Abstract
Objective
Determine the expression and distribution of A Disintegrin And Metalloproteinase with ThromboSpondin motifs-4 (ADAMTS-4), its substrates aggrecan and versican, and their binding partner hyaluronan in laminae of healthy horses as a step towards determining the role of ADAMTS-4 in laminar pathology.
Sample population
Front hoof laminae from 8 healthy horses.
Procedures
Real-time quantitative polymerase chain reaction was used for gene expression analysis. Hyaluronidase, chondroitinase and keratanase digestion of lamina extracts together with sodium dodecylsulfate polyacrylamide gel electrophoresis and Western blotting were used for protein and proteoglycan analysis. Immunofluorescent and immunohistochemical staining of tissue sections were used for protein and hyaluronan localization.
Results
Genes encoding ADAMTS-4, aggrecan, versican and hyaluronan synthase II are expressed in laminae. ADAMTS-4 is predominantly present as a 51 kDa protein bearing a catalytic site neoepitope indicative of active enzyme and in situ activity is inferred from the presence of aggrecan and versican fragments bearing ADAMTS-4 cleavage neoepitopes in laminar protein extracts. Aggrecan, versican and hyaluronan localize to basal epithelial cells within the secondary dermal laminae. ADAMTS-4 also localizes to these cells, but in addition, is present in some cells in the dermal laminae.
Conclusions and clinical relevance
Within the digital laminae, versican exclusively and aggrecan primarily localizes within basal epithelial cells and both are constitutively cleaved by ADAMTS-4 which therefore contributes to their turnover. Based on known properties of these proteoglycans, it is possible that they protect the basal epithelial cells from biomechanical and concussive stress.
Keywords: Equine, Digital Laminae, ADAMTS-4, Aggrecan, Versican, Hyaluronan
Introduction
The equine digital laminae span between the surface of the distal phalanx and the inner hoof wall. Laminae are composed of 500 to 600 vertical folds of keratinized epidermal tissue, the primary epidermal laminae, which are contiguous with the inner hoof wall, and interdigitated folds of connective tissue, the primary dermal laminae, which are contiguous with the distal phalanx1. Each fold of primary epidermal and dermal laminae has 150 to 200 interdigitated folds of secondary laminae resulting in a greatly expanded contact area between these tissue layers. The secondary epidermal and dermal laminae join at a basement membrane, which is a meshwork of collagen fibers and laminins cross-linked and anchored via hemidesmosomes to basal epithelial cells residing at the boundary of the epidermal laminae2. The basement membrane is tethered to tensile collagen fibers of the dermal laminae, which are bound by a variety of extracellular matrix components to each other and to integrins expressed by cells on both sides of the membrane3, thus ensuring integrity of the two layer structure. The basal epithelial cells, or stem cells within this population, give rise to keratinocytes which move outward towards the hoof wall and, analogous to skin epidermal cells, increase their keratin content, generate a cornified cell envelope and undergo terminal differentiation4,5.
The digital laminae resist force imposed by the deep flexor tendon through the distal phalanx to the dermal attachments to the bone6–8. They also support the vertical load of the horse and accommodate compression and stretch deformation created by the flexing, twisting and tilting of the hoof capsule under different loading conditions. In addition, the laminae absorb a portion of the concussive shock imposed when the hoof strikes a solid surface. Concussive shock most likely dissipates in the hoof by freedom of movement of hydrated keratin fibers within the epidermal laminae and transfer of load from the hoof wall to the dermal connective tissue and distal phalanx through the basal epithelial cell layer. Basal epithelial cells are therefore subjected to biomechanical stress and concussion waves of varying amplitude and frequency.
Analyses in model systems show that cells adapt to constant and discontinuous mechanical stress through mechanoreceptor signaling9,10. In this regard, chondrocyte explants that are subjected to dynamic compression within a tolerable range respond by elevated extracellular matrix production9 including elevated production of large polysulfated proteoglycans. Since the equine digital laminae are also subject to dynamic compression/stretching they may also have a highly specialized extracellular matrix that is rich in polysulfated proteoglycans. Indeed, the gene encoding ADAMTS-4, which is a secreted enzyme that regulates turnover of large polysulfated proteoglycans in the extracellular matrix, is expressed in laminae of healthy horses11, which suggests that its substrates may also be present in the tissue.
The main substrates of ADAMTS-4 in peripheral tissues are aggrecan and versican12. These proteoglycans have glycosaminoglycan attachment domains that can be heavily substituted with chondroitin sulfate side chains, and, in the case of aggrecan, also keratan sulfate side chains13–15. Both aggrecan and versican have a hyaluronan binding site in the N terminal G1 domain, which facilitates assembly of massive and highly charged macromolecular complexes in the extracellular matrix. The anionic groups on the glycosaminoglycans carry with them positively charged counter ions, such as Na+, creating an osmotic gradient and drawing water into the tissue. The resulting hydrated gel protects tissue from compression deformation13,16,17. Given that laminae express the gene encoding ADAMTS-4 and are subjected to biomechanical and concussion stress, it was of interest to determine expression and localization patterns of ADAMTS-4 and its proteoglycan substrates in the laminae. Here we report that ADAMTS-4 and its substrates are indeed present in healthy laminae, but, unexpectedly, localize within basal epithelial cells of the secondary epidermal laminae rather than in the extracellular matrix.
Materials and Methods
Animals, Tissue Collection and immunization
Horses
Front hoof laminae were collected from 8 healthy horses as previously described18,19. Briefly, healthy horses were placed under general anesthesia, their distal right forelimbs detached at the metacarpophalangeal joint and the animals euthanized immediately thereafter under protocols approved by the IACUC of the University of Missouri College of Veterinary Medicine. Excised hoofs were sectioned using a band saw and segments of lamina (approximately 5 mm × 5 mm × 5 mm) spanning from the inner hoof wall to the outer face of the distal phalanx were dissected using a sharp scalpel and flash frozen in liquid nitrogen for molecular and biochemical analyses. For immunofluorescent microscopy, segments were embedded in a commercial preparation of water soluble glycols and resinsa and frozen over dry ice (6 of the 8 laminae only). All segments of laminae were processed or frozen within 5 minutes of detachment of the distal right forelimb.
Rabbits
Two female New Zealand White rabbits were immunized with an equine ADAMTS-4-specific peptide (described below) conjugated to keyhole limpet hemocyanin (KLH) and emulsified in an adjuvantb1. Briefly, each rabbit received 500 μg protein dispersed over 10 sites on the back and was boosted three times over the course of 10 weeks with an additional 250 μg peptide-KLH emulsified in an adjuvantb2. The animals were exanguinated 10 days after the last immunization, blood collected for serum preparation and serum stored at −20°C until use. All procedures were carried out under protocols approved by the IACUC of the University of Massachusetts. The peptide, “NTPEDSDPDHFD,” corresponded to amino acids 300–311 of the equine ADAMTS-4 sequence and was synthesizedc with an N-terminal cysteine residue to facilitate conjugation to the KLH. The peptide was chosen from an area of ADAMTS-4 predicted to be a loop structure in the metalloproteinase domain based on homology to the human protein sequence, for which the crystal structure has been solved. Peptide specific antibody was affinity purified from rabbit serum using sepharose beadsd conjugated to the “NTPEDSDPDHFD” peptide. The concentration of purified antibody was determined by spectrophotometrye.
RNA extraction
RNA was collected from three separate sections of dorsal lamina from each horse using an RNA extraction kitf1. Briefly, flash frozen tissue was pulverized in a pre-chilled (on dry ice) slammerg and homogenized in the lysis buffer provided, then passed through the column provided and washed accordingly. The purity and concentration of RNA were determinede and extracted RNA was used for cDNA preparation only when the A260:A280 and the A260:A230 ratios were very close to 2.0. Integrity of isolated RNA was confirmed by electrophoresis on a 1.0% agarose gel and staining with a proprietary polynucleotide gel stainh.
