Abstract
Alcohol use disorders affect millions of individuals. However, the genes and signaling pathways involved in behavioral ethanol responses and addiction are poorly understood. Here we identify a conserved biochemical pathway that underlies the sedating effects of ethanol in Drosophila. Mutations in the Arf6 small GTPase signaling pathway cause hypersensitivity to ethanol-induced sedation. We show that Arf6 functions in the adult nervous system to control ethanol-induced behavior. We also find that the Drosophila Arfaptin protein directly binds to the activated forms of Arf6 and Rac1 GTPases, and mutants in Arfaptin also display ethanol sensitivity. Arf6 acts downstream of Rac1 and Arfaptin to regulate ethanol-induced behaviors, and we thus demonstrate that this conserved Rac1/Arfaptin/Arf6 pathway is a major mediator of ethanol-induced behavioral responses.
Introduction
Alcohol abuse disorders are highly prevalent in many cultures yet come at great individual and societal cost (World Health Organization, 2004). Numerous genes have been found associated with increased alcohol consumption in humans (Gelernter and Kranzler, 2009; Schumann et al., 2011), but our understanding of the genetic risk factors and biochemical signaling pathways involved is far from complete. Therefore, Drosophila melanogaster has been developed as a model organism to investigate the genetics of ethanol-induced behaviors (Rodan and Rothenfluh, 2010). As in mammals, low doses of ethanol lead to locomotor activation in Drosophila, whereas high doses are sedating (Wolf et al., 2002). Furthermore, flies develop conditioned place preference during exposure to alcohol as a reinforcing stimulus (Kaun et al., 2011), and they display preference for alcohol consumption in a two-bottle choice paradigm (Devineni and Heberlein, 2009).
In addition to the similarities in the behavioral responses to ethanol displayed by mammals and flies, numerous genes and signaling pathways have also been found to affect alcohol-induced behaviors in both flies and vertebrates. Mouse EPS8 actin-capping protein knock-outs display increased ethanol consumption and preference (Offenhäuser et al., 2006). In primary cultured neurons from these mice, the ethanol-induced reduction of both F-actin staining and NMDA receptor current is markedly suppressed, suggesting a link between the actin cytoskeleton and alcohol-induced behaviors (Offenhäuser et al., 2006). Similarly, flies with mutations in the arouser gene, encoding the EPS8 homolog, also show altered ethanol-induced behavior (Eddison et al., 2011).
Controlled changes in the actin cytoskeleton are crucial for neuronal development (Watabe-Uchida et al., 2006) and synaptic function (Cingolani and Goda, 2008). The Rho family of GTPases is a major regulator of actin dynamics (Heasman and Ridley, 2008). Drosophila RhoGAP18B inactivates members of the Rho family of small GTPases, and white rabbit (whir) mutations in RhoGAP18B cause resistance to ethanol-induced sedation (Rothenfluh et al., 2006) and decreased voluntary ethanol consumption in a two-bottle choice paradigm (Devineni et al., 2011). However, it is unclear how RhoGAP18B-mediated regulation of the actin cytoskeleton modulates ethanol-induced behavior.
To better understand the signaling pathways in which RhoGAP18B participates, we performed a genetic screen to isolate mutations that genetically interact with RhoGAP18B whir mutants. Here we report that the small GTPase Arf6 and Drosophila Arfaptin (Arfip) act together with RhoGAP18B and the Rho-family GTPase Rac1 to control ethanol-induced sedation. We show that both Arf6 and Arfip are required for normal resistance to ethanol-induced sedation. To display wild-type ethanol sedation, flies need Arf6 in the nervous system and in adulthood but not during development. We find that Arfip binds to Arf6 and Rac1 in vivo and in cultured cells, and we also show that Arf6 acts downstream of Arfip and Rac1 to modulate ethanol-induced behavior. Thus, the conserved Rac1/Arfip/Arf6 pathway is necessary for flies to display normal resistance to ethanol-induced sedation.
Materials and Methods
Fly stocks and genetics.
Flies were maintained on regular cornmeal/yeast/molasses at 25°C/65% humidity (unless otherwise specified). Flies were outcrossed for at least five generations to the w1118 Berlin genetic background. The following fly strains were obtained from the Bloomington Drosophila Stock Center: Arf6P2 (EP2612, stock #17076), Arf6KG (KG02753, stock #13763), and Arfipd04253 (stock #19201, the original whir3-interaction strain). Arfip12 was generated by imprecise excision of ArfipUM-8176-3 (Szeged Drosophila Stock Centre) and deletes nucleotides 7059,346–7060,959, including the first two exons and starting codon of Arfip. Arfip71 is a null mutation obtained from the Eaton laboratory and described previously (Chang et al., 2012). Arf6Gal4 was obtained from the Kyoto Stock Center (NP5226, stock #104910) and harbors a Gal4-containing P-element in the first intron.
Behavioral experiments.
With the exception of experiments using whir/+ females, males were used for all experiments. In both cases, they were 2–7 d old. Ethanol exposure and determination of the ST-50 via measuring the flies' loss-of-righting reflex was performed as described previously (Rothenfluh et al., 2006). Briefly, flies unable to right themselves after light tapping were counted every 5 min, and the time for 10 of 20 flies to sedate was determined by linear interpolation for n = 1. Each set of experimental and control flies was assayed in parallel on the same day and repeated at least one time on a different day.
For developmental versus adult rescue, flies carrying a temperature-sensitive allele of Gal80, Tub–Gal80ts, which suppresses Gal4 at 18°C but is inactive at 25°C, were grown at 18°C, shifted to 25°C for 3 d as adults (or vice versa), and then assayed at room temperature. The driver whir3–Gal4/+ was used, because we obtained incomplete suppression of Gal4 expression by Tub–Gal80ts when using either the elavc155–Gal4 or Arf6Gal4 drivers.
Daily locomotion activity was measured with the Drosophila Activity Monitor system (TriKinetics). CO2 sensitivity was measured using a Flowbuddy/ultimate flypad (Genesee Scientific) and a CO2 gun with a 23 gauge needle. Four flies per tube were exposed to CO2 at 5 l/min, and the time was measured for two of four to sedate.
Ethanol absorption.
Ethanol concentration in flies was measured using the ethanol reagent kit (catalog #229-29) from Genzyme Diagnostics. Ethanol concentration in flies was calculated assuming the volume of a fly to be 2 μl. Controls and Arf6 and Arfip mutants (a total of n = 3 per genotype were tested, where n = 1 consisted of 90 flies) were exposed to ethanol vapors (150:0 ethanol/air) for various times points, and sedation was monitored throughout the exposures. At the end of the exposures, flies were frozen in dry ice and homogenized.
Statistical analyses.
Data were analyzed using Prism (GraphPad Software). ANOVAs were performed, followed by Bonferroni's post hoc comparisons of the indicated datasets.
Cell culture and coimmunoprecipitations.
