gadd45, puma, and pig3 (Applied Biosystems/Celera Assays-on-Demand) were combined with Taqman Universal Master Mix (Applied Biosystems) and cDNA. Polymerase chain reaction (PCR) in a 7500 Sequence Detection System (Applied Biosystems) proceeded as follows: one cycle, two minutes, 50°C (122°F); one cycle, 10 minutes, 94°C (201.2°F); and 40 cycles, 15 seconds, 94°C (201.2°F); and one minute, 60°C (140°F).
cDNA cloning
cDNA was combined with Taqman Universal Master Mix and primers flanking mdm2 exons 2 and 12 (forward: TGTGTTCAGTGGCGATTGGAG, reverse: GGGGGATTCATTTCATTGCATG) for PCR in a GeneAmp 5400 (Perkin Elmer, Waltham, Mass) as follows: one cycle, 10 minutes, 95°C (203°F); 35 cycles, 30 seconds, 95°C (203°F), 30 seconds, 59°C (138.2°F), and 2 minutes 72°C (161.6°F); and 10 minutes, 72°C (161.6°F). PCR product was cloned into the pCR II-TOPO plasmid by using the TOPO TA Cloning Kit (Invitrogen, Carlsbad, Calif). Plasmids were isolated with the FastPlasmid Mini Kit (Eppendorf, Hamburg, Germany), digested with EcoRI (New England Biolabs, Ipswich, Mass), and resolved by agarose gel electrophoresis. Approximately 500 ng of plasmid DNA was sequenced by using M13 forward (TGTAAAACGACGGCCAGT) and reverse (CAGGAAACAGCTATGAC) primers using an Applied Biosystems Prism 3730 DNA Analyzer.
Identification of mdm2 Spliced Variants
cDNA sequences were compared with full-length mdm2 sequence (Gen-Bank AF527840.1) by using the BL2SEQ program.13 A PERL script based on Bio::Graphics, a BioPerl (bioperl.org) module, was written to graphically render BL2SEQ results by using bidirectional sequencing output. Genomic sequence was translated to amino acid sequence by using the ExPASy amino acid to protein translation tool (www.expasy.org/tools/dna).
Results
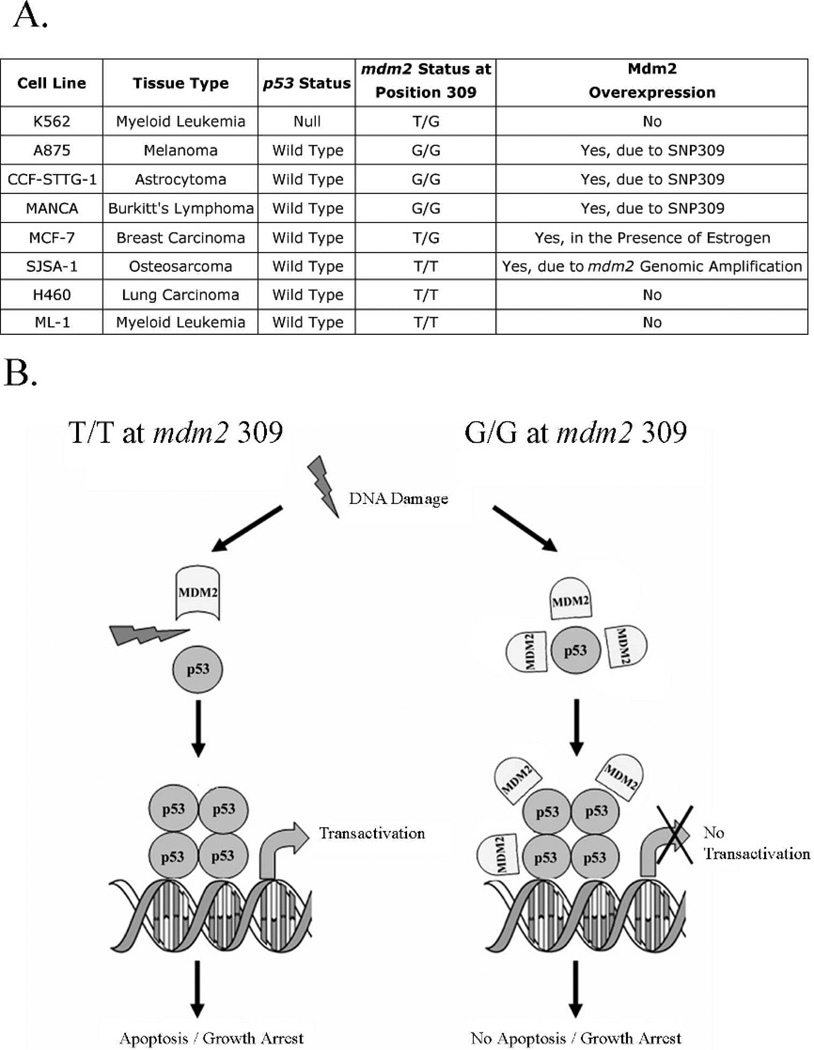