PCR
cDNA was synthesized from isolated RNA using a cDNA synthesis kiti. Relative quantitative real time PCR - Primer sets for ADAMTS-4, aggrecan, versican and hyaluronan synthase II were generated against the equine sequence (Table 1). GAPDH was employed as a housekeeping gene using primers previously described11. Briefly, RT-qPCR reactions were run using a proprietary reaction mixture which contains a high performance reverse transcriptase and reference dyesj according to manufacturer’s instructions and data were read with a thermal cyclerf2 as described11. Non-quantitative PCR- Specific cDNA fragments of four versican isoforms (V0, V1, V2 and V3) were amplified by PCR20 using the primers listed in Table 2. All amplifications were performed for 35 cycles following the conditions: 94°C for 2min, 94°C for 30s, 58°C for 30s, 72°C for 1min and 72°C for 7min using a PCR thermal cyclerk PCR products were visualized after electrophoresis on a 2.0% agarose gel by staining with a proprietary polynucleotide gel stainh, and bands were excised, purified, and sent for sequence confirmation.
Table 1.
Primer sequences utilized in RT qPCR evaluation of gene expression
| Name | Sequence (F-forward 5′; R– reverse 5′) | GenBank ID | Amplicons length | Primer efficiency | R2 |
|---|---|---|---|---|---|
| ADAMTS-4 (C-terminal) | F-gctgggctactattatgtgctg-3′ R-ccacattgttgtatccgtacct-3′ |
100033914 | 227bp | 104% | 0.976 |
| ADAMTS-4 (N-terminal) | F-cagtatcgaggggaccgaact-3′ R-gaaatgctgccatcttgtcat-3′ |
100033914 | 241bp | 92.6% | 1.000 |
| Aggrecan | F-caacaacaatgcccaagactac-3′ R-agttctcaaattgcaaggagtg-3′ |
100033876 | 110bp | 102% | 0.981 |
| Versican | F-cctgcaattaccatctcaccta-3′ R-cagggagttgatttcataacga-3′ |
100065275 | 122bp | 92.1% | 0.988 |
| Hyaluronan synthase 2 | F-cacagacaggctgaggacaa-3′ R-acaggctttggatgatgagg-3′ |
100009708 | 231bp | 94% | 0.930 |
Table 2.
Primer sequences utilized for PCR detection of versican specific isoforms
| Primer | Sequence – GenBank ID 100065275 | Isoform | Primer pair | Expected Product Size (bp) |
|---|---|---|---|---|
| eqV-F1 | 5′-gctgaagaagagtgtgaaaa-3′ | V0 | F2+R1 | 530 |
| eqV-F2 | 5′-tggtgaagaaacaaccagtg-3′ | V1 | F1+R1 | 520 |
| eqV-F3 | 5′-ctcgtgttcctcccactacc-3′ | V2 | F3+R2 | 510 |
| eqV-R1 | 5′-agtggtgactagatgtttcc-3′ | V3 | F1+R2 | 547 |
| eqV-R2 | 5′-tgggcaaagtacttgtagca-3′ |
Protein extracts
NP-40 soluble material
~ 0.35g snap frozen segments of dorsal laminae was pulverized in a pre-chilled (on dry ice) slammerg and immediately homogenized in 10ml of extraction buffer (50mM Tris pH7.0, 150 mM NaCl, 5mM ethylenediaminetetraacetic acid, 0.5% NP-40 containing 10μM E64, 1.5μM pepstatin A and 1 mM phenylmethanesulfonyl fluoride) on ice. The homogenized sample was incubated overnight at 4°C, centrifuged (14,000 g) 15 min in 4°C, supernatant collected and protein in the supernatant precipitated by addition of ice cold absolute ethanol to a final concentration of 80% v/v. The precipitate was washed with ice cold 80% ethanol twice, dried under nitrogen and dissolved in PBS. Protein concentration was determined by a colorimetric assay based on a protein binding dyel1.
Guanidine hydrochloride soluble material
~0.35g snap frozen segments of dorsal laminae was pulverized in a pre-chilled slammer and homogenized in 5ml of cold extraction buffer (0.1 M PBS, with 5 mM iodoacetic acid, 0.1 mM 4-(2-aminoethyl) benzenesulphonyl fluoride, 1% 3-[(cholamidopropyl)-dimethylammonio]-1-propane-sulfonate, 1 μg/ml pepstatin A, 50 mM sodium acetate, 5 mM benzamidine hydrochloride hydrate, 5 mM phenylmethylsulfonyl fluoride, 10 mM N- ethylmaleimide, 4 M guanidine hydrochloride, pH7.6) [additionally supplemented with a proteinase inhibitor cocktailb3] for 30 sec on ice and extracted overnight (~15 hours) at 4°C. The samples were centrifuged at 14,000 g for 30 min at 4°C, a floating layer of insoluble lipids removed and the remaining supernatant collected and precipitated overnight with 4 vol. of ice cold absolute ethanol containing 5 mM sodium acetate. Precipitated molecules were collected by centrifugation at 4°C for 1 hr at 14,000g, dried under nitrogen and the pellet was re-suspended in buffer and digested with 0.06 Units Streptomyces hyaluronidasem/10μg weight of original frozen tissue as described by the manufacturer. After digestion the supernatant solids were precipitated by addition of ice cold absolute ethanol containing 5mM sodium acetate to a final concentration of 80% v/v as above, dried and digested with 0.01 Units Chondroitinase ABCn/10μg weight of original frozen tissue as per manufacturer’s instructions. Supernatant solids were again precipitated and dried as above and digested with 10 μUnits Keratanase IIn/10μg weight of original frozen tissue. The above digestion protocol allowed solubilization of the many molecules that form insoluble macromolecular complexes with hyaluronan and their subsequent analysis by SDS-PAGE.
SDS-PAGE and Western Blotting
An aliquot (30 μg protein content) of extract was boiled in reducing Laemmli (5 mM 2-mercaptoethanol) sample bufferl2 for 5min and subjected to SDS-PAGE in a 4% (w/v) polyacrylamide stacking gel with a 10% (w/v) polyacrylamide gel as previously described21. Proteins were transferred to polyvinylidene fluoride membranes by electroblotting. The membrane was blocked with 5% dry milk in PBS with 0.05% tween-20 for 1 hr, washed with PBS with 0.1% tween-20 for 30 min and then incubated with primary antibodies overnight at 4°C. Antibodies listed in Table 3 were used together with immunoaffinity purified rabbit polyclonal anti-NTPEDSDPDHFD (ADAMTS-4 metalloproteinase domain epitope) described under rabbits above. After incubation with primary antibodies the membranes were washed twice in PBS with 0.1% tween-20 for 30 min and incubated with secondary antibodies conjugated with horseradish peroxidase (Table 3). Detection was performed using enhanced chemiluminescencel3 visualized with a gel imaging and documentation systemo1 and quantification was done using associated softwareo2.
Table 3.
Antibodies used for Western blotanalysis of protein expression
| Primary antibody (dilution used) | Secondary antibody |
|---|---|
| Mouse monoclonal [BC-3] to human aggrecan ARGSVIL (1:2000)r1 | Sheep polyclonal to mouse IgG-H&L (HRP)r2 |
| Rabbit polyclonal to Versican (V0/V1) DPEAAE (within Beta-domain) (1:2000)r3 | Sheep polyclonal to rabbit IgG-H&L (HRP)r4 |
| Rabbit polyclonal to human ADAMTS-4 catalytic neoepitope FASLSRFVET26 (1:500) | Sheep polyclonal to rabbit IgG-H&L (HRP)r4 |
| Mouse monoclonal to beta Actin-Loading Control (1:20,000)r5 | Sheep polyclonal to mouse IgG-H&L (HRP)r2 |
| Rabbit polyclonal to Versican (V0/V2) NIVSFE (within Alpha-domain) (1:2000)r6 | Sheep polyclonal to rabbit IgG-H&L (HRP)r4 |
| Rabbit polyclonal to mouse versican (aGAG domain) aa535–598 of mouse versican V0/V2 (1:1000)s1 | Sheep polyclonal to rabbit IgG-H&L (HRP)r4 |
| Rabbit polyclonal to mouse versican (βGAG domain) aa1360–1439 of mouse versican V1 (1:1000)s2 | Sheep polyclonal to rabbit IgG-H&L (HRP)r4 |
Immunofluorescence
Frozen sections (10 μm thick) were cut from embeddeda tissues and affixed to treated glass slidesp. Immunofluorescent staining was carried out using methods previously described22. Briefly, slides were blocked with 5% BSA in PBS with 0.001% tween-20 and treated with optimal dilutions of primary antibodies, described in table 4, for one hour at room temperature. Sections were washed and treated for one hour at room temperature with secondary antibodies along with a 1:2000 dilution of DAPI. Actin was visualized by incubating tissue sections with a 1:200 dilution of phalloidin-FITCb4 conjugate for one hour at room temperature.