Drosophila S2–Gal4 cells were maintained at 26°C in Schneider's medium (Invitrogen) with 10% fetal bovine serum. Constructs were made using Gateway cloning (T. Murphy, Carnegie Institution for Science, Baltimore, MD) and clonase (Invitrogen) and transfected using standard calcium chloride protocol. Stable transfections were generated with pCoHygro (Invitrogen) and maintained in the presence of 22 mg/ml hygromycin in the medium. For pull-downs, cells were washed in PBS, lysed in immunoprecipitation buffer (50 mm Tris-base, pH 7.4, 50 mm sodium chloride, 1% Triton X-100, 4 mm magnesium chloride, and protease inhibitor mixture; Roche Molecular Biochemicals), incubated for 4 h with FLAG beads (Sigma-Aldrich), and washed in PBS with an equal volume of 2× Laemmli's sample buffer added before Western blot analysis. In vivo coimmunoprecipitation was performed from 100 fly heads homogenized in 800 μl of lysis buffer at 4°C. After centrifugation, the supernatant was transferred to 50 μl of anti-HA affinity matrix (Roche Diagnostics) and incubated for 2 h at 4°C. After three washes with PBS, 50 μl of Laemmli's buffer was added, and 20 μl was run per lane.
Antibody techniques.
Immunohistochemistry was performed on whole-mount brains as described previously (Wu and Luo, 2006), using anti-GFP (1:250; Invitrogen), anti-bruchpilot (nc82, 1:50), and FITC- and TRITC-labeled secondary (1:500; Sigma-Aldrich) antibodies. The nc82 antibody, developed by Erich Buchner, was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by the University of Iowa, Department of Biological Sciences. Western blots were performed using anti-Arf6 antibody (1:1000; Sigma-Aldrich), anti-Arfaptin (1:1000; obtained from the Eaton Laboratory), anti-GFP (1:5000; Invitrogen), anti-FLAG (1:1000; Sigma-Aldrich), anti-actin (1:400; Sigma-Aldrich), anti-Rac1 (1:1000; Millipore), anti-HA (1:5000; Sigma-Aldrich), anti-Cdc42 (1:1000; Santa Cruz Biotechnology), and HRP-coupled secondary (1:5000; Cell Signaling Technology) antibodies and visualized using enhanced chemiluminescence (GE Healthcare).
Results
Mutations in Arfaptin cause sensitivity to ethanol-induced sedation
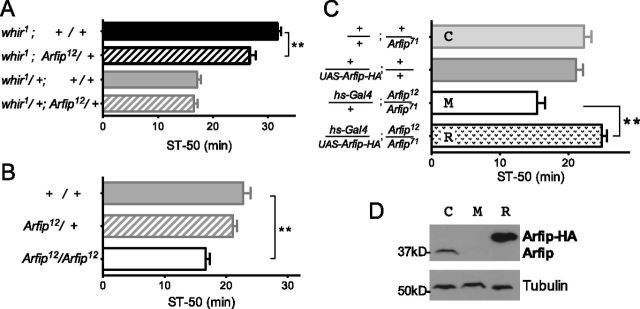
We have shown previously that the Drosophila Rho-family GTPase activating protein RhoGAP18B, encoded by the whir gene, is required for ethanol-induced behavioral responses (Rothenfluh et al., 2006). The strong loss-of-function allele whir3 is semi-lethal, whereas others affecting ethanol responses are fully viable (Rothenfluh et al., 2006). To uncover additional genes involved in RhoGAP18B-mediated signaling, we performed a small-scale genetic screen for modification of whir3 semi-lethality with 300 randomly selected mutants on the third chromosome. We then tested whether mutations that modified this semi-lethal phenotype also modified the ethanol response defects exhibited by whir1. We isolated an interacting mutation in gene CG17184, encoding the sole Drosophila homolog of the two mammalian Arfaptins. Arfaptin2 was originally isolated as a binding partners for GTP-bound Rac1 (Rac1.GTP; Van Aelst et al., 1996), and because RhoGAP18B can act on Rac1 in vitro and acts via Rac or Rho GTPase in vivo (Rothenfluh et al., 2006), we decided to study Arfip (short for Drosophila Arfaptin) in the context of ethanol-induced sedation. We first generated null mutations by imprecise excision of the transposon inserted in Arfip (see Materials and Methods) and tested one of these loss-of-function alleles for behavioral ethanol phenotypes. Heterozygous Arfip12/+ partially suppressed the ethanol resistance of whir1 (Fig. 1A), and homozygous Arfip12 mutants showed behavioral ethanol sensitivity (Fig. 1B). To ascertain that mutation in Arfip was the cause for the observed ethanol sensitivity, we performed a genetic rescue experiment. Expressing HA-tagged Arfip in Arfip mutants using a leaky heat-shock (hs) Gal4 driver rescued both the behavioral ethanol sensitivity (Fig. 1C) and the loss of Arfip protein on Western blots (Fig. 1D). These data indicate that Arfip is required for normal ethanol-induced behavior.
Figure 1.
Arfip is required for behavioral ethanol responses. In this and the following figures, error bars represent SEM. Unless otherwise noted, flies were exposed to 130:20 ethanol/air flow rates, and time was measured for 50% of the flies to sedate (ST-50), as assayed by their loss-of-righting reflex. A, Heterozygous Arfip mutations partially suppress the ethanol-resistance phenotype of whir1 (**p < 0.01, n = 6) but do not alter the wild-type phenotype of whir1/+ (Rothenfluh et al., 2006). B, Homozygous mutant Arfip flies are sensitive to ethanol-induced sedation (**p < 0.001, n = 6–11). C, Expression of an Arfip transgene under control of the Gal4 DNA-binding domain UAS (UAS–Arfip–HA) with the ubiquitously expressed hs–Gal4 driver rescues Arfip12/71 ethanol sensitivity (**p < 0.001, n = 8 per genotype). Flies were reared and kept at 29°C, sufficient for leaky hs–Gal4 and transgene expression. D, Western blot showing restoration of Arfip protein signal in the rescued flies. The genotypes control (C), mutant (M), and rescue (R) are indicated in C.
Loss of Arf6 leads to ethanol sensitivity
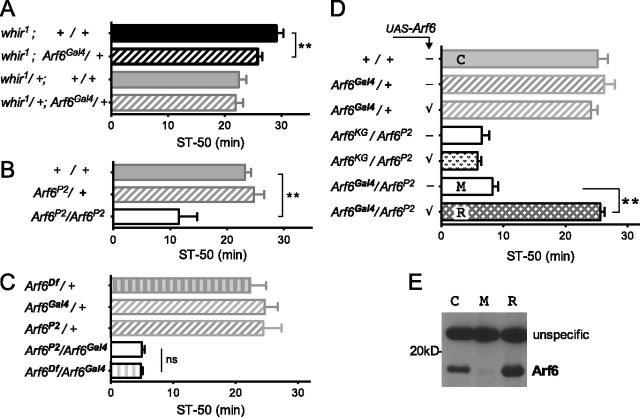
Mammalian Arfaptin2 can bind to both Rac1 and Arf6 small GTPases (Van Aelst et al., 1996; D'Souza-Schorey et al., 1997). We therefore tested Arf6 mutants for their effects on ethanol-induced sedation. Similar to Arfip, an Arf6/+ heterozygous mutant partially suppressed whir1 ethanol resistance (Fig. 2A) and showed ethanol sensitivity when homozygous (Fig. 2B). The ethanol-sensitivity phenotype of the Arf6 transposon-insertion mutants (see Materials and Methods) was unchanged when crossed to a deletion that physically removes the Arf6 locus (Arf6Df; Fig. 2C), indicating that they behave as genetic amorphs, or “nulls.” To confirm that loss of Arf6 was the cause of the observed sensitivity to ethanol, we performed rescue experiments and took advantage of a Gal4-containing transposon insertion in the Arf6 gene, Arf6Gal4, as a driver. Arf6Gal4/Arf6P2 flies showed ethanol sensitivity, and introducing an Arf6 cDNA (UAS–Arf6) into Arf6 mutants rescued that phenotype (Fig. 2D) and also restored Arf6 immunoreactivity on Western blots (Fig. 2E). Rescue was not observed in Arf6KG/Arf6P2;UAS–Arf6 mutant flies lacking the Gal4 transcriptional activator (Fig. 2D). These data show that the small GTPase Arf6 is required for normal ethanol-induced behavior.
