Summary
The homeodomain (HD) protein Bicoid (Bcd) is thought to function as a gradient morphogen that positions boundaries of target genes via threshold-dependent activation mechanisms. Here we analyze 66 Bcd-dependent regulatory elements, and show that their boundaries are positioned primarily by repressive gradients that antagonize Bcd-mediated activation. A major repressor is the pair-rule protein Runt, which is expressed in an opposing gradient, and is necessary and sufficient for limiting Bcd-dependent activation. Evidence is presented that Runt functions with the maternal repressor Capicua and the gap protein Kruppel as the principal components of a repression system that correctly orders boundaries throughout the anterior half of the embryo. These results put conceptual limits on the Bcd morphogen hypothesis, and demonstrate how the Bcd gradient functions within the gene network that patterns the embryo.
Introduction
The “morphogen” hypothesis states that gradients of morphogenetic activities provide spatial information that positions different cell fates along the axes of developing embryos (Wolpert, 1996). Protein gradients exist in many developing systems (reviewed in (Rogers and Schier, 2011), but it is not clear how many concentration thresholds can be provided by a single gradient, or how gradients contribute to the robust systems that ensure body plan consistency within species.
In Drosophila, a network of transcription factors establishes the body plan along the anterior posterior (AP) axis. At the top of this network is the Bicoid (Bcd) gradient, which is formed by diffusion from a localized RNA source near the anterior pole (Driever and Nusslein-Volhard, 1988b; Little et al., 2011). The Bcd protein contains a DNA-binding homeodomain (HD), and activates target genes that are expressed in bands and stripes (Driever and Nusslein-Volhard, 1989; Ochoa-Espinosa et al., 2005; Struhl et al., 1989). Posterior boundaries of these bands and stripes are precisely registered along the AP axis, foreshadowing the organization of the mature body plan.
Several experiments suggest that Bcd functions as a morphogen. For example, increasing bcd gene copy number shifts the posterior boundaries of Bcd-dependent target genes (Gao et al., 1996; Struhl et al., 1989) and the cephalic furrow (a morphological feature) toward the posterior (Driever and Nusslein-Volhard, 1988a). With six or seven copies of bcd, the posterior shifts are very dramatic, but remarkably, the embryos survive to fertile adulthood due to compensation mechanisms (Namba et al., 1997). Thus, the absolute positioning of individual boundaries with respect to the embryonic poles is not essential for development (in the lab).
In recent years, more direct challenges to the Bcd morphogen hypothesis have arisen in the literature. First, boundary shifts caused by copy number changes are less dramatic than predicted by the threshold model (Gao and Finkelstein, 1998; Gao et al., 1996; Houchmandzadeh et al., 2002). Second, there is no strong correlation between boundary position and the Bcd-binding strength of target gene enhancers (Ochoa-Espinosa et al., 2005; Segal et al., 2008). Thus, differential binding sensitivity to Bcd is not the primary design principle that positions Bcd-dependent expression boundaries.
To test the Bcd morphogen hypothesis, we recently used genetic experiments to significantly flatten the Bcd gradient (Ochoa-Espinosa et al., 2009). We further altered Bcd concentration by changing bcd gene copy number. Bcd target genes responded to the flattened gradients in two ways. Three target genes [the head gap genes orthodenticle (otd), empty spiracles (ems), and buttonhead (btd)] formed correctly ordered expression domains with shifted, but well-defined posterior boundaries. Three other target genes [the trunk gap genes giant (gt), hunchback (hb), and Kruppel (Kr)], responded in an all or none fashion to the flattened gradients of Bcd. Very low levels of Bcd could activate zygotic hb and Kr, but approximately twice as much was required for activation of gt (Ochoa-Espinosa et al., 2009). Importantly, all six tested target genes were activated by less Bcd than the amounts present at their boundary positions in wild type embryos. Thus, it was suggested that Bcd concentrations are in excess at every position within the wild type gradient.
Why are target gene boundaries formed in regions where there is more Bcd than the minimal amount required for activation? Perhaps repressors present in middle regions of the embryo can set boundaries of Bcd-dependent expression patterns. For example, the Capicua (Cic) repressor is required for setting the posterior boundaries of Bcd target genes that are expressed in the future cephalic region (Lohr et al., 2009). Also, the Kr repressor sets the posterior boundaries of sloppy-paired 1 (slp1) and even-skipped stripe 2 (Andrioli et al., 2004; Small et al., 1991; Stanojevic et al., 1991). However, it is not clear whether these repressive activities correctly order boundaries of different target genes.
Here we describe a systems approach to better understand the Bcd-dependent patterning system. We use bio-informatics, ChIP-Chip data, and in vivo reporter gene assays to collect and verify 66 Bcd-dependent enhancers, and sequence mining to identify over-represented motifs that correlate with boundary positioning. One prominent motif is a binding site for Runt (Run) protein, which is expressed in a transient graded pattern that directly opposes the Bcd gradient (Gergen and Butler, 1988). We show that Run functions with Cic and Kr to limit Bcd-dependent activation and that repression is required for correctly ordering target gene boundaries.
Results
Identification of 32 novel Bicoid-dependent enhancers
To further understand the relationship between Bcd-binding and embryo patterning, we collected a relatively large set of Bcd-dependent regulatory elements, starting with 34 enhancers that were previously validated in the literature (Suppl. Table I). We used two sets of criteria for predicting new enhancers in genomic sequences: Published ChIP-Chip data for Bcd (Li et al., 2008) and a clustering algorithm that determines Bcd-binding motif density (Lifanov et al., 2003). Candidate enhancers were attached to a lacZ reporter gene and targeted to a specific genomic location selected for its clean background and high activation level [(Bateman et al., 2006); Suppl. Fig. 1].
The ChIP-Chip data show nearly 700 genomic regions that bind Bcd in the embryo with a false discovery rate (FDR) of 1% (Li et al., 2008). Twenty-two of the 34 previously known Bcd-dependent enhancers map to genomic regions that appear in the 50 strongest ChIP-Chip signals. We therefore tested fragments (HC_01 - HC_33) from all remaining regions in the top 50 list (See Experimental Procedures; Fig. 1A). Remarkably, 28 of the 33 tested fragments drove expression during nuclear cycle 14 (NC14) (Fig. 1B; Suppl. Fig. 1). Crossing these reporter genes into embryos lacking bcd showed that the expression patterns of 24 of the 28 were either completely abolished (18) or shifted anteriorly (6), confirming their dependence on Bcd (Suppl. Fig. 2).
Figure 1. Identification of novel Bcd-dependent enhancers.
(A). Testing candidate enhancers from two sources, Bcd ChIP-chip data (Li et al., 2008) and Bcd binding site cluster predictions (Lifanov et al., 2003). There were 34 known Bcd-dependent enhancers (red) when this study started. 37 of 77 tested candidate fragments (blue) drive expression in early NC14 embryos. 32 are Bcd-dependent and 5 (in parentheses) are not altered in bcd mutants. 40 fragments tested negative (green). Note that a single in vivo bound region may contain multiple local binding peaks. Thus, 55 fragments were tested from the top “50” ChIP’d genomic regions. (B). Expression patterns of thirty-two new Bcd-dependent enhancers in early cycle 14 embryos. Embryos here and throughout this paper are oriented with anterior to the left and dorsal up.