Table 4.
Antibodies used for immunofluorescent localization studies
| Primary antibody (dilution) | Secondary antibody |
|---|---|
| Rabbit polyclonal to laminin (1:100)r7 | Goat polyclonal to rabbit IgG H&L – Texas Red conjugater8 Donkey polyclonal to rabbit IgG H&L – conjugated with a flurochrome with Ex/EM* of 493/518nmt1 |
| Goat polyclonal to human versican (T-20) (1:20)u | Donkey polyclonal to goat IgG H&L – conjugated with a fluorochrome with EX/EM* of 591/616nmt2 |
| Rabbit polyclonal to human aggrecan G2 domain peptide (AA 89–106) (1:50)r9 | Donkey polyclonal to rabbit IgG H&L – conjugated with a flurochrome with EX/EM* of 591/616nmt3 |
| Sheep polyclonal to human hyaluronic acid (1:250)r10 | Rabbit polyclonal to sheep IgG H&L – Texas Red conjugater11 |
| Affinity purified rabbit polyclonal to equine ADAMTS- 4 metalloprotease domain peptide (AA 300–311) (1:20; generated as part of this study) | Donkey polyclonal to rabbit IgG H&L – conjugated with a fluorochrome with EX/EM* of 591/616nmt3 |
EX/EM = excitation and emission +/− 4nm in PBS
Specificity of staining was established using enzyme digestion and peptide blocking. Enzymatic digestions were carried out for one hour at 37°C as follows: i) for destruction of hyaluronan epitopes, sections were treated with 0.01Units of chondroitinase ABCb5 in 20 μl 50mM Tris pH 6.8 and 60mM sodium acetate with 0.2% BSA. After digestion, sections were thoroughly washed with PBS with 0.001% tween-20, and reacted with antibodies as described above. ii) For peptide competitions, antibodies were incubated overnight with a 10 fold excess of cognate peptide or antisense peptide in 1% BSA at 4°C prior to incubation with sections as described above. All slides were imaged using an inverted microscope with apotome grid, UV, blue and green excitation light and 20× or 63× objectivesq.
Results
Gene expression
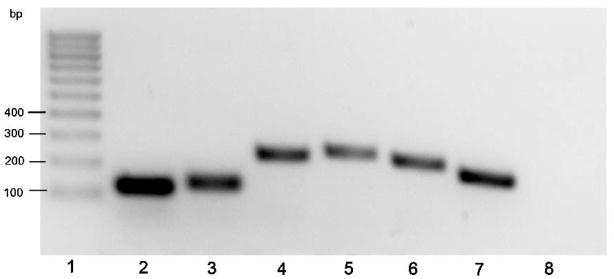
Genes encoding ADAMTS-4, aggrecan, versican and hyaluronan synthase II are expressed in the equine digital laminae of 8 healthy horses as determined by RT-qPCR. Primer sets are described in table 1 and each generated a single product of expected size (Table 1, representative results are shown in Fig 1; Lane 2 aggrecan; Lane 3 versican; Lane 4 hyaluronan synthase II; Lane 5 ADAMTS-4 N terminal domain; Lane 6 ADAMTS-4 C-terminal domain; Lane 7 GAPDH; Lane 8 blank control) and sequence (data not shown). Mean Ct values +/− 1 standard deviation were: GAPDH 21.82+/− 0.88, aggrecan 29.02+/−1.37, versican 25.84+/− 1.20, hyaluronan synthase II 30.34 +/− 0.94, ADAMTS-4 (N-terminal domain primers) 34.12 +/− 1.57, ADAMTS-4 (C-terminal domain primers) 32.45 +/− 1.054. No Ct value was recorded for the blank control.
Figure 1. Analysis ofRT-qPCR products generated using primer pairs specific for aggrecan, versican, hyaluronan synthase II and ADAMTS4, and cDNA generated using RNA from healthy equine laminae.
Lane 1, DNA marker (100 bp ladder); Lane 2 aggrecan; Lane 3 versican; Lane 4 hyaluronan synthase II; Lane 5 ADAMTS-4 N terminal domain; Lane 6 ADAMTS-4 C-terminal domain; Lane 7 GAPDH; Lane 8 blank control.
Versican is composed of 4 domains, namely an N terminal G1 domain, which has the hyaluronan binding site, a αGAG domain which has sites for attachment of chondroitin sulfate side chains, a βGAG domain which also has sites for attachment of chondroitin sulfate side chains and a lectin like C terminal domain, which has 2 epidermal growth factor-like motifs24. Four splice variants (isoforms) of versican are made comprising the V0 isoform which has all 4 domains ordered as above; the V1 isoform which has G1, βGAG and C domains only; the V2 isoform which has G1, α GAG and C domains only; and the V3 isoform which has G1 and C domains only and therefore lacks glycosaminoglycans25. Comparative RT-qPCR analyses performed with primer sets specific for equine versican G1, αGAG, βGAG and C terminal domains showed ratios close to 1:1:1:1 (data not shown) consistent with expression of the full length gene. In addition, RT-PCR analyses performed with primer sets specific for equine V0 versican - αGAG/βGAG (Fig 2, lane 2), V1 versican - G1/βGAG (Fig 2, lane 3), V2 versican - G1/αGAG (Fig 2, lane 4), and V3 versican - G1/C (Fig 2, lane 5) showed that spliced sequence encoding V0 and V1 versican isoforms were abundantly present in laminae while those encoding the V2 isoform were less abundant and spliced sequence encoding the V3 isoform were not detected. Primer specificity was confirmed by sequencing the products (not shown).
Figure 2. Analysis of PCR products generated using primer pairs specific for versican isoforms and cDNA generated using RNA from healthy equine laminae.
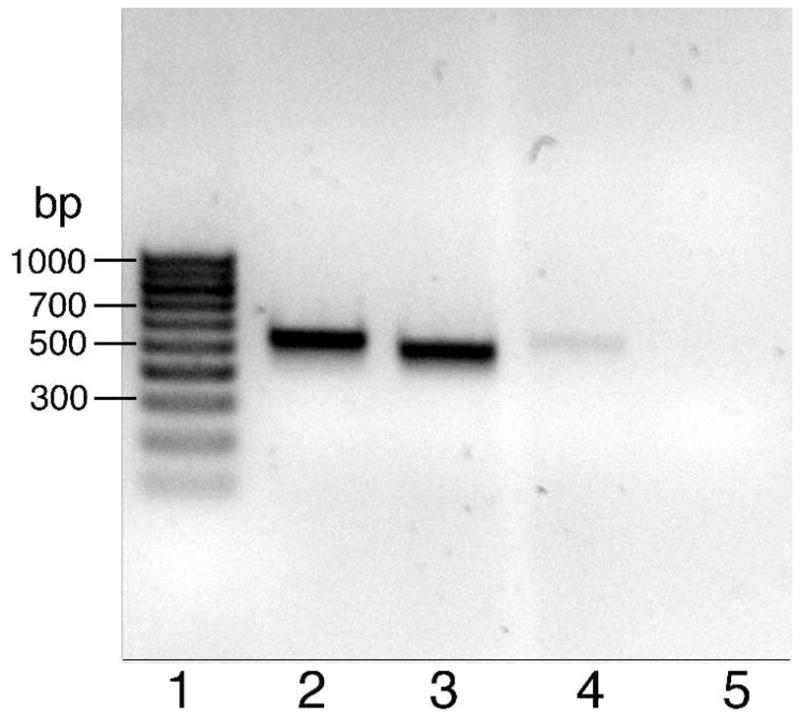
Lane 1, DNA marker (100 bp ladder); Lane 2, V0; Lane 3, V1; Lane 4, V2; Lane 5, V3.
Protein expression
Extracts from the 8 laminae were subjected to Western blotting following SDS-PAGE to determine the protein expression of ADAMTS-4 and its substrates. Representative results are presented in Figure 3.
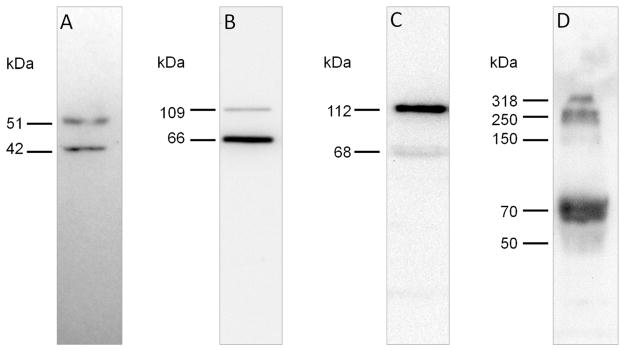
Figure 3. Immunoreactivity of ADAMTS-4 and its substrates in extracts of healthy equine laminae.
