Abstract
The aquaporin family of channels was defined based on the inhibition of water transport by mercurial compounds. Despite the important role of mercurials, little is known about the structural changes involved upon mercury binding leading to channel inhibition. To elucidate the mechanism we designed a mutant, T183C, of aquaporin Z (AqpZ) patterned after the known mercury-sensitive site of aquaporin 1 (AQP1) and determined the x-ray crystal structures of the unbound and mercury blocked states. Superposition of the two structures shows no conformational rearrangement upon mercury binding. In the blocked structure, there are two mercury sites – one bound to Cys183 and occluding the pore, and a second, also bound to the same cysteine but found buried in an interstitial cavity. To test the mechanism of blockade we designed a different mutant, L170C, to produce a more effective mercury block at the pore site. In a dose-response inhibition study, this mutant was 20 times more sensitive to mercury than wild-type AqpZ and 4 times more sensitive than T183C. The x-ray structure of L170C shows four mercury atoms at, or near, the pore site defined in the T183C structure and no structural change upon mercury binding. Thus, we elucidate a steric inhibition mechanism for this important class of channels by mercury.
Keywords: aquaporin, integral membrane protein, x-ray crystallography, membrane protein structure
Introduction
Aquaporins (AQPs) are integral membrane channel proteins that mediate the bi-directional flux of water and selected small amphipathic molecules across cellular membranes.1 In the field of channel biology, selective inhibitors including naturally occurring toxins and organic molecules have played key roles in defining function2, but AQPs have no such specialized inhibitors. Mercurial compounds were found to reduce water transport in the red blood cell membrane to that of a bilayer and so defined the presence of a cellular water channel.3 This selective inhibition subsequently allowed for aquaporin isolation4, cloning5; 6, membrane transport characterization7, and mercury sensitivity mutational analysis.8; 9 The precise mechanism of AQP inhibition by mercury is still undetermined.
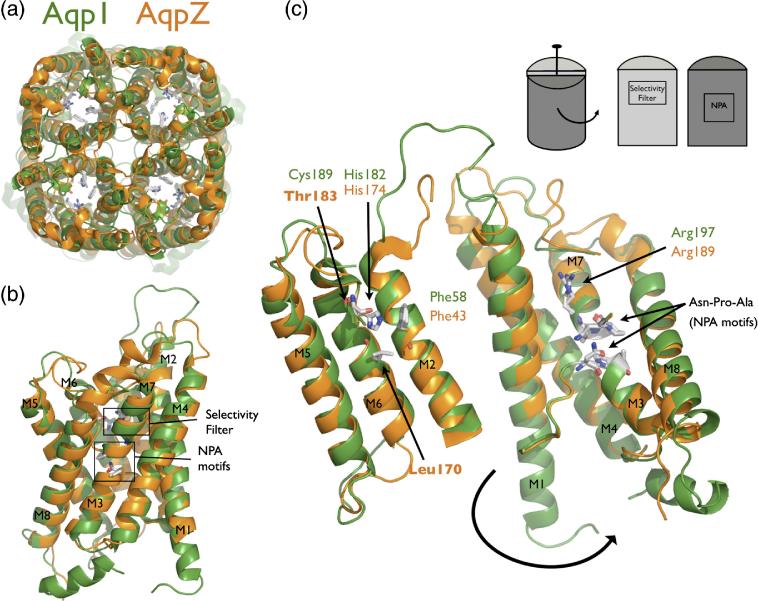
The AQP fold is a right-handed bundle of six transmembrane (TM) helices and two half-spanning helices (named M1-M8) with the pore running through the center of the helical bundle. There is a quasi two-fold symmetric relationship between the N-terminal portion of the polypeptide chain (M1-M4) and the C-terminal portion (M5-M8) from early gene duplication. In the membrane, AQPs occur as homotetramers of four monomer channels related by a four-fold symmetry axis (Figure 1A). The AQP family is divided into two subfamilies, the water selective channels and those with a more promiscuous selectivity for both water and other small amphipathic molecules such as glycerol (aquaglyceroporins).13 A comparison of GlpF, AQP1, and the other recently determined structures, shows that the AQP fold is conserved.14,15,16,17 AQPs are identified by two Asparagine-Proline-Alanine (NPA) sequence motifs located at the ends of the two quasi two-fold related half-spanning helices M3 and M7 (Figure 1B). The selectivity filter, a constricted region formed by four residues near the periplasmic/extracellular entrance, provides distinguishing features that identify the subfamilies (Figure 1B and 1C). In water selective AQPs this region is smaller and more polar and contains a conserved histidine, while in aquaglyceroporins it is larger and more hydrophobic with two conserved aromatic residues 13. Thus, the AQP architecture is conserved and it is the pore side chains that modulate specific functional differences (Figure 1C).
Figure 1.
AqpZ is the bacterial homolog of AQP1. (A) Cartoon representation of the AqpZ (orange) and AQP1 (green) tetramers. Note the presence of the four monomer channels and the hypothetical channel down the tetramer axis. (B) Cartoon representation of the AqpZ and AQP1 monomers. Helices are labeled M1 through M8, and the selectivity filter and NPA motifs are designated with boxes. (C) Monomer opened up showing conservation of the water-selective motif. In this cartoon representation, the monomer is peeled open as shown in the inset schematic. The conserved selectivity filter and NPA motif are shown in sticks. Thr183 and Leu170 in AqpZ are the positions of cysteine mutants in this study. All molecular structure figures were made in Pymol (Delano Scientific).
