Abstract
Membrane Associated Guanylate Kinases (MAGUKs) organize protein complexes at specific cellular sites by regulating interactions with their COOH-terminal Guanylate Kinase-like (GK) domains. Negative regulation of MAGUK GKs by an adjacent SH3 domain is critical for function, yet the mechanism is poorly understood. To gain insight into this process we investigated SH3 regulation of the Discs Large (Dlg) GK domain. Mutational analysis revealed that the binding site of the SH3-inhibited GK ligand GukHolder (GukH) is opposite the SH3 interacting surface indicating that the SH3 domain does not directly occlude GukH binding. We screened for constitutively active SH3GK variants using yeast two-hybrid and a cell polarity/mitotic spindle orientation assay. Residues in both SH3 and GK are required to maintain SH3GK inhibition, including those distant from both the SH3-GK and GK-GukH interaction sites. Activating mutations do not alter the ability of the SH3 and GK to interact in trans. Based on these observations, we propose that the SH3 domain modulates GK domain allostery to control its function.
Cellular structures such as tight junctions and synapses contain specific protein assemblies that include membrane proteins such as receptors and channels, and components involved in signal transduction and cytoskeletal linkage (1, 2). Scaffolding proteins play an essential role in the construction and function of these protein assemblies by linking multiple upstream and downstream components (3-5). Interaction with these components is mediated by a multitude of protein interaction domains that specifically bind individual scaffold ligands (6, 7). Recent evidence indicates that scaffold proteins do not simply play a passive role in these pathways but that they can control both the identity and activity of the proteins they bind (8-10). Thus, a key question in understanding scaffold-mediated organization is how they regulate their protein interaction domains to specify which ligands they bind.
The Membrane-Associated Guanylate Kinases (MAGUKs)1 are scaffold proteins that can regulate the activity of their protein interaction domains (11-15). MAGUKs contain a variable number of PDZ domains that bind cytoskeletal and adhesion proteins followed by an SH3 domain and a domain with homology to the enzyme Guanylate Kinase (16). The Guanylate Kinase-like (GK) domain has no apparent catalytic activity but the fold has been co-opted to mediate interactions with protein ligands (17, 18). In many MAGUKs, the SH3 and GK domains form an intramolecular interaction that regulates GK domain binding and is necessary for function (13, 15, 19-21). For example, the MAGUK Discs large (Dlg) is a tumor suppressor in Drosophila that is required for mitotic spindle orientation in neuronal precursors. In mutants expressing a form of Dlg that lacks the SH3GK intramolecular interaction, the protein localizes correctly but is non-functional (19). As the association of a subset of GK ligands is inhibited when the SH3 is present (13, 15), the essential role of the interaction between MAGUK SH3 and GK domains appears to be to regulate GK ligand binding.
Although the interaction between SH3 and GK domains is an essential component of MAGUK function, remarkably little is known about how the SH3 domain controls GK ligand binding. Because SH3 domains bind proline-rich sequences in target proteins, an initial model for SH3 regulation posited the existence of a cryptic SH3 recognition sequence within the GK (15). However, in structures of the SH3GK module from PSD-95 the PXXP-binding surface on the SH3 is positioned away from the GK and is partially occluded by an insert present in MAGUK SH3s known as the Hook (22, 23). Instead, the core of the interaction is a two-stranded beta sheet formed from a strand that emerges from the SH3 and one following the GK. What has been unclear is how this interaction might participate in regulation.
We have used a combined biochemical, genetic, and cell biological approach to investigate the mechanism of SH3 regulation of MAGUK GKs. To determine if the SH3 domain directly occludes GK ligand binding, we identified the binding surface for the SH3-regulated GK ligand GukHolder (GukH) using a mutational approach and found that it binds to a site distant from the SH3 domain. As the SH3 does not directly occlude GK ligand binding we screened for constitutively active SH3GK modules that are able to bind GukH and identified residues distant from both the SH3 and GukH binding sites. These results suggest that allosteric transitions in the GK domain are important for SH3GK regulation.
Materials and Methods
Expression Constructs
The Drosophila Dlg domain expression vectors were made from the PG isoform and correspond to the following residue numbers: SH3GK (598-975), GK (771-975), GK-F (771-963) and SH3-E (598-784). DNA encoding these constructs was cloned into the pGAD, pGEX or pBH vectors. QuikChange® mutagenesis was used to create single amino acid substitution variants, and two-step PCR was used to create ΔHook (679-766 replaced with 3 GS repeats), ΔI3 (Δ692-737 replaced with 3 GS repeats), and Δ696-701 (replaced with 3 GS repeats) SH3GK variants.