Western blots of laminar extracts (30 μg protein/lane) were probed using: A) antibody against neoepitope FASLSRFVET exposed on ADAMTS-4 catalytic domain after removal of the regulatory propeptide (NP-40 extract); B) antibody against versican V0/V1 neoeptiope DPEAAE generated by ADAMTS-4 cleavage (NP-40 extract); C) antibody against versican V0/V2 neoeptiope NIVSFE generated by ADAMTS-4 cleavage (NP-40 extract); D) antibody against aggrecan neoepitope ARGSVIL (BC-3) generated by ADAMTS-4 cleavage (guanidine hydrochloride extract digested with hyaluronidase, chondroitinase ABC and karatinase III).
ADAMTS-4
Proteins in NP-40 extract of laminae were subjected to SDS-PAGE and ADAMTS-4 was detected by Western blotting using an antibody specific for the catalytic site cleavage neoepitope FASLSRFVET which is revealed only upon removal of the regulatory propeptide26. Results are presented in Figure 3A and show that processed ADAMTS-4 is predominantly present as a ~51 kDa polypeptide accompanied by a ~41 kDa form. The ~51 kDa polypeptide was detected in all 8 laminae extracts analyzed, while the ~41 kDa polypeptide was detected in only 6 of 8 laminae extracts analyzed. The same results were obtained when samples of pulverized laminae were directly solubilized in SDS-sample buffer (data not shown) showing that processing of ADAMTS-4 by propeptide convertase27,28, which exposes the FASLSRFVET catalytic site neoepitope, was not an extraction artifact.
V0/V1 Versican
To explore versican protein expression, NP-40 laminae extracts of laminae were subjected to SDS-PAGE and ADAMTS-4 V0/V1 versican cleavage fragments were revealed by Western blotting using an antibody specific for the C terminal V0/V1 versican β GAG domain neoepitope DPEAAE. Results are presented in Figure 3B and show V0/V1 versican fragments of ~109 kDa and ~66kDa. Detection of the polypeptides was abrogated when anti-DPEAAE antibodies were pre-incubated with sense peptide but not antisense peptide (data not shown). The ~109 kDa polypeptide was detected in 6 of 8 laminae extracts analyzed, while the ~66kDa polypeptide was detected in all 8 laminae extracts analyzed. Polypeptide bands of similar size were detected when pulverized laminae was directly solubilized in SDS-sample buffer (data not shown) showing that processing of V0/V1 versican by ADAMTS-4 was not an extraction artifact,
V0/V2 Versican
NP-40 laminae extracts were subjected to SDS-PAGE and ADAMTS-4 V0/V2 versican cleavage fragments revealed by Western blotting using an antibody specific for the C terminal V0/V2 versican α GAG neoepitope NIVSFE. Results are presented in Figure 3C and show V0/V2 versican fragments of ~112 kDa and ~68 kDa. The polypeptides were detected in all 8 laminae extracts analyzed and detection of bands was abrogated when antibodies were pre-incubated with sense peptide but not antisense peptide (data not shown). Polypeptide bands of similar size were detected when tissue was directly solubilized in SDS-sample buffer (data not shown) showing that processing of V0/V2 versican by ADAMTS-4 was not an extraction artifact.
Aggrecan
SDS-PAGE analysis of ADAMTS-4 fragments of aggrecan required their extraction from pulverized laminae by guanidine hydrochloride buffer, precipitation with ethanol and subsequent digestion with hyaluronidase, chondroitinase ABC and keratanase (data not shown). These treatments were necessary because aggrecan ADAMTS-4 cleavage fragments: i) were not extracted from pulverized laminae by incubation with NP-40 buffer, ii) co-extracted with their binding partner hyaluronan in guanidine hydrochloride buffer, and were held in an insoluble hyaluronan gel upon replacement of guanidine hydrochloride with water or physiological buffer, iii) were heavily glycosylated, particularly with chondroitin sulfate glycosaminoglycans, which prevented migration through the stacking gel in SDS-PAGE in the absence of appropriate digestion. Western blotting performed following SDS-PAGE of the hyaluronidase, chondroitinase ABC and keratanase digested guanidine hydrochloride extracted material using an antibody specific for the aggrecan N terminal ADAMTS-4 cleavage neoepitope ARGSVIL revealed bands at ~318 kDa, ~250 kDa, ~150 kDa and ~70 kDa (a representative result is shown in Fig 3D). These polypeptide bands were detected in the 8 laminae extracts analyzed.
Immunohistological evaluation
To determine the cellular localization of ADAMTS-4 and its substrates, immunohistological studies were performed on flash frozen OCT-embedded laminae (n=6). All samples were analyzed for each staining protocol and representative results are shown below.
General structure – i) Basement membrane
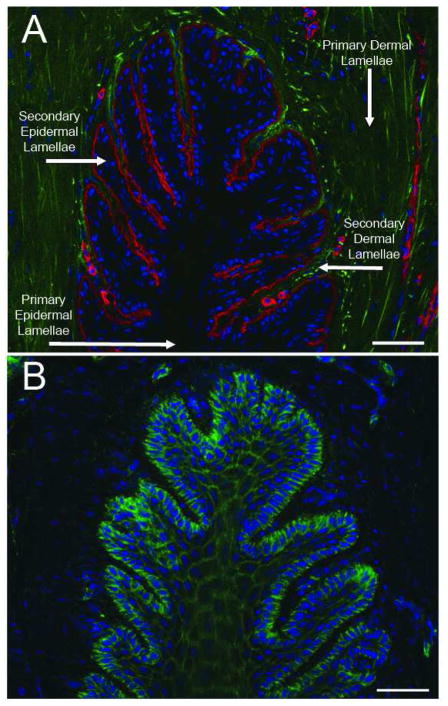
A representative thin transverse section of frozen digital laminae stained with a pan-laminin specific antibody revealed by a texas red-conjugated secondary antibody (red), and with the DNA intercalating dye DAPI (blue), is shown in Figure 4A. Laminin is a component of the basement membrane which lies at the junction between the secondary epidermal and dermal laminae. Laminin is also a component of the basement membrane of small blood vessels which can be seen as small red open circles in the secondary dermal laminae, and of blood vessels in the primary dermal laminae. DAPI (blue) stained cell nuclei can be seen throughout the tissue and those of the basal epithelial cells abut the basement membrane. Some tissue components, putatively collagen fibers in primary and secondary dermal laminae, autofluoresce green at the excitation wavelength (488nm) used in the studies (Fig 4A). This autofluorescent material can also be seen in Figures 5A, 5B, Figure 6A and Figure 7A, has a relatively weak signal which is not digitally recorded when a bright green fluorescent signal is obtained following appropriate specific staining, e.g., after staining thin frozen sections with FITC-phalloidin as shown in Figure 4B and discussed below.
Figure 4. Defining lamellar structure and epidermal/dermal boundaries via localization of laminin and actin.
A) Epidermal/dermal lamellar boundary is visualized by immunofluorescent staining against the basement membrane marker laminin in red. Autofluorescent material (putatively collagen) in green, nuclei stained blue; B) Epidermal cellular boundaries visualized with staining against actin in green, nuclei stained blue. The images are representative of samples from 6 animals analyzed. Scale bars 50μm.