Cancer cells overexpress oncogenic Mdm2 by numerous mechanisms; estrogen receptor positive MCF-7 cells increase Mdm2 expression in the presence of estrogen, which has been proposed to inactivate the p53 checkpoint.14 To study wild-type p53 inactivation by Mdm2 at the cellular and biochemical levels, we compiled human cancer cell lines with variable mdm2 genotypes (Figure 1A).
Fig 1.
mdm2 SNP309 cells have functionally compromised p53 due to a p53-Mdm2 complex. A) Table depicting the different cell lines used in the project, their origins, and mdm2 and p53 status. B) Model demonstrating that in cells homozygous for G/G mdm2 SNP309, p53 is induced after DNA damage and does not dissociate from the excessively expressed Mdm2 protein. This association does not interfere with the ability of p53 to bind to DNA but does impair its transcriptional activity. This model has been slightly modified from one published previously.6
Nutlin-3 Can Reactivate p53 in Cancer Cell Lines with Overexpressed Mdm2 When Activation by Etoposide is Compromised
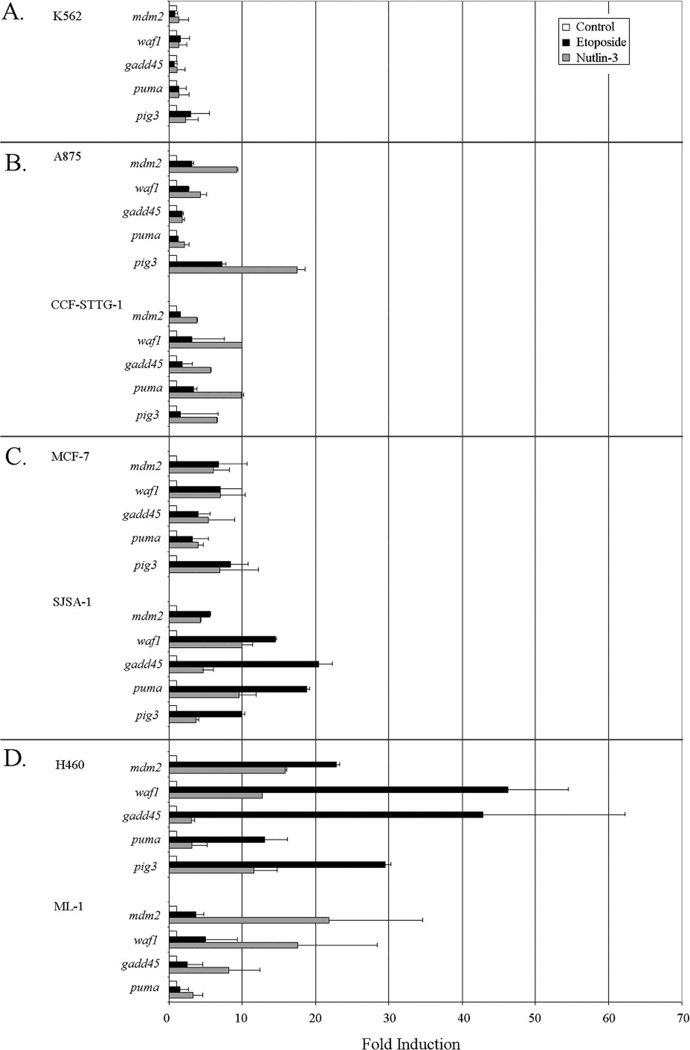
We explored if Nutlin-3, a promising small molecule inhibitor of the p53-Mdm2 interaction, could reactivate functionally compromised p53 in cells overexpressing Mdm2. The cell lines described (Figure 1A) were treated with either Nutlin-3 or etoposide at concentrations previously demonstrated to activate wild-type p53 protein.5,6 Using qRT-PCR, we determined Nutlin-3 treatment induced greater activation of p53 in G/G SNP309 cells than etoposide treatment (Figure 2, panel B). As expected, the p53-null K562 cells showed virtually no induction of p53 target genes in response to either drug treatment (Figure 2, panel A). In A875 cells Nutlin-3 treatment induced >15-fold activation of the p53 target pig3, and in CCF-STTG-1 cells Nutlin-3 mediated activation of p53 targets waf1 and puma reached 10-fold. The other p53 target genes tested in G/G mdm2 SNP309 cell lines were not induced as highly by Nutlin-3 but generally showed