Induced polarity mitotic spindle alignment assay
Cells were polarized and mitotic spindle angle measured as previously described (24). Briefly, Echinoid (Ed):FLAG constructs were made in pMT-V5 (Invitrogen, Carlsbad, CA, USA) replacing the Ed cytoplasmic domain with the FLAG tag and the protein domain(s) of interest at the C-terminus (e.g. Ed:FLAG:GK). Standard methods were used to grow Schneider (S2) cells (Goshima et al., 2007). Cells were seeded at ∼1-3 × 106 cell per well in 6-well culture dishes, transfected with 0.4-1 □g total DNA using Effectene® (Qiagen, Germantown, MD, USA), incubated overnight, and gene expression was induced by the addition of 500 □M CuSO4 for 24-48 hours. Cell clustering was induced by rotation at ∼175 RPM for 1-3 hours. For immunostaining, S2 cells were fixed for 20 minutes in 4% paraformaldehyde, immunostained using standard methods (Goshima et al., 2007), and imaged using a SP2 confocal microscope (Leica) with an oil immersion 60× 1.4 NA objective. Spindle angles were measured using the angle tool in ImageJ, using one vector drawn perpendicular to the center of the Ed crescent and a second vector matching the spindle. Each analysis was performed for at least 20 cells. The results are reported as mean spindle orientation angle, the difference of the random angle (45°) and the observed mean. We have previously measured the standard error of these measurements as ± 3° based on multiple independent trials. The standard deviation differs from the standard error because of natural population variation (i.e. each trial contains a distribution of spindle angles that yields a standard deviation that is typically larger than the variation of the mean from trial to trial).
Yeast Two-Hybrid Screen
A library of SH3GK constructs randomly mutated through MnCl addition to PCR amplification was transformed with linearized pGAD (prey) vector and GUKH (residues 749-1044 from Drosophila GukHolder isoform C) in pGBK (bait) vector into the Y187 strain of S. cerevisiae. Initial survival selection on Leu/Trp-deficient plates ensured homologous recombination of the pGAD vector with a SH3GK construct. In yeast two-hybrid analysis, DNA from colonies that survived when replica plated onto Leu/Trp/His-deficient + 20mM 3-amino triazole (3AT) plates was isolated, transformed into E. coli, and pGAD constructs were selected for sequencing through growth on ampicillin-positive plates. Several plasmids contained multiple mutations. To identify individual mutations that might be responsible for activating the SH3GK, individual point mutations were generated through site-directed mutagenesis of SH3GK, and tested for GukH binding in yeast two-hybrid filter lift assays with X-Gal substrate.
For analysis of GK domain candidate residues and Hook region truncations, the AH109 strain of S. cerevisiae was transformed with GUKH bait, and Dlg prey constructs. Positive clones grew on Leu/Trp/His-deficient + 3AT plates, as analyzed by serial dilution.
Pull-Down Assays
For qualitative in trans GST pull-downs, E. coli cell lysates containing the GST fusion protein of interest were incubated with glutathione-agarose beads and washed three times with binding buffer (100 mM HEPES, pH 7.5, 100 mM NaCl, 1 mM dithiothreitol). A His-tagged fusion of the SH3 domain of Dlg was added to a concentration of 35 μM and agitated with the beads at room temperature for 15 min. The reactions were then washed three times with washing buffer (100 mM HEPES, pH 7.5, 100 mM NaCl, 1 mM dithiothreitol, 0.05% Triton X-100) to remove unassociated proteins. Bound proteins were eluted from the glutathione-agarose beads by the addition of SDS loading buffer and were screened by Western blot analyses using a mouse monoclonal anti-His antibody (1:1000; Qiagen).
Results
The SH3 domain does not sterically block GK ligand binding
No direct structural information is available for any MAGUK GK ligand complex, hampering understanding of GK regulation. The best characterized GK-ligand information is for PSD-95 GK-MAP1A interaction based on NMR chemical shift analysis and mutational studies (18). MAP1A binds to a cleft that corresponds to the guanosine nucleotide-binding (GNB) site in the GK enzyme. We tested if residues within the GNB region of the Dlg GK are required to bind the SH3-regulated GK ligand GukH using yeast two-hybrid, which has been shown to be a reliable assay for this interaction (13). We fused the Dlg GK to the GAL4 activation domain and GukH to the GAL4 DNA binding domain (Figure 1a). To ensure that the mutations did not cause GK unfolding, we also determined if the domains containing the mutations could bind the SH3 domain in trans, which requires a folded GK domain. Mutation of several residues within the GNB (D830, Y831, or Y855) disrupted GukH binding without affecting SH3 binding (Figure 1). Thus, like MAP1A, GukH utilizes the GNB domain of the GK as an interaction surface. As this surface is opposite the SH3 binding site on the GK (Figure 1c), we conclude that the SH3 domain does not regulate GK ligand-binding activity by directly occluding the ligand binding site.