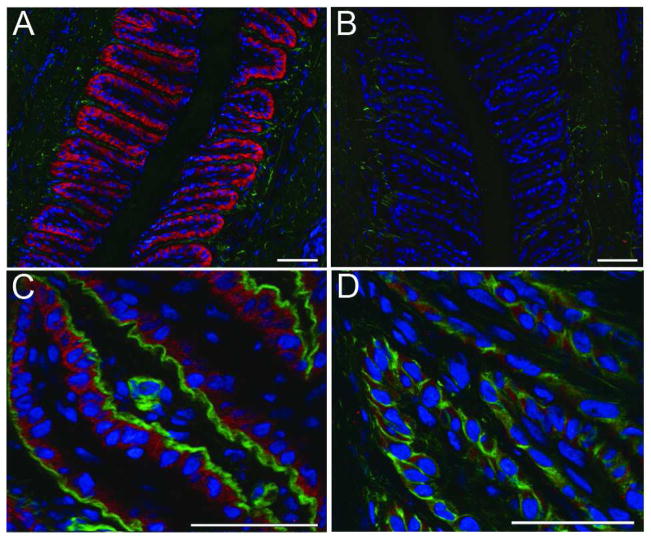
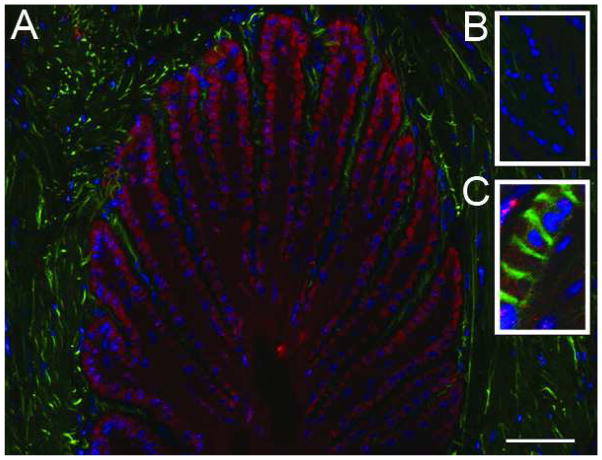
Figure 5. Versican uniquely localizes to the basal epithelia of the secondary epidermal lamellae and is not associated with the basement membrane.
A) Immunofluorescent staining against versican in red, autofluorescent material in green, nuclei stained blue; B) versican staining blocked by pre-incubation of primary antibody with cognate peptide, autofluorescent material in green, nuclei stained blue; C) versican staining in red, basement membrane visualized by staining against laminin in green, nuclei stained blue; D) versican staining in red, epithelial cell boundaries visualized by staining against actin in green, nuclei stained blue. The images are representative of samples from 6 animal analyzed. Scale bars 50μm.
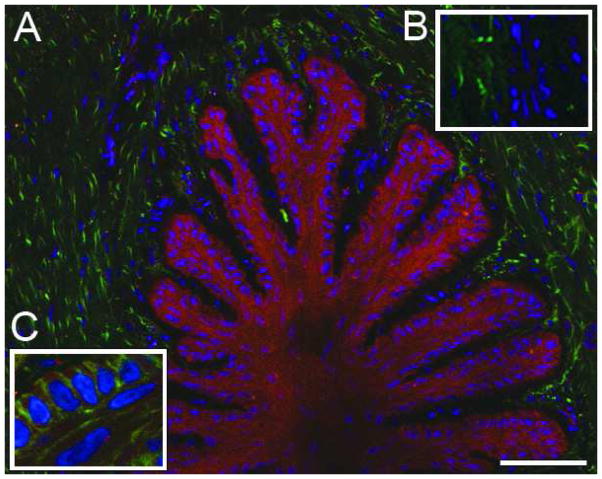
Figure 6. Aggrecan localizes primarily to the secondary epidermal lamellae.
A) Immunofluorescent staining against aggrecan in red, autofluorescent material in green, nuclei stained blue; B) aggrecan staining was blocked by preincubation of the primary antibody with cognate peptide, autofluorescent material in green, nuclei stained blue; C) aggrecan staining in red, epithelial cell boundaries visualized by staining against actin in green, nuclei stained blue. The images are representative of samples from 6 animals analyzed. Scale bars 50μm.
Figure 7. Hyaluronan is present throughout lamellae but enriched in the secondary epidermal lamellae.
A) Immunofluorescent staining against hyaluronan in red, autofluorescent material in green, nuclei stained blue; B) hyaluronan staining abrogated by incubation of tissue section with epitope-digesting enzyme (Chondroitinase ABC pH 6.8), autofluorescent material in green, nuclei stained blue; C) hyaluronan staining in red, epithelial cell boundaries visualized by staining against actin in green, nuclei stained blue. The images are representative of samples from 6 animals analyzed. Scale bars 50μm.
General Structure - ii) Cortical actin
A representative thin transverse section of frozen digital laminae stained with FITC-phalloidin (green) to detect actin bundles and DAPI (blue) to detect DNA in nuclei is shown in Figure 4B. The FITC-phalloidin is most densely associated with the cortical actin skeleton of basal epithelial cells (the outermost cell layer of the secondary epidermal laminae; Fig 4B), but can also be seen defining the cortical actin skeleton of suprabasal cells extending into the primary epidermal laminae (Fig 4B). Note that DAPI (blue) stained nuclei are faded or absent from keratinocytes in the primary epidermal laminae. The contiguous keratinocyte cortical actin network evident throughout the secondary epidermal laminae indicates that there is little room for extracellular matrix in this tissue. The phalloidin-FITC stain is bright and very little exposure time was required to capture the image shown in Figure 4B, accounting for the almost complete absence in the captured image of the green autofluorescent materials seen in Fig 4A.
Versican
A representative thin transverse section of frozen digital laminae stained with antibody specific for the carboxy-terminal domain of all versican isoforms revealed with a secondary antibody conjugated with a proprietary red dye, and with the DNA intercalating dye DAPI (blue), is shown in Figure 5A. It is notable that versican staining is restricted to a single layer of cells, putatively the basal epithelial cells. Specific staining of versican was inhibited by pre-incubation of the antibody with its competing peptide (Fig 5B) but not with an antisense peptide (not shown).
Identification of the versican-stained (red) cell layer as basal epithelial cells was confirmed by showing that the cells abut the basement membrane. This was achieved by staining a thin section of frozen laminae with: i) pan-anti-laminin antibody revealed here by a secondary antibody conjugated with a proprietary green fluorescent dye, ii) anti-versican antibody revealed by red fluorescent dye -conjugated secondary antibody and iii) with DAPI which stains nuclei blue. Results are presented in Figure 5C. The versican (red) does not actually contact (co-localize with) basement membrane laminin (green), which would have given a yellow merged image. To test whether versican is contained with basal epithelial cells, a thin cryosection was co-stained with anti-versican antibody which was detected using a texas red-conjugated secondary antibody (red) and the section was also stained with FITC-phalloidin to detect actin bundles (green) and with DAPI (blue) to detect cell nuclei. Results presented in Figure 5D show that versican (red) is located between the cortical actin of basal epithelial cells (green) and the cell nuclei (blue). In addition, it is clear that versican staining is not pronounced along cell boundaries suggesting little or no accumulation of the proteoglycan in extracellular space.
Aggrecan
A representative thin transverse section of frozen digital laminae was stained with antibody that reacted with an epitope on the G2 domain of aggrecan revealed with a proprietary red fluorescent dye-conjugated secondary antibody, and co-stained with DAPI (blue) (Figure 6A). Aggrecan staining (red) was greatest in basal epithelial cells but was also detected in regions of the epidermal laminae occupied by supra-basal epithelial cells. Aggrecan was largely, if not entirely, absent from primary and secondary dermal laminae. Autofluorescent material (putatively collagen; green) is readily seen in this image. Aggrecan staining was inhibited by pre-incubation of the antibody with its competing peptide (Fig 6, inset B). To further identify the distribution of aggrecan in laminae, a transverse thin frozen section was co-stained with aggrecan specific antibody (red), FITC-phalloidin (green) to detect actin bundles and DAPI (blue) to detect nuclei. Aggrecan staining (red) was present within punctuate bodies contained within boundaries defined by phalloidin stained cortical actin (green) (Fig 6, inset C). A similar tissue distribution was seen in immunohistological sections stained with the antibody against the “ARGSVIL” epitope of aggrecan (data not shown).
Hyaluronan
A representative thin transverse section of frozen digital laminae was stained with antibody specific for hyaluronan polysaccharide revealed with texas red-conjugated secondary antibody (red), and co-stained with DAPI (blue). Results are shown in Figure 7A which shows that hyaluronan (red) is diffusely present in the epidermal laminae and is not pronounced along cell boundaries (Fig 7A). Staining was absent from tissue sections that had been pre-treated with hyaluronidase (Fig 7, inset B). Hyaluronan staining within the secondary epidermal laminae was contained within cell boundaries defined by phalloidin stained cortical actin (green) (Fig 7 inset C).