Mercurials can bind non-selectively to accessible cysteines, but in AQPs, due to a decrease in solvent accessibility from the membrane bilayer, mercury typically binds selectively to residues associated with the pore. In the mutational analysis of AQP1, removal of endogenous cysteines identified Cys189 as the one responsible for mercury sensitivity and predicted its pore location. Confirmation of Cys189 as a pore residue came from the first atomic resolution AQP structure, that of the E. coli glycerol channel (GlpF)10, and the later AQP1 structure11; 12 showed precisely how the cysteine, not present in the GlpF sequence, was oriented. The structure of AQP1 was determined with protein obtained from natural sources so is not conveniently amenable to mutation and mercury was not used in the structural analysis. To understand the mechanism of mercury inhibition in AQPs we focused our efforts on the bacterial homolog of AQP1, AqpZ 18. AqpZ contains the water-selective sequence motif of AQP1 at the selectivity filter, has functionally been described as a water channel19, is not mercury sensitive, can be over-expressed and mutated, and its x-ray structure has previously been determined in our laboratory. AqpZ is thus a useful model system for probing the relationship between structure and function (Figure 1).14,20
Results
Structure of mutant T183C complexed with mercury
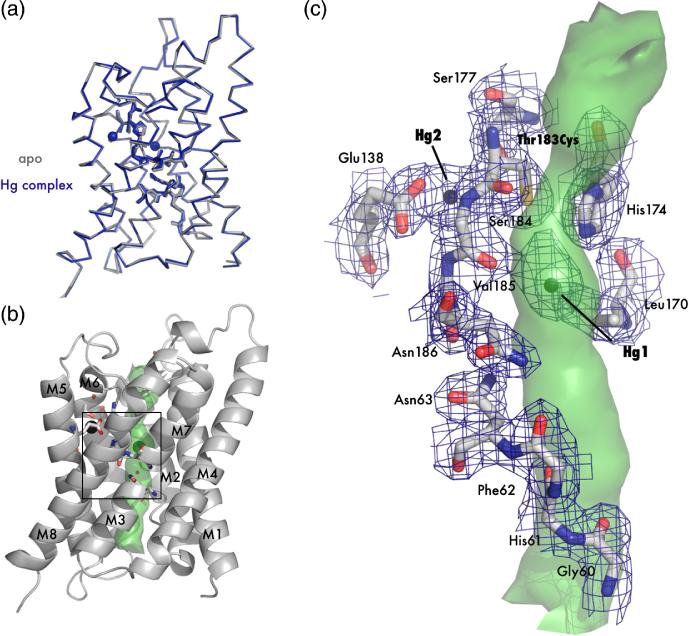
AqpZ and AQP1 are both functionally characterized as water-selective channels and have an identical selectivity filter except that AqpZ lacks the well-described mercury sensitive cysteine of AQP1 (Thr183 in AqpZ, Cys189 in AQP1). Based on this similarity, we postulated that a mutant of AqpZ lacking all endogenous cysteines but including the known mercury sensitive site of AQP1, would serve as a model for AQP1 (Figure 1C). The two endogenous cysteines of AqpZ were replaced with serine by mutagenesis, and the AQP1 mercury sensitive site was introduced via mutation Thr183Cys (protein hereby denoted T183C). This mutant, T183C, was expressed, purified, and crystallized in the presence or absence of HgCl2 to determine the structural implications of mercury binding. The crystals diffracted to atomic resolution, and the two structures were solved by molecular replacement using the previously published structure of WT AqpZ (Protein Data Bank (PDB) code 1RC2)14. The final resolution cutoffs were 2.30 Å and 2.20 Å, and the Rfree statistics for the refined structures were 23.8% and 24.2% for the apo and mercury-bound forms respectively (Table 1). The overall structures of both the apo and complexed forms display the canonical AQP fold (Figures 2A and 2B) 10. Superposition of the structures shows there is no significant conformational change between the two forms, and the root mean square deviation (RMSD) of the main chain α-carbons is 0.27 Å.
Table 1.
Crystallographic Data and Refinement Statistics
| T183C | T183C- Mercury |
L170C | L170C- Mercury |
|
|---|---|---|---|---|
| Data Collection | ||||
| Space Group | P4 | I4 | P4 | I4 |
| Unit Cell | ||||
| a (Å) | 92.4 | 91.1 | 91.3 | 91.2 |
| c (Å) | 78.2 | 77.9 | 77.5 | 77.1 |
| Resolution range (Å)a | 50-2.30 (2.30-2.36) |
50-2.20 (2.20-2.26) |
50-2.55 (2.55-2.62) |
50-1.90 (19.0-1.95) |
| Unique reflections | 28897 | 16199 | 15206 | 23096 |
| Completenessa | 98.3 (93.8) | 99.8 (100.0) | 74.1 (65.5) | 92.8 (66.4) |
| Rsymb(%)a | 7.3 (59.5) | 8.8 (59.5) | 7.5 (46.2) | 6.4 (33.4) |
| I / σ(I) a | 15.3 (1.1) | 15.5 (1.9) | 10.3 (1.2) | 15.4 (1.7) |
| Refinement Statistics | ||||
| Rwork / Rfree (%) | 19.7 / 23.8 | 19.3 / 24.2 | 23.6 / 28.0 | 16.6 / 19.5 |
| RMSD bonds (Å)c | 0.016 | 0.015 | 0.012 | 0.014 |
| RMSD angles (°)c | 1.65 | 1.64 | 1.45 | 1.61 |
| Number of protein atoms | 3356 | 1671 | 3368 | 1696 |
| Number of solvent atoms | 109 | 68 | 43 | 139 |
| Number of Hg | 0 | 3 | 0 | 4 |
| Average B-factor (Å2) | 37.7 | 36.4 | 24.9 | 23.2 |
|
| ||||
| PDB code | 209D | 209E | 209F | 209G |
values in parenthesis refer to the highest-resolution shell
∑|I–<I>|/∑I, where I equals observed intensity and <I> equals average intensity for symmetry-related reflections
Root-mean-square deviation of bond lengths and angles from ideal values
Figure 2.
Crystal structure of apo T183C and mercury bound T183C mutants. (A) Main chain overlay of the apo (gray) and Hg-complex (blue) with an RMSD (Cα) of 0.27 Å. Bound Hg2+ atoms are displayed as spheres with a van der Waals radius of 1.10 Å. (B) Cartoon representation of T183C. Transmembrane helices are labeled M1-M8 and the interior surface of the channel is drawn as a green surface. The black square denotes the area of interest depicted in panel C. (C) Structure of the blocked channel. Amino acids involved with water binding in AQPs are shown as sticks and with 2Fo-Fc electron density mapped contoured at 1.2σ drawn in blue. Mercury atoms are shown as spheres. In this orientation it can be seen that T183C-Hg1 sterically blocks the pore (green surface).