Figure 1.
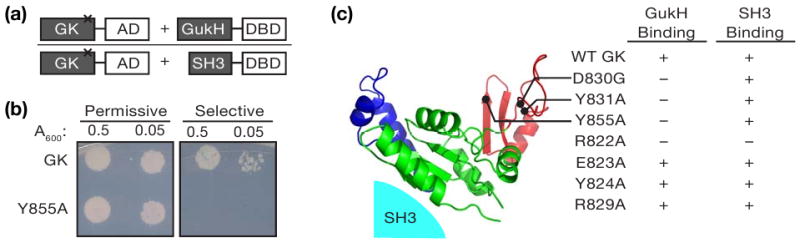
GukH binding site on the Dlg GK. (a) Schematic representation of yeast two-hybrid bait and prey constructs. AD = activation domain, DBD = DNA binding domain. (b) Representative results from yeast two-hybrid assay. Serial dilution onto plates of permissive (-LT) or selective (-L-T-H + 3AT) medium starting with A600 of 0.5 shows loss of interaction between GukH and the Dlg GK domain with tyrosine 855 mutated to alanine. (c) Summary of yeast two-hybrid results. Yeast growth indicates interaction (+) while no growth indicates no interaction (-).Mutations that repress GukH binding but not SH3 binding are mapped onto the PSD-95 GK structure (22; PDB code: 1kjw). The Lid, Core, and GBD domains are colored in blue, green, and red, respectively. The location of the SH3 domain is shown.
Identification of SH3GK mutations that restore ligand binding and function
How might the SH3 domain regulate GK activity given that it does not directly block ligand binding? To gain insight into SH3GK regulated complex assembly, we devised a screen to identify mutations that render the Dlg SH3GK module constitutively activated, interacting with GK ligands even when the SH3 domain is present. As GukH binds the isolated GK domain but not the SH3GK, we generated a library of SH3GK point mutants by error-prone PCR and screened the library for sequences that bind GukH in the yeast two-hybrid assay. Using this screen, we identified six individual point mutations that allow the SH3GK to bind GukH (Figure 2a). These mutated residues vary in their degree of conservation (Figure 2b) suggesting that some might be general features of SH3GK regulation, while others may be specific to GukH regulation.
Figure 2.
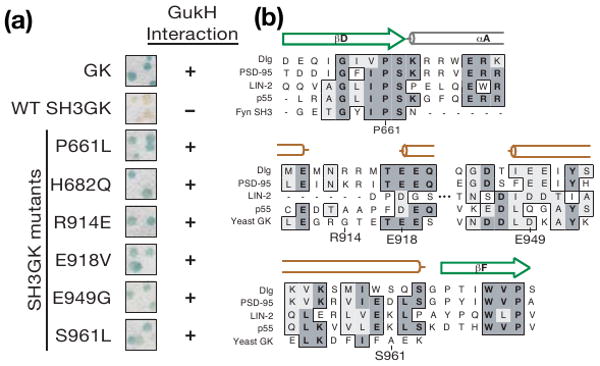
SH3GK activation screen. (a) SH3GK mutations identified that allow binding to GukH in yeast two-hybrid survival screening are shown in a blue/white colony filter lift assay. GK and wild-type SH3GK were used as positive and negative controls, respectively, to determine GukH interaction (+) and no interaction (-). (b) CLUSTALW alignment of Dlg residues required for SH3GK repression with the corresponding regions of both MAGUK (R. norvegicus PSD-95, C. elegans LIN-2, H. sapiens MPP3) and non MAGUK (H. sapiens Fyn, and S. cerevisiae Guanylate Kinase) sequences. Amino acid identities are shaded and similarities are boxed. Secondary structure elements from the SH3 and GK domains of PSD-95 are shown above the sequences with β-sheets that fold into the SH3 domain as green arrows and α-helicies from the Hook region and GK domain as gray and orange cylinders respectively.
Although the binding assay we used in our screen suggests that the point mutations lead to constitutive SH3GK activity, we also sought to test them in a functional context. While the intramolecular interaction between MAGUK SH3 and GK domains has been known for some time, few assays have been developed to examine its function. Dlg plays a role in positioning the mitotic spindle apparatus, possibly through interactions between its GK domain and the kinesin-like protein Khc73/GAKIN (25, 26). We recently found that the Dlg GK domain can orient the spindle when polarized along the cortex of cultured Drosophila S2 cells (24). Here we use this assay to test whether the GK domain is SH3-regulated, and the effect of SH3GK point mutants on SH3GK function in a cellular context.