greater activation than seen with etoposide. In MCF-7 cells with estrogen-induced overexpression of Mdm2, etoposide and Nutlin-3 treatment resulted in similar p53 target gene activation (Figure 2, panel C). Analysis of these target genes in SJSA-1 cells with wild-type p53 and wild-type Mdm2 gene amplification (Figure 2, panel C) or in H460 and ML-1 cells with normally expressed wild-type Mdm2 (Figure 2, panel D) showed variable activation phenotypes after etoposide treatment, as compared to Nutlin-3. Etoposide provoked greater p53 target gene activation than Nutlin-3 in SJSA-1 and H460 cells, while in ML-1 cells Nutlin-3 was more effective (Figure 2, panels C and D). In the H460 cell line, etoposide caused activation of waf1, gadd45, and pig3 of ≥30-fold. Nutlin-3 activation levels for both H460 and ML-1 cells were slightly higher but comparable to those seen in mdm2 SNP309 cells (Figure 2, panel D). The Mdm2 overexpressing and non-overexpressing cell lines responded similarly to Nutlin-3, suggesting Nutlin-3 can activate the p53 pathway across different strata of p53-Mdm2 cellular phenotypes, while DNA damage by etoposide results in cell type variability.
Fig 2.
Nutlin-3 can reactivate p53 in cancer cell lines with overexpressed Mdm2 when activation by etoposide is compromised. Quantitative reverse transcription–polymerase chain reaction was used to detect fold induction of the mdm2, waf1, gadd45, puma, and pig3 transcripts in K562 (panel A), A875 and CCF-STTG-1 (panel B), SJSA-1 and MCF-7 (panel C), and H460 and ML-1 (panel D) cells. Exponentially growing cells were either left untreated (white bars), treated with 8 µM etoposide (50 µM for MCF-7 cells) (black bars), or 10 µM Nutlin-3 (gray bars), for 24 hours. All results were normalized to untreated samples and gapdh values. Results are representative of three independent experiments, and error bars indicate standard deviation.