Figure 2.
Arf6 is required for behavioral ethanol responses. A, Heterozygous Arf6 mutations partially suppress the ethanol-resistance phenotype of whir1 (*p < 0.05, n = 8–10) but does not alter the wild-type phenotype of whir1 /+. B, Arf6 mutants are sensitive to ethanol-induced sedation (**p < 0.001, n = 4–9). Note that all Arf6 alleles used, Arf6P2, Arf6KG, and Arf6Gal4, showed the same sensitivity phenotype (see also C, D). C, Arf6 mutations are genetic amorphs, i.e., nulls, because Arf6P2 mutation with a physical deletion of the Arf6 locus (Arf6Df) is no more sensitive than Arf6P2/Arf6Gal4 (NS, p = 0.73, n > 8). D, Gal4 expression from the Arf6Gal4 mutant allele rescues the Arf6 sensitivity phenotype when a UAS–Arf6 transgene is introduced (**p < 0.001, n = 6–7, checkmarks indicate the presence of the UAS transgene). E, Restoration of Arf6 signal on a Western blot from extracts of behaviorally rescued flies. The genotypes control (C), mutant (M), and rescue (R) are indicated in D.
Arf6 and Arfip mutants had wild-type ethanol absorption and metabolism (Fig. 3A), indicating that the ethanol sensitivity of these mutants (Fig. 3B) was not caused by a pharmacokinetic effect but reflected a pharmacodynamic change in Arf6 and Arfip mutant animals. We also tested the mutants for their reaction to the commonly used fly anesthetic CO2: it took Arf6 flies 24.9 ± 0.4 s, Arfip flies 26.4 ± 0.4 s, and control flies 27.9 ± 0.5 s to sedate under CO2 (n = 13 each). The differences were significant (p < 0.05), but the mutants showed a smaller change from wild type compared with their ethanol-sensitivity phenotype. To test whether the mutants were subvital or were generally weak and had locomotor defects, we tested them for their spontaneous locomotion activity over the span of 5 d. They showed no difference in their total activity counts per day (791 ± 62 for Arf6, 1018 ± 66 for Arfip, and 985 ± 77 for controls, p > 0.06, n = 16 each), indicating that the mutants were not generally weak and unhealthy.
Figure 3.
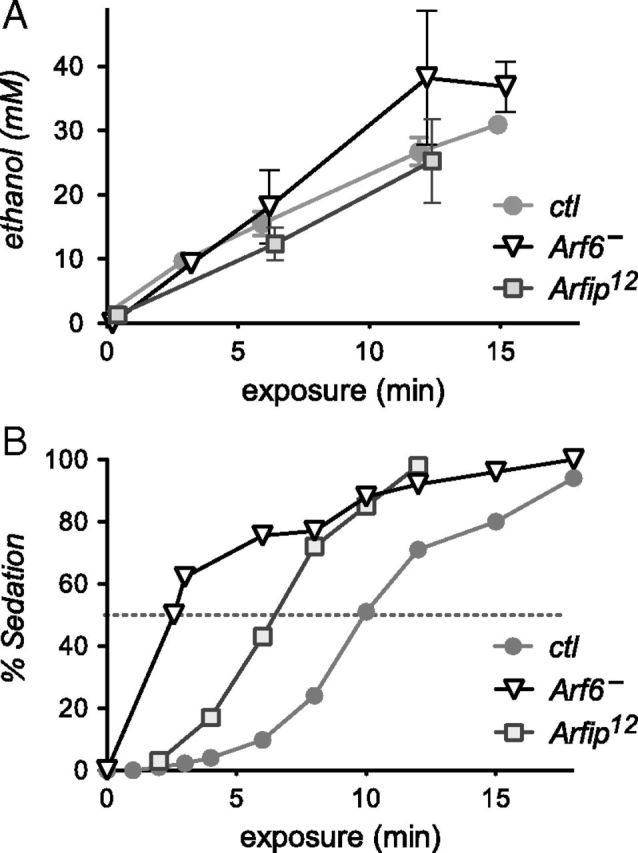
No change in ethanol absorption and metabolism in Arf6 or Arfip mutant flies. A, Flies were exposed to 150:0 ethanol/air and flash frozen, and their internal ethanol concentration was measured. Two-way ANOVA indicates significant ethanol increase over exposure time (p < 0.001, n = 3–6 per time and genotype) but no effect of genotype (p > 0.21). The Arf6− mutant genotype is Arf6P2/Gal4. B, Fraction of sedated flies from A. The intersection of the sedation curves with the stippled line indicates the ST-50 (at 150:0 ethanol/air). ctl, Control w Berlin flies.
Arf6 is required in the nervous system for normal ethanol-induced responses
To learn where in the animal Arf6 was required for wild-type ethanol behavior, we asked whether neural expression from Arf6Gal4 was necessary for rescue. To test this, we used elav–Gal80, which suppresses Gal4 activity specifically in the nervous system (Yang et al., 2009). Figure 4A shows that Arf6 mutant flies containing the Arf6Gal4 driver and elav–Gal80 did not rescue behavioral ethanol sensitivity, indicating that neural expression of Arf6 is required for normal ethanol-induced behavior. We then used the weak neural driver elav3E1–Gal4 to ask whether exclusive nervous system expression of Arf6 was sufficient to rescue the Arf6 mutant ethanol sensitivity. Figure 4B shows that selective Arf6 expression in neurons was indeed sufficient for behavioral rescue, arguing that Arf6 functions in the nervous system to regulate ethanol-induced behavior. Neural expression promoted by the Arf6Gal4 driver includes the mushroom bodies and neurosecretory cells (Fig. 4C), both known to be involved in ethanol-induced behavior (Rodan et al., 2002; King et al., 2011). Note that Arfip is expressed predominantly in the CNS in Drosophila embryos (Chang et al., 2012), consistent with a role of Arfip and Arf6 in the nervous system.
Figure 4.