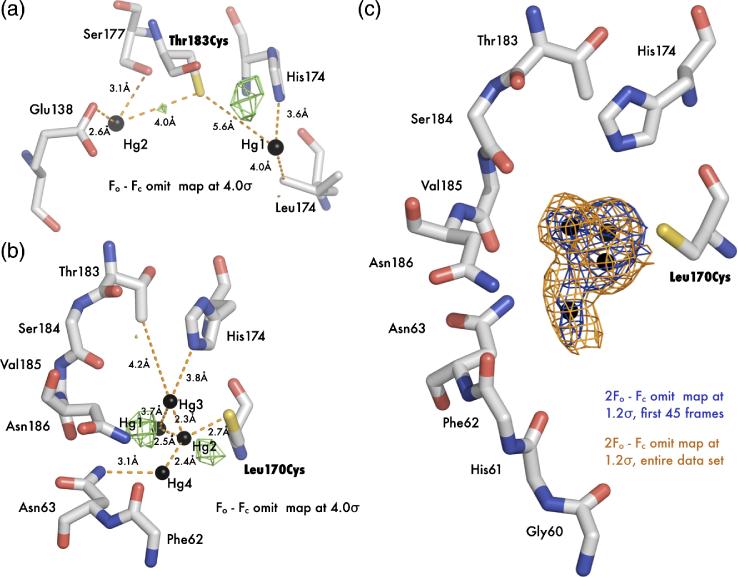
A calculation of the channel surface (Figure 2C) using van der Waals radii with the program HOLE221 reveals a 20Å long pore resembling the WT structure. The most striking feature of the complex structure is the two large electron density peaks of the mercury atoms and their unexpected location – one is located directly in the pore and one is interstitially bound in a cavity just outside the pore (Figure 2C). T183C-Hg1 (nomenclature is mutant followed by mercury atom number), the mercury in the pore, is located roughly halfway between the NPA region and the narrow selectivity filter. It makes favorable electrostatic contacts with the main chain carbonyl of S184 (3.3 Å) and the imidazole ring of His174 (3.7 Å). Somewhat surprisingly, T183C-Hg1 is approximately 5.6 Å away from Cys183 and is 3.9 Å to the closest water. T183C-Hg2, the mercury outside of the pore, is bound to Cys183 (distance of 4.0 Å) and resides in a hydrophilic pocket formed by conserved Glu138 and Ser177 where it makes favorable electrostatic interactions of 2.6 Å and 3.1 Å respectively.
The thiol-mercury bond distances are considerably longer than the ideal length of 2.5 Å and noise in difference maps suggests that both mercury atoms are disordered and at partial occupancy. We therefore carried out joint occupancy and anisotropic B-factor refinement in SHELX.22 In this calculation, T183C-Hg1 and T183C-Hg2 refine to occupancies of 0.24 and 0.32 respectively, suggesting a higher affinity or less disorder at the interstitial site. The anisotropic displacement parameters for T183C-Hg1 suggests disorder parallel to the channel axis, while T183C-Hg2 is disordered between residues Glu138 and Cys183. This disorder can also be observed in the 2Fo – Fc electron density (Figure 2C).
Based on these results, we hypothesized that the mercury site in the pore produces a steric block of the channel. To test this hypothesis we designed an optimized mercury-binding site in the pore, to provide a ‘switch’ as a probe of conductance in AqpZ. This new mutation, Leu170Cys in the cysteine-less background, was intended to bind mercury only at the site within the pore. We next biophysically characterized WT, T183C, and L170C to determine their relative mercury sensitivities.
Kinetics of Water Flux and Mercury Inhibition
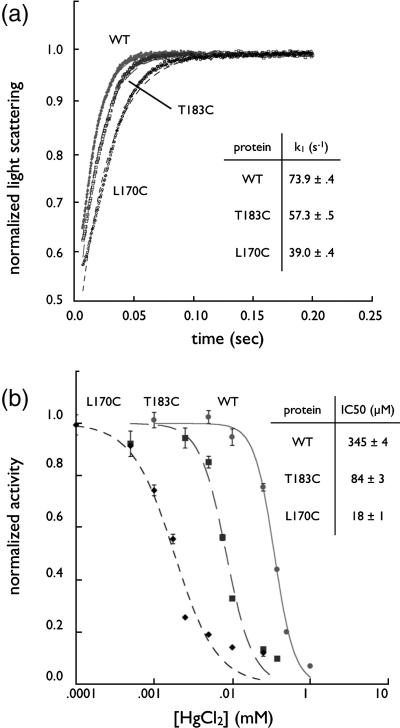
Flux through the AQP channel was assayed in osmotically-driven liposome permeability experiments19. AqpZ is a water-selective channel with high rates of conduction and we first sought to determine the activity of WT, T183C, and L170C. Purified proteins were reconstituted into liposomes and kinetics assayed in a stopped-flow device by mixing proteoliposomes with a hyperosmolar reconstitution buffer to drive water efflux. The resulting proteoliposome shrinkage was measured by light scattering and the curve fit to an exponential equation with a single rate constant. Raw light scattering data are provided in Figure 3A, along with the fitted curves. We measured the rates of WT, T183C, and L170C to be 73.9 ± 0.4 (standard deviation) s−1, 57.3 ± 0.5 s−1 and 39.0 ± 0.4 s−1 respectively, indicating that all proteins are functional water channels. Thus, WT has the fastest rates of conduction, T183C is slightly slower, and L170C much slower. Control liposomes without protein had rates of 4.6 ± 0.1 s−1.
Figure 3.
Kinetic Studies of Aquaporin Z. (A) Water conduction of WT, T170C, and T183C. Proteins were reconstituted in liposomes, challenged with a higher osmotic gradient in a stopped-flow device, and liposome shrinkage measured by light scattering at 440 nm. Plots were fit to a single exponential and the resulting rates are shown in the inset table (n=5). (B) Dose-response curve of proteoliposomes incubated with HgCl2. After incubation with HgCl2 proteoliposomes were assayed as in panel A and the rates (normalized to maximum rate for each mutant, n=5) were fit to a sigmoid dose-response curve in Kaleidagraph (Synergy Software). IC50 values are shown in the inset table.
In order to ascertain the role of HgCl2 as an inhibitor we determined the half maximal inhibitory concentration (IC50) using a dose-response relationship. In this experiment proteoliposomes were incubated with various concentrations of HgCl2 and then assayed for water conduction as described above. The results (Figure 3B) demonstrate that WT is the most sensitive to inhibition by mercury. The mutants, as expected, are inhibited at progressively lower concentrations with IC50s of 345μM, 84μM, and 18μM for WT, T183C and L170C respectively,19 and thus, L170C is the most sensitive to mercury. HgCl2 did not affect control liposomes and mercury inhibition was reversible with the addition of 5 mM 2-mercaptoethanol (BME) following mercury incubation (data not shown).