In the S2 spindle orientation assay, the GK domain is polarized by attachment to the cytoplasmic region of the adhesion protein Echinoid (Ed). Clusters of cells expressing Ed-GK have enriched “crescents” of the protein at sites of cell-cell contact and the mitotic spindle aligns with this crescent (24). We assess spindle-orienting activity by measuring the angle between the spindle and the crescent for a large number of mitotic, clustered cells (Figure 3a; see methods). Whereas Ed has no influence on the spindle angle by itself (24), the Ed-GK fusion has a mean spindle orientation (degrees from the random position of 45°) of 18 ± 20° (1 SD; standard error = ± 3°; see methods) indicating that it is able to orient the spindle relative to the Ed-GK crescent.
Figure 3.
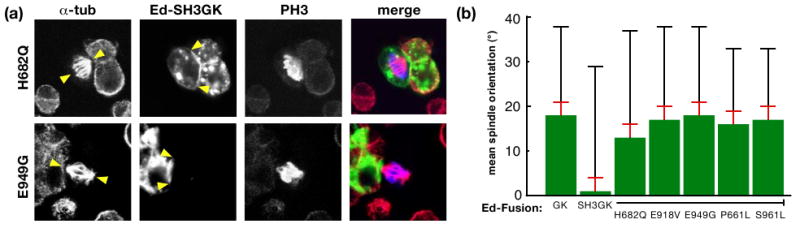
The SH3 domain regulates GK domain function. (a) Example cells from spindle orientation assay. Mitotic S2 cells were stained with antibodies against α-tubulin (red) to identify the mitotic spindle, the FLAG tag (green) to identify Ed-SH3GK with either H682Q or E949G point mutations, and Phospho-Histone H3 (blue) to identify DNA. Arrows denote spindle position (in α-tubulin channel) or edges of Ed-fusion crescent (in Ed channel). Merged images show mitotic spindle alignment to fusion protein crescents. (b) Spindle orienting ability of Ed-Dlg fusions. The mean spindle orientation angle is shown for Ed fusions to Dlg-GK or Dlg-SH3GK (wild type and those harboring putative activating mutations). The mean spindle orientation angle is the difference between the random angle (45°) and the mean observed angle for a particular Ed fusion. The standard error (calculated by measuring the mean angle from several independent experiments; see methods) is shown as an orange bar whereas the standard deviation (due to the natural variation of the population of cells) is shown as a black bar.
To determine if the spindle-orienting activity of the Dlg GK is regulated by the intramolecular interaction, we compared Ed-GK spindle orientation to that of Ed-SH3GK. We found that, whereas the GK domain exhibits robust spindle orientation consistent with microtubule attachment during prophase, the SH3GK has no detectable activity with a mean spindle orientation of 1° ± 28° (Figure 3b). We conclude that the SH3 domain regulates the spindle orienting activity of the GK domain, presumably through the intramolecular interaction, further emphasizing the important role of this interaction and providing a functional context for assessing SH3GK regulation.
We used the polarized Ed-Dlg spindle orientation assay to determine if several of the point mutations identified in the screen break SH3GK regulation, allowing it to orient the spindle. While the wild-type SH3GK has no spindle orienting activity, SH3GKs that bind GukH in the yeast two-hybrid assay are able to orient the spindle when polarized in S2 cells (Figures 3a,b). The point mutations identified in the screen render the SH3GK fully active as they allow the SH3GK to orient the spindle at a level indistinguishable from Ed-GK. Thus, both in interaction and functional assays, these mutations lead to constitutive SH3GK activity. In the next several sections, we examine these mutations to understand how they bypass SH3GK regulation.
SH3GK activation does not require breaking ‘E’ and ‘F’ strand interactions
The interaction between MAGUK SH3 and GK domains was originally identified based on their ability to interact intermolecularly (27). Isolated SH3 domains containing the ‘E’-strand interact with the ‘F’-strand that follows the GK domain and both strands are required for the interaction (Figure 4a). These two strands pair in the intact SH3GK structure to form a β-sheet between the two domains and pairing is required both for interaction between isolated SH3 and GK domains, as well as for the intact SH3GK module (22). For example, the dlgsw allele encodes a protein with intact SH3 and GK domains but a truncated F-strand that disrupts the intramolecular interaction. Interestingly, Dlgsw localizes properly but is non-functional (19). As the E-F pairing appears to be a core element of the SH3GK interaction, activation might require disruption of the two strands. Alternatively the β-sheet could serve as a structural motif that is present in both repressed and activated MAGUK states.
