Abstract
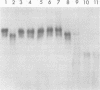
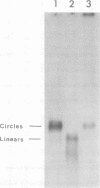
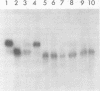
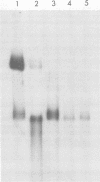
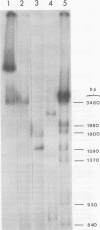
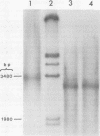
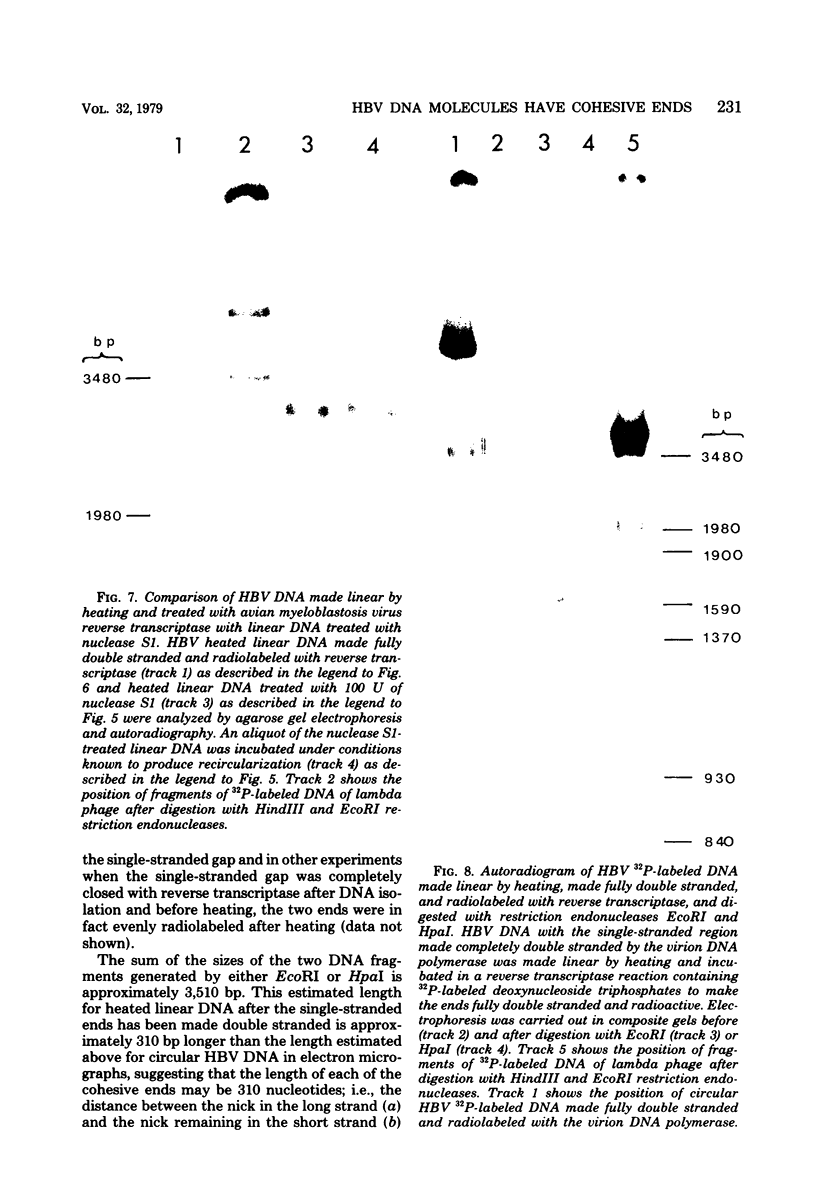
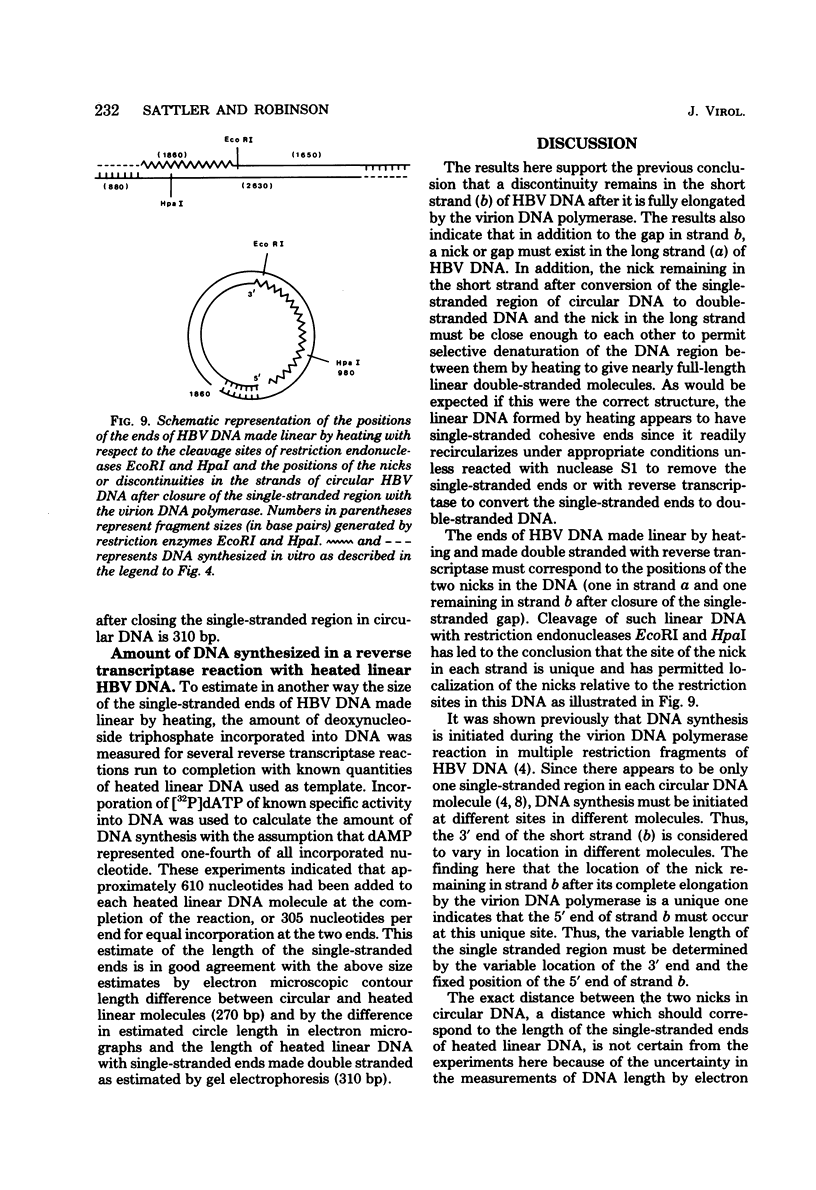
Hepatitis B virus DNA made fully double stranded by a virion DNA polymerase reaction could be converted from circular to linear molecules by heating in 10 mM NaCl at 77°C or in 100 mM NaCl at 90°C for 15 min. Heat-generated linear hepatitis B virus DNA was reannealed to circular molecules by incubating in higher salt concentrations. The identity of the molecular forms was established by their electrophoretic mobility and appearance in electron micrographs. Recircularization was blocked by reacting linear molecules with nuclease S1 or avian myeloblastosis virus reverse transcriptase. These results suggest that the heated linear DNA had single-stranded ends with complementary nucleotide sequences. It also suggests that a discontinuity or nick is present in each strand of the circular DNA molecule after the single-stranded region is made double stranded by the virion DNA polymerase reaction. The difference in contour length by electron microscopy of circular and linear molecules spread under aqueous conditions suggested that the discontinuities in the two strands were about 270 base pairs apart. The amount of nucleotide incorporated into the ends of heat-generated linear hepatitis B virus DNA by reverse transcriptase suggested that the single-stranded ends were about 305 bases in length. This fully double-stranded linear DNA was cleaved with EcoRI or HpaI restriction endonuclease. The sum of the two fragments generated by each totaled 3,510 base pairs, 310 base pairs greater than the contour length of circular hepatitis B virus DNA which represents a third estimate of the distance between the discontinuities in the two DNA strands of circular DNA. Restriction endonuclease cleavage also indicated that the ends of heated linear DNA which correspond to the discontinuities in the two strands of the circular DNA are at unique sites in the DNA with respect to the restriction sites.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Clayton D. A., Davis R. W., Vinograd J. Homology and structural relationships between the dimeric and monomeric circular forms of mitochondrial DNA from human leukemic leukocytes. J Mol Biol. 1970 Jan 28;47(2):137–153. doi: 10.1016/0022-2836(70)90335-9. [DOI] [PubMed] [Google Scholar]