Structure of mutant L170C blocked by mercury
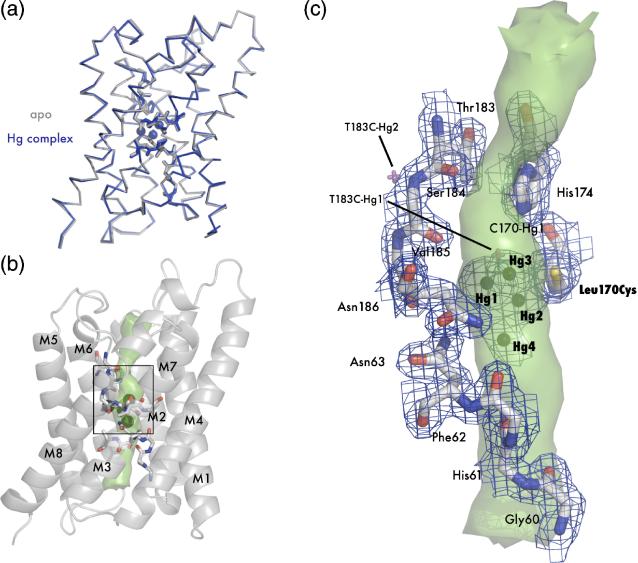
Since L170C has a heightened sensitivity to mercury we determined the x-ray structure of the apo and complex forms. L170C was overexpressed, purified, and crystallized with and without mercury using HgCl2 as an additive. Diffraction data were collected to 2.55 Å and 1.90 Å for the apo and complex proteins respectively, and the structures were solved by molecular replacement. As described in Materials and Methods, the model was built and refined iteratively to an Rfree of 28.0% and 19.5% for the apo and complex forms respectively (Table 1). Like T183C, L170C displays the same AQP canonical fold, and also like T183C, shows very little structural difference between the apo and complex form. The superposition (RMSD 0.27 Å) is shown in Figure 4A.
Figure 4.
Crystal structure of apo L170C and mercury bound L170C. (A) Main chain overlay of the apo (gray) and Hg-complex (blue) with an RMSD of 0.27 Å. Bound mercury atoms are displayed as spheres with a van der Waals radius of 1.10 Å. (B) Cartoon representation of L170C. Transmembrane helices are labeled M1-M8 and the interior surface of the channel is drawn as a green surface. The black square denotes the area of interest depicted in panel C. (C) Structure of the blocked channel. Amino acids classically involved with water binding in AQPs are shown as sticks and with 2Fo-Fc - electron density mapped contoured at 1.2 σ drawn in blue. Mercury are shown as spheres. Superposition of mercury atoms from the T183C structure are shown as magenta crosses. In this orientation it can be seen that all three mercury atoms sterically block the pore (green surface).
As we predicted, introduction of a cysteine residue at position 170 increases the affinity at the T183C-Hg1 site, and we were able to locate four mercury atoms (named L170C-Hg1, L170C-Hg2, L170C-Hg3, and L170C-Hg4) near the NPA region between Cys170 and His174. These atoms are clustered together, as indicated in the channel pathway of Figure 4B and 4C. L170C-Hg2, L170C-Hg3, and L170C-Hg4 lie directly in the pore, while L170C-Hg1 is at the edge (Figure 4C). The occupancies of L170C-Hg1, L170C-Hg2, L170C-Hg3, and L170C-Hg4 were refined to 0.40, 0.23, 0.20, and 0.18 respectively. L170C-Hg2 is covalently bound to Cys170 at the distance expected for a sulfur-mercury bond length (2.6 Å). L170C-Hg2 is also at the proper Hg dinuclear complex distance from L170C-Hg1 (2.5 Å), L170C-Hg3 (2.3 Å), and L170C-Hg4 (2.4 Å) and there is continuous electron density between all mercury atoms at 1.5 σ in a 2Fo-Fc map. L170C-Hg2 may therefore mediate the binding of a second mercury in the pore at any of the three other positions. L170C-Hg3 lies directly in the pore at the same site as T183C-Hg1 (magenta double cross in Figure 4C) and may interact with the imidazole ring of H174 (3.8 Å) in a similar manner.
Discussion
Aquaporins and mercury inhibition
Even after the knowledge of the extreme toxicity of mercury and its various compounds, it continued to find use in disinfectants, cosmetics, and a suite of so-called medicines.23 Mercurials attack the reactive thiol moiety of cysteine found in nearly all proteins and are known to have a host of side effects including polyuria induced by AQP2 inhibition in the apical membrane of the kidney collection tubule.24 Due to its affinity for thiols, mercury has been useful in chemical probes of protein-mediated biological processes, as it was in defining the water channels as proteins. Mercury as a pore blocker has been instrumental in characterizing the AQP channel and revealing the role of AQPs in membrane transport for numerous tissues6; 7.
Steric inhibition by mercury
We located two mercury atoms in the T183C-mercury complex x-ray structure. These atoms, T183C-Hg1 and T183C-Hg2, are located in the pore and at an interstitial site respectively. Given that T183C is four times more sensitive to mercury than WT, we sought to establish the relevance of each site to this important functional result and therefore determine whether the mechanism is steric or rooted in conformational rearrangement.
The most obvious explanation for inhibition is that T183C-Hg1, due to its location, directly blocks the pore and inhibits through a steric mechanism. This is further supported by the fact that there is little structural change between the apo and mercury-complexed forms (Figure 2A). Divalent mercury has a small ionic radius of 1.10 Å which would explain the lack of structural perturbation and its ability to bind deep within the protein (T183C-Hg2). But, while steric blockage is both supported by the data and simple, can we rule out conformational change?
Occupancy refinement revealed that both T183C-Hg1 and T183C-Hg2 are present at significantly less than unity (0.24 and 0.32 respectively). Since the mercury sites are of low occupancy, they may occur (i) alternately in different channels, i.e. they may be exclusive of one another, or (ii) they may both be bound, either cooperatively or uncooperatively, and have a lower occupancy for reasons outline below. In either case, any changes in the protein around a partially occupied site would also be of low (~.3) occupancy, and so be difficult to refine as multiple conformers. This could be particularly important for T183C-Hg2, bound to the highly conserved Glu138. Rearrangement of Glu138 could perturb the essential water-coordinating carbonyls and potentially disrupt water flux. However, inspection of Fo - Fc difference maps reveals an increased noise level (Figures 6A and 6B), but no obvious alternate conformer. Also the potentially cleaner Fo(apo) - Fo(Hg) maps between observations per se were not possible due to the degree of lack of isomorphism (Table 1). Thus, distinguishing between a steric mechanism and one involving a conformational change of the protein is difficult.