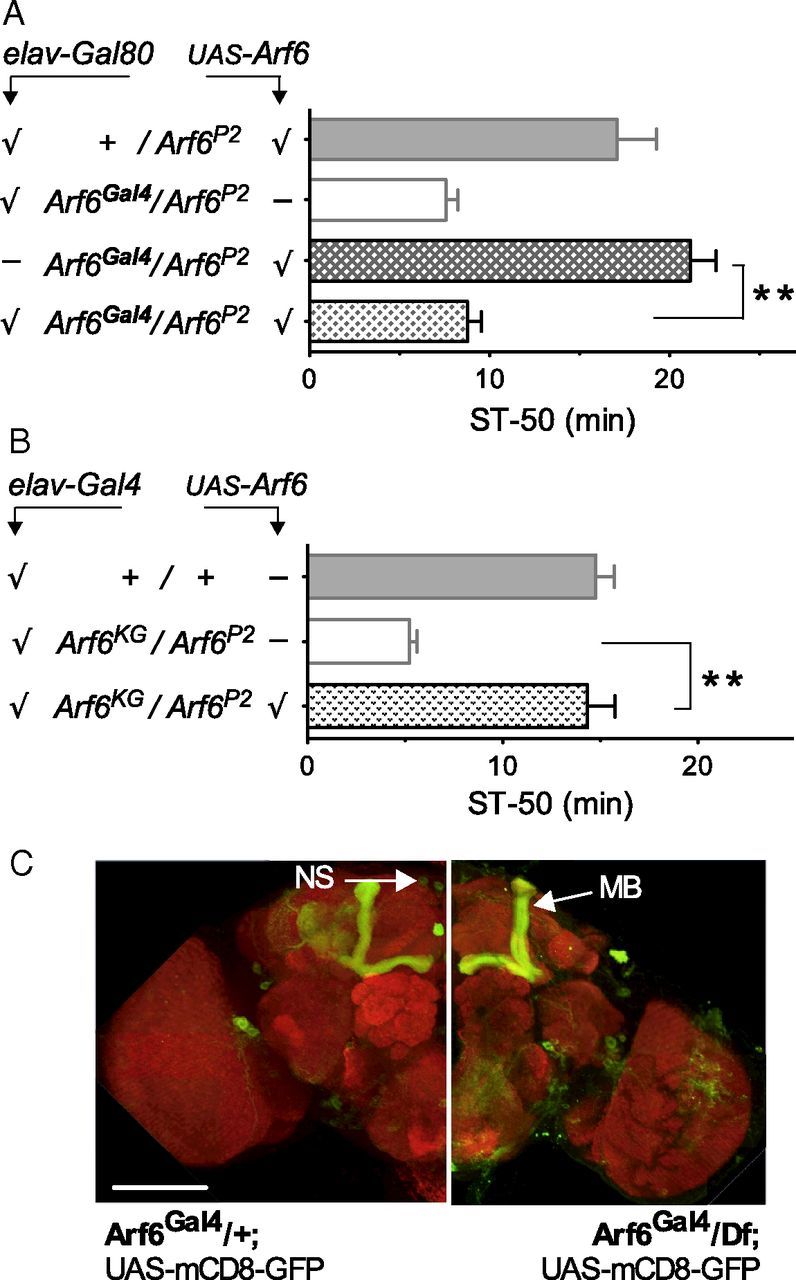
Arf6 mediates normal ethanol responses in the nervous system. A, Gal4 expression in the nervous system from Arf6Gal4 is necessary for rescue, because neuronal expression of the Gal4 inhibitor Gal80 (elav–Gal80) suppresses rescue (**p < 0.001, n = 7–8, checkmarks indicate transgene presence). B, Neuronal expression of UAS–Arf6 (with the weak elav3E1–Gal4 driver) is sufficient to rescue the Arf6 mutant ethanol sensitivity (**p < 0.001, n = 5–6). C, Expression of Gal4 from the Arf6Gal4 allele in wild-type Arf6Gal4/+ (left) and Arf6Gal4/Df mutant (right) brain visualized with anti-GFP (green). The brain neuropil was stained with anti-Brp antibody (also known as nc82, red). Regions stained include the mushroom bodies (MB) and neurosecretory cells (NS), which are known to be involved in behavioral ethanol responses. No gross morphological aberrations are obvious in the mutant (see Results). Scale bar (bottom left), 100 μm.
Adult expression of Arf6 is necessary and sufficient for normal ethanol responses
Arf6 is involved in many processes in the nervous system, including neurite development (Hernández-Deviez et al., 2002) and synapse function (Scholz et al., 2010). For normal ethanol-induced sedation, Arf6 could be required for sedation-circuit assembly or, alternatively, Arf6 might be required for normal function in adult neurons. In mutant Arf6Gal4 flies, neither gross brain anatomy nor the morphology of the Gal4-expressing neurons showed obvious abnormalities (Fig. 4C), suggesting that neural development was essentially normal in Arf6 mutant flies. Because we might have missed subtle developmental defects in this experiment, we wanted to directly test the requirement for Arf6 in adults, using Gal80ts, which allows for temperature-dependent suppression of Gal4-driver activity (McGuire et al., 2003). Expressing UAS–Arf6 in Arf6 mutants throughout development and then shutting off expression in the adult did not rescue the ethanol-sensitivity phenotype of Arf6 mutants (Fig. 5A). In contrast, adult-specific UAS–Arf6 expression was sufficient to rescue Arf6 mutant ethanol sensitivity to wild-type levels (Fig. 5B). This indicates that Arf6 is not required during development for normal ethanol-induced sedation but is consistent with an acute, adult function of Arf6 in ethanol-induced behavior. For technical reasons (see Materials and Methods), we used the whir3–Gal4/+ driver in this experiment, which is active in RhoGAP18B-expressing neurons. Note that whir3–Gal4/+ flies show wild-type ethanol responses and that the whir3–Gal4-driven expression is sufficient to rescue both the whir mutant phenotype [when providing UAS–RhoGAP18B (Rothenfluh et al., 2006)], as well as the Arf6 mutant phenotype [when providing UAS–Arf6 (Fig. 5B)], further suggesting a functional link between RhoGAP18B and Arf6.
Figure 5.
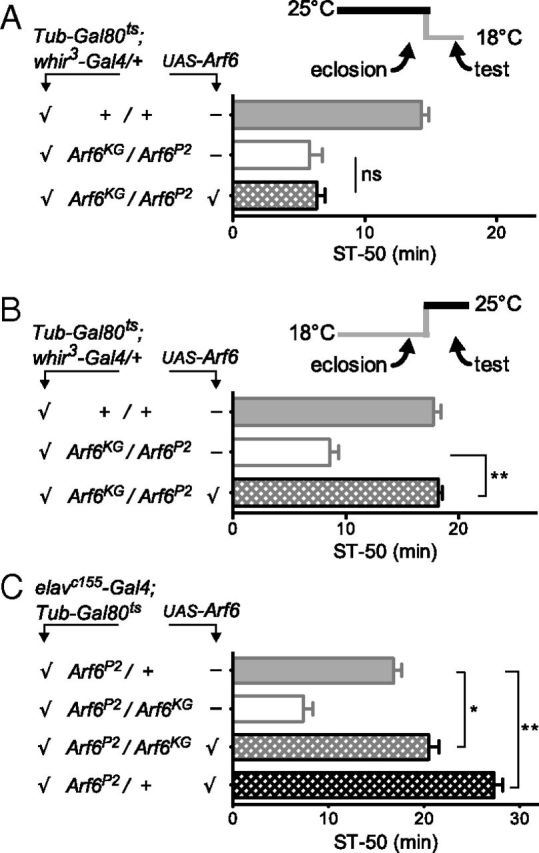
Adult, but not developmental, Arf6 expression is required for normal ethanol-induced behavior. UAS–Arf6 expression was suppressed using ubiquitous Gal80ts, which inhibits Gal4 at 18°C but not at 25°C. The flies were shifted to experimental temperature for 3 d after eclosion, as indicated by the small schematics. All flies in each panel underwent the same temperature shifts. A, Expression of UAS–Arf6 during development, using whir3–Gal4/+ as a driver, does not rescue ethanol sensitivity of Arf6 mutants (NS, p = 0.63, n = 11 per genotype). B, UAS–Arf6 expression in the adult only completely rescues the Arf6 ethanol sensitivity (**p < 0.001, n = 8). C, Strong nervous system expression of UAS–Arf6 (with the elavc155–Gal4 driver) causes lethality. When expressed in the adult only (same temperature regimen as in B), it rescues mutant Arf6 flies beyond wild type (light stippled bar, *p < 0.05) and causes strong ethanol resistance when overexpressed in normal flies (dark stippled bar, **p < 0.01, n > 7).