The homotypic clustering model (Experimental Procedures) identified 221 genomic regions with high densities of Bcd-site motifs. Of the 34 previously known enhancers, this model correctly predicted 15, but 5 of these do not overlap with the top 50 ChIP-Chip signals (Fig. 1A). Thus, we used the clustering model to filter the list of 1% FDR ChIP-Chip signals below the top 50, and tested 25 fragments contained in both sets (HC_34 - HC_58). Eight of these elements drove Bcd-dependent expression in the early embryo, bringing the total number of known Bcd-dependent enhancers to 66. We also tested 19 elements predicted by the clustering model that do not appear in the ChIP-Chip list (HC_59 - HC_77). None directed expression in the early embryo, despite having what appear to be strong clusters of Bcd-binding sites (see Discussion).
A common sequence motif in enhancers with shared boundary positions
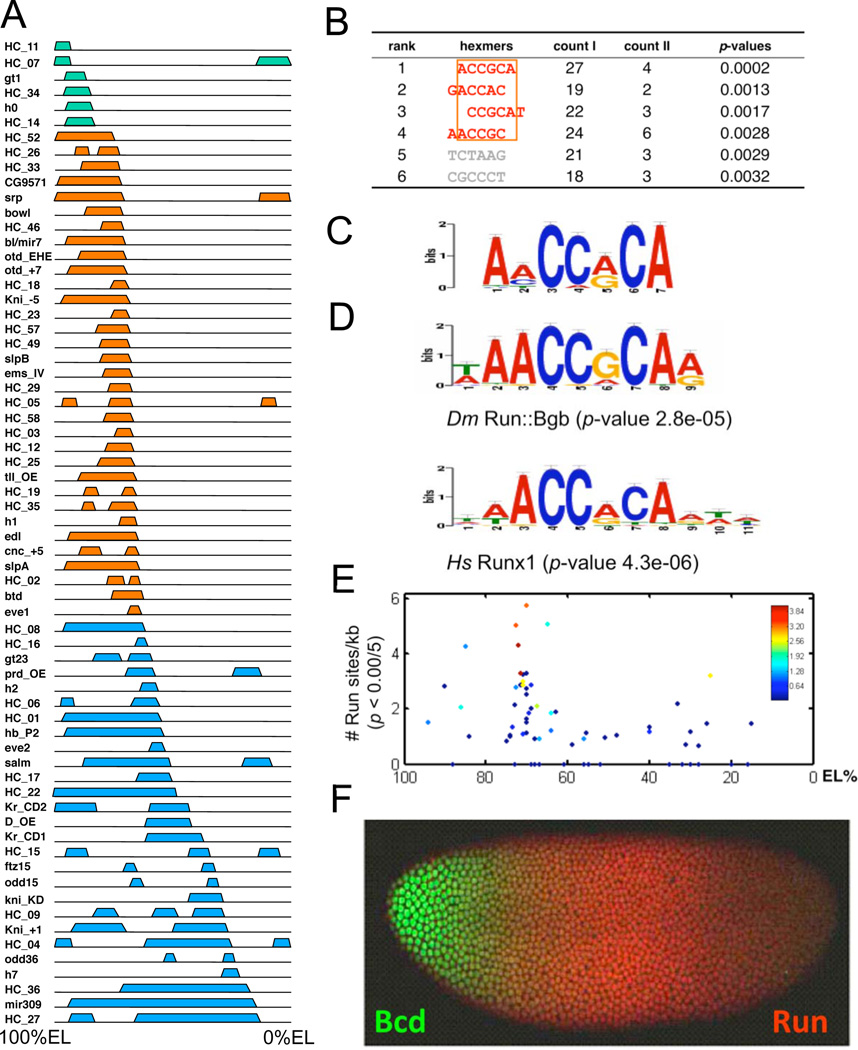
During early NC14, the 66 Bcd-dependent enhancers drive expression patterns with boundaries at a large number of positions along the AP axis (Fig. 2A). Six enhancers (Type 0) drive reporter expression patterns (green) that are limited to the anterior-most 25% of the embryo (100–75% embryo length (EL); 100% = the anterior pole), which corresponds to the unsegmented region near the anterior pole. 33 enhancers (Type I) establish expression boundaries (orange) in the region between 75 and 65% EL (Fig. 2A). The posterior-most boundary in this group is formed by the eve stripe 1 enhancer, which marks the cephalic furrow, the division between the presumptive head and thorax. Finally, 27 enhancers (Type II) form boundaries in embryonic regions posterior to the cephalic furrow (blue, Fig. 2A).
Figure 2. Over-represented Run-binding sites in Type I enhancers.
(A). Schematics of expression patterns driven by 66 known Bcd-dependent enhancers. Colored blocks above each line show the expression patterns driven by individual enhancers in early NC14 embryos. The left end of each line represents the anterior tip of the embryo (100%EL). The enhancers are classified by the positions of their Bcd-dependent posterior boundaries (see text). Type 0 is in green, Type I in orange, and Type II in blue. (B). Actual counts of specific hexamers in Type I and Type II enhancers. p-values were calculated using an exact binomial test (Experimental Procedures). The top four over-represented hexamers can be aligned (orange rectangle). (C). Motif derived from Type I enhancers using a discriminative MEME algorithm and the Type II sequences as negative filter(Bailey et al., 2010). This motif is very similar to Runt domain transcription factor binding motifs Drosophila Runt and human Runx1 (D). P-values were determined by the TOMTOM program(Gupta et al., 2007). (E). Correlation between Run-binding site number (y-axis) and posterior boundary position (x-axis) for all Bcd dependent enhancers. Each enhancer is represented by a single diamond, and aggregate binding strength (estimated by the STUBB algorithm) is represented by the color of each diamond according to the heat map shown. (F). Bcd and Run protein expression patterns in an early NC14 embryo.
If repressors present in middle regions of the embryo help set boundary positions of Type I patterns, their binding sites might be over-represented in Type I enhancers. By contrast, these same binding sites should be lacking or under-represented in Type II enhancers, which drive expression in regions where repressors might be present. We used an enumerative method to evaluate the distribution of all possible hexamers in the sequences of Type I and Type II enhancers. Then, we used an exact binomial test (Experimental Procedures) to assess significance of hexamer count differences between the two types. At a stringent cutoff p-value (0.005), only six hexamers are significantly overrepresented in Type I enhancers compared to Type II enhancers (Fig. 2B). The top four can also be aligned, producing a composite hexamer ACCRCA (Fig. 2B). We also applied a discriminative motif discovery tool (DREME) to search Type I enhancers, using Type II enhancers as negative filter (Experimental Procedures). A motif that contains the ACCRCA core was discovered (Fig. 2C), which shows a very high similarity to canonical binding motifs for Runt domain proteins (Melnikova et al., 1993), including Drosophila Runt (Run) and human Runx1 (Fig. 2D).
We then checked the distribution of Run binding sites in all Bcd-dependent enhancers using the Drosophila Run binding motif (Fig. 2E). High affinity Run binding sites (with matching p<0.0005) are significantly enriched in Type I enhancers (p<10−7; one-tail Poisson) and under-represented in Type II (p=.0026). We also calculated a cut-off-free index (Stubb score;(Sinha et al., 2006)), which represents integrated input over all potential Run sites. Type I enhancers show significantly higher Stubb scores than Type II enhancers (Fig. 2E). Finally, we checked the ChiP-Chip data, which shows Run binding to 82% of Type I enhancers, but only to 50% of Type II, with generally weaker signals (Suppl. Fig. 3).