Figure 4.
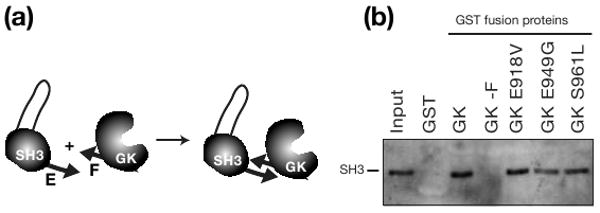
Mutations that allow SH3GK to bind GukH do not disrupt ‘E’-‘F’ strand interaction (a) Schematic representation of “E”-strand containing SH3 and “F”-strand containing GK domains used in GST pull-down assays. (b) GK domains with activating mutations interact with SH3. Western Blot to visualize His-tagged SH3 shows no difference in pull-down between wild-type GK and GK with an activating mutation. Pull-down of SH3 by immobilized GST-GK fusion protein. Lanes show input (20%) and protein bound by GST-alone control and GST-GK fusion proteins.
The mutations identified in the SH3GK activating screen allowed us to test the hypothesis that the E-F strand interaction must be disrupted to allow SH3GK to bind ligands. We constructed GK domains containing individual activating mutations and measured their ability to interact in trans, which requires the E-F β-sheet. As shown in Figure 4b, although these mutations activate the SH3GK, they do not disrupt the E-F interaction. We conclude that relieving SH3 repression of the GK does not require breaking the E-F pairing interaction. Furthermore, the ability of the active variants to interact in trans indicates that the intermolecular interaction assay is not a reliable indicator of MAGUK activation state.
SH3 and Hook residues required for regulation
Of the mutations that allow the SH3GK to bind GukH, P661L and H682Q reside in the SH3 domain. P661 is highly conserved and is directly NH2-terminal to the Hook region, which is an insertion in MAGUK SH3 domains that consists of a conserved α-helix followed by a variable region (Figure 5a). Multiple Hook configurations are present in SH3GK crystal structures (23), suggesting that it can undergo hinge-type movements. Mutating P661 may change the orientation of the Hook region relative to the GK. H682, which is mutated to a glutamine in an activation mutant, lies directly in the Hook in an extended region not present in PSD-95.
Figure 5.
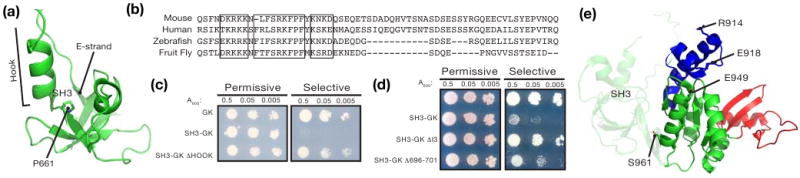
SH3GK residues required for regulation. (a) Ribbon representation of the Dlg SH3 domain including the Hook region and the E Strand. Proline 661, a residue that activates SH3GK when mutated, is shown by a stick representation. (b) CLUSTALW alignment of I3 regions from Dlg-like MAGUKs shows boxed regions of high conservation. (c) The Hook is required for SH3 regulation of GK. Serial dilutions onto plates of permissive (-LT) or selective (-L-T-H + 3AT) medium starting with A600 of 0.5 show restoration of interaction between GUKH and SH3GK with a truncated Hook region. (d) Example of yeast two-hybrid assay. Serial dilutions onto plates of permissive (-LT) or selective (-L-T-H + 3AT) medium starting with A600 of 0.5 show restoration of interaction between GUKH and SH3GK with I3 region truncations. (E) GK activating mutations are mapped onto the PSD-95 GK structure (22; PDB code: 1kjw). The Lid, Core, and GBD domains are colored in blue, green, and red, respectively.
To further explore the role of the Hook in regulating SH3GK activity, we generated additional variants and tested if they bind GukH. We made two Hook truncations: the first (ΔHook) reduced the Hook to just 24 amino acids and added a 6 residue linker, and the second truncation removed a smaller section of the Hook found in numerous MAGUKs known as insert 3 (ΔI3; Figure 5b). SH3GK proteins with either ΔHook or ΔI3 truncations bind GukH indicating that the Hook and the I3 region within are required for SH3 inhibition of GK ligand binding (Figure 5c,d). To further narrow the specific requirements for I3-mediated regulation, we made an additional SH3GK variant, replacing a highly basic segment within the I3 (residues 696-701). As shown in Figure 5d, this region is partially required for SH3GK repression. Thus, we conclude that SH3 residues outside of its interface with the GK, particularly those within the Hook insert, are required for inhibition of GK ligand binding.
