Abstract
Zinc finger proteins comprise the largest class of eukaryotic transcription factors. The metal binding sites in these proteins have been proposed as plausible targets for exchange reactions between zinc and toxic metal ions that lead to the alteration of function of the proteins in gene transcription. According to the present work, both Cd2+ and Pb2+ displace Zn2+ from transcription factor IIIA (TFIIIA). Neither product binds to the internal control region (ICR) of the 5 S rRNA gene, the normal binding site for Zn-TFIIIA. Furthermore, the adduct of Zn-TFIIIA with ICR is also reactive with Cd2+ and Pb2+, leading to the dissociation of the DNA–protein complex. Cd-TFIIIA reacts with apometallothionein (apoMT) to form Cd-MT and apoTFIIIA. Similarly, Cd2+ and Zn2+ can be exchanged in the reaction of Cd-TFIIIA with Zn-MT. Zn-finger 3 of TFIIIA has also been examined to compare the reactivity of a single finger motif with fingers in the holoprotein. Zn-finger 3 reacts with much faster kinetics than the holoprotein.
Keywords: Zinc-finger, Cadmium, Lead TFIIIA, Metallothionien
The most common protein DNA binding motif among transcription factors in eucaryotes is the Zn finger structure that is stabilized through binding of a Zn2+ ion to two imidazole nitrogen (N) and two cysteine sulfhydryl (S) ligands. In the absence of Zn2+ its conformational integrity is lost and the domain no longer associates with DNA. Because transcription factors play such a central role in cell regulation, Zn finger proteins have attracted much attention.
The prototypical Zn finger transcription factor is transcription factor IIIA (TFIIIA), isolated from the immature ovary of Xenopus laevis. It binds to the internal control region (ICR) of the 5 S ribosomal RNA gene and stimulates its transcription. The product 5 S rRNA competes with the ICR for binding Zn-TFIIIA to inhibit its own synthesis. TFIIIA largely comprises nine consecutive Zn finger domains which differentially interact with the ICR DNA or with 5 S rRNA.
Studies with TFIIIA and other Zn finger structures suggest that the Zn2+ is not bound as tightly in these molecules as in numerous other types of Zn-metalloproteins. Thus, when N2S2 Zn finger proteins are prepared, buffers commonly contain Zn2+ to insure that the isolated proteins are saturated with metal ion (Del Rio & Setzer, 1991). Furthermore, competing small ligands such as EDTA rapidly inactivate TFIIIA and other Zn fingers, whereas, such reactions involving other Zn-metalloproteins commonly occur very slowly (Hanas, Hazuda, Bogenhager, Wu & Wu, 1983; Petering, Krezoski, Chen, Pattanaik & Shaw, 1991). For these and other reasons, investigators have speculated that cellular control of gene regulation might include control by alteration in accessibility of Zn finger proteins to intracellular Zn2+. Furthermore, these same properties make Zn finger sites attractive targets for toxic metal ions to bind in competition with Zn2+ (Makowski & Sunderman, 1992).
A protein where both of these possibilities may occur is in MTF-1, the transcriptional control protein for induction of metallothionein (MT) synthesis by Zn2+. MTF-1 contains six Zn finger domains, is required for Zn2+ responsive MT induction in vivo, and binds to its DNA metal response element in a Zn dependent fashion in vitro (Radtke et. al., 1993). It has also been assumed that MTF-1 is the binding site for Cd2+ which results in MT induction (Andrews, 1990).
The present study examined interactions of Zn2+ and Cd2+ or Pb2+ with TFIIIA and reactions of these structures with MT to explore the chemical possibilities for reaction of Zn finger proteins with toxic metal ions and competing metal binding ligands.
TFIIIA was isolated from E. coli containing the cloned gene for Xenopus laevis TFIIIA (Del Rio & Setzer, 1991). The 106 base pair ICR was also obtained from a plasmid grown in E. coli and used as a 32P-labelled DNA binding site for TFIIIA in a standard electrophoretic mobility shift assay (Romaniuk, 1990). Finger 3 of TFIIIA (F3) was made by peptide synthesis and used for comparison with the native structure.