Runt-binding activity is sufficient for repression of Type I enhancers
run was identified as a pair-rule mutation (Gergen and Wieschaus, 1985), and is expressed in a seven-striped pattern in NC14 embryos (Gergen and Butler, 1988). However, run is first expressed ubiquitously in NC10 embryos, and then repressed in anterior-most regions in NC12, forming a broad domain in middle regions that lasts until very early in NC14 (Fig. 2F). Run protein at the anterior edge of this domain forms a gradient that directly opposes the Bcd gradient, consistent with a potential role as a repressor. To test this, we used a hs-run transgene to induce ubiquitous expression in early NC14 (Tsai and Gergen, 1994). This caused a strong repression of reporter gene expression driven by all 15 tested Type 1 enhancers (Fig. 3B, data not shown). In contrast, patterns driven by all tested Type 0 (2) and Type II (7) enhancers were not detectably altered in embryos with ubiquitously expressed Run (Fig. 3A, C, data not shown).
Figure 3. Runt is sufficient to antagonize Bcd-dependent activation.
(A–C). Expression patterns of reporter genes containing Type 0 (A), Type I (B), and Type II enhancers (C) in embryos also carrying a hs-run construct. The left embryo of each pair was raised at room temperature (wt); the right embryo was heat-shocked to induce ubiquitous expression of Run (hs-run). (D–H). Expression patterns of a reporter gene carrying the wild type hb P2 enhancer (D) or mutated forms of the enhancer with one (E), two (F), or three introduced Run-binding motifs (G). Patterns are shown in wild type (wt) and run3 mutants as indicated. (H). Expression of a mutated form of the hb P2 reporter in which each inserted Run motif in (G) (green box) was mutated to a different unrelated sequence (a, b, c). Posterior boundary positions (% EL; mean ± std) and sample sizes (n) are shown for each experiment.
To test whether Run-binding sites are sufficient for establishing posterior boundaries, we introduced 1, 2, or 3 Run-binding sites into a Type II enhancer from the hb gene (hbP2-lacZ, Fig. 3D–H; Suppl. Table 2). The hbP2 enhancer contains four high affinity Bcd binding sites (p<.0005), but no native Run-binding sites, and forms a posterior boundary at ~55% EL. Adding Run sites to this enhancer progressively shifted the posterior boundary of hbP2-lacZ expression toward the anterior (Fig. 3 E–G). We also constructed a hbP2-lacZ reporter gene in which the nucleotides we changed to insert Run sites were mutated to other nucleotides (Fig. 3H). The posterior boundary formed by this reporter appears at a position very similar to that formed by the wild type enhancer (Fig. 3D). Finally, we crossed all hbP2-lacZ reporter genes into run mutants(Fig. 3D–G, data not shown). This caused posterior expansions of patterns driven by reporters containing Run sites (Fig. 3E–G), but no change in the patterns driven by the wild type and the scrambled site reporter. Finally, we inserted Run-binding sites into a second Type II enhancer (HC_01, Fig. 1B, Suppl. Table 2), which caused an anterior shift of the posterior boundary similar to that seen with the hbP2-lacZ reporter (Suppl. Fig. 4). Together these experiments show that direct Run-binding can set Bcd-dependent boundaries.
Runt is required for boundary positioning of multiple Bcd target genes
We next tested whether endogenous Run sets posterior boundaries of real Bcd target genes by examining expression patterns in run mutant embryos. Consistent with previous results, loss of run caused ectopic expression of the head gap gene otd in the posterior part of the embryo (Fig. 4A). Careful measurements revealed that the posterior boundary of the anterior otd expression domain also shifts posteriorly by ~3% in run mutants. This shift was more significant in early NC14 embryos [68.9 ± 1.8% EL in run (n=10) compared to 72.2 ± 1.6 EL in wild type (n=48)], but was still detectable in mid-NC14 embryos (Fig. 4A). Other anterior genes regulated by Type I enhancers, including empty spiracles (ems; Fig. 4B), sloppy paired 1 and 2 (slp1 and slp2), also showed similar boundary shifts in run− embryos (Suppl. Fig. 5).
Figure 4. Runt is required for boundary positioning of Bcd target genes regulated by Type I enhancers.
(A, B). otd (A) and ems (B) expression in wild type embryos (wt) and mutants lacking run mRNA (run3). Arrow in A shows the posterior duplication of otd in run3. (C). Expression of an otd-lacZ reporter gene in a wild type embryo (top), in a run3 embryo (middle), and when Run sites within the otd enhancer were mutated (bottom). To orient the embryos precisely along the DV axis, they were co-stained with sna, which labels the ventral-most region of the embryos. PBP measurements for each experiment are shown in the plots on the right. Each horizontal blue line represents the expression pattern in a specific region (75–65% EL) of a single embryo, with the expression boundary shown as the point at the end of the line. Means ± standard deviations and sample sizes (n) are shown for each experiment; mean boundary positions are also marked by red bars. p-values were calculated using a one-tail t-test. All plots are colinear with the scale at bottom right.
We also examined a reporter gene containing the otd EHE enhancer in run mutants (Fig. 4C). The pattern driven by this enhancer showed an expansion similar to that seen for the endogenous otd gene. This enhancer contains seven Run-binding sites; we mutated these sites (Suppl. Table 2), which caused a posterior shift of the expression boundary similar to that seen for the wild type enhancer in run mutants (Fig. 4C).
Combinatorial repression establishes the correct order of Bcd-dependent expression boundaries
The boundary shifts of endogenous expression patterns observed in run mutants (Fig. 4, Suppl. Fig. 5) are modest (~3% EL) compared to the dramatic shift (~15% EL) observed when the hbP2-lacZ reporter gene with three inserted Run sites was crossed into the same mutants. One explanation is that the otd EHE contains weak Bcd binding sites that do not support activation in more posterior regions. Alternatively, Type I enhancers may receive inputs from other transcription repressors, in particular Cic and/or Kr. We tested this by genetically removing both run and maternal Cic, which also causes a severe reduction in Kr levels (Ajuria et al., 2011). This double mutant caused more dramatic expansions of head gap patterns than observed in either single mutant (Suppl. Fig. 5).
To test whether the correct registry of gene expression boundaries is affected in these mutants, we performed multiplex FISH experiments to simultaneously detect three Bcd-dependent expression patterns. We compared otd, slp1, and gt (anterior domain), which have well-separated posterior boundaries (Fig 5A, E). In single run or cic mutants, there were noticeable posterior shifts of all three boundaries, but no change in their spatial order (Fig. 5B, C, F, G). In the double mutant, however, the increased expansions of otd and slp1 extended to a point where they completely overlapped with the gt posterior boundary (Fig. 5D, H).
Figure 5. The correct order of Bcd-dependent boundaries is established by combinatorial repression.
Left column. Triple FISH of otd, slp1 and gt in wild type (A), run− (B), cic− (C) and run− cic− double mutant (D) embryos. White rectangles indicate the regions used for image quantification. Middle column. Quantification of otd, slp1 and gt in wild type (E), run− (F), cic− (G) and run− cic− double mutant (H) embryos. Y-axis represents normalized intensities. Right column. engrailed (en) mRNA patterns in stage 10–11 embryos. In wild type embryos (I), en is expressed in five head domains that foreshadow the ocular (oc), antennal (an), intercalary (ic), mandibular (md), and maxillary (mx) segments. In run− (J) and cic− (K) mutant embryos, the head region appears enlarged, and all cephalic en domains are still visible. In double mutant embryos (L), only a single cephalic en domain (oc) is observed. Arrow in (L) points to the oc on the opposite side of the embryo.