GK domain residues required for regulation
We also identified several residues within the GK domain that are required for repression of ligand binding activity by SH3: R914, E918, E949, and S961. These residues map to one face of the GK and form a straight path from the Lid to the Core domain (Figure 1c, 5e). In addition, they are solvent-exposed and only one of them, S961, is in close proximity to the SH3 domain such that it could mediate direct interactions between SH3 and GK. R914 and E918 are part of the Lid domain that is opposite the GBD that binds GukH. E949 is in the Core domain between the Lid and the COOH-terminus of the GK near S961. The three charged residues form salt bridges on the surface of the GK that may be important for stabilizing the inactive conformation. Thus, several surface residues distant from both the SH3 domain and GukH binding site are required for SH3GK regulation.
Discussion
MAGUK scaffold proteins are critical elements of many signaling pathways, particularly those that involve the construction and maintenance of membrane specializations such as adhesions and synapses. MAGUKs are defined by the GK, a unique protein interaction domain originating from an enzyme. An SH3 domain adjacent to the GK inhibits the binding of certain ligands (some GK ligands are not inhibited by the SH3) and this regulation is a critical element of function (19). The SH3 forms an intramolecular interaction with the GK via a β-sheet that is formed from individual strands following each domain. Although SH3 regulation of the GK is required for MAGUK function, very little is known about the mechanism of GK regulation.
SH3 regulation of the GK domain is a form of autoinhibition, a common method of regulating multidomain signaling proteins in which a regulatory domain within the protein inhibits the activity of another domain (28). In one mechanism for autoinhibition, termed “modular allostery”, the regulatory domain interacts such that activity is inhibited through a steric mechanism (i.e. interaction and active sites overlap). This type of regulation is distinguished from classical allostery in which regulation involves conversion between inactive and active states of an individual domain. The elements of modular allostery important for regulation are the interface between the two domains and the linker connecting the domains (the domains themselves may act as rigid bodies and it is the linker that undergoes a conformational change).
We have tested for the presence of modular allostery in the SH3GK module by determining the binding site of regulated GK ligand GukH and comparing it to the SH3 binding site, as determined from the PSD-95 SH3GK crystal structure. GukH binds to the site that corresponds to the GMP binding site in the GK enzyme. As this site is opposite the SH3 binding site, the SH3GK is unlikely to use modular allostery for regulation.
How then might the SH3 regulate the GK if not by modular allostery? We propose that regulation takes advantage of classical allostery within the GK domain itself. The GK enzyme undergoes a large conformational change as it transitions from unliganded to the adenosine and guanosine nucleotide bound form(29, 30). The SH3 bound GK adopts a conformation very similar to the unliganded form. However, MAGUK GKs appear to undergo dynamics similar to their enzyme counterparts based on NMR measurements of the PSD-95 GK interaction with MAP1A (18). We hypothesize that the SH3 alters the energetic landscape of the GK, reducing its ability to adopt conformations with high ligand affinity. Our observation that mutation of GK residues distant from both the SH3 and GukH binding sites is consistent with this hypothesis. Thus, not only do MAGUK proteins co-opt the GK enzyme fold as a protein interaction domain, but may also use similar allosteric hinge motions present in the enzyme's catalytic pathway to regulate ligand binding.
Hook regulation of GK activity
Our results implicate the Hook as a required element for SH3 repression of binding to the GK. The Hook is the most variable region among the SH3GK module of MAGUK proteins (16). We have found that the I3 region within the Dlg Hook is required for GK regulation, and this has also been demonstrated for the mammalian MAGUK SAP97 (15). As the I3 insert is in an alternatively spliced exon, it's possible that isoforms of these proteins are expressed that contain constitutively active SH3GK modules. Furthermore, the Dlg Hook is a binding site for FERM domain proteins and Calmodulin raising the possibility that binding to the Hook could allow the GK to interact with regulated ligands. Further research will be required to test these possibilities.
Activation of the SH3GK
Our results also allow us to comment on the mechanism of SH3GK activation. Because the interaction between SH3 and GK domains is based on pairing of the ‘E’ and ‘F’ strands and disruption of this interaction leads to SH3GK activation, it is possible that activation requires interrupting the E-F interaction. However, we have found this is not the case: activation of the Dlg SH3GK can occur while the E-F interaction is intact. The mechanism by which physiological factors activate SH3GK will require further investigation.