We also sought to rescue the Arf6 mutant phenotype by specifically expressing UAS–Arf6 in the adult nervous system. Figure 5C shows that mutant Arf6 flies expressing UAS–Arf6 with the neural driver elavc155–Gal4 displayed ethanol-induced sedation rescued beyond wild type, i.e., they were slightly more resistant than wild-type flies. This suggested that the strong nervous system driver elavc155–Gal4 might not just restore Arf6 levels to wild type but cause overexpression of Arf6, leading to resistance. We directly tested whether too much Arf6 in wild-type flies leads to ethanol resistance and found that neuronal overexpression of UAS–Arf6 in adult wild-type flies caused strong ethanol resistance (Fig. 5C). Keeping the same flies at Gal4-suppressing 18°C caused neither behavioral ethanol resistance nor Arf6 overexpression (as assessed by Western blots; data not shown). Together, these data are consistent with the interpretation that Arf6 modulates adult neuronal function and that too little Arf6 protein leads to ethanol sensitivity, whereas too much Arf6 causes ethanol resistance.
Drosophila Arfaptin functionally connects activated Rac1.GTP to Arf6.GTP
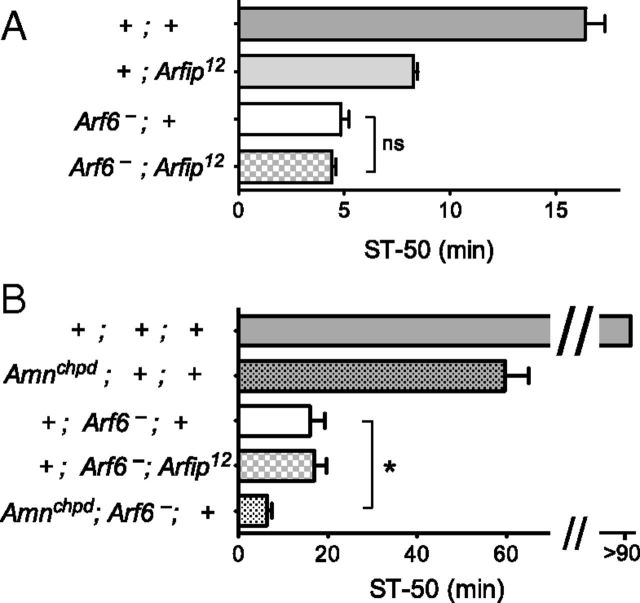
Mammalian Arf6 directly binds to Arfaptin2 (Tarricone et al., 2001), suggesting that these proteins act in the same signaling pathway. To genetically test this, we assayed the phenotype of Arf6;Arfip double mutants and found that introducing Arfip12 into an Arf6 mutant genotype did not enhance Arf6 ethanol sensitivity (Fig. 6A). In contrast, when we introduced the unrelated ethanol-sensitive mutation cheapdate (Moore et al., 1998), we observed an additional increase of ethanol sensitivity compared with Arf6 single mutants (Fig. 6B). These data suggest that Drosophila Arf6 and Arfip regulate behavioral ethanol sedation in the same pathway in vivo.
Figure 6.
Arf6 and Arfip mediate ethanol resistance in the same genetic pathway. A, Mutations in Arfip do not increase the ethanol sensitivity of Arf6− mutant flies (NS, p = 0.30, n = 9–14 per genotype). B, To ascertain that this was not attributable to a floor effect of maximal ethanol sensitivity, the ethanol/air flow was reduced to 20:130, which does not sedate wild-type flies (gray bar). Introducing the unrelated ethanol-sensitive cheapdate allele of the Amnesiac gene (dotted bars), but not Arfip12, further decreased the ethanol sensitivity of Arf6 mutant flies (*p < 0.05, n = 5 per genotype). Arf6−, Arf6P2/Gal4.
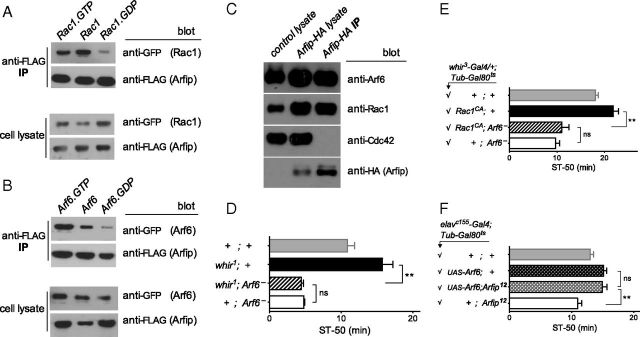
We next wanted to know whether, and how, Drosophila Arfip interacts with Arf6 and Rho-family GTPases. Tarricone et al. (2001) suggested that mammalian Arfaptin2 binds directly to either Arf or Rac GTPases. The interaction between Arfip and Arf6 has consistently been found to depend on GTP loading of Arf6 (D'Souza-Schorey et al., 1997; Shin and Exton, 2001). Conversely, binding to Rac1 has been observed with either GDP-bound (Shin and Exton, 2001) or GTP-bound (D'Souza-Schorey et al., 1997) Rac1. To determine which form of the GTPases fly Arfip interacted with, we cotransfected Drosophila Schneider cells with FLAG–Arfip and various GTPases tagged with yellow fluorescent protein (YFP). Figure 7A shows that Arfip preferentially interacted with GTP-locked Rac1G12V and only weakly with GDP-bound Rac1T17N. As with mammalian Arfaptin2, fly Arfip showed preferential interaction with GTP-locked Arf6Q67L over GDP-bound Arf6T44N (Fig. 7B). Thus, Arfip binds to the activated form of both Arf6 and Rac1, and all three proteins are required for normal resistance to ethanol sedation (Figs. 1, 2) (Rothenfluh et al., 2006).
Figure 7.