Cell Cycle Analysis of Human Cancer Cells Treated with Etoposide or Nutlin-3
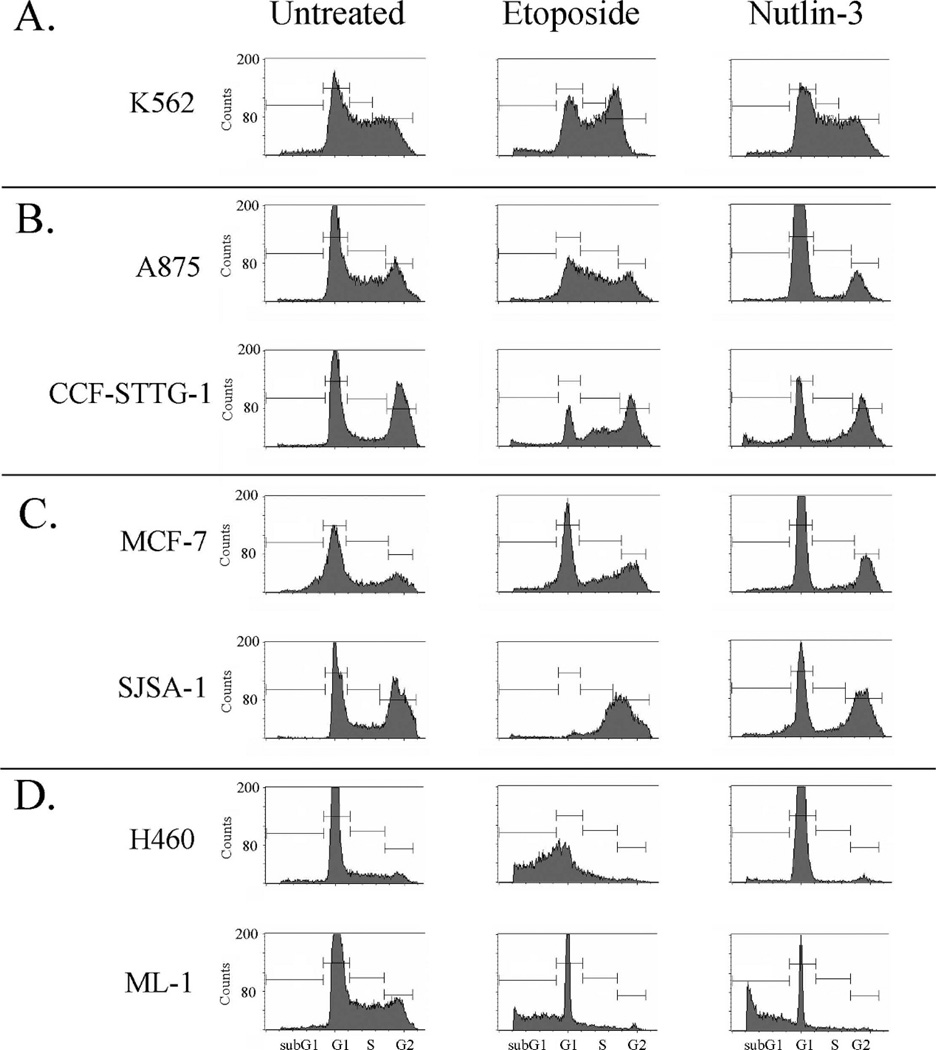
Using FACS analysis, we compared the influence of Nutlin-3 and etoposide on the growth of these cell lines. Nutlin-3 treatment of K562 cells (p53-null) showed no change in the cell cycle profile (Figure 3, panel A). In agreement with published Nutlin-3 data,12 we saw cell cycle arrest as the predominant outcome after Nutlin-3 treatment in wild-type p53 cancer cell lines (Figure 3). Nutlin-3-mediated G1 arrest occurred in G/G mdm2 SNP309 cells (Figure 3, panel B), the Mdm2 estrogen-induced overexpresser and the mdm2 genomic amplification overexpresser (Figure 3, panel C), and one wild-type Mdm2 normal expresser (Figure 3, panel D, H460). The notable exception was ML-1 cells (normal wild-type Mdm2 expression) with increased sub-G1 DNA content after Nutlin-3 treatment, indicating apoptosis (Figure 3, panel D, ML-1). The etoposide-provoked cell cycle profiles showed a profound change in cancer cells with wild-type p53 and normal wild-type mdm2 expression (Figure 3, panel D), with a major increase in sub-G1 DNA content for both. A875 and CCF-STTG-1 cells were resistant to etoposide-induced G1 arrest or apoptosis but were sensitive to induction of G1 arrest by Nutlin-3. Compared to etoposide, data suggest that Nutlin-3 treatment can provoke a more consistent activation of the p53-mediated checkpoints, with a trend towards cell cycle arrest even in the presence of a homozygous G/G mdm2 SNP309 genotype.
Fig 3.