In conclusion, we have begun to elucidate the mechanism of regulated scaffolding in MAGUK proteins. Future work will be directed at understanding how the SH3 domain might modulate GK conformation and dynamics.
Acknowledgments
We thank Rhonda Newman and members of the Prehoda lab for helpful comments.
Footnotes
Funding was provided by a Damon Runyon postdoctoral fellowship to C.A.J., NIH grant GM068032 to K.E.P., and the Howard Hughes Medical Institute to C.Q.D.
Abbreviations: MAGUK, Membrane Associated Guanylate Kinase; GK, Guanylate Kinase-like domain; Dlg, Discs large; SH3, Src Homology 3; GukH, GukHolder; GNB, guanosine nucleotide-binding.
References
- 1.Jamora C, Fuchs E. Intercellular adhesion, signalling and the cytoskeleton. Nat Cell Biol. 2002;4:E101–108. doi: 10.1038/ncb0402-e101. [DOI] [PubMed] [Google Scholar]
- 2.Yamada S, Nelson WJ. Synapses: sites of cell recognition, adhesion, and functional specification. Annu Rev Biochem. 2007;76:267–294. doi: 10.1146/annurev.biochem.75.103004.142811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dimitratos SD, Woods DF, Stathakis DG, Bryant PJ. Signaling pathways are focused at specialized regions of the plasma membrane by scaffolding proteins of the MAGUK family. Bioessays. 1999;21:912–921. doi: 10.1002/(SICI)1521-1878(199911)21:11<912::AID-BIES3>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 4.Lajoie P, Goetz JG, Dennis JW, Nabi IR. Lattices, rafts, and scaffolds: domain regulation of receptor signaling at the plasma membrane. J Cell Biol. 2009;185:381–385. doi: 10.1083/jcb.200811059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang M, Wang W. Organization of signaling complexes by PDZ-domain scaffold proteins. Acc Chem Res. 2003;36:530–538. doi: 10.1021/ar020210b. [DOI] [PubMed] [Google Scholar]
- 6.Garner CC, Nash J, Huganir RL. PDZ domains in synapse assembly and signalling. Trends Cell Biol. 2000;10:274–280. doi: 10.1016/s0962-8924(00)01783-9. [DOI] [PubMed] [Google Scholar]
- 7.Pawson T, Scott JD. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 8.Gardoni F, Polli F, Cattabeni F, Di Luca M. Calcium-calmodulin-dependent protein kinase II phosphorylation modulates PSD-95 binding to NMDA receptors. Eur J Neurosci. 2006;24:2694–2704. doi: 10.1111/j.1460-9568.2006.05140.x. [DOI] [PubMed] [Google Scholar]
- 9.Lalonde D, Bretscher A. The scaffold protein PDZK1 undergoes a head-to-tail intramolecular association that negatively regulates its interaction with EBP50. Biochemistry. 2009 doi: 10.1021/bi802089k. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang M. Scaffold proteins as dynamic switches. Nat Chem Biol. 2007;3:756–757. doi: 10.1038/nchembio1207-756. [DOI] [PubMed] [Google Scholar]
- 11.Fukunaga Y, Matsubara M, Nagai R, Miyazawa A. The interaction between PSD-95 and Ca2+/calmodulin is enhanced by PDZ-binding proteins. J Biochem. 2005;138:177–182. doi: 10.1093/jb/mvi107. [DOI] [PubMed] [Google Scholar]
- 12.Massimi P, Narayan N, Cuenda A, Banks L. Phosphorylation of the discs large tumour suppressor protein controls its membrane ocalization and enhances its susceptibility to HPV E6-induced degradation. Oncogene. 2006;25:4276–4285. doi: 10.1038/sj.onc.1209457. [DOI] [PubMed] [Google Scholar]
- 13.Qian Y, Prehoda KE. Interdomain interactions in the tumor suppressor discs large regulate binding to the synaptic protein GukHolder. J Biol Chem. 2006;281:35757–35763. doi: 10.1074/jbc.M607057200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sabio G, Arthur JS, Kuma Y, Peggie M, Carr J, Murray-Tait V, Centeno F, Goedert M, Morrice NA, Cuenda A. p38gamma regulates the ocalization of SAP97 in the cytoskeleton by modulating its interaction with GKAP. Embo J. 2005;24:1134–1145. doi: 10.1038/sj.emboj.7600578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu H, Reissner C, Kuhlendahl S, Coblentz B, Reuver S, Kindler S, Gundelfinger ED, Garner CC. Intramolecular interactions regulate SAP97 binding to GKAP. Embo J. 2000;19:5740–5751. doi: 10.1093/emboj/19.21.5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Funke L, Dakoji S, Bredt DS. Membrane-associated guanylate kinases regulate adhesion and plasticity at cell junctions. Annu Rev Biochem. 2005;74:219–245. doi: 10.1146/annurev.biochem.74.082803.133339. [DOI] [PubMed] [Google Scholar]