Arfaptin binds to activated GTPases and transduces a signal from Rac1 to Arf6. A, Arfip preferentially binds to the GTP-locked forms of Rac1 and Arf6 in B. Drosophila S2 cells, stably expressing Arfip–FLAG, were transiently transfected with Rac1–YFP or Arf6–YFP. GTPases pulled down with anti-FLAG beads were then detected with anti-GFP antibody, and representative pull-downs of multiple independent ones are shown. Mutants used were Rac1G12V (GTP-locked), Rac1T19N (GDP-locked), Arf6Q67L (GTP-locked), and Arf6T44N (GDP-locked). C, Arfip–HA interacts with Rac1 and Arf6 but not Cdc42 GTPase in vivo. Western blots of control lysate, Arfip–HA lysate (expressing UAS–Arfip–HA in the nervous system with elavc155–Gal4), and Arfip-HA pulled down with anti-HA were probed with anti-GTPase or anti-HA antibody. The lysate lanes represent 5% of the immunoprecipitation (IP) pull-down input. D, E, Double mutants of ethanol-sensitive Arf6− with ethanol-resistant whir1 (in D) or activated Rac1 (in E, UAS–Rac1CA, driven with whir3--Gal4/+ in the adult only as in Fig. 5B using Tub–Gal80ts to avoid developmental lethality) are no different from ethanol-sensitive Arf6− mutants alone, indicating that Arf6 acts downstream of RhoGAP18B and Rac1 (NS, p = 0.25, **p < 0.001, n > 7 per genotype for D; NS, p = 0.44, **p < 0.001, n > 6 per genotype for E). Arf6− is Arf6KG/P2 for both panels. F, The ethanol-resistant UAS–Arf6 overexpression phenotype is unchanged when ethanol-sensitive Arfip12 is introduced, indicating that Arf6 acts downstream of Arfip (NS, p = 0.85, **p < 0.001, n > 6 per genotype; pan-neuronal elavc155–Gal4/+ was used to drive UAS–Arf6 in the adult only as in Fig. 5C).
To test whether Arfip would also interact with Arf6 and Rac1 in vivo, we immunoprecipitated neuronally expressed Arfip–HA from (elavc155–Gal4;UAS–Arfip-HA/+) fly heads. The proteins pulled down together with Arfip–HA included Arf6 and Rac1 but not another small GTPase, Cdc42 (Fig. 7C). This indicates that Arfip exists in physical complexes with both Arf6 and Rac1.
We then wanted to understand the hierarchical relationship between Arf6, Arfip, and Rac1 and how signaling is transduced from one component to the next. In the induction of membrane ruffling, Rac1 acts downstream of Arf6 (Franco et al., 1999), whereas it acts upstream of Arf6 in inducing dendrite branching (Hernández-Deviez et al., 2002). To determine the relationship of Rac1, Arf6, and Arfip in the control of ethanol-induced behavior, we performed genetic epistasis experiments. Adding ethanol-resistant whir1 or activated Rac1G12V mutations to ethanol-sensitive Arf6 mutants did not change the ethanol sensitivity of Arf6 mutants alone (Fig. 7D,E). This suggests that Arf6 acts downstream of both RhoGAP18B and Rac1 in the control of ethanol-induced sedation. We also found that the ethanol-sensitive mutant Arfip12 was unable to change ethanol resistance caused by UAS–Arf6 overexpression (Fig. 7F), indicating that Arf6 also acts downstream of Arfip. Together, these data suggest that Arf6, Arfip, and Rac1 all act to promote ethanol resistance in wild-type flies and that Arf6 acts downstream of Arfip and Rac1 to mediate normal ethanol-induced behavior.
Discussion
Arf6 function in behavior
We have shown previously that RhoGap18B and Rac1, proteins involved in the dynamic regulation of the actin cytoskeleton, are required for normal ethanol-induced behavioral responses (Rothenfluh et al., 2006). To better understand the molecular mechanisms involved in this regulation of behavior, we performed a genetic interaction screen. Here, we show that Arf6 is a critical regulator of behavioral responses to ethanol and that flies lacking Arf6 are highly sensitive to ethanol-induced sedation. This is the first report showing an involvement of Arf6 in adult behavior. Drosophila and mammalian Arf6 are highly conserved, with 96% identity at the amino acid level. Mice with Arf6 knocked out survive throughout early development but die midgestation or shortly after birth with small, abnormal livers (Suzuki et al., 2006). In flies, the only phenotype described so far for Arf6 mutants is male sterility (Dyer et al., 2007; Huang et al., 2009). Molecularly, Arf6 is involved in actin organization and membrane trafficking at the plasma membrane (Donaldson, 2003; Schweitzer et al., 2011), and, in the nervous system, Arf6 has been implicated in numerous processes, including developmental ones, such as dendrite establishment (Hernández-Deviez et al., 2002; Choi et al., 2006), or functional ones, such as long-term depression (LTD) (Scholz et al., 2010). We show that strong loss-of-function mutations in Arf6 cause flies to be very sensitive to ethanol-induced sedation, whereas other behaviors, such as baseline locomotion, are unaffected. We find that Arf6 is required in the adult nervous system, but not during development, to control ethanol-induced behavior. This suggests that Arf6 is not required to set up the neuronal circuits controlling ethanol-induced behavior but rather functions in the adult nervous system, perhaps acutely modulating neuronal function or plasticity, as it does in LTD-mediated AMPA receptor endocytosis (Scholz et al., 2010).
Ethanol-induced behavior and Arfaptin
The Drosophila Arfip protein physically interacts with Arf6 in cultured cells and in vivo, and our genetic experiments indicate that these two genes regulate behavioral ethanol sensitivity in the same pathway. We show that Arfip mutants are also sensitive to ethanol-induced sedation, for the first time showing a function of this protein in adult behavior. Mammalian Arfaptin2 dimers form a curved BAR domain (Tarricone et al., 2001), which are thought to induce, or bind to, curved membranes (Frost et al., 2009). This suggests that Arfaptin2 may promote vesicle formation, and Arfaptin2 can indeed tubulate liver liposomes in vitro (Peter et al., 2004) and trans-Golgi structures when overexpressed in HeLa cells (Man et al., 2011).
Arfaptin2 binds to Arf6 and Rac1 proteins in vitro and in cultured cells (D'Souza-Schorey et al., 1997; Shin and Exton, 2001), and we show that fly Arfip binds to these two GTPases in vivo, using coimmunoprecipitation assays from fly head extracts. Arfip acts synergistically with Arf6 to cause actin reorganization at the cell periphery (D'Souza-Schorey et al., 1997), suggesting that these two proteins act together in the same pathway and direction. Conversely, overexpressed Arfaptin1 inhibited the activation of the Arf effector phospholipase D, thereby antagonizing the activity of Arf GTPase (Williger et al., 1999). Our findings argue that Arfaptin acts together with GTPase signaling in the same direction. We show that loss of either Arfip or Arf6 function causes sensitivity to ethanol-induced sedation, and we have shown previously that that reduced Rac1 signaling causes ethanol sensitivity (Rothenfluh et al., 2006). Together, these findings suggest that Arf6, Arfip, and Rac1 all act together to control peripheral actin and membrane organization and thereby promote behavioral ethanol resistance.
Role of Rac/Arfaptin/Arf6 signaling in ethanol responses
Previous experiments have suggested that Arf6 could act downstream of Rac1. This was shown by suppressing Rac1-induced membrane ruffling with dominant-negative Arf6 (Radhakrishna et al., 1999) and by suppressing increased dendrite branching, induced by dominant-negative Rac1, with constitutive-active Arf6 (Hernández-Deviez et al., 2002). Our genetic data indicate that Arf6 controls ethanol-induced behavior downstream of Rac1 and Arfip (Fig. 7). Canonically, this would be interpreted as Rac1 activating Arfip, which then activates Arf6. However, because Arfip contains neither a CRIB (Cdc42/Rac-interactive binding protein) Rac effector nor an ArfGEF GTP-loading domain, we suggest an alternative model wherein activated Arf6 recruits Arfip, which helps in vesicle formation, and also recruits Rac1, which is required for controlling actin dynamics to permit vesicle transport from, or to, the plasma membrane. This would allow for spatial and temporal coordination of these two essential functional components of membrane trafficking. Disrupting the actin cytoskeleton does indeed interfere with both endocytosis (Galletta and Cooper, 2009) and redistribution of Arf6 and receptors from the recycling endosomes to the plasma membrane (Radhakrishna and Donaldson, 1997). In this model, Arf6 would be required for Rac1 recruitment, and Rac1 would be required for Arf6 redistribution. This might explain why dominant-negative Rac1 inhibited actin reorganization induced by the Arf6 activator EFA6 (arguing that Rac1 is downstream of Arf6) (Franco et al., 1999).