ADAMTS-4
Thin transverse sections of frozen digital laminae were stained with an antibody specific to the peptide “NTPEDSDPDHFD” corresponding to amino acids 300–311 of the metalloprotease domain of equine ADAMTS-4. ADAMTS-4 staining was revealed with a red fluorescent dye-conjugated secondary antibody and nuclei counterstained with DAPI (blue). Antibody specificity was confirmed by western blotting, which revealed similar band patterns to those obtained using the “FASLSRFVET” ADAMTS-4 antibody (shown in Fig 3a) and by inhibition of Western blotting with sense but not antisense peptide (data not shown).
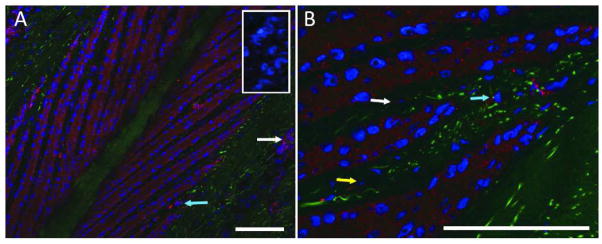
Staining was diffusely present throughout the epithelial cells of the secondary epidermal lamellae (Figs 8A and B), and was abrogated by pre-incubation of the antibody with its immunizing peptide (Fig 8A, inset) but not with a non-competing peptide (data not shown). Punctate staining was also present in cells associated with the vasculature (Fig 8A, white arrow), mononuclear cells of the dermal lamellae (Figs 8A and B, blue arrows), and dermal fibroblasts (Fig 8B, yellow arrows).
Figure 8. ADAMTS-4 is present in epithelial cells of the secondary epidermal lamellae.
A) Immunofluorescent staining against ADAMTS-4 “NTPEDSDPDHFD” epitope in red, autofluorescent material in green, nuclei stained blue. Staining blocked by cognate peptide in inset. B) Higher magnification image, ADAMTS-4 visualized in red, autofluorescent material in green, nuclei in blue. Images representative of samples from 6 animals analyzed. Scale bars 50μm. White arrows indicate vascular endothelia, blue arrows indicate mononuclear cells of the dermal lamellae, yellow arrow indicates a dermal fibroblast.
Discussion
Using RT-qPCR and validated primers we show that equine digital laminae express genes encoding ADAMTS-4, aggrecan, versican (processed to V0, V1 and V2 splice isoforms) and hyaluronan synthase II. Using SDS-PAGE followed by Western blotting with antibodies specific for conserved peptides in horse and human ADAMTS-4, aggrecan and versican, and validated by peptide competition, we confirmed that gene expression is accompanied by protein expression and that the core proteins of the large polysulfated proteoglycans are subject to constitutive ADAMTS-4 cleavage in vivo. Using immunofluorescent and immunohistochemical staining of thin sections of laminae with specific antibodies validated by peptide competition or targeted epitope digestion, we show that aggrecan, versican and hyaluronan are predominantly present within basal epithelial cells of the secondary epidermal laminae, while ADAMTS-4 is present within these cells and also in cells of the dermal laminae.
In all laminar extracts, ADAMTS-4 was present predominantly as a ~51 kDa form bearing the conserved FASLSRFVET neoepitope, which is exposed upon removal of regulatory propeptide by furin propeptide convertase in the trans golgi27. A minor ~61 kDa form bearing the neoepitope was also detected in all samples but staining was too weak to be visible in the images presented. In addition, a ~42kDa form of ADAMTS-4 bearing the FASLSRFVET neoepitope was detected in extracts from 75% of samples tested. Processed ADAMTS-4 bearing the FASLSRFVET neoepitope has a MW between ~68 kDa and 70 kDa in mice and humans and a portion of the material undergoes autoproteolysis to generate ~61 kDa, 51 kDa and 40 kDa polypeptides17,29,30 corresponding to the ~61 kDa, ~51 kDa, ~42 kDa laminae material reported here. The absence of the ~68 kDa– 70 kDa enzyme from extracts of laminae suggests that autoproteolytic activity of equine ADAMTS-4 may be greater than that of murine and human ADAMTS-4.
In addition to cleaving the large polysulfated proteoglycans, autoproteolysed ADAMTS-4 has been shown to cleave low molecular weight leucine rich proteoglycans which bind collagens and fibronectin, affecting fiber formation and network organization30,31. Thus, in the equine digital laminae, ADAMTS-4 may cause the re-organization of a broad range of ECM components. This increased cleavage capability may have implications for the development of laminitis.
Aggrecan is a highly glycosylated multidomain protein16. The core protein of ~220 kDa can be decorated with up to 100 chondroitin sulfate side chains each of ~20 kDa positioned between globular domains 2 and 3. Aggrecan can also be decorated with up to 60 keratin sulfate side chains each of ~5 kDa to ~15 kDa which are more widely distributed on the molecule than the chondroitin sulfate glycosaminoglycans. In addition, a variable number of O- and N-linked oligosaccharide side chains can also be added. ADAMTS-4 cleaves aggrecan between the G1 and G2 domains at the Glu373-Ala374 bond yielding a large glycosaminoglycan rich fragment with an N terminal ARGSVI cleavage neoepitope and a G1 fragment with a NITEGE neoepitope. Antibodies specific for ARGSVIL detected polypeptides of ~318 kDa, ~250 kDa, ~150 kDa and ~70 kDa, present in guanidine hydrochloride extracts of laminae that were further subjected to hyaluronidase/chondroitinase ABC digestion. The largest fragments most likely correspond to N or O glycosylated material. This presence of polypeptides bearing the ADAMTS-4 cleavage neoepitope in lamina extracts indicates that endogenous ADAMTS-4 is active in the laminae.
Cleavage of aggrecan between Glu373-Ala374 by ADAMTS-4 and ADAMTS-5 in cartilage allows large glycosaminoglycan rich fragments, which are no longer anchored to hyaluronan, to diffuse into synovial fluid32. However, constitutive cleavage of aggrecan in the laminae did not allow the ARGSVI positive glycosaminoglycan-rich fragments to easily detach from the tissue, inferred from our inability to extract the fragments into NP-40 homogenization buffer. Rather, overnight incubation of pulverized laminae in a 4M guanidine hydrochloride extraction buffer was required to extract the cleavage fragments suggesting that they are held in the lamellar tissue by interactions with other as yet unidentified molecules.
NP-40 and direct SDS sample buffer extracts of pulverized laminae contain V0/V1 versican fragments of ~109 kDa and ~66 kDa bearing the C terminal β GAG domain ADAMTS-4 cleavage neoepitope DPEAAE. The MWs of these fragments are unaffected by digestion with hyaluronidase or chondroitinase ABC and hence the fragments do not have attached hyaluronan or chondroitin sulfate glycosaminoglycans. It is likely that the ~66 kDa fragment described here is equivalent to the ~70 kDA DPEAAE positive G1 domain of versican V1 reported to be present in ADAMTS-4 treated human aorta33. The extracts of laminae also contained ~112 kDa and ~68 kDa V0/V2 versican fragments bearing the C terminal ADAMTS-4 α GAG domain cleavage neoepitope NIVSFE. ADAMTS-4 cleavage of fully deglycosylated human aorta V0/V2 versican has been shown to yield an NIVSFE positive fragment of ~64 kDa which is equivalent to the glial hyaluronate binding protein of human brain V2 versican34. Although we digested extracts of laminae with chondroitinase ABC we did not digest with sialidase or O-glycanase perhaps accounting for the larger sized (~68 kDa) putative versican G1- α GAG domain fragment detected in our studies. The ~112 kDa material is however too large to be comprised solely of the ADAMTS-4 cleaved versican G1 and α GAG core protein domains, suggesting that it may be heavily N-glycosylated. The ~112 kDa and ~68 kDa polypeptides bearing the NIVSFE neoepitope were also detected (data not shown) by antibodies raised against the N-terminal 13-residue peptide sequence of human versican (LF-9935, a gift from Dr. Larry Fisher, NIH) which is 87% homologous to matched equine sequence further supporting their identify as fragments of versican.