Cell cycle analysis of human cancer cells treated with etoposide or Nutlin-3. Fluorescence-activated cell sorting (FACS) analysis of K562 (panel A), A875 and CCF-STTG-1 (panel B), SJSA-1 and MCF-7 (panel C), and H460 and ML-1 (panel D) cells. Exponentially growing cells were either left untreated (left panel), treated with 8 µM etoposide (50 µM for MCF-7 cells) (center panel), or treated with 10 µM Nutlin-3 (right panel) for 48 hours. Cells were harvested, fixed in 30% ethanol, and cellular DNA was stained with propidium iodide before FACS analysis. Cell cycle phase is indicated on the graphs in the bottom panel.
Cloning of mdm2 cDNA from SNP309 Cancer Cells Shows Large Deletions
Many alternatively spliced forms of mdm2 exons 1–12 are produced in cancer cells,15 and DNA damage can facilitate changes in splicing patterns of mdm2 mRNA.16 The mdm2 gene has two well-characterized promoters (P1 and P2), and when transcription begins from the P2 promoter, the traditional mRNA includes exons 2 through 12.17 We are working to determine the predominant spliced variants in G/G SNP309 cells. Using nested PCR primer sets for mdm2 we observed that products of different sizes were more commonly generated in SNP309 cells (data not shown).
We utilized TA cloning of RT-PCR products to construct mdm2 cDNA clones, with primers flanking exons 2 and 12. Using 2–12 cDNA clones from the cell lines in Figure 1A, we have begun to examine spliced variants resulting from the mdm2 P2 promoter (which is more active than P1 in G/G SNP309 cells). Two spliced variants we isolated were similar to those previously associated with tumor formation, MDM2-F18 and MDM2-C.19 MDM2-C has transforming ability and is frequently detected in human cancer. The cDNA clones we obtained are missing exons 5–9 and have been named P2 MDM2-C because they are derived from the P2 promoter. The frequency of P2 MDM2-C clones obtained suggests that Mdm2-spliced variants related to SNP309 require further investigation, as the region spliced out is required for Mdm2-mediated p53 degradation.20 Other clone sequences of the high-frequency, or novel splicing, were analyzed with the ExPASy nucleic acid to protein translation tool (www.expasy.org/tools/dna). Figure 4 shows that these clones can produce in-frame truncated Mdm2 products. The spliced variants shown in Figure 4 have retained the same amino acids as the full-length protein, thus indicating their potential to produce product in vivo. The sequences of the P2 promoter clones obtained have been submitted to the National Center for Biotechnology Information database and can be retrieved (accession numbers: P2-MDM2-C1 EU07674, P2-MDM2-10 EU076747, P2-MDM2-C EU076748, P2-MDM2-FL EU076749).
Fig 4.
Spliced variants have the potential to produce in-frame truncated Mdm2 products. cDNA was prepared by using reverse transcription–polymerase chain reaction (RT-PCR). Using specific primers flanking exons 2 and 12 of mdm2, PCR products were generated, cloned, and sequenced. The resulting nucleotide sequences were translated by using the translation tool found at www.ExPASy.org. The output amino acid sequences were compared to the Mdm2 full-length protein as depicted above, where letters represent amino acids.
Discussion
Standard DNA-damaging drugs do not adequately disrupt the p53-Mdm2 chromatin associated complex in G/G SNP309 cells (Figure 1B).6 To explore an alternative option, we used a human cancer cell line-based study to investigate the effectiveness of Nutlin-3 in activating the p53 tumor suppressor pathway in cell lines that do not respond adequately to standard chemotherapeutics. Nutlin-3 has shown promise in cell culture and mouse model studies. This is the first study to examine its effects in G/G mdm2 SNP309 cells and in breast cancer cells with Mdm2 overexpressed because of prolonged estrogen exposure.
The inactivation of wild-type p53 by oncogenic proteins is a common phenomenon in human cancers. Oncogenic Mdm2 can inactivate p53 by at least two pathways: E3 ubiquitin-mediated proteolysis and inhibition of p53 transcription by formation of a chromatin associated complex. Conventional DNA-damaging chemotherapeutics inadequately release the p53-Mdm2 chromatin complex in G/G mdm2 SNP309 cells. We have shown by qRT-PCR that Nutlin-3 treatment of mdm2 G/G SNP309 cells causes p53 target gene activation that is comparable to activation in cancer cell lines with low Mdm2 protein levels, while etoposide treatment does not (Figure 2). The ability of Nutlin-3 to activate p53 transcriptional activity was similar in cell lines with Mdm2 overexpressed from G/G SNP309, estrogen exposure, gene amplification, or in normally expressing wild-type mdm2 cells.