Isolation of Zn-TFIIIA in the absence or presence of Zn2+ resulted in protein samples that varied in Zn to protein stoichiometry from 2 to 9±2, confirming the qualitative view that Zn2+ is readily lost from the protein or difficult to acquire. One can reproducibly bind nine Zn2+, Cd2+, or Pb2+ ions per mol of TFIIIA by titrating the apoprotein with metal ion at pH 7.4.
Titration data have been used to measure metal ion binding constants for F3. The results in Table 1 show that Zn2+ does not bind strongly to F3, and both Cd2+ and Pb2+ display larger formations constants, indicating that these metal ions bind preferentially to F3 in comparison with Zn2+. Available information for two other finger peptides is also shown in Table 1. It is seen that there can be a large variation in binding affinity of Zn2+ for related finger structures. In the case of CP1, peptide association with Zn2+ is much more favorable than with Cd2+. These results show first that individual Zn finger sites will be differentially sensitive to the concentration of cellular Zn2+ and second that certain Zn finger proteins will be preferentially susceptible to exchange of Zn2+ with Cd2+ and, presumably, also with Pb2+.
Table 1.
Log formation constants for M2+-finger complexes at pH 7.4 and 25°CF3 of TFIIIA and other finger peptidesa
| F3 | Sp1 (F3) | CP1 | |
|---|---|---|---|
| Co2+ | 5.1 | 6.5 | 7.2 |
| Zn2+ | 7.4 | 9.2 | 11.2 |
| Cd2+ | 8.3 | 8.7 | |
| Pb2+ | 8.7 |
Abbreviations and references: Sp1 (F3), finger 3 of Sp1 transcription factor (Posewitz & Wilcox, 1995), CP1, consensus peptide derived from available sequences of N2S2 domains (Krizek, Merkle & Berg, 1993).
Cd2+ and Pb2+ stoichiometrically and rapidly displace Zn2+ from Zn-F3, consistent with the results shown in Table 1. Similarly, using an ultrafiltration method to separate TFIIIA from unbound metal ions, it was determined that these metal ions react rapidly with Zn-TFIIIA to form Cd- or Pb-TFIIIA. It had previously been shown that binding of Cd2+ inhibited subsequent association of TFIIIA with its ICR DNA binding site (Hanas & Gun, 1996). It was important to find out whether the Zn-TIIIA-ICR adduct was also reactive with these metal ions. An electrophoretic mobility shift assay was done on reaction mixtures of Cd2+ or Pb2+ with the preformed Zn-TFIIIA-ICR adduct. With 13 μM Zn2+ in the buffer to help maintain Zn-saturated TFIIIA, four- and seven-fold excesses of Pb2+ and Cd2+, respectively, were sufficient to completely dissociate the preformed adduct of Zn-TFIIIA-ICR. Thus, binding of Zn-TFIIIA to the ICR does not protect it from metal ion exchange reactions, which destroy its ability to specifically associate with DNA.
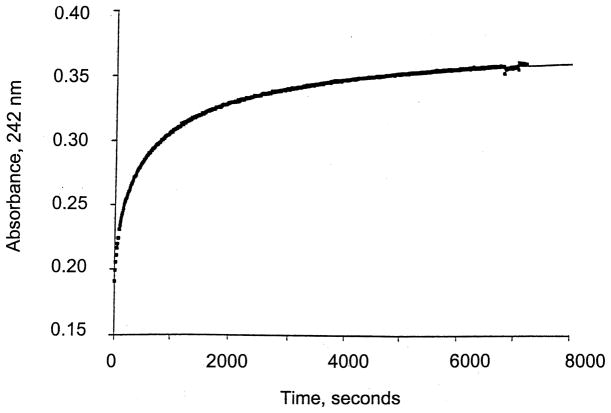
Previous experiments showed that apoMT can remove Zn2+ from TFIIIA, either in the absence or presence of the ICR (Petering et al., 1999; Zeng, Vallee & Kägi, 1991). In Fig. 1, a reaction of apoMT with Cd-TFIIIA is shown, indicating that Cd-MT and apoTFIIIA form efficiently with a modest rate constant of 40 s−1 M−1. In the cell this reaction might rescue the Cd-TFIIIA molecule. The metal ion exchange reaction between Cd-TFIIIA and Zn-MT, which produces Zn-TFIIIA and Cd-MT, was also observed by ultrafiltration but took place at a slower rate. The same reaction of Cd-F3 with Zn-MT occurred rapidly, indicative of the greater kinetic reactivity of the peptide than the holoprotein.