To test whether overlapping boundaries disrupt A–P segment formation in the cephalic region, we assayed expression of engrailed (en), a segment polarity gene expressed in five anterior domains that mark anlage for cephalic segments (Fig. 5I). In single run or cic mutants, all five expression domains were still detectable (Fig. 5J, K). However, in double mutant embryos, only a single expression pattern was observed (Fig. 5L). These results suggest that the double mutant causes drastic head defects, consistent with the loss of normally registered expression boundaries. Also, since the amplitude and shape of the Bcd gradient appear unchanged from wild type in all mutant embryos (Suppl. Fig. 6), we conclude that the formation of specific head segments, and the spatial registration of multiple Bcd-dependent expression boundaries are formed primarily by antagonistic repression of the Bcd gradient.
Patterned run expression persists in embryos with flattened Bcd gradients
Repression by Run and Cic provides a mechanism that is consistent with the idea that boundaries of Bcd target genes are normally set in positions where Bcd concentrations are in excess of those required for activation. We previously showed that boundaries of some target genes (the Type I genes otd, ems and btd) are correctly ordered in double mutant [exuperantia vasa (exu vas)] embryos that flatten the Bcd gradient(Ochoa-Espinosa et al., 2009). Are Run and/or Cic involved in setting these boundaries?
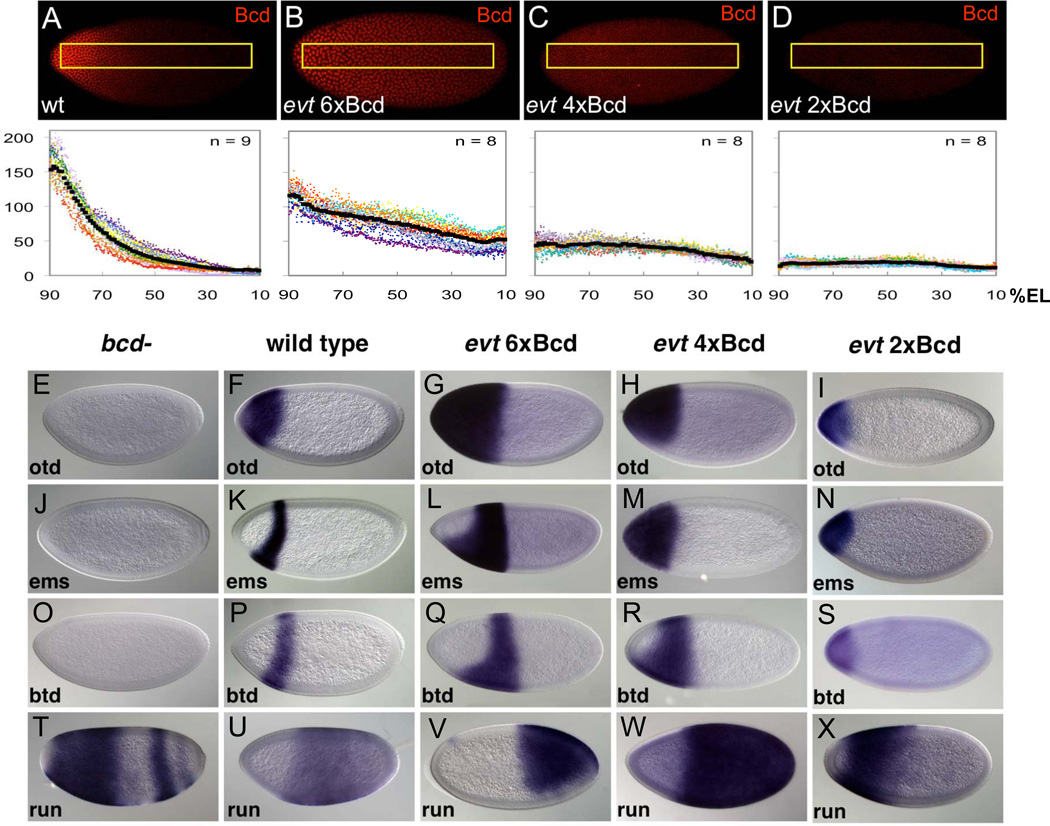
In addition to the anterior expression domains of otd, ems, and btd, exu vas mutant embryos show posterior stripes of all three genes, arranged with anterior boundaries in mirror image to the order of boundaries in anterior regions(Ochoa-Espinosa et al., 2009). Because the terminal system functions through a graded phosphorylation cascade at both poles (Furriols and Casanova, 2003), it could in principle account for the observed mirror image expression patterns (Gao et al., 1996). To test this, we used a triple mutant chromosome (evt) that contains exu, vas, and a null mutation of torso (tor), which encodes the terminal system receptor, and varied bcd copy number (Fig. 6A–D). The additional removal of the terminal system completely abolished the posterior otd, ems, and btd stripes observed in exu vas embryos, but each target gene was still expressed in an anterior domain with a sharp posterior expression boundary (Fig. 6G–I, L–N, Q–S). In embryos with 6 copies of bcd, boundaries were established very near the center of the embryo (Fig. 6G, L, Q); reducing bcd copy number shifted these boundaries to more anterior positions (Fig. 6H–I, M–N, R–S).
Figure 6. Boundary positioning in embryos with flattened Bcd gradients.
(A–D). Bcd gradient profiles in wild type embryos (A) and exu vas tor (evt) triple mutant embryos with six (B), four (C) or two copies (D) of bcd. Boxes in top panels represent regions used for quantification (bottom panels: Y Axis = raw intensities). Each color represents data extracted from a single embryo. Mean expression profiles for each expression pattern (Experimental Procedures) are depicted by black lines. (E–X). An exu vas tor (evt) triple mutant chromosome was used to make embryos with flattened Bcd gradients, and bcd copy numbers were varied to provide different levels of Bcd. Individual genes assayed are indicated on each panel.
Since the gradient of Cic repressive activity is abolished in terminal system mutants (Jimenez et al., 2000), it is unlikely that Cic-mediated repression is involved in forming these boundaries. Thus, we assayed the run RNA expression pattern in evt embryos. run is indeed expressed, and the anterior run boundary, normally positioned at ~65%EL (Fig. 6U), shifts with bcd copy number in the evt mutant background, always forming at a position that abuts the posterior boundaries of the head gap genes (Fig. 6V–X). This strongly supports the hypothesis that Run-mediated repression is critical for positioning these boundaries.
Discussion
Bcd-dependent enhancers
In this paper we identified 32 novel enhancers that respond to Bcd-dependent activation and form expression boundaries at different positions along the AP axis of fly embryos. To our knowledge, adding these elements to the 34 previously known enhancers constitutes the largest dataset of in vivo tested and confirmed enhancers regulated by a specific transcription factor in all of biology.
The 32 new confirmed enhancers were identified among 77 tested genomic fragments, which were selected because they showed in vivo binding activity (Li et al., 2008), or they conformed to a stringent homotypic clustering model for predicted Bcd-binding sites (Lifanov et al., 2003), or both. All seven previously unknown fragments showing in vivo binding and a predicted site cluster directed Bcd-dependent transcription in the early embryo. Other fragments from the top 50 ChIP-Chip signals (which do not conform to the clustering model) were also very likely (21/26) to test positive in the in vivo test, but this likelihood drops significantly (9/25) in a set of fragments from lower on the list of ChIP-Chip fragments. Interestingly, of 19 tested fragments that contain clusters of predicted sites, but no in vivo binding activity, not one tested positive in vivo. These results suggest that in vivo binding assays are much better predictors of regulatory function than simple site clustering algorithms alone.