In addition to activating p53 transcription in G/G mdm2 SNP309 cells, Nutlin-3 also induced growth arrest, evident by increases in G1 and G2 phases and a decrease in S phase (Figure 3). While Nutlin-3 activated the p53 G1 cell cycle checkpoint in G/G SNP309 cells, the DNA damage-inducing drug etoposide did not. However, in SJSA-1 cells that overexpressed Mdm2 (gene amplification) at a higher level than in the G/G SNP309 cells (Arva and Bargonetti, unpublished data), etoposide activated the G1 growth arrest p53 pathway (Figures 2 and 3), suggesting either the Mdm2 protein produced in G/G SNP309 cells differs from that produced from the amplification genotype or that Mdm2 works in combination with cell type-specific factors and the DNA damage pathway in order to activate the p53 G1 arrest checkpoint in SJSA-1 cells. It is interesting that etoposide caused the most robust activation of p53 targets in H460 cells, even as compared to ML-1 cells, which like H460 are also wild-type for p53 and T/T for mdm2 SNP309. H460 cells may be more sensitive to the induction of double strand breaks, or there may be other genotypic differences between these cells that drive particular drug responses.
Because transcription factor loading can influence splicing,21 it would not be surprising if SNP309-induced transcription from the mdm2 P2 promoter prompted changes in mdm2 splicing. Additionally, DNA damage-induced stress changes mdm2 splicing.16 Importantly, our data from G/G SNP309 mdm2 cDNA clones and p53 activity analyses suggest alternative forms of Mdm2 are produced from the G/G SNP309 genotype. The region we see spliced out in cDNA clones includes exon 9, an important region for ubiquitin-mediated p53 degradation,20 which coincides with the fact that the Mdm2 protein in G/G SNP309 cells does not inhibit p53 by increased proteolysis but inhibits p53 by chromatin-mediated inhibition.6 While we have identified that cells overexpressing Mdm2 often produce alternatively spliced mdm2 mRNA from the P2 promoter, these messages are co-expressed with other spliced variants in the cell population. It would be interesting to determine if the expression of these clones in different human cell types will affect growth patterns.
In conclusion, we see that Nutlin-3 can activate p53 in G/G SNP309 cells by disrupting the p53-Mdm2 complex to allow for p53 target gene transcription and G1 cell cycle arrest as the predominant outcome after treatment.10 Thus, Nutlin-3 can activate the p53 pathway in a situation where DNA damage cannot.
Implications for Improving Health Disparities
Individuals homozygous for G/G mdm2 SNP309 have higher cancer incidence, develop cancer earlier, and have more recurrent tumors. Additionally, some of these cancers are hormone sensitive and can be provoked by estrogen. The increased tumor incidence in minority populations likely results from gene-environment interactions, and the mdm2 SNP309-estrogen interaction may associate with certain health disparities. The use of Nutlin-3 as a chemotherapeutic agent could allow for better across-the-board treatment of different cancers.