Figure 6.
Mercury disorder in electron density maps. (A) Fo-Fc electron density map (green) of mercury bound T183C structure contoured at 4 σ. (B) Fo-Fc electron density map (green) of mercury bound L170C structure contoured at 4 σ. (C) 2Fo-Fc omit electron density map solved with the first 45 frames of data (blue) and the entire dataset (orange). Both maps are contoured at 1.2 σ around the three mercury atoms.
Favoring the more simple steric mechanism, we set out to show blockage occurs at the T183C-Hg1 site through creation of a new cysteine mutant. To differentiate between the steric and conformational mechanism, such a mutant should bind mercury only at the pore site. Furthermore, the lower occupancy of T183C-Hg1 and longer thiol-mercury distances (5.6 Å versus 4.0 Å) relative to T183C-Hg2 suggest the T183C-Hg1 interaction with Cys183 is not ideal. We can thus hypothesize a structurally optimized mutant would also be more sensitive to mercury. In this region, the pore is formed mostly by main chain water-coordinating carbonyls and side chains from selectivity filter residues. Among the few side chains that project into the pore near T183C-Hg1, Leu170 is positioned closely to the site (Figure 4C). Thus, mutant L170C should be functionally sensitive to mercury, only bind mercury at the pore site, and therefore support a steric blockage model.
In inhibition assay we determined that L170C, with a cysteine at the proposed steric blockage site, is actually the most sensitive to mercury with respect to WT and T183C. Furthermore, this increase in sensitivity is also echoed in the structure. As with T183C, there is little evidence of conformational change upon mercury binding. Instead, there is complete occlusion of the pore and importantly, there are no interstitially bound mercury atoms to disturb the water-coordinating carbonyls. There are four mercury atom positions in the pore clustered around the introduced cysteine that refine to occupancies of 0.40, 0.23, 0.20 and 0.18 for L170C-Hg1, L170C-Hg2, L170C-Hg3, and L170C-Hg4 respectively. It is therefore possible these sites actually represent one mercury atom that is blocking the pore and is statistically disordered throughout the crystal lattice. Or, because of the low occupancy values, these sites may reflect mercury conformations that exist alternately in different blocked channels. Based on bond lengths, an attractive model is that L170C-Hg2 would be directly bound to Cys170 and then form a dinuclear mercury complex with one of three other mercury atoms. In either case, we conclude that although there may be multiple conformations of a blocked channel in L170C, the mechanism is a steric one. Finally, because the mercury atom sites in the L170C structure are at the same position as T183C-Hg1, T183C (and by analogy AQP1) may also be blocked in a steric manner.
Water permeability of AqpZ reconstituted into liposomes
Using a proteoliposome-based assay we measured the water conduction rates and determined that both mutants were active at lower conductances than for WT. While this decreased permeability may result from perturbation of the protein native state due to mutation, the decrease also appears to correlate with the introduction of a more polar cysteine side chain into the hydrophobic pore. T183C shows a moderate decrease in rate and the cysteine rotamer points away from the pore, while L170C shows a much larger decrease and the side chain is in the pore (Figures 2C and 4C). AQPs most likely achieve high conduction rates by partially stabilizing the subtrate, similar to an idea suggested for ion channels27. In this case water is stabilized as a line of molecules against eight carbonyls, the guanidium group of Arg189, and Nδ2 moieties from the two conserved asparagines of the NPA motifs, in an otherwise hydrophobic channel. Introduction of a polar pore residue therefore increases channel water affinity and decreases flux.
Mercury and WT AqpZ
We unexpectedly discovered that WT AqpZ is also inhibited by HgCl2. The WT protein contains two (Cys9 and Cys20) endogenous cysteines, which can presumably bind mercury but attempts at co-crystallizing WT with mercury to justify this were unsuccessful. Problems with co-crystallization suggests non-specific binding, so the pore itself may have some low affinity for mercury due to the presence of histidine and other polar side chains. We measured the IC50 of a mutant in which both endogenous cysteines were mutated to serine to be roughly 1mM (data not shown). In light of this result it may be inappropriate to classify AQPs as either distinctively mercury sensitive or insensitive. The ionic radius of Hg2+ is even smaller than water (1.10 Å) and so regardless of the presence of a pore cysteine, it may become partially stabilized by the partial negative charge on the carbonyl oxygens and the imidazole ring of His174. Therefore, at higher (i.e. mM) mercury concentrations all AQPs may be inhibited. This also suggests there may be other charged inhibitors, possibly by the mechanism outlined above.28; 29
Mercury and the tetrameric axis
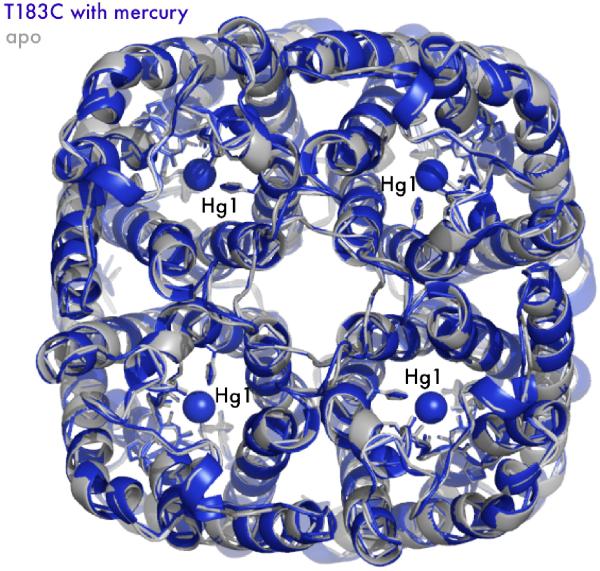
A steric mechanism with no significant structural change also validates the use of mercury to distinguish between conduction through the tetramer axis and the monomer pore. As described above, the arrangement of the helical bundle monomer creates a four-fold symmetric channel running parallel to the monomer channel (Figure 4). This four-fold axis is hydrophobic, of larger dimensions than the monomer channel, and previous x-ray structures indicate the presence of some as yet undetermined molecules. Both experimental and computational studies have investigated possible substrates, including water, ions25, and CO2.26 The mercury-bound structure of T183C, along with a simple steric blockage at the proposed site, suggests conduction studies using mercury are solely inhibiting the monomer channel.