One explanation for the failure of these predicted site clusters to bind Bcd in vivo is that they lie in heterochromatic regions of the genome that prevent site access. However, since they fail to function when taken out of their normal context (in reporter genes), whatever is preventing activation must be a property of the fragment itself and not its location in the genome. Interestingly, a number of Bcd site cluster-containing fragments drive expression later in development (Xu et al, in prep.). We propose that these fragments fail to bind Bcd because they lack sites for co-factors that facilitate Bcd-binding. In preliminary experiments, we have observed that Bcd-activated fragments contain on average more binding sites for the ubiquitous activator protein Zelda (Zld) than those that fail to activate (Xu et al., in prep.). Zld has been shown to be critical for timing the zygotic expression of hundreds of genes in the maternal to zygotic transition (Liang et al., 2008; Struffi et al., 2011).
The role of repressors in registering Bcd-dependent expression boundaries
Our results suggest strongly that a gradient of Run protein plays a major role in limiting Bicoid-dependent activation. Run seems to work as part of a repression system that also includes Cic and possibly Kr (Fig. 7A–E). Expression boundaries in the region anterior to the presumptive cephalic furrow shift toward the posterior in run and cic mutants, and the double mutant causes boundaries that are normally well separated to collapse into a single position (Fig. 5D, H).
Figure 7. A network of repressors registers Bcd-dependent posterior expression boundaries.
(A–D). Expression patterns of Bcd (A), Kr (B), Run (C), and Cic (D) in early NC14 embryos. Boxes represent regions used for quantification. (E). Average expression profiles of Bcd (n=8), Kr (n=6), Run (n=8) and Cic (n=8). (F). Model of gene expression boundary registration in the anterior half of the embryo. The Cic repression gradient is established via Tor down-regulation of Cic at the anterior tip. Bcd activates slp1 and gt, which encode repressors that set the anterior boundaries of run and Kr transcription. The Cic, Run, and Kr gradients repress Bcd-dependent activation of target genes, including slp1 and gt themselves.
The use of multiple repressors permits flexibility in binding site architecture within enhancers that establish boundaries at similar positions. For example, Type I enhancers show over-representations of both Run (Fig. 2) and Cic sites (data not shown), but 27% lack strong matches to the Cic PWM, and 12% lack strong matches to the Run PWM (Suppl. Fig. 7). Importantly, however, all Type I enhancers lacking Cic sites contain Run sites, and those lacking Run sites contain Cic sites. We observe multiple Kr sites in a large number of Bcd-dependent enhancers (Suppl. Fig. 7), which suggests that Kr is also a major component of the repression system that orders Bcd-dependent expression boundaries. Taken together, our data suggest that antagonistic repression of Bcd-mediated activation is a key design principle of the system that organizes the AP body plan. The repressors identified so far (Run, Cic, and Kr) are expressed in overlapping domains with gradients at different positions (Fig. 7B–E), consistent with the formation and ordering of a relatively large number of boundaries throughout the anterior half of the embryo.
The close linkage between repressor sites and Bcd sites within discrete enhancers suggests that repression occurs via short-range interactions that interfere directly with Bcd-binding or activation. Interestingly, Cic also shows repressive effects that seem to be binding-site independent. For example, some Type I enhancers do not contain recognizable Cic sites (Suppl. Fig. 7), but their expression boundaries expand posteriorly in cic mutants (data not shown). This could be caused by the reduced expression of run (Suppl. Fig. 5) and Kr (Ajuria et al., 2011) in cic mutants. However, genetically removing both Kr and run causes a less dramatic expansion than that seen in the absence of cic (data not shown). This suggests that Cic binds these enhancers via sub-optimal sites or that it is required for the correct patterning of another unknown repressor. Another possibility is that these expansions are caused indirectly by changing the balance of MAPK phosphorylation events that control terminal patterning (Kim et al., 2011; Ronchi et al., 1993).
Integrating the Bcd morphogen hypothesis with the AP patterning network
Our results do not strictly falsify the Bcd morphogen hypothesis, but they support the idea that the Bcd gradient can establish only a “rough framework that is elaborated by the interaction of the zygotic segmentation genes”(Driever and Nusslein-Volhard, 1988a). What is the nature of this framework, and what role does it play in the network that precisely positions target gene boundaries?
One component of the system, the Cic repression gradient, is maternally produced and formed by down-regulation at the poles via the terminal patterning system (Jimenez et al., 2000). This gradient is formed independently of Bcd, but is critical for establishing boundaries of Bcd-dependent target genes(Lohr et al., 2009); Fig. 5; Suppl. Fig. 5). In contrast, Bcd is involved in activating the expression patterns of run and Kr, and in repressing them in anterior regions. Both run and Kr expand anteriorly in bcd mutants [(Hoch et al., 1990); Fig. 6T]. There is no evidence that Bcd functions directly as a transcriptional repressor, so these repressive activities are probably indirect. Previous work showed that the Bcd target gene gt is involved setting the anterior Kr boundary (Wu et al., 1998; Yu and Small, 2008), and we hypothesize that another Bcd target gene, slp1, encodes a forkhead domain (FKH) protein that sets the anterior boundary of the early run pattern. slp1 is expressed in a pattern reciprocal to the run pattern, and was previously shown to position the anterior boundaries of several pair-rule gene stripes including run stripe 1 (Andrioli et al., 2004).
These results suggest that a major function of the Bcd gradient is the differential positioning of two repressors, Slp1 and Gt, which set the positions of the Run and Kr repression gradients, which then feedback to repress Bcd-dependent target genes (Fig.7F). How are slp1 and gt differentially positioned? One possibility is that slp1 and gt enhancers respond to specific concentrations within the Bcd gradient, consistent with the original model for morphogen activity. However, the fact that the slp1 and gt expression domains form boundaries at the same positions in embryos lacking the Cic and Run repressors argues against this model for these genes.
We also show that Bcd target genes normally expressed in cephalic regions form and correctly position posterior boundaries in embryos containing flattened Bcd gradients((Ochoa-Espinosa et al., 2009); Fig. 6). Run is still expressed in these embryos, specifically in a domain that consistently abuts the boundaries of the anterior Bcd target genes, regardless of copy number (Fig. 6). This suggests that a mutually repressive interaction between Slp1 and Run is maintained in these embryos, but does not explain how these boundaries are consistently oriented perpendicularly to the AP axis. The answer might lie in the fact that the flattened Bcd gradients in these embryos are not completely flat (Fig. 6B–D), but are present as a shallow gradient with slightly higher levels in anterior regions. In these embryos, the slight changes in concentration along the AP axis might cause a bias that enables the orientation of the mutual repression interaction. In wild type embryos, Bcd is much more steeply graded, which makes this bias stronger and the boundary between these mutual repressors more robust.
Antagonistic repression: a general mechanism for refining morphogen activities?