- Hruska J. F., Clayton D. A., Rubenstein J. L., Robinson W. S. Structure of hepatitis B Dane particle DNA before and after the Dane particle DNA polymerase reaction. J Virol. 1977 Feb;21(2):666–672. doi: 10.1128/jvi.21.2.666-672.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan P. M., Greenman R. L., Gerin J. L., Purcell R. H., Robinson W. S. DNA polymerase associated with human hepatitis B antigen. J Virol. 1973 Nov;12(5):995–1005. doi: 10.1128/jvi.12.5.995-1005.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landers T. A., Greenberg H. B., Robinson W. S. Structure of hepatitis B Dane particle DNA and nature of the endogenous DNA polymerase reaction. J Virol. 1977 Aug;23(2):368–376. doi: 10.1128/jvi.23.2.368-376.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy V. B., Thimmappaya B., Dhar R., Subramanian K. N., Zain B. S., Pan J., Ghosh P. K., Celma M. L., Weissman S. M. The genome of simian virus 40. Science. 1978 May 5;200(4341):494–502. doi: 10.1126/science.205947. [DOI] [PubMed] [Google Scholar]
- Robinson W. S., Clayton D. A., Greenman R. L. DNA of a human hepatitis B virus candidate. J Virol. 1974 Aug;14(2):384–391. doi: 10.1128/jvi.14.2.384-391.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson W. S., Greenman R. L. DNA polymerase in the core of the human hepatitis B virus candidate. J Virol. 1974 Jun;13(6):1231–1236. doi: 10.1128/jvi.13.6.1231-1236.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sninsky J. J., Siddiqui A., Robinson W. S., Cohen S. N. Cloning and endonuclease mapping of the hepatitis B viral genome. Nature. 1979 May 24;279(5711):346–348. doi: 10.1038/279346a0. [DOI] [PubMed] [Google Scholar]
- Summers J., O'Connell A., Millman I. Genome of hepatitis B virus: restriction enzyme cleavage and structure of DNA extracted from Dane particles. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4597–4601. doi: 10.1073/pnas.72.11.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai H. T., Smith C. A., Sharp P. A., Vinograd J. Sequence heterogeneity in closed simian virus 40 deoxyribonucleic acid. J Virol. 1972 Feb;9(2):317–325. doi: 10.1128/jvi.9.2.317-325.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]