Fig. 1.
Kinetics of Cd2+ transfer between Cd-TFIIIA (42 μM Cd) and apoMT (6 μM). Formation of Cd-MT was followed at 242 nm. Buffer: 20 mM HEPES, pH 7, 25°C.
In summary, these results demonstrate how differential binding of toxic metal ions to Zn finger transcription factors can occur and show that even when bound to their DNA binding sites, Zn finger structures can be readily reactive with Cd2+ or Pb2+. The fact that both metal ions cause dissociation of TFIIIA from the 5 S rDNA ICR indicates that these metal ions alter the finger domain conformation needed to specifically interact with DNA. In particular, this result brings into question the hypothesis that Cd2+ binds to MTF-1 and activates its binding to DNA. Finally, the much larger reactivity of the individual Zn finger, Zn-F3, than the holoprotein Zn-TFIIIA, suggests that tertiary structural features play a significant role in its reactions.
Acknowledgments
This research was supported by NIH grants ES-04026 and ES-04184.
References
- Andrews GK. Prog Food Nutr Sci. 1990;14:193–258. [PubMed] [Google Scholar]
- Del Rio S, Setzer DR. Nucl Acid Res. 1991;19:6197–6203. doi: 10.1093/nar/19.22.6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanas JS, Gun CG. Nucl Acid Res. 1996;24:924–930. doi: 10.1093/nar/24.5.924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanas JS, Hazuda DJ, Bogenhager DF, Wu FYH, Wu CW. J Biol Chem. 1983;258:14120–14125. [PubMed] [Google Scholar]
- Krizek BA, Merkle DL, Berg JM. Inorg Chem. 1993;32:937–940. [Google Scholar]
- Makowski GS, Sunderman FW., Jr J Inorg Biochem. 1992;48:107–119. doi: 10.1016/0162-0134(92)80020-v. [DOI] [PubMed] [Google Scholar]
- Petering DH, Krezoski S, Chen P, Pattanaik A, Shaw CF., III . In: Metallothioneins: synthesis, structure and properties of metallothioneins, phy-tochelatins and metal-thiolate complexes. Stillman MJ, Shaw CF III, Suzuki KT, editors. New York: VCH Publishers; 1991. pp. 164–185. [Google Scholar]
- Petering DH, Dughish M, Huang M, Krezoski S, Krull S, Lewand D, Munñoz A, Ren L, Venkatesh S, Blumenthal S, Shaw CF., III . In: Metallothionein IV. Klaassen CD, editor. Basel: Birkhäuser Verlag; 1999. pp. 459–466. [Google Scholar]
- Posewitz MC, Wilcox DE. Chem Res Toxicol. 1995;8:1020–1028. doi: 10.1021/tx00050a005. [DOI] [PubMed] [Google Scholar]
- Radtke F, Heuchel R, Georgiev O, Hergersberg M, Gariglio M, Dembic Z, Schaffner W. EMBO J. 1993;12:1355–1361. doi: 10.1002/j.1460-2075.1993.tb05780.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romaniuk PJ. J Biol Chem. 1990;265:17593–17600. [PubMed] [Google Scholar]
- Zeng J, Vallee BL, Kägi JH. Proc Natl Acad Sci, USA. 1991;88:9984–9988. doi: 10.1073/pnas.88.22.9984. [DOI] [PMC free article] [PubMed] [Google Scholar]