Our results suggest that antagonistic repression precisely orders Bcd-dependent expression boundaries. However, repression may not be required for the activity of all morphogens. For example, the extracellular signal activin has been shown to activate target genes in a threshold-dependent manner in isolated animal caps from frog embryos(Gurdon et al., 1998). Also, a gradient of the transcription factor Dorsal (Dl) is critical for setting boundaries between different tissue types along the dorsal-ventral (DV) axis of the fly embryo(Roth et al., 1989). It is thought that the major mechanism in Dorsal-specific patterning is threshold-dependent activation(Hong et al., 2008; Jiang and Levine, 1993), which is quite different from the system described in this paper. One major difference between Bcd and Dl is the number of boundaries specified: three for Dl, and more than ten for Bcd (Fig. 2A). We propose that the robust ordering of more boundaries simply requires a more complex system.
In general, though, it seems that antagonistic mechanisms are involved in controlling the establishment or interpretation of most morphogen activities (reviewed in Rogers and Schier, 2011). For example, in the Drosophila wing disc, the TGF-β signal Dpp forms an activity gradient that is refined by interactions with multiple extracellular factors (Affolter and Basler, 2007). Also, in vertebrates, the signaling activity of the extracellular morphogen Sonic hedgehog (Shh) is affected by positive and negative interactions with specific molecules on the surfaces of receiving cells(Allen et al., 2011; Allen et al., 2007; Jeong and McMahon, 2005).
There is some evidence that transcriptional repression is also used for refining the patterning activities of extracellular molecules. Dpp acts as a long-range morphogen that activates two major target genes [optomotor blind (omb) and spalt (sal)] in nested patterns with boundaries at different positions with respect to the source of Dpp (Lecuit et al., 1996; Nellen et al., 1996). Although these boundaries could in theory be formed by differential responses to the morphogen, it is clear that the transcriptional repressor Brinker (Brk), which is expressed in an oppositely oriented gradient, also plays an important role (Campbell and Tomlinson, 1999; Jazwinska et al., 1999; Minami et al., 1999). The Brk gradient is itself positioned by Dpp activity in a manner analogous to positioning of the Run and Kr repressor gradients by Bcd (Muller et al., 2003). Also, a similar transcriptional network functions in Sonic Hedgehog (Shh)-mediated patterning of the vertebrate neural tube, where a series of spatially oriented repressors feeds back to limit the expression boundaries of Shh-mediated cell fate decisions (Balaskas et al., 2012).
Conceptually, these more complex systems are reminiscent of the reaction-diffusion model proposed by Turing (Turing, 1952), in which a localized activator would activate a repressor, which would diffuse more rapidly than the activator, and feed back on its activity. These systems strongly suggest that the patterning activity of a single monotonic gradient is insufficiently robust for establishing precise orders of closely positioned expression boundaries. By integrating gradients with repressive mechanisms that refine gradient shape or influence outputs, systems are generated that ensure consistency in body plan establishment while still maintaining the flexibility required for complex systems to evolve.
Experimental Procedures
Fly strains, genetics, and heat shock experiments
Fly strains and genetic manipulations are described in the Extended Experimental Procedures. For heat shock experiments, 1–3 hour embryos carrying one copy of hs-run and one copy of a given reporter gene were collected and transferred immediately into a 37°C incubator for 20 minutes, allowed to recover at 25°C for 40 minutes, and fixed immediately. The same embryos without the heat shock treatment and heat-shocked yw embryos lacking the hs-run construct were used as controls. Control embryos showed essentially wild type patterns. In other experiments, yw1118 flies were used as wild-type controls if not specifically mentioned.
Transcription factor binding motifs
We used the Bcd-binding motif extracted from DNase I footprinted sites (Lifanov et al., 2003). The Cic, Kr and Run motifs are from B1H experiments (Noyes et al., 2008). To assay for differentially represented hexamers among enhancer Types, we used an exact binomial test to calculate the significance of a word being more over-represented in one enhancer type compared to another(Robin et al., 2007). DREME was run using the following parameters: distribution of motif occurrences: 0 or 1 per sequence, motif width: 6–9; second order background(Bailey et al., 2010). The derived motif in Fig. 2C was compared to all known TF binding motifs in TransAct and Jaspar databases using TOMTOM(Gupta et al., 2007). Also see detailed information in the Extended Experimental Procedures.
Candidate enhancers
Fragments containing in vivo Bcd-binding signals (top 50 for Bcd antibody 1; (Li et al., 2008)) were selected by searching the regions from 550 bp upstream to 550 bp downstream of the primary binding peaks for Bcd motifs (cutoff PWM score 4.2). Primers were designed to amplify the minimal sequences that include all Bcd sites. For some bound regions, secondary peaks were tested separately. Fragments containing clusters of predicted Bcd-binding sequence motifs were selected using a homotypic clustering model (Lifanov et al., 2003). See the Extended Experimental Procedures for details. Primer sequences for all tested fragments are listed in Supplementary Table 1.
Vector construction and transgenesis
To construct reporter genes, a 4.0 kb HindIII-SphI fragment from the pEl1 vector, which includes the even-skipped (eve) basal promoter, the lacZ coding region, and the 3’ UTR from the α-tubulin gene(Lawrence et al., 1987), was fused with an attB backbone derived from a 3.5 kb HindIII SphI fragment from the piB-GFP vector (Bateman et al., 2006) to generate the empty vector piB-HC-lacZ. Amplified candidate enhancers (HC_01-HC_77) were digested with BglII (or BamHI) and AscI, and inserted into the piB-HC-lacZ vector. Enhancers containing new Run sites and enhancer containing mutated Run sites (Supplementary Table II) were chemically synthesized (Integrated DNA Technologies, Inc.) and inserted into the piB-HC-lacZ vector as described above. ΦC31 integrase-mediated cassette exchange was used to insert all transgenes into the 38F1 landing site (Bateman et al., 2006). Detailed information about constructing the artificial enhancers and generating transgenic reporter lines are available in the Extended Experimental Procedures.
In situ hybridizations, immunohistochemistry, and image processing
Enzymatic in situ hybridizations were performed using digoxigenin (DIG) labeled antisense RNA probes and the alkaline phosphotase assay(Small, 2000). Embryos were imaged at 200X on a Zeiss Axioskop, and analyzed using MATLAB (Mathworks). See the Extended Experimental Procedures for details of measuring the posterior boundary positions (PBPs). Triple-color FISH was performed as previously described using DIG-labeled, fluorescein (FITC)-labeled, and biotin (BIO)-labeled probes (Kosman et al., 2004). Detection and quantification of protein profiles for Bcd, Run, Kr and Cic was described previously (Ochoa-Espinosa et al., 2009; Yu and Small, 2008). Also see the Extended Experimental Procedures for the antibodies used.
Supplementary Material
Research highlights.
-
*
A Runt protein gradient represses Bicoid-dependent activation.
-
*
Runt and two other repressors spatially order Bicoid-dependent boundaries.
-
*
The repressors bind directly to all 66 known Bicoid-dependent enhancers.
-
*
The repression system puts conceptual limits on the Bicoid morphogen hypothesis.