- 17.Kuhlendahl S, Spangenberg O, Konrad M, Kim E, Garner CC. Functional analysis of the guanylate kinase-like domain in the synapse-associated protein SAP97. Eur J Biochem. 1998;252:305–313. doi: 10.1046/j.1432-1327.1998.2520305.x. [DOI] [PubMed] [Google Scholar]
- 18.Reese ML, Dakoji S, Bredt DS, Dotsch V. The guanylate kinase domain of the MAGUK PSD-95 binds dynamically to a conserved motif in MAP1a. Nat Struct Mol Biol. 2007;14:155–163. doi: 10.1038/nsmb1195. [DOI] [PubMed] [Google Scholar]
- 19.Newman RA, Prehoda KE. Intramolecular interactions between the SH3 and GK domains of discs large regulate its function in asymmetric cell division. J Biol Chem. 2009;284:12924–12932. doi: 10.1074/jbc.M809304200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shin H, Hsueh YP, Yang FC, Kim E, Sheng M. An intramolecular interaction between Src homology 3 domain and guanylate kinase-like domain required for channel clustering by postsynaptic density-95/SAP90. J Neurosci. 2000;20:3580–3587. doi: 10.1523/JNEUROSCI.20-10-03580.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woods DF, Hough C, Peel D, Callaini G, Bryant PJ. Dlg protein is required for junction structure, cell polarity, and proliferation control in Drosophila epithelia. J Cell Biol. 1996;134:1469–1482. doi: 10.1083/jcb.134.6.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGee AW, Dakoji SR, Olsen O, Bredt DS, Lim WA, Prehoda KE. Structure of the SH3-guanylate kinase module from PSD-95 suggests a mechanism for regulated assembly of MAGUK scaffolding proteins. Mol Cell. 2001;8:1291–1301. doi: 10.1016/s1097-2765(01)00411-7. [DOI] [PubMed] [Google Scholar]
- 23.Tavares GA, Panepucci EH, Brunger AT. Structural characterization of the intramolecular interaction between the SH3 and guanylate kinase domains of PSD-95. Mol Cell. 2001;8:1313–1325. doi: 10.1016/s1097-2765(01)00416-6. [DOI] [PubMed] [Google Scholar]
- 24.Johnston CA, Hirono K, Prehoda KE, Doe CQ. Identification of an Aurora-A/PinsLINKER/Dlg Spindle Orientation Pathway using Induced Cell Polarity in S2 Cells. Cell. 2009;138:1150–1163. doi: 10.1016/j.cell.2009.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siegrist SE, Doe CQ. Microtubule-induced Pins/Gαi cortical polarity in Drosophila neuroblasts. Cell. 2005;123:1323–1335. doi: 10.1016/j.cell.2005.09.043. [DOI] [PubMed] [Google Scholar]
- 26.Albertson R, Doe CQ. Dlg, Scrib and Lgl regulate neuroblast cell size and mitotic spindle asymmetry. Nat Cell Biol. 2003;5:166–170. doi: 10.1038/ncb922. [DOI] [PubMed] [Google Scholar]
- 27.McGee AW, Bredt DS. Identification of an intramolecular interaction between the SH3 and guanylate kinase domains of PSD-95. J Biol Chem. 1999;274:17431–17436. doi: 10.1074/jbc.274.25.17431. [DOI] [PubMed] [Google Scholar]
- 28.Dueber JE, Yeh BJ, Bhattacharyya RP, Lim WA. Rewiring cell signaling: the logic and plasticity of eukaryotic protein circuitry. Curr Opin Struct Biol. 2004;14:690–699. doi: 10.1016/j.sbi.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 29.Blaszczyk J, Li Y, Yan H, Ji X. Crystal structure of unligated guanylate kinase from yeast reveals GMP-induced conformational changes. J Mol Biol. 2001;307:247–257. doi: 10.1006/jmbi.2000.4427. [DOI] [PubMed] [Google Scholar]
- 30.Sekulic N, Shuvalova L, Spangenberg O, Konrad M, Lavie A. Structural characterization of the closed conformation of mouse guanylate kinase. J Biol Chem. 2002;277:30236–30243. doi: 10.1074/jbc.M204668200. [DOI] [PubMed] [Google Scholar]