Mercury dynamics in the pore
Mercury, due to its 80 electrons and aggressive thiol attacking nature, is one of the most common heavy atom derivatives for de novo phasing of x-ray crystal structures via isomorphous replacement and, to a lesser extent, anomalous diffraction (AD).30 The lack of successful mercury based AD experiments can be partially attributed to the success of other approaches, such as selenomethionine incorporation, but one emerging reason for the failure of mercury in AD experiments is the labile nature of the thiol-mercury bond under x-ray radiation exposure.31; 32
It was our initial intent to solve the mercury-complex structures with unbiased experimental phases using either isomorphous replacement or AD methods, as this would allow unambiguous identification of mercury sites. Mercury bound crystals were not isomorphous to the native dataset (Table 1) so we adopted a multiwavelength AD strategy. We were unable to obtain interpretable experimental maps and subsequently solved the structure by molecular replacement. Mercury sites in both structures were located by a combination of anomalous difference and omit maps combined with thorough investigation of each putative site’s chemical environment. During refinement of both mercury bound structures, it became clear there was significant motion in mercury atoms bound to the introduced cysteines. Fo-Fc difference maps (Figure 6A and 6B), which show differences between the observed data and model, show small (less than three electrons) “noisy” positive peaks after anisotropic refinement of the individual mercury sites. Based on their small size and proximity to adjacent Hg sites, we attribute these peaks to radiation-induced change of the mercury sites and alternate states of the protein as described above.
The difficulty in using mercury as an anomalous scatterer is due in part to the relative ease with which the mercury-thiol bond is cleaved, so we postulate that motion in the mercury peaks also reflects radiation damage induced cleavage of the mercury atoms’ interaction with cysteine. A previous radiation damage study on the problems associated with using mercury has seen solvent exposed mercury atoms “escaping” over time, while those that are buried are more likely to remain bound to their respective sulfur moiety.32 Using the L170C structure as a test case, we refine the mercury-bound structure against the data derived only from the first 45 frames (72.9% complete; scaled to the entire dataset) and the resulting omit map is shown in Figure 6C. A comparison with omit maps calculated from all of the data, shows there is indeed a change in the occupancies of sites. This change in occupancy can be seen as diffusion of mercury out of the solvent accessible channel while the more deeply buried L170C-Hg1 shows little motion.
The consequence of such dynamics is not understood, but it does highlight the need for determining exactly what mercury does upon binding. In the case of T183C, new mutants may be used to probe the role of the interstial site in the absence of mercury bound in the pore. In L170C we know, at least under x-ray exposure, that mercury is dynamic and influenced by the solvent-filled channel. Here, higher-resolution structures will allow more accurate refinement of occupancies and distances, and functional analysis of new mutants (e.g. double cysteine mutants) can be used to further probe the effect of mercury binding. It will also be important to structurally characterize the role of larger mercurials such as p-chloromercuriphenylsulfonic acid, which are extensively used in physiology studies and may have a more complex inhibition mode. Moving away from mercurials, structural studies should be extended to include the recently discovered cationic charged inhibitors, to understand the more general principles of aquaporin inhibition. Such studies may open up new avenues for the discovery of specific small molecule aquaporin inhibitors for clinical use. The mechanisms we have elucidated and outlined here are a first step towards this important goal.
Materials and Methods
Expression and Purfication
Mutants of AqpZ were generated by site-directed-mutagenesis of the pET28b-AqpZ construct used in the original structure paper. 14 Prior to mutagenesis, endogenous cysteines were removed via the mutations Cys9Ser and Cys20Ser. The E. coli strain C43(DE3) was transformed, grown to 0.6-1 OD at 600 nm at 37°C in 2 x LB media, 0.5% glycerol (v/v), 1x M9 salts, and 25 mg / L kanamycin, and induced with 1 mM isopropyl -D-thiogalactoside (Anatrace).33; 34
All purification was carried out at 4°C or on ice as necessary. Cells from six L of culture were harvested and lysed by a microfludizer in 20 mM Tris pH 7.4, 100 mM NaCl, .5 mM phenylmethylsulforyl fluoride, and 5 mM BME. Membranes were recovered from supernatant by 100,000 x g centrifugation for 2 hours. AqpZ was solubilized from membranes by agitation in 20 mM Tris pH 7.4, 100 mM NaCl, 5 mM BME, 10% glycerol (v/v), and 270 mM n-Octyl-β-D-glucopyranoside (OG) (Anatrace) for 12-16 hours at 4°C. Solubilized protein was bound in batch to Ni-NTA resin (Qiagen) for 1 hour, washed with 25 resin volumes of 20mM Tris pH 7.4, 100 mM NaCl, 5 mM BME, 10% glycerol, 40 mM OG, and 20 mM imidazole, and eluted with 20 mM Tris pH 7.4, 100 mM NaCl, 5 mM BME, 10% glycerol, 40 mM OG, and 250 mM imidazole. Imidazole was removed using a Biorad Econo-Pac 10DG desalting column and the 6xHis tag was removed by digestion with 5 μg of trypsin for 12 hours at 4°C. Trypsin was removed by passing over a benzamidine-sepharose matrix (GE Healthcare), and the protein sample was injected onto a Pharmacia Superose 12 gel filtration column running a mobile phase of 20 mM Tris pH 7.4, 100 mM NaCl, 2 mM dithiothreitol, 10% glycerol, and 40 mM OG. Except as noted, all materials were purchased from Sigma or Fisher. The sample was judged pure and homogeneous by both gel filration chromatography and coomassie-stained denaturing gels. Final yields were approximately ten, seven, and three mg of protein / L culture for WT, T183C, and L170C respectively.
Crystalization and Data Collection
Following gel filtration chromatography, the protein was concentrated to 25 mg / ml using a 30 kDa cutoff Amicon Ultra-15 Centrifugal Filter. Crystals were grown by hanging drop vapor diffusion at room temperature by 1:1 addition of protein and 25-30% polyethylene glycol (PEG) monomethyl ether 2000 (Fluka), 100 mM sodium cacodylate pH 6.5, and 50-100 mM MgCl2. For co-crystallization, divalent mercury in the form of 1-3 mM HgCl2 was added in batch to the protein sample before mixing with precipitant. In general, the best co-crystals were obtained at slightly lower (2-4%) PEG concentrations than in apo crystallization. Crystals grew to roughly 300 μm x 300 μm x 150 μm over the course of several days and were flash frozen in liquid nitrogen following a brief washing in the mother liquor plus 15% glycerol for cryoprotection. Diffraction intensities were collected on Advanced Light Source Beamline 8.3.1 using an ADSC Quantum-Q210 CCD detector. Prior to data collection on mercury-complex crystals, fluorescence energy scans of the LIII mercury edge were taken to verify mercury substitution.