Aggrecan and versican have been extensively studied in articular cartilage, tendons, and atherosclerotic plaque where they associate with hyaluronan in the extracellular matrix (ECM)12,16,36. Hyaluronan is a non-sulfated glycosaminoglycan that is synthesized at the plasma membrane by one of three distinct hyaluronan synthases37 and extruded into the extracellular space. Hyaluronan synthase II, which was shown here to be expressed in the laminae, makes the longest hyaluronan chains, which can be larger than 2000 kDa. Hyaluronan binds a large number of aggrecan monomers with binding stabilized by link protein. These complexes can reach several hundred millions of Daltons13. The large negatively charged complexes of aggrecan and hyaluronan attract and hold water in tissues, forming a hydrated gel. This is packaged in collagen fibers in articular cartilage and tendon, accounting for the high resistance of these tissues to compression deformation16. Versican is also a hyaluronan-binding proteoglycan. It is expressed in fast growing cells of many tissues including the skin, the media of the aorta and in developing chicken limb buds38, where it is implicated in regulating cell proliferation and migration as well as in expanding the ECM and increasing its viscoelasticity. Given these data, it was expected that aggrecan, versican and hyaluronan in the digital laminae would also be associated with ECM as well as present in producer cells. However, the materials were not observed to be present in the ECM. Furthermore the secondary epidermal laminae had little to no discernable ECM.
Aggrecan was detected in punctuate bodies contained within cortical actin boundaries throughout the basal and suprabasal epithelial cells of the secondary epidermal laminae. Hyaluronan staining was also present in punctuate bodies contained within cortical actin boundaries throughout the secondary epidermal laminae. In contrast, versican staining was restricted to basal epithelial cells only. Versican was not detected in any suprabasal epithelial cells in the secondary epidermal laminae. Thus, our studies suggest that versican, aggrecan and hyaluronan may be useful markers for defining specialization within epidermal keratinocytes.
In addition to aggrecan, versican and hyaluronan, basal epithelial cells also contained ADAMTS-4. Because versican fragments bearing an ADAMTS-4 neoepitope were readily extracted from laminae into SDS sample buffer, which prevents post extraction processing, it can be concluded that a portion if not all versican and ADAMTS-4 share an intracellular compartment that permits ADAMTS-4 activity. Furthermore, because aggrecan fragments bearing an ADAMTS-4 neoepitope could also be extracted from laminae, it is likely that at least a portion of aggrecan and ADAMTS-4 also share an intracellular compartment that permits ADAMTS-4 activity. Thus, ADAMTS-4 contributes to processing, and most likely turnover of aggrecan and versican within laminar basal epithelial cells.
The biological roles of aggrecan, versican, hyaluronan and ADAMTS-4 in equine digital laminae are not directly illuminated by the above studies. However, based on known compression-resistance and cell signaling properties of the large polysulfated proteoglycans it is reasonable to propose that the proteoglycans affect development and maintenance of the basal epithelial cell layer, and may cushion basal epithelial cells against the severe biomechanical stresses associated with their anatomical location.
Acknowledgments
The study was supported by D08EQ-054 from the Morris Animal Foundation, by 2007-35204-18313 from USDA NRI CSREES and by MAS00907 from USDA CSREES to SJB. D. Alfandari The authors thank Drs. Tortorella and Malfait for providing antibody specific for the ADAMTS-4 active site cleavage neoepitope FASLSRFVET, Dr. Andria Cogswell for co-ordinating was supported by NIH DE016289, Erica Pawlak was supported by a Lotta M. Crabtree Fellowship in Agriculture and. Almaz Taye was supported by a fellowship from the Northeast Alliance for Graduate Education and the Professoriate. collection of laminae, Ms. Aileen Thomas for rabbit immunizations and Dr. Hannah Galantino-Homer for helpful discussion.
Abbreviations
- ADAMTS-4
A Disintegrin And Metalloproteinase with ThromboSpondin motifs-4
- BSA
bovine serum albumin
- DAPI
4′,6-diamidino-2-phenylindole
- ECM
extracellular matrix
- FITC
fluorescein isothiocyanate
- GAPDH
Glyceraldehyde 3-phosphate dehydrogenase
- GAG
glycosaminoglycan, also refers to glycosaminoglycan domain of proteins e.g., α GAG and β GAG
- HRP
horse radish peroxidase
- IgG H&L
heavy and light chains of immunoglobulin of the IgG class
- KLH
keyhole limpet hemocyanin
- NP-40
Tergitol-type NP-40, which is nonyl phenoxylpolyethoxylethanol
- PCR
polymerase chain reaction
- PBS
phosphate buffered saline pH 7.2
- SDS-PAGE
sodium dodecylsulfate polyacrylamide gel electrophoresis
- RT-qPCR
real-time quantitative polymerase chain reaction
Footnotes
Tissue-Tek O.C.T.; Sakura Finetek USA, Inc., Torrance, CA
Freund’s adjuvant, complete;
Freund’s adjuvant, incomplete;
Sigma Fast Protease Inhibitor Cocktail;
Phalloidin-FITC;
Chondroitinase ABC; Sigma-Aldrich, St. Louis, MO
GenScript USA, Inc., Piscataway, NJ
NHS-Activated Sepharose Fast Flow Beads, GE Healthcare Biosciences, Pittsburgh, PA
NanoDrop 1000; Thermo Scientific, Wilmington, DE.
Stratagene Absolutely RNA kit;
Stratagene MX 3005p; Stratagene, La Jolla, CA.
Biospec Products, Inc., Bartlesville, OK
SYBRSafe DNA Gel Stain; Molecular Probes, Eugene, OR.
Quanta q Script cDNA Synthesis Kit; Quanta BioSciences, Gaithersburg, MD
SYBR Premix Ex Taq; Applied Biosystems, Foster City, CA.
PTC-100PCR System; MJ Research Inc., Waltham, MA
Bradford Assay;
Laemmli Reducing Sample Buffer;
E.C.L; Bio Rad Life Sciences, Hercules, CA
Calbiochem, EMD, Merck KGaA, Darmstadt, Germany
Seikagaku, Tokyo, Japan.
G : Box ;
Gene Tools ; Syngene, Frederick, MD
Fisher Superfrost Plus ; Fisher Scientific, Fair Lawn, NJ
Zeiss MOT200 with Zeiss Apotome ; Carl Zeiss MicroImaging, Inc., Thornwood, NY
#ab3773;
#ab6808;
#ab19345;
#ab6795;
#ab8226;
#ab28671;
#ab11575;
#ab6719;
#ab16320 (no longer available);
#ab53842;
#ab6745 AbCam, Cambridge, MA
#AB1032;
#AB1033; Millipore, Billerica, MA
#711-485-152;
#705-295-147;
#711-515-152 ; Jackson ImmunoResearch Laboratories, Inc., West Grove, PA
#26706; Santa Cruz Biotechnologies, Santa Cruz, CA
Presented in part at the American Association of Equine Practitioners Foundation Equine Laminitis Workshop, West Palm Beach, Nov. 2009 and at the Conference for Research Workers in Animal Diseases, Chicago, Nov. 2010.