Acknowledgements
We thank Jerry Huang for assistance with computer searches for clusters of Bcd-binding motifs, Hsiao-lan Liang for run expression data, and Peter Gergen, Stas Shvartsman, Jack Bateman, and Gary Struhl for fly stocks. We thank Claude Desplan, Justin Blau, Jens Rister, and Rhea Datta for suggestions that improved the manuscript, and Thomas Gregor, Eric Wieschaus, Jim Jaynes, and Christine Rushlow for insightful discussions. This work was supported by NIH grant RO1 GM51946 to SS, and was conducted in a facility constructed with support from Research Facilities Improvement Grant C06 RR-15518-01 from the National Center for Research Resources, National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Affolter M, Basler K. The Decapentaplegic morphogen gradient: from pattern formation to growth regulation. Nat Rev Genet. 2007;8:663–674. doi: 10.1038/nrg2166. [DOI] [PubMed] [Google Scholar]
- Ajuria L, Nieva C, Winkler C, Kuo D, Samper N, Andreu MJ, Helman A, Gonzalez-Crespo S, Paroush Z, Courey AJ, et al. Capicua DNAbinding sites are general response elements for RTK signaling in Drosophila. Development. 2011;138:915–924. doi: 10.1242/dev.057729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen BL, Song JY, Izzi L, Althaus IW, Kang JS, Charron F, Krauss RS, McMahon AP. Overlapping roles and collective requirement for the coreceptors GAS1, CDO, and BOC in SHH pathway function. Dev Cell. 2011;20:775–787. doi: 10.1016/j.devcel.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen BL, Tenzen T, McMahon AP. The Hedgehog-binding proteins Gas1 and Cdo cooperate to positively regulate Shh signaling during mouse development. Genes Dev. 2007;21:1244–1257. doi: 10.1101/gad.1543607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrioli LP, Oberstein AL, Corado MS, Yu D, Small S. Groucho-dependent repression by Sloppy-paired 1 differentially positions anterior pair-rule stripes in the Drosophila embryo. Dev Biol. 2004;276:541–551. doi: 10.1016/j.ydbio.2004.09.025. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Whitington T, Machanick P. The value of position-specific priors in motif discovery using MEME. BMC Bioinformatics. 2010;11:179. doi: 10.1186/1471-2105-11-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaskas N, Ribeiro A, Panovska J, Dessaud E, Sasai N, Page KM, Briscoe J, Ribes V. Gene regulatory logic for reading the sonic hedgehog signaling gradient in the vertebrate neural tube. Cell. 2012;148:273–284. doi: 10.1016/j.cell.2011.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman JR, Lee AM, Wu CT. Site-specific transformation of Drosophila via phiC31 integrase-mediated cassette exchange. Genetics. 2006;173:769–777. doi: 10.1534/genetics.106.056945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell G, Tomlinson A. Transducing the Dpp morphogen gradient in the wing of Drosophila: regulation of Dpp targets by brinker. Cell. 1999;96:553–562. doi: 10.1016/s0092-8674(00)80659-5. [DOI] [PubMed] [Google Scholar]
- Driever W, Nusslein-Volhard C. The bicoid protein determines position in the Drosophila embryo in a concentration-dependent manner. Cell. 1988a;54:95–104. doi: 10.1016/0092-8674(88)90183-3. [DOI] [PubMed] [Google Scholar]
- Driever W, Nusslein-Volhard C. A gradient of bicoid protein in Drosophila embryos. Cell. 1988b;54:83–93. doi: 10.1016/0092-8674(88)90182-1. [DOI] [PubMed] [Google Scholar]
- Driever W, Nusslein-Volhard C. The bicoid protein is a positive regulator of hunchback transcription in the early Drosophila embryo. Nature. 1989;337:138–143. doi: 10.1038/337138a0. [DOI] [PubMed] [Google Scholar]
- Furriols M, Casanova J. In and out of Torso RTK signalling. EMBO J. 2003;22:1947–1952. doi: 10.1093/emboj/cdg224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q, Finkelstein R. Targeting gene expression to the head: the Drosophila orthodenticle gene is a direct target of the Bicoid morphogen. Development. 1998;125:4185–4193. doi: 10.1242/dev.125.21.4185. [DOI] [PubMed] [Google Scholar]
- Gao Q, Wang Y, Finkelstein R. Orthodenticle regulation during embryonic head development in Drosophila. Mech Dev. 1996;56:3–15. doi: 10.1016/0925-4773(96)00504-7. [DOI] [PubMed] [Google Scholar]
- Gergen JP, Butler BA. Isolation of the Drosophila segmentation gene runt and analysis of its expression during embryogenesis. Genes Dev. 1988;2:1179–1193. doi: 10.1101/gad.2.9.1179. [DOI] [PubMed] [Google Scholar]
- Gergen JP, Wieschaus E. The localized requirements for a gene affecting segmentation in Drosophila:Analysis of larvae mosaic for runt. Developmental Biology. 1985;109:321–335. doi: 10.1016/0012-1606(85)90459-2. [DOI] [PubMed] [Google Scholar]
- Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS. Quantifying similarity between motifs. Genome Biol. 2007;8:R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon JB, Dyson S, St Johnston D. Cells' perception of position in a concentration gradient. Cell. 1998;95:159–162. doi: 10.1016/s0092-8674(00)81747-x. [DOI] [PubMed] [Google Scholar]
- Hoch M, Schroder C, Seifert E, Jackle H. cis-acting control elements for Kruppel expression in the Drosophila embryo. Embo J. 1990;9:2587–2595. doi: 10.1002/j.1460-2075.1990.tb07440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JW, Hendrix DA, Papatsenko D, Levine MS. How the Dorsal gradient works: insights from postgenome technologies. Proc Natl Acad Sci U S A. 2008;105:20072–20076. doi: 10.1073/pnas.0806476105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houchmandzadeh B, Wieschaus E, Leibler S. Establishment of developmental precision and proportions in the early Drosophila embryo. Nature. 2002;415:798–802. doi: 10.1038/415798a. [DOI] [PubMed] [Google Scholar]
- Jazwinska A, Kirov N, Wieschaus E, Roth S, Rushlow C. The Drosophila gene brinker reveals a novel mechanism of Dpp target gene regulation. Cell. 1999;96:563–573. doi: 10.1016/s0092-8674(00)80660-1. [DOI] [PubMed] [Google Scholar]
- Jeong J, McMahon AP. Growth and pattern of the mammalian neural tube are governed by partially overlapping feedback activities of the hedgehog antagonists patched 1 and Hhip1. Development. 2005;132:143–154. doi: 10.1242/dev.01566. [DOI] [PubMed] [Google Scholar]
- Jiang J, Levine M. Binding affinities and cooperative interactions with bHLH activators delimit threshold responses to the dorsal gradient morphogen. Cell. 1993;72:741–752. doi: 10.1016/0092-8674(93)90402-c. [DOI] [PubMed] [Google Scholar]
- Jimenez G, Guichet A, Ephrussi A, Casanova J. Relief of gene repression by torso RTK signaling: role of capicua in Drosophila terminal and dorsoventral patterning. Genes Dev. 2000;14:224–231. [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Andreu MJ, Lim B, Chung K, Terayama M, Jimenez G, Berg CA, Lu H, Shvartsman SY. Gene regulation by MAPK substrate competition. Dev Cell. 2011;20:880–887. doi: 10.1016/j.devcel.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosman D, Mizutani CM, Lemons D, Cox WG, McGinnis W, Bier E. Multiplex detection of RNA expression in Drosophila embryos. Science. 2004;305:846. doi: 10.1126/science.1099247. [DOI] [PubMed] [Google Scholar]