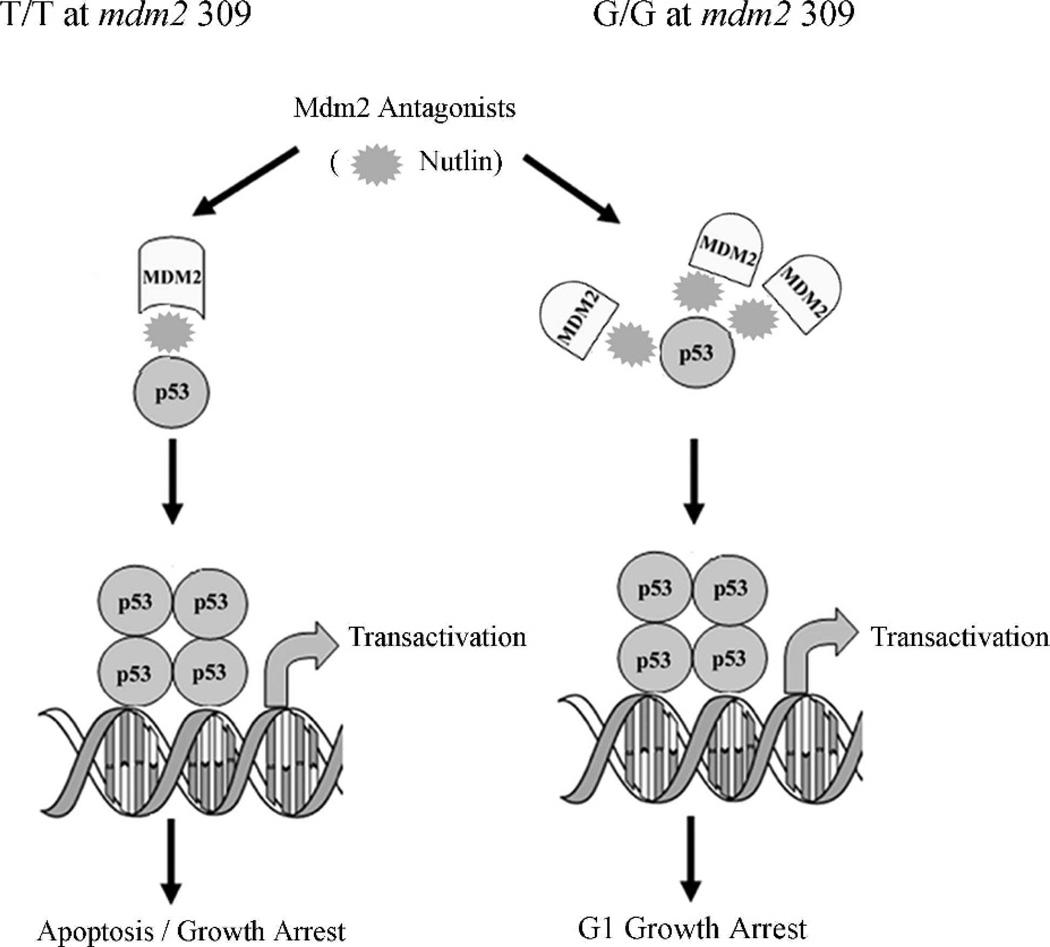
Fig 5.
Nutlin-3 can release the p53-Mdm2 complex in mdm2 SNP309 cells. A model illustrating that in cells homozygous for G/G mdm2 SNP309, p53 transcriptional activity is blocked through a chromatin-bound inhibitory complex that might contain an alternative Mdm2 isoform. DNA damage is unable to release p53 from this complex; however, the p53-Mdm2 antagonist, Nutlin-3, activates p53 function.
Acknowledgments
This work was supported by grants from The National Science Foundation (MCB-0212761) and The Breast Cancer Research Foundation to JB and was facilitated by an NIH National Center for Research Resources award (RR03037) to Hunter College. DO was supported by a Minority Access to Research Careers grant from the NIH/NIGMS (5T34GM007823-27). We thank Albana Thomaraj and Aashish Jethra for their help in screening and sequencing clones, Cindy Puente for her help in summarizing and organizing clone data, and Sandy Gamss for her help in editing the manuscript.