References
- 1.Pollitt CC. The Anatomy and Physiology of the Suspensory Apparatus of the Distal Phalanx. Veterinary Clinics of North America: Equine Practice. 2010;26:29–49. doi: 10.1016/j.cveq.2010.01.005. Advances in Laminitis, Part I. [DOI] [PubMed] [Google Scholar]
- 2.French KR, Pollitt CC. Equine laminitis: loss of hemidesmosomes in hoof secondary epidermal lamellae correlates to dose in an oligofructose induction model: an ultrastructural study. Equine Vet J. 2004;36:230–235. doi: 10.2746/0425164044877125. [DOI] [PubMed] [Google Scholar]
- 3.Black SJ. Extracellular matrix, leukocyte migration and laminitis. Vet Immunol Immunopathol. 2009;129:161–163. doi: 10.1016/j.vetimm.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 4.Alonso L, Fuchs E. Stem cells of the skin epithelium. Proc Natl Acad Sci U S A. 2003;100 (Suppl 1):11830–11835. doi: 10.1073/pnas.1734203100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Budras KD, Hullinger RL, Sack WO. Light and electron microscopy of keratinization in the laminar epidermis of the equine hoof with reference to laminitis. Am J Vet Res. 1989;50:1150–1160. [PubMed] [Google Scholar]
- 6.Bertram JE, Gosline JM. Functional design of horse hoof keratin: the modulation of mechanical properties through hydration effects. J Exp Biol. 1987;130:121–136. doi: 10.1242/jeb.130.1.121. [DOI] [PubMed] [Google Scholar]
- 7.Kasapi MA, Gosline JM. Micromechanics of the equine hoof wall: optimizing crack control and material stiffness through modulation of the properties of keratin. J Exp Biol. 1999;202:377–391. doi: 10.1242/jeb.202.4.377. [DOI] [PubMed] [Google Scholar]
- 8.Willemen MA, Jacobs MW, Schamhardt HC. In vitro transmission and attenuation of impact vibrations in the distal forelimb. Equine Vet J Suppl. 1999;30:245–248. doi: 10.1111/j.2042-3306.1999.tb05227.x. [DOI] [PubMed] [Google Scholar]
- 9.Buschmann MD, Kim YJ, Wong M, et al. Stimulation of aggrecan synthesis in cartilage explants by cyclic loading is localized to regions of high interstitial fluid flow. Arch Biochem Biophys. 1999;366:1–7. doi: 10.1006/abbi.1999.1197. [DOI] [PubMed] [Google Scholar]
- 10.Momberger TS, Levick JR, Mason RM. Mechanosensitive synoviocytes: a Ca2+-PKCalpha-MAP kinase pathway contributes to stretch induced hyaluronan synthesis in vitro. Matrix Biol. 2006;25:306–316. doi: 10.1016/j.matbio.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 11.Coyne MJ, Cousin H, Loftus JP, et al. Cloning and expression of ADAM-related metalloproteases in equine laminitis. Vet Immunol Immunopathol. 2009;129:231–241. doi: 10.1016/j.vetimm.2008.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salter RC, Ashlin TG, Kwan AP, et al. ADAMTS proteases: key roles in atherosclerosis? J Molec Med. 2010;88:1203–1211. doi: 10.1007/s00109-010-0654-x. [DOI] [PubMed] [Google Scholar]
- 13.Kiani C, Chen L, Wu YJ, et al. Structure and function of aggrecan. Cell Res. 2002;12:19–32. doi: 10.1038/sj.cr.7290106. [DOI] [PubMed] [Google Scholar]
- 14.Kenagy RD, Plaas AH, Wight TN. Versican degradation and vascular disease. Trends Cardiovasc Med. 2006;12:92–101. doi: 10.1016/j.tcm.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heinegard D. Proteoglycans and more – from molecules to biology. Int J Exp Pathol. 2009;90:575–586. doi: 10.1111/j.1365-2613.2009.00695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roughley PJ. The structure and function of cartilage proteoglycans. Eur Cell Mater. 2006;12:92–101. doi: 10.22203/ecm.v012a11. [DOI] [PubMed] [Google Scholar]
- 17.Miwa HE, Gerken TA, Huynh TD, et al. Mammalian expression of full-length bovine aggrecan and link protein: formation of recombinant proteoglycan aggregates and analysis of proteolytic cleavage by ADAMTS-4 and MMP-13. Biochim Biophys Acta. 2006;1760:472–486. doi: 10.1016/j.bbagen.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 18.Johnson PJ, Kreeger JM, Keeler M, et al. Serum markers of lamellar basement membrane degradation and lamellar histopathological changes in horses affected with laminitis. Equine Vet J. 2000;32:462–468. doi: 10.2746/042516400777584695. [DOI] [PubMed] [Google Scholar]
- 19.Loftus JP, Johnson PJ, Belknap JK, et al. Leukocyte-derived and endogenous matrix metalloproteinases in the lamellae of horses with naturally acquired and experimentally induced laminitis. Vet Immunol Immunopathol. 2009;129:221–230. doi: 10.1016/j.vetimm.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 20.Soma T, Tajima M, Kishimoto J. Hair cycle-specific expression of versican in human hair follicles. J Dermatol Sci. 2005;39:147–154. doi: 10.1016/j.jdermsci.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 21.Wyckoff M, Rodbard D, Chrambach A. Polyacrylamide gel electrophoresis in sodium dodecyl sulfate-containing buffers using multiphasic buffer systems: properties of the stack, valid Rf-measurement, and optimized procedure. Anal Biochem. 1977;78:459–482. doi: 10.1016/0003-2697(77)90107-5. [DOI] [PubMed] [Google Scholar]
- 22.Hayes A, Hughes CE, Caterson B. Antibodies and immunohistochemistry in extracellular matrix research. Methods. 2008;45:10–21. doi: 10.1016/j.ymeth.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 23.Nuovo GJ, Elton TS, Nana-Sinkam P, et al. A methodology for the combine in situ analysis of the precursor and mature forms of microRNAs and correlation with their putative targets. Nat Protoc. 2009;4:107–115. doi: 10.1038/nprot.2008.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu YJ, La Pierre DP, Wu J, et al. The interaction of versican with its binding partners. Cell Res. 2005;15:483–494. doi: 10.1038/sj.cr.7290318. [DOI] [PubMed] [Google Scholar]
- 25.Rahmani M, Wong BW, Ang L, et al. Versican: signaling to transcriptional control pathways. Can J Physiol Pharmacol. 2006;84:77–92. doi: 10.1139/y05-154. [DOI] [PubMed] [Google Scholar]
- 26.Tortorella MD, Arner EC, Hills R, et al. ADAMTS-4 (aggrecanase-1): N-terminal activation mechanisms. Arch Biochem Biophys. 2005;444:34–44. doi: 10.1016/j.abb.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 27.Wang P, Tortorella M, England K, et al. Proprotein convertase furin interacts with and cleaves pro-ADAMTS4 (Aggrecanase-1) in the trans-Golgi network. J Biol Chem. 2004;279:15434–15440. doi: 10.1074/jbc.M312797200. [DOI] [PubMed] [Google Scholar]
- 28.Malfait AM, Arner EC, Song RH, et al. Proprotein convertase activation of aggrecanases in cartilage in situ. Arch Biochem Biophys. 2008;478:43–51. doi: 10.1016/j.abb.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 29.Gao G, Westling J, Thompson VP, et al. Activation of the proteolytic activity of ADAMTS4 (aggrecanase-1) by C-terminal truncation. J Biol Chem. 2002;277:11034–11041. doi: 10.1074/jbc.M107443200. [DOI] [PubMed] [Google Scholar]
- 30.Kashiwagi M, Enghild JJ, Gendron C, et al. Altered proteolytic activities of ADAMTS-4 expressed by C-terminal processing. J Biol Chem. 2004;279:10109–10119. doi: 10.1074/jbc.M312123200. [DOI] [PubMed] [Google Scholar]
- 31.Gendron C, Kashiwagi M, Lim NH, et al. Proteolytic activites of human ADAMTS-5: comparative studies with ADAMTS-4. J Biol Chem. 2007;282:18294–18306. doi: 10.1074/jbc.M701523200. [DOI] [PubMed] [Google Scholar]
- 32.Sandy JD, Verscharen C. analysis of aggrecan in human knee cartilage and synovial fluid indicates that aggrecanase (ADAMTS) activity is responsible for the catabolic turnover and loss of whole aggrecan whereas other protease activity is required for C-terminal processing in vivo. Biochem J. 2001;358:615–626. doi: 10.1042/0264-6021:3580615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sandy JD, Westling J, Kenagy RD, et al. Versican V1 proteolysis in human aorta in vivo occurs at the Glu441-Ala442 bond, a site that is cleaved by recombinant ADAMTS-1 and ADAMTS-4. J Biol Chem. 2001;276:13372–13378. doi: 10.1074/jbc.M009737200. [DOI] [PubMed] [Google Scholar]
- 34.Westling J, Gottschall PE, Thompson VP, et al. ADAMTS4 (aggrecanase-1) cleaves human brain versican V2 at Glu405-Gln406 to generate glial hyaluronate binding protein. Biochem J. 2004;377:787–795. doi: 10.1042/BJ20030896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fisher LW, Stubbs JT, 3rd, Young MF. Antisera and cDNA probes to human and certain animal model bone matrix noncollagenous proteins. Acta Orthop Scand Suppl. 1995;266:61–65. [PubMed] [Google Scholar]
- 36.Yoon JH, Halper J. Tendon proteoglycans: biochemistry and function. J Musculoskelet Neuronal Interact. 2005;5:22–34. [PubMed] [Google Scholar]
- 37.Itano N, Kimata K. Mammalian hyaluronan synthases. IUBMB Life. 2002;54:195–199. doi: 10.1080/15216540214929. [DOI] [PubMed] [Google Scholar]
- 38.Wight TN. Versican: a versatile extracellular matrix proteoglycan in cell biology. Curr Opin Cell Biol. 2002;14:617–623. doi: 10.1016/s0955-0674(02)00375-7. [DOI] [PubMed] [Google Scholar]