What cellular processes could this signaling cascade be involved in to control ethanol-induced sedation? Endogenous Arf6 and Rac1 colocalize at the plasma membrane and on recycling endosomes, and stimulation of Arf6 relocates both Arf6 and Rac1 to the plasma membrane (Radhakrishna et al., 1999). These GTPases thus act in concert to regulate structural changes of peripheral actin and membrane (Palamidessi et al., 2008), and, given our finding that Arfip binds to GTP-bound forms of Arf6 and Rac1, Arfip may serve as a physical link between these two regulators of the actin cytoskeleton and membrane dynamics. In primary hippocampal neurons, Arf6 and Rac1 are both required for the maturation of dendritic filapodia, which is associated with synaptic maturation (Raemaekers et al., 2012). Increases in synapse number in adult flies has been shown to cause increased ethanol sensitivity (Eddison et al., 2011), and one intriguing possibility is that the Rac1/Arfip/Arf6 pathway might be involved in Drosophila synapse maturation, regulating the strength or number of connections. A second possibility is that Arf6 plays a role in trafficking of receptors important for ethanol-induced behavior, similar to the role of Arf6 in μ-opioid receptor trafficking, which is involved in opiate addiction (Rankovic et al., 2009). Future experiments will investigate these possibilities and help to elucidate the processes regulated by Rac1/Arfip/Arf6 signaling to control ethanol-induced behaviors.
Footnotes
This work was supported by National Institute of Health T32 Fellowship Grant DA7290 (R.L.P.y.C.d.P.) and Grant 5R01AA019526 (A.R.). A.R. is the Effie Marie Cain Scholar in Biomedical Research at University of Texas Southwestern. We thank Geetha Kalahasti and Antonio Lopez for help with experiments, Chris Cowan, Robin Hiesinger, Mike Buszczak, Helmut Krämer, and the Rothenfluh laboratory for helpful discussion and critical comments on this manuscript. We thank Elizabeth Chen, Yuh Nung Jan, and the Bloomington, Szeged, and Kyoto stock centers for fly strains.
The authors declare no competing financial interests.
References
- Chang L, Davison H, Kreko T, Cusmano T, Wu Y, Rothenfluh A, Eaton B. Genetic dissection of synaptic pathologies in dynactin mutants identifies Arfaptin, a specifier of dynactin complex function during synapse growth. Mol Biol Cell. 2012 doi: 10.1091/mbc.E12-09-0697. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S, Ko J, Lee JR, Lee HW, Kim K, Chung HS, Kim H, Kim E. ARF6 and EFA6A regulate the development and maintenance of dendritic spines. J Neurosci. 2006;26:4811–4819. doi: 10.1523/JNEUROSCI.4182-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani LA, Goda Y. Actin in action: the interplay between the actin cytoskeleton and synaptic efficacy. Nat Rev Neurosci. 2008;9:344–356. doi: 10.1038/nrn2373. [DOI] [PubMed] [Google Scholar]
- Devineni AV, Heberlein U. Preferential ethanol consumption in Drosophila models features of addiction. Curr Biol. 2009;19:2126–2132. doi: 10.1016/j.cub.2009.10.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devineni AV, McClure KD, Guarnieri DJ, Corl AB, Wolf FW, Eddison M, Heberlein U. The genetic relationships between ethanol preference, acute ethanol sensitivity and ethanol. Fly. 2011;5:191–199. doi: 10.4161/fly.5.3.16987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson JG. Multiple roles for Arf6: sorting, structuring, and signaling at the plasma membrane. J Biol Chem. 2003;278:41573–41576. doi: 10.1074/jbc.R300026200. [DOI] [PubMed] [Google Scholar]
- D'Souza-Schorey C, Boshans RL, McDonough M, Stahl PD, Van Aelst L. A role for POR1, a Rac1-interacting protein, in ARF6-mediated cytoskeletal rearrangements. EMBO J. 1997;16:5445–5454. doi: 10.1093/emboj/16.17.5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer N, Rebollo E, Domínguez P, Elkhatib N, Chavrier P, Daviet L, González C, González-Gaitán M. Spermatocyte cytokinesis requires rapid membrane addition mediated by ARF6 on central spindle recycling endosomes. Development. 2007;134:4437–4447. doi: 10.1242/dev.010983. [DOI] [PubMed] [Google Scholar]
- Eddison M, Guarnieri DJ, Cheng L, Liu CH, Moffat KG, Davis G, Heberlein U. arouser reveals a role for synapse number in the regulation of ethanol sensitivity. Neuron. 2011;70:979–990. doi: 10.1016/j.neuron.2011.03.030. [DOI] [PubMed] [Google Scholar]
- Franco M, Peters PJ, Boretto J, van Donselaar E, Neri A, D'Souza-Schorey C, Chavrier P. EFA6, a sec7 domain-containing exchange factor for ARF6, coordinates membrane recycling and actin cytoskeleton organization. EMBO J. 1999;18:1480–1491. doi: 10.1093/emboj/18.6.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost A, Unger VM, De Camilli P. The BAR domain superfamily: membrane-molding macromolecules. Cell. 2009;137:191–196. doi: 10.1016/j.cell.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletta BJ, Cooper JA. Actin and endocytosis: mechanisms and phylogeny. Curr Opin Cell Biol. 2009;21:20–27. doi: 10.1016/j.ceb.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR. Genetics of alcohol dependence. Hum Genet. 2009;126:91–99. doi: 10.1007/s00439-009-0701-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Bio. 2008;9:690–701. doi: 10.1038/nrm2476. [DOI] [PubMed] [Google Scholar]
- Hernández-Deviez DJ, Casanova JE, Wilson JM. Regulation of dendritic development by the ARF exchange factor ARNO. Nat Neurosci. 2002;5:623–624. doi: 10.1038/nn865. [DOI] [PubMed] [Google Scholar]
- Huang J, Zhou W, Dong W, Watson AM, Hong Y. From the Cover: Directed, efficient, and versatile modifications of the Drosophila genome by genomic engineering. Proc Natl Acad Sci U S A. 2009;106:8284–8289. doi: 10.1073/pnas.0900641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaun KR, Azanchi R, Maung Z, Hirsh J, Heberlein U. A Drosophila model for alcohol reward. Nat Neurosci. 2011;14:612–619. doi: 10.1038/nn.2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King I, Tsai LT, Pflanz R, Voigt A, Lee S, Jäckle H, Lu B, Heberlein U. Drosophila tao controls mushroom body development and ethanol-stimulated behavior through par-1. J Neurosci. 2011;31:1139–1148. doi: 10.1523/JNEUROSCI.4416-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man Z, Kondo Y, Koga H, Umino H, Nakayama K, Shin HW. Arfaptins are localized to the trans-Golgi by interaction with Arl1, but not Arfs. J Biol Chem. 2011;286:11569–11578. doi: 10.1074/jbc.M110.201442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302:1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- Moore MS, DeZazzo J, Luk AY, Tully T, Singh CM, Heberlein U. Ethanol intoxication in Drosophila: genetic and pharmacological evidence for regulation by the cAMP signaling pathway. Cell. 1998;93:997–1007. doi: 10.1016/s0092-8674(00)81205-2. [DOI] [PubMed] [Google Scholar]