REFERENCES
- 1.Reihsaus E, Kohler M, Kraiss S, Oren M, Montenarh M. Regulation of the level of the oncoprotein p53 in non-transformed and transformed cells. Oncogene. 1989;5:137–145. [PubMed] [Google Scholar]
- 2.Bargonetti J, Manfredi JJ. Multiple roles of the tumor suppressor p53. Curr Opin Oncol. 2002;14:86–91. doi: 10.1097/00001622-200201000-00015. [DOI] [PubMed] [Google Scholar]
- 3.Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 4.Mendrysa SM, McElwee MK, Michalowski J, O’Leary KA, Young KM, Perry ME. mdm2 Is critical for inhibition of p53 during lymphopoiesis and the response to ionizing irradiation. Mol Cell Biol. 2003;23:462–472. doi: 10.1128/MCB.23.2.462-473.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.White DE, Talbott KE, Arva NC, Bargonetti J. Mouse double minute 2 associates with chromatin in the presence of p53 and is released to facilitate activation of transcription. Cancer Res. 2006;66:3463–3470. doi: 10.1158/0008-5472.CAN-05-1381. [DOI] [PubMed] [Google Scholar]
- 6.Arva NC, Gopen TR, Talbott KE, et al. A chromatin associated and transcriptionally inactive p53-MDM2 complex occurs in MDM2 SNP 309 homozygous cells. J Biol Chem. 2005;280:26776–26787. doi: 10.1074/jbc.M505203200. [DOI] [PubMed] [Google Scholar]
- 7.Bond GL, Hu W, Bond EE, et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004;119:591–602. doi: 10.1016/j.cell.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 8.Bond GL, Hirshfield KM, Kirchhoff T, et al. MDM2 SNP309 accelerates tumor formation in a gender-specific and hormone-dependent manner. Cancer Res. 2006;66:5104–5110. doi: 10.1158/0008-5472.CAN-06-0180. [DOI] [PubMed] [Google Scholar]
- 9.Minsky N, Oren M. The RING domain of Mdm2 mediates histone ubiquitylation and transcriptional repression. Mol Cell. 2004;16:631–639. doi: 10.1016/j.molcel.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 10.Tovar C, Rosinski J, Filipovic Z, et al. Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: implications for therapy. Proc Natl Acad Sci U S A. 2006;103:1888–1893. doi: 10.1073/pnas.0507493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shieh SY, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 12.Vassilev LT, Vu BT, Graves B, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 13.Tatusova T, Madden TL. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- 14.Phelps M, Darley M, Primrose JN, Blaydes JP. p53-independent activation of the hdm2-P2 promoter through multiple transcription factor response elements results in elevated hdm2 expression in estrogen receptor alpha-positive breast cancer cells. Cancer Res. 2003;63:2616–2623. [PubMed] [Google Scholar]
- 15.Bartel F, Harris LC, Wurl P, Taubert H. MDM2 and its splice variant messenger RNAs: expression in tumors and down-regulation using antisense oligonucleotides. Mol Cancer Res. 2004;2:29–35. [PubMed] [Google Scholar]
- 16.Chandler DS, Singh RK, Caldwell LC, Bitler JL, Lozano G. Genotoxic stress induces coordinately regulated alternative splicing of the p53 modulators MDM2 and MDM4. Cancer Res. 2006;66:9502–9508. doi: 10.1158/0008-5472.CAN-05-4271. [DOI] [PubMed] [Google Scholar]
- 17.Barak Y, Gottlieb E, Juven-Gershon T, Oren M. Regulation of mdm2 expression by p53: alternative promoters produce transcripts with nonidentical translation potential. Genes Dev. 1994;8:1739–1749. doi: 10.1101/gad.8.15.1739. [DOI] [PubMed] [Google Scholar]
- 18.Tamborini E, Della Torre G, Lavarino C, et al. Analysis of the molecular species generated by MDM2 gene amplification in liposarcomas. Int J Cancer. 2001;92:790–796. doi: 10.1002/ijc.1271. [DOI] [PubMed] [Google Scholar]
- 19.Sigalas I, Calvert AH, Anderson JJ, Neal DE, Lunec J. Alternatively spliced mdm2 transcripts with loss of p53 binding domain sequences: transforming ability and frequent detection in human cancer. Nat Med. 1996;2:912–917. doi: 10.1038/nm0896-912. [DOI] [PubMed] [Google Scholar]
- 20.Wallace M, Worrall E, Pettersson S, Hupp TR, Ball KL. Dual-site regulation of MDM2 E3-ubiquitin ligase activity. Mol Cell. 2006;23:251–263. doi: 10.1016/j.molcel.2006.05.029. [DOI] [PubMed] [Google Scholar]
- 21.Calvo O, Manley JL. Strange bedfellows: polyadenylation factors at the promoter. Genes Dev. 2003;17:1321–1327. doi: 10.1101/gad.1093603. [DOI] [PubMed] [Google Scholar]