Phasing and Model Refinement
Data were processed with Elves35 and CCP436 (using MOSFLM37) and the structures were solved by molecular replacement with the published WT structure (Protein Data Bank Code 1RC2) using Phaser38. The models were refined with iterative cycles of manual building with Coot39 and restrained refinement with individual B-factor refinement in Refmac540. After several initial rounds of refinement, mercury atoms were located unambiguously by inspecting anomalous difference maps, Fo-Fc omit maps (peaks of 15-7 σ), and the local chemical environment. Following refinement, the appearance of negative density in Fo-Fc difference maps indicated that occupancy for mercury atoms was not unity. Occupancies for mercury atoms, along with their anisotropic B-factors were refined using least-squares refinement in SHELXL.22
Proteoliposome Reconstitution
Before removing the 6xHis tag with trypsin, aliquots of protein were set aside for proteoliposome reconstitution. E. coli polar lipids were sonicated to clarity and the reconstitution cocktail was prepared by sequentially adding 100 mM MOPS pH 7.5, 51.3 mM OG (1.5%, w/v), 50 μg / ml of purified protein, and 10 mg / ml E. coli polar lipids (Avanti).20 To reduce oxidation, lipid stocks were stored in 2 mM BME and all buffers were under argon atmosphere. Following cocktail incubation for one hour at room temperature (RT), proteoliposomes were formed by diluting the mixture 50-fold into a running buffer of 20 mM Hepes pH 7.5 and harvested by centrifugation at 100,000 x g for two hours. Pelleted liposomes were resuspended into one ml of running buffer (20 mM Hepes pH 7.5) and stored at 4°C. Liposome monodispersity was verified by dynamic light scattering with a mean diameter of 90 nm.
To analyze the kinetics of water conduction through the channel, we subjected the proteoliposomes to an osmotic gradient by mixing 1:1 proteoliposomes (final AqpZ monomer concentration of 0.27 μM) and running buffer with osmolyte (20 mM Hepes pH 7.5, 570 mM sucrose) and measured water efflux (liposome shrinkage) by light scattering in a stopped-flow device at 440 nm. Resulting curves were fit to a single-exponential rate constant (k1) as a measure of conduction to use in comparison between mutants and with inhibitor (n=5, same liposome preparation). Inhibition of the mutants was accomplished by incubating the resuspended proteoliposomes with the appropriate concentration of HgCl2 for one hour at RT prior to stopped-flow analysis (n=5, same liposome preparation). Data were consistent across multiple protein and liposome preparations.
Figure 5.
Mercury blocks the monomer channel. While AQPs are tetramers in the membrane, the monomer is the functional unit. By imposing crystal symmetry on both the apo (grey) and complex structures (blue), T183C is drawn as a tetramer in cartoon representation. Mercury-Hg1, with its proper van der Waals radius, is drawn as a sphere blocking the channel. Note there is almost no structural change to the tetrameric axis.
Acknowledgements
We thank Franklin A. Hays, Pascal Egea, Janet Finer-Moore and Zach Newby for helpful manuscript suggestions. D.F. Savage was supported by a Burroughs Wellcome Trainee Fellowship. Research was supported by grant GM 24485 from the NIGMS.
Abbreviations
- AQP1
aquaporin 1
- AqpZ
aquaporin Z
- TM
transmembrane
- GlpF
glycerol facilitator
- WT
wild-type
- IC50
half maximal inhibitory concentration
- RMSD
root mean square deviation
- BME
2-mercaptoethanol
- AD
anomalous diffraction
- RT
room temperature
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Borgnia M, Nielsen S, Engel A, Agre P. Cellular and molecular biology of the aquaporin water channels. Annu Rev Biochem. 1999;68:425–58. doi: 10.1146/annurev.biochem.68.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Hille B. Ion channels of excitable membranes. 3rd edit Sinauer; Sunderland, Mass: 2001. [Google Scholar]
- 3.Macey RI. Transport of water and urea in red blood cells. Am J Physiol. 1984;246:C195–203. doi: 10.1152/ajpcell.1984.246.3.C195. [DOI] [PubMed] [Google Scholar]
- 4.Zeidel ML, Ambudkar SV, Smith BL, Agre P. Reconstitution of functional water channels in liposomes containing purified red cell CHIP28 protein. Biochemistry. 1992;31:7436–40. doi: 10.1021/bi00148a002. [DOI] [PubMed] [Google Scholar]
- 5.Preston GM, Carroll TP, Guggino WB, Agre P. Appearance of water channels in Xenopus oocytes expressing red cell CHIP28 protein. Science. 1992;256:385–7. doi: 10.1126/science.256.5055.385. [DOI] [PubMed] [Google Scholar]
- 6.Fushimi K, Uchida S, Hara Y, Hirata Y, Marumo F, Sasaki S. Cloning and expression of apical membrane water channel of rat kidney collecting tubule. Nature. 1993;361:549–52. doi: 10.1038/361549a0. [DOI] [PubMed] [Google Scholar]
- 7.Javot H, Maurel C. The role of aquaporins in root water uptake. Ann Bot (Lond) 2002;90:301–13. doi: 10.1093/aob/mcf199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Preston GM, Jung JS, Guggino WB, Agre P. The mercury-sensitive residue at cysteine 189 in the CHIP28 water channel. J Biol Chem. 1993;268:17–20. [PubMed] [Google Scholar]
- 9.Kuang K, Haller JF, Shi G, Kang F, Cheung M, Iserovich P, Fischbarg J. Mercurial sensitivity of aquaporin 1 endofacial loop B residues. Protein Sci. 2001;10:1627–34. doi: 10.1110/ps.5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu D, Libson A, Miercke LJ, Weitzman C, Nollert P, Krucinski J, Stroud RM. Structure of a glycerol-conducting channel and the basis for its selectivity. Science. 2000;290:481–6. doi: 10.1126/science.290.5491.481. [DOI] [PubMed] [Google Scholar]