- Offenhäuser N, Castelletti D, Mapelli L, Soppo BE, Regondi MC, Rossi P, D'Angelo E, Frassoni C, Amadeo A, Tocchetti A, Pozzi B, Disanza A, Guarnieri D, Betsholtz C, Scita G, Heberlein U, Di Fiore PP. Increased ethanol resistance and consumption in Eps8 knockout mice correlates with altered actin dynamics. Cell. 2006;127:213–226. doi: 10.1016/j.cell.2006.09.011. [DOI] [PubMed] [Google Scholar]
- Palamidessi A, Frittoli E, Garré M, Faretta M, Mione M, Testa I, Diaspro A, Lanzetti L, Scita G, Di Fiore PP. Endocytic trafficking of Rac is required for the spatial restriction of signaling in cell migration. Cell. 2008;134:135–147. doi: 10.1016/j.cell.2008.05.034. [DOI] [PubMed] [Google Scholar]
- Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- Radhakrishna H, Donaldson JG. ADP-ribosylation factor 6 regulates a novel plasma membrane recycling pathway. J Cell Biol. 1997;139:49–61. doi: 10.1083/jcb.139.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishna H, Al-Awar O, Khachikian Z, Donaldson JG. ARF6 requirement for Rac ruffling suggests a role for membrane trafficking in cortical actin rearrangements. J Cell Sci. 1999;112:855–866. doi: 10.1242/jcs.112.6.855. [DOI] [PubMed] [Google Scholar]
- Raemaekers T, Peric A, Baatsen P, Sannerud R, Declerck I, Baert V, Michiels C, Annaert W. ARF6-mediated endosomal transport of telencephalin affects dendritic filopodia-to-spine maturation. EMBO J. 2012;31:3252–3269. doi: 10.1038/emboj.2012.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankovic M, Jacob L, Rankovic V, Brandenburg LO, Schröder H, Höllt V, Koch T. ADP-ribosylation factor 6 regulates mu-opioid receptor trafficking and signaling via activation of phospholipase D2. Cell Signal. 2009;21:1784–1793. doi: 10.1016/j.cellsig.2009.07.014. [DOI] [PubMed] [Google Scholar]
- Rodan AR, Rothenfluh A. The genetics of behavioral alcohol responses in Drosophila. Int Rev Neurobiol. 2010;91:25–51. doi: 10.1016/S0074-7742(10)91002-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodan AR, Kiger JA, Jr, Heberlein U. Functional dissection of neuroanatomical loci regulating ethanol sensitivity in Drosophila. J Neurosci. 2002;22:9490–9501. doi: 10.1523/JNEUROSCI.22-21-09490.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenfluh A, Threlkeld RJ, Bainton RJ, Tsai LT, Lasek AW, Heberlein U. Distinct behavioral responses to ethanol are regulated by alternate RhoGAP18B isoforms. Cell. 2006;127:199–211. doi: 10.1016/j.cell.2006.09.010. [DOI] [PubMed] [Google Scholar]
- Scholz R, Berberich S, Rathgeber L, Kolleker A, Köhr G, Kornau HC. AMPA receptor signaling through BRAG2 and Arf6 critical for long-term synaptic depression. Neuron. 2010;66:768–780. doi: 10.1016/j.neuron.2010.05.003. [DOI] [PubMed] [Google Scholar]
- Schumann G, Coin LJ, Lourdusamy A, Charoen P, Berger KH, Stacey D, Desrivières S, Aliev FA, Khan AA, Amin N, Aulchenko YS, Bakalkin G, Bakker SJ, Balkau B, Beulens JW, Bilbao A, de Boer RA, Beury D, Bots ML, Breetvelt EJ, et al. Genome-wide association and genetic functional studies identify autism susceptibility candidate 2 gene (AUTS2) in the regulation of alcohol consumption. Proc Natl Acad Sci U S A. 2011;108:7119–7124. doi: 10.1073/pnas.1017288108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer JK, Sedgwick AE, D'Souza-Schorey C. ARF6-mediated endocytic recycling impacts cell movement, cell division and lipid homeostasis. Semin Cell Dev Biol. 2011;22:39–47. doi: 10.1016/j.semcdb.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin OH, Exton JH. Differential binding of arfaptin 2/POR1 to ADP-ribosylation factors and Rac1. Biochem Biophys Res Commun. 2001;285:1267–1273. doi: 10.1006/bbrc.2001.5330. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Kanai Y, Hara T, Sasaki J, Sasaki T, Kohara M, Maehama T, Taya C, Shitara H, Yonekawa H, Frohman MA, Yokozeki T, Kanaho Y. Crucial role of the small GTPase ARF6 in hepatic cord formation during liver development. Mol Cell Biol. 2006;26:6149–6156. doi: 10.1128/MCB.00298-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarricone C, Xiao B, Justin N, Walker PA, Rittinger K, Gamblin SJ, Smerdon SJ. The structural basis of Arfaptin-mediated cross-talk between Rac and Arf signalling pathways. Nature. 2001;411:215–219. doi: 10.1038/35075620. [DOI] [PubMed] [Google Scholar]
- Van Aelst L, Joneson T, Bar-Sagi D. Identification of a novel Rac1-interacting protein involved in membrane ruffling. EMBO J. 1996;15:3778–3786. [PMC free article] [PubMed] [Google Scholar]
- Watabe-Uchida M, Govek EE, Van Aelst L. Regulators of Rho GTPases in neuronal development. J Neurosci. 2006;26:10633–10635. doi: 10.1523/JNEUROSCI.4084-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williger BT, Ostermann J, Exton JH. Arfaptin 1, an ARF-binding protein, inhibits phospholipase D and endoplasmic reticulum/Golgi protein transport. FEBS Lett. 1999;443:197–200. doi: 10.1016/s0014-5793(98)01707-4. [DOI] [PubMed] [Google Scholar]
- Wolf FW, Rodan AR, Tsai LT, Heberlein U. High-resolution analysis of ethanol-induced locomotor stimulation in Drosophila. J Neurosci. 2002;22:11035–11044. doi: 10.1523/JNEUROSCI.22-24-11035.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. Global status report on alcohol. Geneva: World Health Organization; 2004. Economic and social costs of alcohol use; pp. 65–55. [Google Scholar]
- Wu JS, Luo L. A protocol for dissecting Drosophila melanogaster brains for live imaging or immunostaining. Nat Protoc. 2006;1:2110–2115. doi: 10.1038/nprot.2006.336. [DOI] [PubMed] [Google Scholar]
- Yang CH, Rumpf S, Xiang Y, Gordon MD, Song W, Jan LY, Jan YN. Control of the postmating behavioral switch in Drosophila females by internal sensory neurons. Neuron. 2009;61:519–526. doi: 10.1016/j.neuron.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]