- Lawrence PA, Johnston P, Macdonald P, Struhl G. Borders of parasegments in Drosophila embryos are delimited by the fushi tarazu and evenskipped genes. Nature. 1987;328:440–442. doi: 10.1038/328440a0. [DOI] [PubMed] [Google Scholar]
- Lecuit T, Brook WJ, Ng M, Calleja M, Sun H, Cohen SM. Two distinct mechanisms for long-range patterning by Decapentaplegic in the Drosophila wing. Nature. 1996;381:387–393. doi: 10.1038/381387a0. [DOI] [PubMed] [Google Scholar]
- Li XY, MacArthur S, Bourgon R, Nix D, Pollard DA, Iyer VN, Hechmer A, Simirenko L, Stapleton M, Luengo Hendriks CL, et al. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol. 2008;6:e27. doi: 10.1371/journal.pbio.0060027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang HL, Nien CY, Liu HY, Metzstein MM, Kirov N, Rushlow C. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature. 2008;456:400–403. doi: 10.1038/nature07388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifanov AP, Makeev VJ, Nazina AG, Papatsenko DA. Homotypic regulatory clusters in Drosophila. Genome Res. 2003;13:579–588. doi: 10.1101/gr.668403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little S, Tkacik G, Kneeland T, Wieschaus E, Gregor T. The Formation of the Bicoid Morphogen Gradient Requires Protein Movement from Anteriorly Localized mRNA. PLoS Biol. 2011;9:e1000596. doi: 10.1371/journal.pbio.1000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr U, Chung HR, Beller M, Jackle H. Antagonistic action of Bicoid and the repressor Capicua determines the spatial limits of Drosophila head gene expression domains. Proc Natl Acad Sci U S A. 2009;106:21695–21700. doi: 10.1073/pnas.0910225106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikova IN, Crute BE, Wang S, Speck NA. Sequence specificity of the core-binding factor. J Virol. 1993;67:2408–2411. doi: 10.1128/jvi.67.4.2408-2411.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami M, Kinoshita N, Kamoshida Y, Tanimoto H, Tabata T. brinker is a target of Dpp in Drosophila that negatively regulates Dpp- dependent genes [In Process Citation] Nature. 1999;398:242–246. doi: 10.1038/18451. [DOI] [PubMed] [Google Scholar]
- Muller B, Hartmann B, Pyrowolakis G, Affolter M, Basler K. Conversion of an extracellular Dpp/BMP morphogen gradient into an inverse transcriptional gradient. Cell. 2003;113:221–233. doi: 10.1016/s0092-8674(03)00241-1. [DOI] [PubMed] [Google Scholar]
- Namba R, Pazdera TM, Cerrone RL, Minden JS. Drosophila embryonic pattern repair: how embryos respond to bicoid dosage alteration. Development. 1997;124:1393–1403. doi: 10.1242/dev.124.7.1393. [DOI] [PubMed] [Google Scholar]
- Nellen D, Burke R, Struhl G, Basler K. Direct and long-range action of a DPP morphogen gradient. Cell. 1996;85:357–368. doi: 10.1016/s0092-8674(00)81114-9. [DOI] [PubMed] [Google Scholar]
- Noyes MB, Meng X, Wakabayashi A, Sinha S, Brodsky MH, Wolfe SA. A systematic characterization of factors that regulate Drosophila segmentation via a bacterial one-hybrid system. Nucleic Acids Res. 2008;36:2547–2560. doi: 10.1093/nar/gkn048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa-Espinosa A, Yu D, Tsirigos A, Struffi P, Small S. Anterior-posterior positional information in the absence of a strong Bicoid gradient. Proc Natl Acad Sci U S A. 2009;106:3823–3828. doi: 10.1073/pnas.0807878105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa-Espinosa A, Yucel G, Kaplan L, Pare A, Pura N, Oberstein A, Papatsenko D, Small S. The role of binding site cluster strength in Bicoid-dependent patterning in Drosophila. Proc Natl Acad Sci U S A. 2005;102:4960–4965. doi: 10.1073/pnas.0500373102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin S, Schbath S, Vandewalle V. Statistical tests to compare motif count exceptionalities. BMC Bioinformatics. 2007;8:84. doi: 10.1186/1471-2105-8-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers KW, Schier AF. Morphogen Gradients: From Generation to Interpretation. Annu Rev Cell Dev Biol. 2011;27:28.21–28.31. doi: 10.1146/annurev-cellbio-092910-154148. [DOI] [PubMed] [Google Scholar]
- Ronchi E, Treisman J, Dostatni N, Struhl G, Desplan C. Down-regulation of the Drosophila morphogen bicoid by the torso receptor-mediated signal transduction cascade. Cell. 1993;74:347–355. doi: 10.1016/0092-8674(93)90425-p. [DOI] [PubMed] [Google Scholar]
- Roth S, Stein D, Nusslein-Volhard C. A gradient of nuclear localization of the dorsal protein determines dorsoventral pattern in the Drosophila embryo. Cell. 1989;59:1189–1202. doi: 10.1016/0092-8674(89)90774-5. [DOI] [PubMed] [Google Scholar]
- Segal E, Raveh-Sadka T, Schroeder M, Unnerstall U, Gaul U. Predicting expression patterns from regulatory sequence in Drosophila segmentation. Nature. 2008;451:535–540. doi: 10.1038/nature06496. [DOI] [PubMed] [Google Scholar]
- Sinha S, Liang Y, Siggia E. Stubb: a program for discovery and analysis of cis-regulatory modules. Nucleic Acids Res. 2006;34:W555–W559. doi: 10.1093/nar/gkl224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small S. In vivo analysis of lacZ fusion genes in transgenic Drosophila melanogaster. Methods Enzymol. 2000;326:146–159. doi: 10.1016/s0076-6879(00)26052-7. [DOI] [PubMed] [Google Scholar]
- Small S, Kraut R, Hoey T, Warrior R, Levine M. Transcriptional regulation of a pair-rule stripe in Drosophila. Genes Dev. 1991;5:827–839. doi: 10.1101/gad.5.5.827. [DOI] [PubMed] [Google Scholar]
- Stanojevic D, Small S, Levine M. Regulation of a segmentation stripe by overlapping activators and repressors in the Drosophila embryo. Science. 1991;254:1385–1387. doi: 10.1126/science.1683715. [DOI] [PubMed] [Google Scholar]
- Struffi P, Corado M, Kaplan L, Yu D, Rushlow C, Small S. Combinatorial activation and concentration-dependent repression of the Drosophila even skipped stripe 3+7 enhancer. Development. 2011;138:4291–4299. doi: 10.1242/dev.065987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G, Struhl K, Macdonald PM. The gradient morphogen bicoid is a concentration-dependent transcriptional activator. Cell. 1989;57:1259–1273. doi: 10.1016/0092-8674(89)90062-7. [DOI] [PubMed] [Google Scholar]
- Tsai C, Gergen JP. Gap gene properties of the pair-rule gene runt during Drosophila segmentation. Development. 1994;120:1671–1683. doi: 10.1242/dev.120.6.1671. [DOI] [PubMed] [Google Scholar]
- Turing AM. The chemical basis of morphogenesis. Philosophical Transactions of Royal Society in London. 1952;B-237:37–84. [Google Scholar]
- Wolpert L. One hundred years of positional information. Trends Genet. 1996;12:359–364. doi: 10.1016/s0168-9525(96)80019-9. [DOI] [PubMed] [Google Scholar]
- Wu X, Vakani R, Small S. Two distinct mechanisms for differential positioning of gene expression borders involving the Drosophila gap protein giant. Development. 1998;125:3765–3774. doi: 10.1242/dev.125.19.3765. [DOI] [PubMed] [Google Scholar]
- Yu D, Small S. Precise registration of gene expression boundaries by a repressive morphogen in Drosophila. Curr Biol. 2008;18:868–876. doi: 10.1016/j.cub.2008.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.