- 11.Sui H, Han BG, Lee JK, Walian P, Jap BK. Structural basis of water-specific transport through the AQP1 water channel. Nature. 2001;414:872–8. doi: 10.1038/414872a. [DOI] [PubMed] [Google Scholar]
- 12.Walz T, Hirai T, Murata K, Heymann JB, Mitsuoka K, Fujiyoshi Y, Smith BL, Agre P, Engel A. The three-dimensional structure of aquaporin-1. Nature. 1997;387:624–7. doi: 10.1038/42512. [DOI] [PubMed] [Google Scholar]
- 13.Park JH, Saier MH., Jr. Phylogenetic characterization of the MIP family of transmembrane channel proteins. J Membr Biol. 1996;153:171–80. doi: 10.1007/s002329900120. [DOI] [PubMed] [Google Scholar]
- 14.Savage DF, Egea PF, Robles-Colmenares Y, O’Connell JD, 3rd, Stroud RM. Architecture and selectivity in aquaporins: 2.5 a X-ray structure of aquaporin Z. PLoS Biol. 2003;1:E72. doi: 10.1371/journal.pbio.0000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harries WE, Akhavan D, Miercke LJ, Khademi S, Stroud RM. The channel architecture of aquaporin 0 at a 2.2-A resolution. Proc Natl Acad Sci U S A. 2004;101:14045–50. doi: 10.1073/pnas.0405274101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tornroth-Horsefield S, Wang Y, Hedfalk K, Johanson U, Karlsson M, Tajkhorshid E, Neutze R, Kjellbom P. Structural mechanism of plant aquaporin gating. Nature. 2006;439:688–94. doi: 10.1038/nature04316. [DOI] [PubMed] [Google Scholar]
- 17.Lee JK, Kozono D, Remis J, Kitagawa Y, Agre P, Stroud RM. Structural basis for conductance by the archaeal aquaporin AqpM at 1.68 A. Proc Natl Acad Sci U S A. 2005;102:18932–7. doi: 10.1073/pnas.0509469102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calamita G, Bishai WR, Preston GM, Guggino WB, Agre P. Molecular cloning and characterization of AqpZ, a water channel from Escherichia coli. J Biol Chem. 1995;270:29063–6. doi: 10.1074/jbc.270.49.29063. [DOI] [PubMed] [Google Scholar]
- 19.Borgnia MJ, Kozono D, Calamita G, Maloney PC, Agre P. Functional reconstitution and characterization of AqpZ, the E. coli water channel protein. J Mol Biol. 1999;291:1169–79. doi: 10.1006/jmbi.1999.3032. [DOI] [PubMed] [Google Scholar]
- 20.Borgnia MJ, Agre P. Reconstitution and functional comparison of purified GlpF and AqpZ, the glycerol and water channels from Escherichia coli. Proc Natl Acad Sci U S A. 2001;98:2888–93. doi: 10.1073/pnas.051628098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smart OS, Goodfellow JM, Wallace BA. The pore dimensions of gramicidin A. Biophys J. 1993;65:2455–60. doi: 10.1016/S0006-3495(93)81293-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheldrick GM, Schneider TR. SHELXL: High Resolution Refinement. Methods in Enzymology. 1997;277:319–343. [PubMed] [Google Scholar]
- 23.Emsley J. Nature’s building blocks : an A-Z guide to the elements. Oxford University Press; Oxford ; New York: 2001. [Google Scholar]
- 24.Zalups RK. Molecular interactions with mercury in the kidney. Pharmacol Rev. 2000;52:113–43. [PubMed] [Google Scholar]
- 25.Yu J, Yool AJ, Schulten K, Tajkhorshid E. Mechanism of gating and ion conductivity of a possible tetrameric pore in aquaporin-1. Structure. 2006;14:1411–23. doi: 10.1016/j.str.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 26.Cooper GJ, Boron WF. Effect of PCMBS on CO2 permeability of Xenopus oocytes expressing aquaporin 1 or its C189S mutant. Am J Physiol. 1998;275:C1481–6. doi: 10.1152/ajpcell.1998.275.6.C1481. [DOI] [PubMed] [Google Scholar]
- 27.Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 28.Niemietz CM, Tyerman SD. New potent inhibitors of aquaporins: silver and gold compounds inhibit aquaporins of plant and human origin. FEBS Lett. 2002;531:443–7. doi: 10.1016/s0014-5793(02)03581-0. [DOI] [PubMed] [Google Scholar]
- 29.Yool AJ, Brokl OH, Pannabecker TL, Dantzler WH, Stamer WD. Tetraethylammonium block of water flux in Aquaporin-1 channels expressed in kidney thin limbs of Henle’s loop and a kidney-derived cell line. BMC Physiol. 2002;2:4. doi: 10.1186/1472-6793-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blundell TL, Johnson LN. Protein crystallography. Academic Press; New York: 1976. [Google Scholar]
- 31.Ji X, Blaszczyk J, Chen X. The absorption edge of protein-bound mercury and a double-edge strategy for HgMAD data acquisition. Acta Crystallogr D Biol Crystallogr. 2001;57:1003–7. doi: 10.1107/s0907444901006370. [DOI] [PubMed] [Google Scholar]
- 32.Ramagopal UA, Dauter Z, Thirumuruhan R, Fedorov E, Almo SC. Radiation-induced site-specific damage of mercury derivatives: phasing and implications. Acta Crystallogr D Biol Crystallogr. 2005;61:1289–98. doi: 10.1107/S0907444905022316. [DOI] [PubMed] [Google Scholar]
- 33.Miroux B, Walker JE. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol. 1996;260:289–98. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 34.Mohanty AK, Wiener MC. Membrane protein expression and production: effects of polyhistidine tag length and position. Protein Expr Purif. 2004;33:311–25. doi: 10.1016/j.pep.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 35.Holton J, Alber T. Automated protein crystal structure determination using ELVES. Proc Natl Acad Sci U S A. 2004;101:1537–42. doi: 10.1073/pnas.0306241101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–3. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 37.Leslie AG. The integration of macromolecular diffraction data. Acta Crystallogr D Biol Crystallogr. 2006;62:48–57. doi: 10.1107/S0907444905039107. [DOI] [PubMed] [Google Scholar]
- 38.Read RJ. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr D Biol Crystallogr. 2001;57:1373–82. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- 39.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 40.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]